Abstract
Endoplasmic reticulum–mitochondrial contacts (EMCs) regulate multiple critical cellular activities, dysregulation of which correlates with various human maladies such as neurodegenerative diseases. A new study makes use of the ascorbate peroxidase proximity-labeling proteomics approach to scrutinize the components of EMCs in live cells, leading to the identification of reticulon 1A as a novel promoter of EMCs.
Introduction
Subcellular organelles are critical sites within cells for specialized metabolism, storage, and signaling processes. These functions can be highly compartmentalized but are also often linked up to other organelles via membrane contact sites created and regulated by specific protein components, and the resultant communication is important for maintaining cellular homeostasis (1). Among organelle cross-talk, the endoplasmic reticulum (ER)3–mitochondrial contact (EMC) has drawn wide attention due to its pivotal roles in multiple biological processes. Dysregulation of EMCs has been associated with various illnesses, including neurodegenerative diseases, cancers, and metabolic diseases (2). However, little is known about the key protein components of EMCs that may play important roles in physiological and disease states. In this issue, Cho et al. (3) tackle this question by applying a proximity-labeling approach using ascorbate peroxidase (APEX) to identify the proteome of EMCs and validate one of the identified proteins reticulon 1a (RTN1A) as promoting EMC formation.
Previous studies have shown that EMCs facilitate Ca2+ and phospholipid exchange between the ER and mitochondria and play pivotal roles in mitochondrial biogenesis. In addition, EMCs have been linked to autophagy, apoptosis and the ER stress response (3, 4). Some previously defined EMC components, mutations of which either promote or impair EMCs, thus disrupting cellular homeostasis (3), have been reported to be associated with neurodegenerative diseases. Therefore, thorough study of the components of EMCs and their biological functions can both improve our understanding of cell biology processes and inform therapeutic strategies against such diseases. These studies have been relatively straightforward to conduct in yeast through genetic screening, which has resulted in the identification of components of the ER–mitochondrial encounter structure complex. However, no orthologs of these components exist in mammalian cells. Cho et al. (3) therefore adopted a different strategy of using “proximity proteomics” to systematically identify the components of mammalian EMCs. Proximity proteomics refers to a group of methods that employ enzymes fused to a protein of interest or an organelle-targeting sequence (5). The enzymes generate reactive molecules from fed substrates, and these enzymatic products then label proteins within 10–50 nm of the protein of interest due to their short half-life. Subsequent affinity purification using the introduced label followed by mass spectrometric analysis results in a snapshot of local proteins that can be used as leads for further experimental analysis.
In the current work, Cho et al. (3) used the mitochondria-targeting sequence derived from mouse mitochondrial AKAP1 (protein kinase A anchor protein 1) to direct APEX to the outer membrane of mitochondria (named Mit-APEX) and then treated cells with biotin-phenol and hydrogen peroxide to generate the reactive biotin labels. This system can only label proteins within ∼20 nm of the enzyme's location. Both ER and mitochondrial proteins are labeled at the EMC sites, whereas only mitochondrial proteins are labeled when ER is not in close contact with mitochondria. Because the proteins of interest are labeled with biotin followed by affinity purification, any endogenous biotin-binding proteins and the proteins nonspecifically bound to the affinity media will also be identified (6, 7). To distinguish the proteins of interest from the nonspecific binders, Cho et al. (3) adopted a quantitative proteomics approach called SILAC (stable isotope labeling by amino acids in cell culture) (8), in which heavy isotope–labeled amino acids are incorporated into proteins during cell culture. The authors first labeled the Mit-APEX-overexpressed HEK293T cells with heavy amino acids, whereas mock-transfected cells were cultured with light amino acids. The cells were pooled, lysed, and centrifuged to enrich the microsomal fraction containing ER material, and then affinity-purified using streptavidin magnetic beads, followed by quantitative mass spectrometric analysis. Endogenous biotin-binding proteins and proteins nonspecifically bound to the affinity media (streptavidin-magnetics beads) would have equal abundance between Mit-APEX-overexpressed cells and the mock-transfected cells, whereas the proteins specifically labeled by Mit-APEX would show a different ratio. A reverse-labeling experiment was also conducted for enhanced reliability. Approximately 1100 proteins were commonly identified in the duplicate experiments, and 405 proteins were selected as specific targets of Mit-APEX using a 2-fold cutoff threshold (Fig. 1). Gene ontology analysis was conducted, and the proteins were classified based on cellular compartment definition, resulting in 88 proteins that could be annotated as localizing to the ER, including several well-known ER–mitochondria-tethering proteins.
Figure 1.
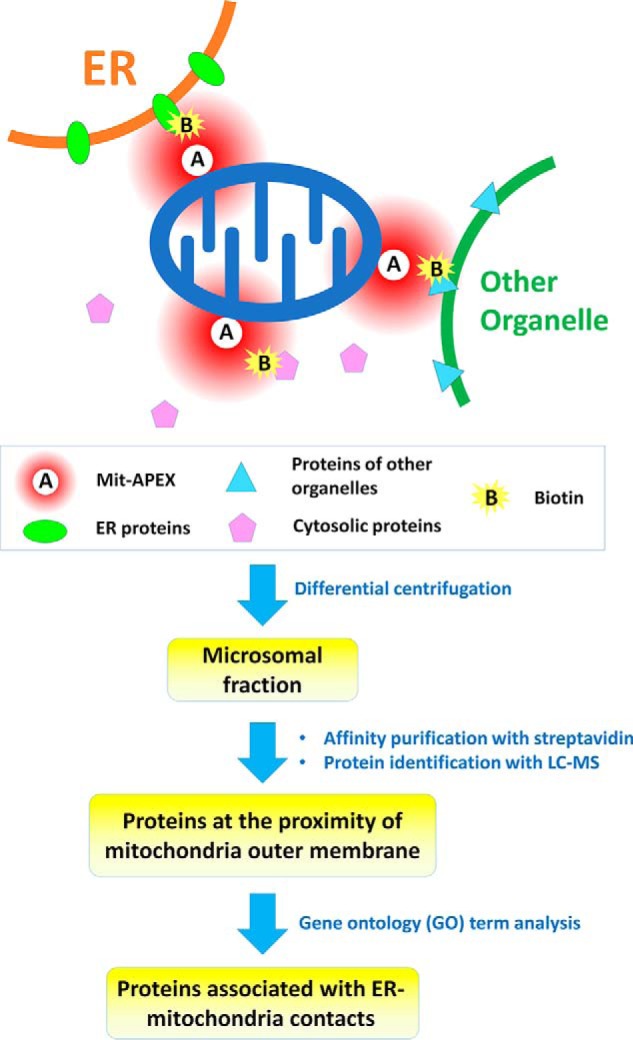
Schematic of identification of protein components of ER–mitochondrial contacts through proximity labeling. Ascorbate peroxidase is expressed on the mitochondria outer membrane (Mit-APEX) so that it can label proteins in its proximity (∼20 nm) with biotin, using biotin phenol and H2O2 as substrates. The microsomal fraction containing ER materials was isolated through differential centrifugation, and the biotin-labeled proteins at the proximity of mitochondrial outer membrane were affinity-purified and identified with LC-MS. The list of identified proteins was subjected to gene ontology term analysis, and the proteins annotated to the ER were assigned as the proteins of ER–mitochondrial contacts.
Among the identified proteins, atlastin (ATL2, ATL3) and reticulon (RTN1, RTN3) were particularly enriched. These proteins are known to play roles in forming and stabilizing tubular ER structures, suggesting to the authors that they might also be well suited to participate in the formation of EMCs. They conducted a split Renilla luciferase (Rluc8) complementation assay to test whether these proteins could enhance the formation of EMCs, in which the N-terminal part of Rluc8 was expressed on the outer membrane of mitochondria, and the C-terminal part of Rluc8 was expressed on the ER membrane. If the ER and mitochondria form EMCs, luminescence signals can be detected. Using this assay, Cho et al. (3) found that RTNs, especially RTN1A, promote the formation of EMCs, whereas ALTs do not have such a function. The authors further showed that the EMC-promoting activity relies on the N-terminal part of RTN1A by creating hybrid proteins using the C-terminal part of other RTN family members.
The current study thus comprehensively identified the proteome of the EMCs and discovered a novel key player RTN1A that can promote the formation of EMCs. Subsequent studies could examine whether mutations of RTN1A are associated with ailments such as neurodegenerative diseases or metabolic diseases through modulation of EMC formation. If so, RTN1A could be a novel target for the development of new therapeutic regimens. Other ER proteins in the identified list should also be investigated with the split Rluc8 complementation assay or other techniques to further explore their potential roles in EMC formation. The current study also identified a comprehensive list of proteins from other organelles, which may play important roles in the contacts of mitochondria with these organelles. These identified proteins provide a wealth of information for future investigation.
This work was supported in part by a Tier1 grant (R -154-000-A15-114) from the Ministry of Education and NMRC CBRG-0067-2014 from the National Medical Research Council, Singapore (to Y.-C. L.). The authors declare that they have no conflicts of interest with the contents of this article.
- ER
- endoplasmic reticulum
- EMC
- ER–mitochondrial contact
- APEX
- ascorbate peroxidase
- RTN1A
- reticulon 1a
- Rluc8
- Renilla luciferase.
References
- 1. Schrader M., Godinho L. F., Costello J. L., and Islinger M. (2015) The different facets of organelle interplay-an overview of organelle interactions. Front. Cell Dev. Biol. 3, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Filadi R., Theurey P., and Pizzo P. (2017) The endoplasmic reticulum-mitochondria coupling in health and disease: Molecules, functions and significance. Cell Calcium 62, 1–15 [DOI] [PubMed] [Google Scholar]
- 3. Cho I.-T., Adelmant G., Lim Y., Marto J. A., Cho G., and Golden J. A. (2017) Ascorbate peroxidase proximity labeling coupled with biochemical fractionation identifies promoters of endoplasmic reticulum–mitochondrial contacts. J. Biol. Chem. 292, 16382–16392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rowland A. A., and Voeltz G. K. (2012) Endoplasmic reticulum-mitochondria contacts: Function of the junction. Nat. Rev. Mol. Cell Biol. 13, 607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rees J. S., Li X. W., Perrett S., Lilley K. S., and Jackson A. P. (2015) Protein neighbors and proximity proteomics. Mol. Cell Proteomics 14, 2848–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tytgat H. L., Schoofs G., Driesen M., Proost P., Van Damme E. J., Vanderleyden J., and Lebeer S. (2015) Endogenous biotin-binding proteins: an overlooked factor causing false positives in streptavidin-based protein detection. Microb. Biotechnol. 8, 164–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zheng W., Li G., and Li X. (2015) Affinity purification in target identification: The specificity challenge. Arch. Pharm. Res. 38, 1661–1685 [DOI] [PubMed] [Google Scholar]
- 8. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., and Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]


