Abstract
Glycosylation changes associated with cellular transformation can facilitate the growth and progression of tumors. Previously we discovered that the gene Mgat3 encoding the glycosyltransferase GnT-III is elevated in epithelial ovarian carcinomas (EOCs) and leads to the production of abnormal truncated N-linked glycan structures instead of the typical bisected forms. In this study, we are interested in discovering how these abnormal glycans impact the growth and progression of ovarian cancer. We have discovered using stable shRNA gene suppression that GnT-III expression controls the expansion of side-population cells, also known as cancer stem cells. More specifically, we found that GnT-III expression regulates the levels and activation of the heavily glycosylated Notch receptor involved in normal and malignant development. Suppression of GnT-III in EOC cell lines and primary tumor-derived cells resulted in an inhibition of Notch signaling that was more potent than pharmacologic blockage of Notch activation via γ-secretase inhibition. The inhibition resulted from the redirection of the Notch receptor to the lysosome, a novel mechanism. These findings demonstrate a new role for bisecting glycosylation in the control of Notch transport and demonstrate the therapeutic potential of inhibiting GnT-III as a treatment for controlling EOC growth and recurrence.
Keywords: cancer, glycosylation, N-linked glycosylation, Notch pathway, ovarian cancer
Introduction
Epithelial ovarian cancer (EOC)3 is the second most common gynecologic malignancy in the United States and is the fifth leading cause of cancer deaths among women with over 14,000 deaths reported each year (1). Among the histological subtypes of EOC, serous ovarian cancer is the most prevalent subtype and accounts for the majority of deaths (2). Although the war on cancer has led to improved survival rates for certain cancers, survival rates for ovarian cancer patients have not shown much improvement over a 30-year period (1). This fact highlights the need to focus our efforts on the development of new therapeutic strategies for this disease. Increasing our knowledge of the key drivers of ovarian cancer growth and metastasis may to lead to new therapeutics.
Over 80% of the cell membrane is composed of glycoconjugates such as glycolipids and glycoproteins. Previously, we discovered that a specific glycosyltransferase known as GnT-III is up-regulated in the two most prevalent subtypes of EOC, serous and endometrioid (3, 4). GnT-III transfers a β1,4-linked N-acetylglucosamine to the mannosyl core of N-linked glycans to produce a structure commonly referred to as a bisecting glycan (5) (see Fig. 1A). The presence of bisecting glycans affects the overall conformation of the growing N-linked glycan in the Golgi and often suppresses further branching (6). We isolated N-linked glycans from membrane proteins that were enriched using the lectin E-PHA from primary serous and endometrioid ovarian carcinoma tissues and discovered a very unusual bisecting glycan structure that is not only missing further branching, it is also missing galactose capping and sialic acid extensions (4) (see Fig. 1A). The role of GnT-III expression and bisecting glycosylation has been variable in the literature. In breast cancer where the glycosyltransferase GnT-V dominates, the expression of GnT-III suppresses tumor burden. In fact, loss of heterozygosity in the chromosomal region where GnT-III is located, 22q13.1, correlates with breast cancer (7, 8). However, in leukemia, GnT-III levels and bisecting glycans are increased and associated with blast crisis (9). Therefore, the cell type and overall levels of different branching glycosyltransferases may play roles in determining the overall bisecting glycan structure and functional outcomes.
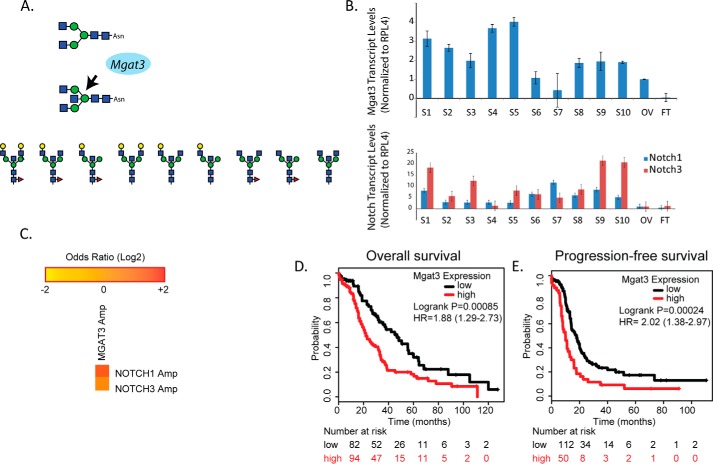
Figure 1.
GnT-III expression levels are elevated in high-grade serous ovarian carcinoma tissues and correlate with reduced survival. A, Mgat3 encodes for the glycosyltransferase GnT-III that catalyzes the transfer of N-acetylglucosamine (blue box) to the trimannosyl core in β 1,4 linkage to form the bisecting N-linked glycan structure. Shown below are examples of the abnormal truncated bisecting N-linked structures previously identified in ovarian cancer tissues (4). B, Mgat3, NOTCH1, and NOTCH3 expression levels are increased in primary high grade serous ovarian carcinoma tissues. The data are ± S.E., n = 3 experiments using primers and analysis as previously described (3). Sample pathology, grade, and origin of tumors S1–S10 are indicated in supplemental Table S1. OV and FT correspond to normal ovary and normal fallopian tube. C, the multidimensional cancer genome browser, cBioportal, was used to analyze trends in mutual exclusivity or co-occurrence between NOTCH1, NOTCH3, and Mgat3 in the TCGA Ovarian Serous Cystadenocarcinoma database (10, 11). **, p values were derived using a Fisher exact test; Logs odds ratio was utilized to quantify how robustly the presence or absence of gene A alterations associated with the presence or absence of gene B alterations from selected tumors. The co-occurrence of Mgat3 with Notch 3 has a p value of 0.007 and a log odds ratio of 1.4, whereas Mgat3 with Notch 1 has a p value of 0.019 and a log odds ratio of 1.6. D, Kaplan–Meier analysis (http://kmplot.com/analysis/index) of OS was plotted for stage IV ovarian cancer patients (n = 176). The OS was significantly longer in the Mgat3 low expression group than in the Mgat3 high expression group. A cutoff value of 140 was chosen by auto select in the analysis configuration, with the expression value of the probe (209764_at) ranging from 4 to 587. E, Kaplan–Meier analysis of PFS was plotted for stage IV ovarian cancer patients (n = 162). The PFS was significantly longer in the Mgat3 low expression group than in the Mgat3 high expression group. A cutoff value of 183 was chosen by auto select in the analysis configuration, with the expression value of the probe (209764_at) ranging from 4 to 700. HR, hazard ratio.
In this study, we are assessing for the first time the role of GnT-III expression and bisecting glycosylation on the growth and progression of EOC. We report that GnT-III expression significantly increases ovarian cancer growth and metastasis and increases the expansion of the ovarian cancer stem cell (CSC) population. Furthermore, glycosylation mediated by GnT-III increases Notch receptor levels and activity. We find that suppression of GnT-III results in a relocation of Notch receptor to the lysosome, resulting in reduced Notch receptor levels and an impairment of γ-secretase–mediated activation. Our data provide insights into a novel molecular mechanism to inhibit the Notch pathway in EOC and cancers with elevated GnT-III expression.
Results
GnT-III expression is increased in high grade serous ovarian cancers and correlates with decreased patient survival
We have previously discovered that the glycosyltransferase known as GnT-III is elevated in primary ovarian carcinoma tissues leading to increased bisecting glycosylation on glycoproteins (3, 4). We find that Mgat3 transcript levels are elevated in 8 of 10 high grade ovarian carcinoma tumors relative to non-malignant ovary and non-malignant fallopian tube tissues (Fig. 1B and supplemental Table S1). Our previous glycoproteomic study led to the discovery that a number of proteins with bisecting glycosylation had potential roles in the Notch pathway (4). Therefore, we have analyzed the levels of Notch receptors and find that 9 of the 10 high grade ovarian carcinoma tissues have elevated levels of NOTCH1 and NOTCH3 compared with non-malignant ovarian tissue and fallopian tube tissue (Fig. 1B). We have used the multidimensional cancer genome browser cBioportal to analyze trends in mutual exclusivity or co-occurrence between NOTCH1, NOTCH3, and Mgat3 in the TCGA Ovarian Serous Cystadenocarcinoma database (10, 11) Our results demonstrate a significant tendency toward co-occurrence with a log odds ratio for Mgat3 and NOTCH1 at 1.6 and a log odds ratio of Mgat3 and NOTCH3 at 1.4 (Fig. 1C). The prognostic value of GnT-III levels were evaluated in high grade serous and endometrioid ovarian carcinomas (Fig. 1, D and E). Survival data for patients indicate that patients with high GnT-III levels had significantly reduced overall survival (Fig. 1D) and progression-free survival (Fig. 1E) compared with patients with low GnT-III levels. The distribution of Mgat3 levels across patients is included in a bee-swarm plot (supplemental Fig. S1)
GnT-III expression and bisecting glycosylation expand side-population cells and enhance spheroid formation
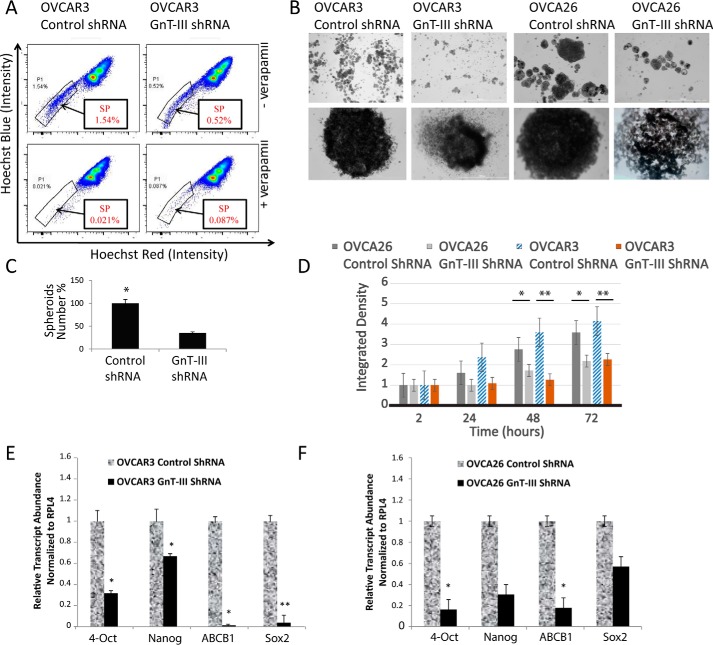
Our previous glycoproteomic study suggests that glycoproteins that receive bisecting glycans in ovarian cancer participate in stem cell signaling pathways (4). Side-population (SP) cells have been demonstrated to be a source of CSCs and progenitor cells in several malignancies including ovarian cancer (12, 13). SP cells are isolated based on their ability to efflux the Hoechst 33342 fluorescent dye. To determine the effect of bisecting glycosylation on the SP cells, we measured the SP in OVCAR3 cells with stable expression of control (scrambled) short hairpin RNA (control shRNA) or short hairpin RNA targeting GnT-III (GnT-III shRNA). Stable suppression of GnT-III and rescue of this phenotype have been previously described (4). In FACS experiments, we consistently observe (n = 3) a significant reduction in the percentage of SP cells in OVCAR3 GnT-III shRNA cells (Fig. 2A) (0.52%) versus OVCAR3 control shRNA cells (1.54%).
Figure 2.
GnT-III expression maintains the SP and promotes spheroid formation in ovarian cancer cells. A, stable OVCAR3 control shRNA and OVCAR3 GnT-III shRNA cells were labeled with Hoechst 33342 dye and analyzed by flow cytometry before and after verapamil treatment. Representative results shown from n = 3 experiments. B, OVCAR3 and OVCA26 cell lines generated 3D independent, self-renewing spheroid cells under stem cell selection medium. Upper row, lower magnification; lower row, zoom in on individual spheroid. Bar, 100 μm. C, cumulative spheroids formed were counted, and data from n = 3 experiments are shown as a percentage. *, p < 0.05. D, spheroid growth and formation were evaluated by dispersing cells from spheroids that had been grown in ultralow adhesion plates in stem cell selective media for 3 weeks. Spheroid density was calculated using ImageJ from five fields for each sample at 2, 24, 48, and 72 h after seeding. The data shown are results from three independent experiments with the density of the OVCA26 GnT-III ShRNA spheroids being set at 1.0. *, p < 0.05; **, p < 0.005. E, total RNA isolated from OVCAR3 spheroid cells were examined for expression of various stemness genes as indicated (primer sequences in supplemental Table S2). Relative expression levels normalized to RPL4 are shown for GnT-III shRNA levels compared with control shRNA set to 1.0 for comparison. The data are means ± S.E., n = 3. *, p < 0.05; **, p < 0.01. F, total RNA isolated from OVCA26 spheroid cells were examined for expression of various stemness genes as indicated (primer sequences in supplemental Table S2). Relative expression levels normalized to RPL4 are shown for GnT-III shRNA levels compared with control shRNA set to 1.0 for comparison. The data are means ± S.E., n = 3. *, p < 0.05.
Previous studies have shown that ovarian cancer cell lines including OVCAR3 have sphere-forming and self-renewal ability when grown in stem cell-selective medium (14). To further examine the role of GnT-III expression on sphere formation, we established stable expression of control and GnT-III shRNA in a patient-derived ovarian cancer cell line known as OVCA26. OVCA26 cells express significant levels of ALDH1 and CD133, two markers associated with cancer stem cell populations. OVCAR3 and OVCA26 cells expressing control shRNA demonstrate greater (p < 0.001) sphere-forming ability compared with GnT-III shRNA cells (Fig. 2, B and C). Interestingly, GnT-III shRNA-expressing cells cluster together in irregularly shaped cell clumps compared with control shRNA cells. Serial passaging of the cells revealed that control cells maintain sphere-forming ability over multiple passages, whereas the number of cell aggregates formed in GnT-III shRNA cells were reduced in number and density after passage 3 (Fig. 2, C and D). The difference in spheroid densities can be observed as early as 24 h for OVCA26 and OVCAR3 GnT-III ShRNA cells (Fig. 2D). Based on these data, we investigated the molecular profile of the spheroids, focusing on genes important for stem cell renewal and expansion. The expression levels of Oct-4, Nanog, ABCB1, and Sox-2 were significantly decreased in OVCAR3 GnT-III shRNA cells compared with OVCAR3 control shRNA cells (Fig. 2E). Similar results were obtained with OVCA26 cells (Fig. 2F).
Suppression of GnT-III reduces ovarian tumor growth and metastasis in vivo
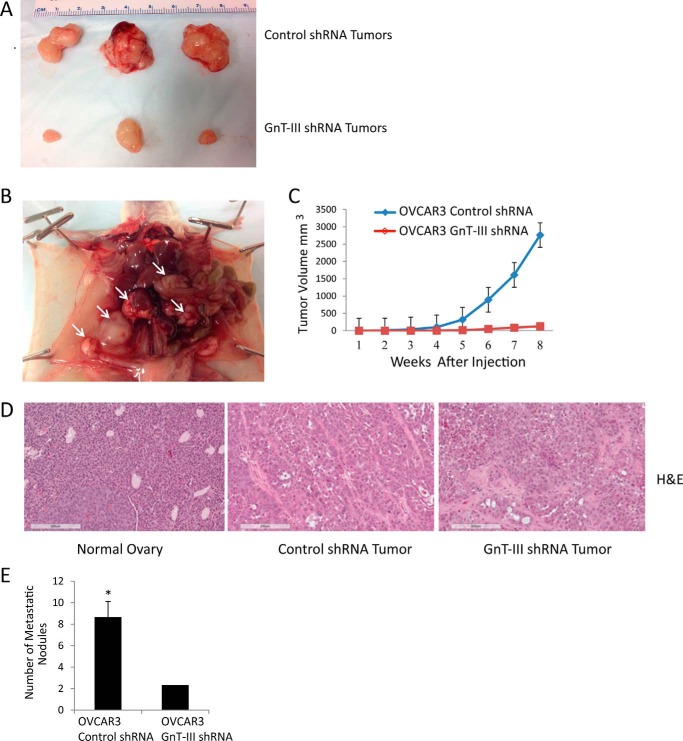
A xenograft model was used to examine the effect of GnT-III expression and bisecting glycosylation on tumor growth. OVCAR3 control shRNA cells displayed robust tumor initiation compared with OVCAR3 GnT-III shRNA cells. Tumor growth was noticed in 6 of 6 animals at 8 weeks following injection of control cells, whereas only 3 of 6 mice injected with an equal number of GnT-III shRNA cells had tumors at 8 weeks (Fig. 3A, representative tumors). Control cell tumors were more invasive following i.p. injections (Fig. 3B, white arrows identify multiple masses in the peritoneum and mesentery), whereas GnT-III shRNA tumors were less vascularized (Fig. 3, A and D compare vessel number in control shRNA tumor (middle) with GnT-III shRNA tumor (right)) and never invaded surrounding tissues. The i.p. injections of control shRNA cells resulted in the development of bloody ascites in 3 of 4 mice with tumor latencies of 70–90 days (supplemental Fig. S2A), whereas the same number of GnT-III shRNA cells never resulted in bloody ascites (supplemental Fig. S3A). We also observed a significant increase in ovarian masses and metastasis (Fig. 3E) in control shRNA cells compared with GnT-III shRNA cells (p < 0.05) (supplemental Figs. S2B and 3B, respectively).
Figure 3.
GnT-III suppression inhibits tumor growth and metastasis in a xenograft model. A, OVCAR3 control shRNA or GnT-III shRNA were injected subcutaneously in the right and left flanks of NOD/SCID mice. The sizes of selected resected tumors are shown. B, control OVCAR3 tumor invasion into the abdominal cavity with multiple masses on the peritoneum and mesentery are marked with white arrows. C, cumulative growth curves for OVCAR3 control shRNA and OVCAR3 GnT-III shRNA xenografts. D, hematoxylin and eosin (H&E) staining for normal ovary (left), control shRNA xenograft tumor (middle), and GnT-III shRNA xenograft tumor (right). Bar, 200 μm. E, cumulative number of metastatic nodules for control siRNA and GnT-III siRNA xenografts. n = 12 per cell line. *, p < 0.05.
GnT-III expression and bisecting glycosylation regulate Notch levels and activity
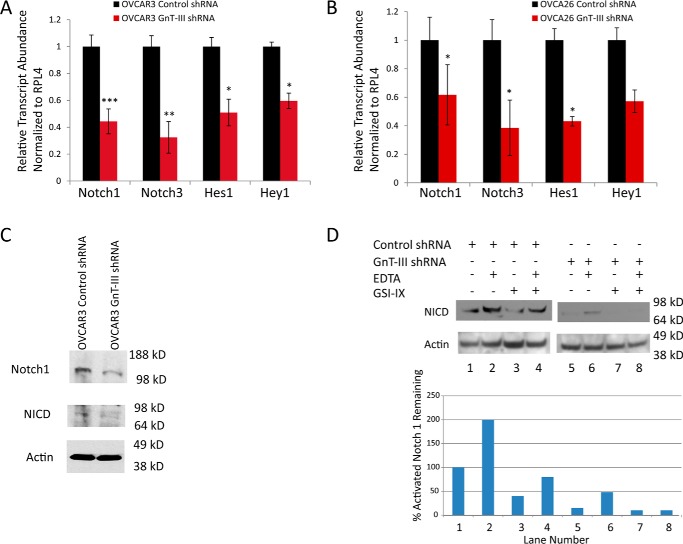
Several of the glycoproteins that were previously identified in our glycoproteomic study have roles in the Notch signaling pathway (4). Notch receptors have been shown to be important regulators of the ovarian CSC population (15). To evaluate the effect of GnT-III expression on Notch signaling, we measured the levels of Notch receptors and downstream effectors of Notch using RNA extracted from spheroids of OVCAR3 and OVCA26 stable cell lines. Control shRNA levels were set to 1.0 for comparison. We observed a significant reduction in the levels of Notch 1 and Notch 3 in the RNA from spheroids of both OVCAR3 and OVCA26 GnT-III shRNA cells compared with control cells (p < 0.001, p < 0.01, and p < 0.05, respectively) (Fig. 4, A and B). The levels of downstream effectors of the Notch signaling pathway HES-1 and Hey-1 were also significantly reduced in both OVCAR3 and OVCA26 GnT-III shRNA cells compared with control (p < 0.05) (Fig. 4, A and B). To confirm that the reduction in transcript levels in spheroids corresponds to protein reduction, an antibody to the 120-kDa Notch 1 transmembrane domain of the S1-cleaved heterodimeric Notch 1 detected Notch 1 in OVCAR3 GnT-III shRNA cells at 40% the level of control cells (Fig. 4C). Furthermore, we find a reduction in the levels of activated Notch 1 measured using an antibody that recognizes the cleaved Notch intracellular domain (NICD) (Fig. 4C).
Figure 4.
GnT-III expression regulates Notch levels and activity. A, Notch 1, Notch 3, Hes 1, and Hey 1 expression levels were significantly reduced in OVCAR3 GnT-III shRNA cells compared with control shRNA levels. The data are means ± S.E., n = 3 experiments. ***, p < 0.001; **, p < 0.01; *, p < 0.05, respectively. B, Notch 1, Notch 3, and Hes 1 levels were significantly reduced in OVCA26 GnT-III shRNA cells compared with control cells. The data are means ± S.E., n = 3 experiments. *, p < 0.05. C, Western blot analysis of Notch 1 and active NICD for OVCAR3 cell lines. D, Western blot analysis of cleaved NICD in OVCAR3 control shRNA and OVCAR3 Gnt-III shRNA cells treated with EDTA and or GSI-IX. The graph represents the percentage of activated Notch remaining following normalization to actin levels; the blot shown is representative of n = 2 experiments.
The γ-secretase cleavage of Notch to release the NICD is an essential step in the activation of Notch signaling. To determine whether there is any effect of GnT-III expression and bisecting glycosylation on γ-secretase cleavage, the cells were treated with γ-secretase inhibitor IX (GSI-IX). Exposure of cells to EDTA stimulates detachment of the Notch extracellular domain, rendering the residual transmembrane fragment a substrate for γ-secretase cleavage and Notch activation (16). Cells were treated with EDTA prior to returning them to regular medium for incubation with or without GSI-IX. Control OVCAR3 shRNA cells treated with EDTA show an increase in activated Notch 1 NICD (Fig. 4F, compare lanes 1 and 2), as expected. GnT-III shRNA cells also show an increase over basal when treated with EDTA (Fig. 4F, compare lanes 5 and 6). OVCAR3 GnT-III shRNA cells without any treatment show an inhibition of activated Notch 1 greater than that of control cells treated with GSI-IX (Fig. 4F, compare lanes 3 and 5). Treatment of OVCAR3 GnT-III shRNA cells with GSI-IX results in further inhibition of activated Notch 1 when compared with control cells (Fig. 4F, compare lanes 3 and 7). These results indicate that GnT-III expression promotes Notch receptor cleavage.
GnT-III suppression alters Notch receptor localization
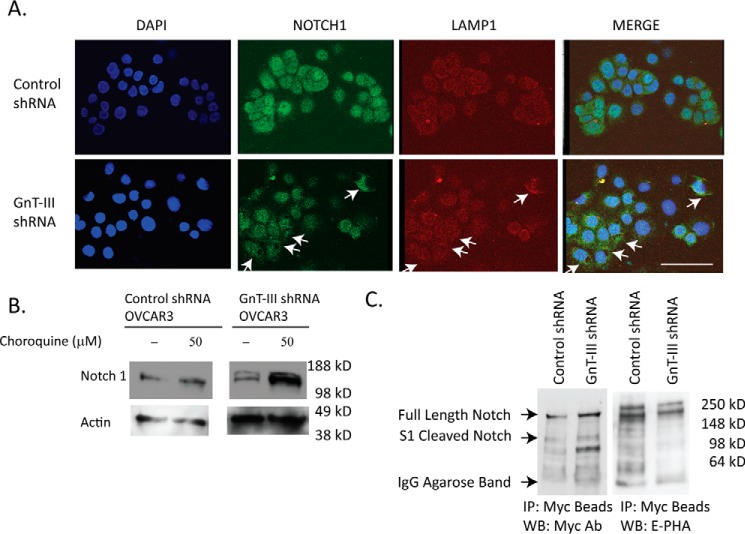
Autophagy has been shown to regulate the Notch pathway in normal physiology and disease (17). Therefore we performed immunocytochemistry to determine the localization of Notch 1 in OVCAR3 cells. Notch 1 is observed in intracellular vesicles and the nucleus in control shRNA OVCAR3 cells and does not co-localize with LAMP1 (lysosomal associated membrane protein 1; Fig. 5A). However, in GnT-III shRNA OVCAR3 cells, Notch 1 is primarily localized with the late endosome/lysosome marker LAMP1, indicating that a larger percentage of Notch 1 is in this fraction (Fig. 5A, white arrows). To further confirm these findings, we treated cells with chloroquine to raise the pH of this cellular compartment and inhibit lysosomal enzymes (18). We found no effect on Notch 1 levels in control cells treated with chloroquine, whereas GnT-III shRNA cells show a significant increase in Notch 1 levels when lysosomal enzymes are inhibited (Fig. 5B). These results indicate that Notch 1 is localized outside of the late endosome/lysosome compartment in control OVCAR3 cells, and inhibition of GnT-III causes a relocation of Notch 1 to the late endosome/lysosome.
Figure 5.
Notch 1 receptor has altered localization and glycosylation following GnT-III suppression. A, immunofluoresnce staining of Notch 1 (green) and LAMP1 (red) in OVCAR3 control shRNA and GnT-III shRNA cells. Bar, 100 μm. Significant areas of co-localization are shown with arrows. B, Western blot analysis of Notch 1 from lysates of cells in the absence or presence of the lysosome inhibitor chloroquine overnight. C, to analyze bisecting glycosylation of Notch 1 in ovarian cancer, mNotch 1/6× Myc was transfected into OVCAR3 control and GnT-III ShRNA cells. The mNotch 1 was isolated using anti-Myc-agarose resin prior to Western blot (WB) detection using Myc antibody or E-PHA. IP, immunoprecipitation.
Notch 1 is modified with bisecting glycosylation in OVCAR3 cells
We previously conducted a glycoproteomic analysis of membrane proteins isolated from primary ovarian cancer tumor tissues (4). In this study Notch was not identified as a substrate for GnT-III glycosylation. These results may have been influenced by the high levels of O-linked glycans on Notch peptides that would suppress ionization efficiency in the mass spectrometer. In this study, we can confirm that Notch 1 reacts with the lectin E-PHA, indicating the presence of bisecting glycans (Fig. 5C). OVCAR3 control shRNA and GnT-III shRNA cells were transfected with Myc-tagged mNotch 1. Notch 1/6× Myc was captured using beads covalently coupled with Myc antibody. We observe several Notch 1/6× Myc bands, including the full-length Notch 1 and the S1 cleaved Notch 1 with a predicted molecular mass of ∼100 kDa (19) (Fig. 5C). There are additional lower molecular mass bands that may be due to proteolysis. All of these bands are present in both the control and the GnT-III shRNA lanes at approximately the same levels, confirming equivalent transfection and expression of mNotch 1/6× Myc fusion proteins (Fig. 5C, blot to the left). The 6× Myc-tagged Notch 1 bands are E-PHA–positive in the control shRNA lane with complete absence or significant reduction in the GnT-III shRNA lane (Fig. 5C, blot to the right). The reduction in reactivity of these bands with the lectin E-PHA in the GnT-III shRNA lane confirms the presence of bisecting glycans on mNotch 1/6× Myc fusion proteins.
Discussion
Research over the past few years has focused on the identification of glycan changes specific for ovarian cancer. The next step is to investigate the functional impact of these glycosylation changes for ovarian cancer progression. In this manuscript, we have examined the influence of a specific form of N-linked glycosylation that is abnormally elevated in ovarian cancer and report novel mechanisms that GnT-III–mediated glycosylation exerts on ovarian cancer progression.
Disease recurrence and chemoresistance are major clinical issues facing clinicians and ovarian cancer patients that have been linked to CSC levels. Our data indicate that GnT-III expression levels have a significant impact on patient survival (Fig. 1). Furthermore, GnT-III expression promotes the CSC population and the expression of required stemness genes (Fig. 2). Suppression of GnT-III dramatically reduced ovarian tumor growth and metastasis in a xenograft model (Fig. 3 and supplemental Figs. S1 and S2). These findings suggest that bisecting glycosylation is a significant factor promoting the growth and metastasis of ovarian cancers.
The Notch signaling pathway is important for the activation and maintenance of stem cell progenitors in normal tissue development that can be abnormally activated in ovarian cancers. Recent studies support a role for the Notch pathway in ovarian cancer progression and chemoresistance (15, 20). We report, for the first time, that bisecting glycans increase Notch receptor levels and activity (Fig. 4). The influence of GnT-III expression on Notch signaling involves increased Notch receptor levels, increased levels of NICD, and enhanced γ-secretase cleavage (Fig. 4). We find that the use of GSI with GnT-III suppression resulted in additional inhibition, indicating that targeting GnT-III expression may be a new mechanism to inhibit Notch signaling in ovarian cancer that may augment GSI therapy. The effects of GnT-III expression levels on γ-secretase activity in ovarian cancer are novel and have not previously been reported. The mechanism for γ-secretase activation by GnT-III may be linked to control of Notch receptor trafficking. There is a precedent for bisecting glycosylation in the control of protein localization in the cell. A recent study found that mice deficient for GnT-III in an Alzheimer's model show a change in intracellular localization of the protein BACE1. BACE1 with bisecting glycosylation is localized to early endosomes, whereas BACE1 in mice lacking GnT-III was localized to the late endosome/lysosome (21).
The significance of N-linked glycosylation present on the Notch receptor has not been extensively studied in human cancers. It has been show that Notch has occupied N-linked glycosylation in CHO cells (22). Our data suggest that the Notch receptor has bisecting glycosylation in ovarian cancer. Notch 1 and Notch 3 signaling are active in ovarian cancer (15, 23). Our results confirm a role for N-linked bisecting glycosylation in the activation of Notch 1. Our finding that inhibition of GnT-III blocks Notch activation suggests that targeting GnT-III or blocking bisecting glycosylation sites on Notch receptor may lead to new therapeutics for ovarian cancer. The impact of N-linked glycosylation on O-fucose glycosylation of Notch is not known. The N-linked sites within Notch are located in close proximity to O-fucose modified sites. O-Fucose glycosylation of Notch EGF repeats has been shown to create a code to either inhibit or activate Notch ligand-dependent activation (24). It will be important in the future to determine how the bisecting glycan structures on Notch in ovarian cancer alter this O-fucose code. Overall, the evaluation of GnT-III expression levels could have clinical utility for identifying patients that would benefit from Notch inhibitor therapies.
Experimental procedures
Primary tissue samples and cell lines
Tumor samples from patients with ovarian cancer were obtained from the ovarian cancer patient repository of the University of Pennsylvania (Philadelphia, PA) (supplemental Table S1). All specimens were collected under a university institutional review board–approved protocol, and written consent from each donor was obtained. The EOC cell lines OVCAR3 and the primary patient cell line OVCA26 were grown in RPMI 1640 (Invitrogen) supplemented with 10% FBS (Gemini Bioproducts, West Sacramento, CA) and 100 mg/ml streptomycin (Invitrogen). The OVCA26 patient-derived cell line was produced under an approved institutional review board protocol by Dr. Martin Cannon at the University of Arkansas for Medical Sciences. The cell lines were maintained at 37 °C in a humidified atmosphere with 5% CO2. Lentiviral transduction to establish stable GnT-III suppression was performed as described previously (4). Spheroids were generated under stem cell–selective medium (Stem Cell Technologies, Vancouver, Canada) in ultralow adhesion dishes for 3 weeks. SP analysis was performed using Hoechst 33342 fluorescent dye in the Flow Cytometry Core at University of Arkansas for Medical Sciences, and data were analyzed using FlowJo software.
Kaplan–Meier plot analysis
The prognostic value of the MGAT3 gene in ovarian cancer was analyzed using the Kaplan–Meier plotter (http://kmplot.com/analysis/) (25–27).4 For analysis, patients with stage IV serous and endometrioid ovarian cancer were divided into two groups according to higher and lower MGAT3 expression levels (specifics on the cutoffs are included in the figure legend). The overall survival (OS) and progression-free survival (PFS) were then compared between the two groups. Log rank p values and hazard ratios with 95% confidence intervals were then calculated.
Quantitative RT-PCR
Samples (50 μl of packed cells) were extracted using TRIzol (Invitrogen) according to the manufacturer's instructions. After DNase treatment, RNA (2 μg) was reverse-transcribed using Superscript III (Invitrogen) with random hexamers and oligo(dT). Primers were validated with respect to primer efficiency and single product detection. The primer sequences are shown in supplemental Table S2. Quantitative real-time PCR was performed in a Bio-Rad CFX96 C1000 instrument using RT SYBR Green/Fluorescein qPCR Master Mix (Bio-Rad). Relative expression of target genes was calculated using the comparative cycle threshold method with genes normalized to RPL4.
Immunochemistry studies
Ovarian cancer cells were plated in chamber slides or on ultralow adhesion plates at 50% confluence prior to immunofluorescence staining. The cells were fixed in 3% formaldehyde, 1× PBS and blocked in 1× PBS, 1% BSA prior to incubation with primary antibodies, anti-Notch 1 antibody (Abcam), or anti LAMP-1 antibody (Abcam) in 1× PBS, 1% BSA, 0.2% Tween 20 for 1 h. Alexa Fluor 488– or 594–conjugated secondary antibodies were incubated for 30 min in 1× PBS, 1% BSA, 0.2% Tween 20 for 30 min prior to counterstain with DAPI (Vector Laboratories, Burlingame, CA) and mounting in Vectashield.
In vivo xenograft mouse model
All procedures were performed following the guidelines adopted by the Animal Care and Use Committee at the University of Arkansas for Medical Sciences. Female immunodeficient mice were obtained from Jackson Labs (NSG, NOD scid γ/8 weeks old). Five million OVCAR3 control or OVCAR3 GnT-III shRNA cells in PBS were inoculated subcutaneously into the right and left flank, respectively, of each mouse (n = 6). Five million OVCAR3 control or OVCAR3 GnT-III shRNA cells were injected intraperitoneal (n = 6 mice/cell line). The mice were monitored daily for 8 weeks prior to necropsy and tumor extraction. Tumor volume in mg is calculated using the following formula: volume = (length × width2/2).
Notch immunoprecipitation and Western blot analysis
Control or GnT-III shRNA cells were transfected with mNotch 1/6× Myc pCS2+ plasmid (gift from Robert Haltiwanger) (28). The cells were lysed in 1× TBS, 1% Triton X-100 supplemented with protease inhibitors. Lysates (500 μg) were precleared before the capture of Myc-tagged Notch using Myc antibody agarose beads (Santa Cruz Biotechnologies). The beads with captured Notch 1/6× Myc were washed with 1× TBS/1% Triton X-100 before proteins were separated on Tris-glycine gel prior to detection using Myc antibody at 1:500 dilution followed by anti-species secondary conjugated to HRP and chemiluminescent detection. The blot was stripped using Restore (Thermo Fisher) and detected using biotinylated E-PHA (Vector Labs) diluted 1:3,000 in 3% BSA/TBS with Tween 20 followed by streptavidin HRP (Vector Labs) diluted 1:10,000 in 3% BSA/TBS with Tween 20 prior to chemiluminescent detection.
Author contributions
H. A. and B. P. J. conducted most of the experiments, analyzed results, and wrote segments of the manuscript. M. Z. and Z. L. conducted the experiments to examine the localization of Notch and the glycosylation of Notch. M. J. C. derived the OVCA26 cell line and provided valuable insight in the course of the study. K. L. A. conceived of the idea for the project and worked with H. A., B. P. J., M. Z., and Z. L. on all experiments and wrote the manuscript with H. A. and B. P. J.
Supplementary Material
Acknowledgments
We thank Dr. Carolyn Bertozzi and Dr. Robert Haltiwanger for helpful discussions during the course of this study.
This work was supported by NCI, National Institutes of Health Grant UO1CA168870 (to K. L. A.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Tables S1 and S2 and Figs. S1–S3.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- EOC
- epithelial ovarian carcinoma
- E-PHA
- phaseolus vulgaris erythroagglutinin
- CSC
- cancer stem cell
- SP
- side-population
- NICD
- Notch intracellular domain
- OS
- overall survival
- PFS
- progression-free survival.
References
- 1. Siegel R., Ma J., Zou Z., and Jemal A. (2014) Cancer statistics, 2014. CA Cancer J. Clin. 64, 9–29 [DOI] [PubMed] [Google Scholar]
- 2. McCluggage W. G. (2011) Morphological subtypes of ovarian carcinoma: a review with emphasis on new developments and pathogenesis. Pathology 43, 420–432 [DOI] [PubMed] [Google Scholar]
- 3. Abbott K. L., Nairn A. V., Hall E. M., Horton M. B., McDonald J. F., Moremen K. W., Dinulescu D. M., and Pierce M. (2008) Focused glycomic analysis of the N-linked glycan biosynthetic pathway in ovarian cancer. Proteomics 8, 3210–3220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Allam H., Aoki K., Benigno B. B., McDonald J. F., Mackintosh S. G., Tiemeyer M., and Abbott K. L. (2015) Glycomic analysis of membrane glycoproteins with bisecting glycosylation from ovarian cancer tissues reveals novel structures and functions. J. Proteome Res. 14, 434–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Narasimhan S. (1982) Control of glycoprotein synthesis. UDP-GlcNAc:glycopeptide β 4-N-acetylglucosaminyltransferase III, an enzyme in hen oviduct which adds GlcNAc in β1–4 linkage to the β-linked mannose of the trimannosyl core of N-glycosyl oligosaccharides. J. Biol. Chem. 257, 10235–10242 [PubMed] [Google Scholar]
- 6. Schachter H. (1986) Biosynthetic controls that determine the branching and microheterogeneity of protein-bound oligosaccharides. Biochem. Cell Biol. 64, 163–181 [DOI] [PubMed] [Google Scholar]
- 7. Iida A., Kurose K., Isobe R., Akiyama F., Sakamoto G., Yoshimoto M., Kasumi F., Nakamura Y., and Emi M. (1998) Mapping of a new target region of allelic loss to a 2-cM interval at 22q13.1 in primary breast cancer. Genes Chromosomes Cancer 21, 108–112 [PubMed] [Google Scholar]
- 8. Nagahata T., Hirano A., Utada Y., Tsuchiya S., Takahashi K., Tada T., Makita M., Kasumi F., Akiyama F., Sakamoto G., Nakamura Y., and Emi M. (2002) Correlation of allelic losses and clinicopathological factors in 504 primary breast cancers. Breast Cancer 9, 208–215 [DOI] [PubMed] [Google Scholar]
- 9. Yoshimura M., Nishikawa A., Ihara Y., Nishiura T., Nakao H., Kanayama Y., Matuzawa Y., and Taniguchi N. (1995) High expression of UDP-N-acetylglucosamine: β-d mannoside β-1,4-N-acetylglucosaminyltransferase III (GnT-III) in chronic myelogenous leukemia in blast crisis. Int. J. Cancer 60, 443–449 [DOI] [PubMed] [Google Scholar]
- 10. Cancer Genome Atlas Research Network (2011) Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discovery 2, 401–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang S., Balch C., Chan M. W., Lai H. C., Matei D., Schilder J. M., Yan P. S., Huang T. H., and Nephew K. P. (2008) Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res. 68, 4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Szotek P. P., Pieretti-Vanmarcke R., Masiakos P. T., Dinulescu D. M., Connolly D., Foster R., Dombkowski D., Preffer F., Maclaughlin D. T., and Donahoe P. K. (2006) Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc. Natl. Acad. Sci. U.S.A. 103, 11154–11159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang L., Mezencev R., Bowen N. J., Matyunina L. V., and McDonald J. F. (2012) Isolation and characterization of stem-like cells from a human ovarian cancer cell line. Mol. Cell. Biochem. 363, 257–268 [DOI] [PubMed] [Google Scholar]
- 15. McAuliffe S. M., Morgan S. L., Wyant G. A., Tran L. T., Muto K. W., Chen Y. S., Chin K. T., Partridge J. C., Poole B. B., Cheng K. H., Daggett J. Jr., Cullen K., Kantoff E., Hasselbatt K., Berkowitz J., et al. (2012) Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc. Natl. Acad. Sci. U.S.A. 109, E2939–E2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. van Tetering G., van Diest P., Verlaan I., van der Wall E., Kopan R., and Vooijs M. (2009) Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem. 284, 31018–31027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu X., Fleming A., Ricketts T., Pavel M., Virgin H., Menzies F. M., and Rubinsztein D. C. (2016) Autophagy regulates Notch degradation and modulates stem cell development and neurogenesis. Nat. Commun. 7, 10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Steinman R. M., Mellman I. S., Muller W. A., and Cohn Z. A. (1983) Endocytosis and the recycling of plasma membrane. J. Cell Biol. 96, 1–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nye J. S., Kopan R., and Axel R. (1994) An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development 120, 2421–2430 [DOI] [PubMed] [Google Scholar]
- 20. Alvero A. B., Chen R., Fu H. H., Montagna M., Schwartz P. E., Rutherford T., Silasi D. A., Steffensen K. D., Waldstrom M., Visintin I., and Mor G. (2009) Molecular phenotyping of human ovarian cancer stem cells unravels the mechanisms for repair and chemoresistance. Cell Cycle 8, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kizuka Y., Kitazume S., Fujinawa R., Saito T., Iwata N., Saido T. C., Nakano M., Yamaguchi Y., Hashimoto Y., Staufenbiel M., Hatsuta H., Murayama S., Manya H., Endo T., and Taniguchi N. (2015) An aberrant sugar modification of BACE1 blocks its lysosomal targeting in Alzheimer's disease. EMBO Mol. Med. 7, 175–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Moloney D. J., Shair L. H., Lu F. M., Xia J., Locke R., Matta K. L., and Haltiwanger R. S. (2000) Mammalian Notch1 is modified with two unusual forms of O-linked glycosylation found on epidermal growth factor-like modules. J. Biol. Chem. 275, 9604–9611 [DOI] [PubMed] [Google Scholar]
- 23. Rose S. L., Kunnimalaiyaan M., Drenzek J., and Seiler N. (2010) Notch 1 signaling is active in ovarian cancer. Gynecol. Oncol. 117, 130–133 [DOI] [PubMed] [Google Scholar]
- 24. Kakuda S., and Haltiwanger R. S. (2017) Deciphering the fringe-mediated notch code: identification of activating and inhibiting sites allowing discrimination between ligands. Dev. Cell 40, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Györffy B., Lanczky A., Eklund A. C., Denkert C., Budczies J., Li Q., and Szallasi Z. (2010) An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 123, 725–731 [DOI] [PubMed] [Google Scholar]
- 26. Gyorffy B., Lánczky A., and Szállási Z. (2012) Implementing an online tool for genome-wide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocrine-related Cancer 19, 197–208 [DOI] [PubMed] [Google Scholar]
- 27. Szász A. M., Lánczky A., Nagy Á., Förster S., Hark K., Green J. E., Boussioutas A., Busuttil R., Szabó A., and Gyrffy B. (2016) Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget 7, 49322–49333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rampal R., Arboleda-Velasquez J. F., Nita-Lazar A., Kosik K. S., and Haltiwanger R. S. (2005) Highly conserved O-fucose sites have distinct effects on Notch1 function. J. Biol. Chem. 280, 32133–32140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.