Abstract
Multiple reaction monitoring-MS analysis of lipid extracts from human carotid endarterectomy and carotid artery samples from young individuals consistently demonstrated the presence of bacterial serine dipeptide lipid classes, including Lipid 654, an agonist for human and mouse Toll-like receptor (TLR)2, and Lipid 430, the deacylated product of Lipid 654. The relative levels of Lipid 654 and Lipid 430 were also determined in common oral and intestinal bacteria from the phylum Bacteroidetes and human serum and brain samples from healthy adults. The median Lipid 430/Lipid 654 ratio observed in carotid endarterectomy samples was significantly higher than the median ratio in lipid extracts of common oral and intestinal Bacteroidetes bacteria, and serum and brain samples from healthy subjects. More importantly, the median Lipid 430/Lipid 654 ratio was significantly elevated in carotid endarterectomies when compared with control artery samples. Our results indicate that deacylation of Lipid 654 to Lipid 430 likely occurs in diseased artery walls due to phospholipase A2 enzyme activity. These results suggest that commensal Bacteriodetes bacteria of the gut and the oral cavity may contribute to the pathogenesis of TLR2-dependent atherosclerosis through serine dipeptide lipid deposition and metabolism in artery walls.
Keywords: atherosclerosis, phospholipase A2, Toll like receptors, mass spectrometry
Microorganisms of the phylum Bacterioidetes are prevalent in the human intestinal flora and within this phylum, members of the Bacteroides genera represent approximately one-third of the cultivable microbial flora of the human intestinal microbiome (1, 2). Periodontal diseases are also associated with increased percentages of specific Bacteroidetes species at periodontal disease sites. Among the oral Bacteroidetes, the genera Porphyromonas, Prevotella, Tannerella, and Capnocytophaga predominate. Porphyromonas gingivalis is considered to be a primary pathogen for chronic destructive periodontal disease (3). P. gingivalis has also been implicated in the development of atherosclerosis in experimental animals (4, 5) and P. gingivalis genomic products have been identified in a limited percentage of human atherosclerotic artery samples (6–9). Periodontal Bacteroidetes pathogens have been shown to invade human arterial endothelial cells in culture (10, 11). In contrast to the atherogenic members of the oral flora, little is known regarding the capacity of intestinal organisms, particularly intestinal Bacteroidetes organisms, to contribute to the development of atherosclerosis.
Serine dipeptide lipids are produced by common oral and intestinal Bacteroidetes bacteria (12) and the serine dipeptide lipids produced by P. gingivalis engage human and mouse Toll-like receptor (TLR)2 (12). The serine lipids of P. gingivalis are comprised of two classes. One class is termed Lipid 430 and contains a single hydroxyl fatty acid linked to a serine-glycine dipeptide. The other class, termed Lipid 654, contains two fatty acids. The second fatty acid of Lipid 654 is ester-linked to the core hydroxy fatty acid of Lipid 430 (12). The Lipid 654 class was originally isolated from Flavobacterium, a member of the Bacteroidetes phylum, and was termed flavolipin (13, 14). Flavolipin was originally reported to be a TLR4 ligand, the canonical receptor for lipopolysaccharide. However, our work has shown that Lipid 654 engages TLR2 and not TLR4 (12). Using ESI-multiple reaction monitoring (MRM), we have demonstrated that human blood sera samples contain detectable levels of Lipid 654 (15) and lipid extracts of diseased periodontal tissues also contain Lipid 654 (12). Therefore, accumulation of Lipid 654 in human tissues represents the presence of an exogenous TLR2 ligand produced by organisms of either the oral cavity or intestinal tract. TLR2 has been shown in experimental animal models to be an important innate immune receptor in the development of atherosclerosis (16–18) and TLR2 expression is upregulated in human endothelial cells exposed to turbulent fluid flow (19, 20). Other than bacterial peptidoglycan (21), no other exogenous TLR2 ligands of bacterial origin have been identified in human atheromas.
The first goal of this investigation was to determine whether Lipid 654 is recovered in lipid extracts of common intestinal and oral Bacteroidetes, as well as in lipid extracts of human carotid artery tissue, brain, and blood samples. Evaluation of Lipid 654 in endarterectomies included the fractionation of pooled total lipid extracts from carotid endaraterectomy samples using normal phase HPLC with neutral solvent followed by acidic refractionation of the combined fractions containing Lipid 654. Mass spectrometric analysis was used to confirm fractions containing Lipid 654. In order to evaluate metabolic breakdown of Lipid 654 to Lipid 430, ESI-MRM was used to examine lipid extracts of common oral and intestinal Bacteroides bacteria, and atherosclerotic or healthy carotid arteries and serum samples for the relative levels of Lipid 430 and Lipid 654. The median Lipid 430/Lipid 654 ratio for each sample category was calculated from electronically integrated MRM scans. To further investigate the physiological mechanism for conversion of Lipid 654 to Lipid 430 in carotid atheromas, we investigated the capacity of common lipases to cause deacylation of Lipid 654. Common phospholipase (PL) enzyme preparations as well as LPL were investigated for their capacity to convert Lipid 654 to Lipid 430. Because PLA2 was the only enzyme class within our survey that converted Lipid 654 to Lipid 430, the last goal of this investigation was to evaluate the capacity of PLA2 enzyme preparations to hydrolyze Lipid 654 to Lipid 430.
MATERIALS AND METHODS
Reagents
BBL biosate peptone, trypticase peptone, yeast extract and brain heart infusion (BHI), and HPLC solvents were obtained from Fisher Scientific. Porcine pancreatic (PP) PLA2 (≥600 U/mg protein), honey bee venom (HBV) PLA2 (Apis mellifera, 600–2,400 U/mg protein), PLA1 from Thermomyces lanuginosus, PLC (Clostridium perfringens, 10–50 U/mg protein), PLD (Arachis hypogaea, 300–700 U/mg protein), LPL (Pseudomonas sp., ≥50,000 U/mg protein), and diisopropylethylamine were obtained from Sigma. For HBV PLA2 and PP PLA2, 1 U is the amount of enzyme required to hydrolyze 1 μmol of soybean L-α-phosphatidylcholine to L-α-lysophosphatidylcholine per minute at pH 8.0 and 37°C. Human recombinant secretory (Rs) PLA2 (type V, 9.9 U/mg, ≥95% purity by SDS-PAGE) and human recombinant lipoprotein (RLp) PLA2 (platelet-activating factor acetylhydrolase, 4.36 U/mg, ≥95% purity by SDS-PAGE) were obtained as N-terminal hexahistadine-tagged proteins purified from Escherichia coli (Cayman Chemical, Ann Arbor, MI). One unit is the amount of recombinant enzyme required to release 1 μmol of 2-nitro-5-thiobenzoic acid per minute at 37°C in 25 mM Tris buffer (pH 7.5), 10 mM CaCl2, 100 mM KCL with 0.3 mM Triton X-100, 1 mg/ml BSA, 1 mM 1,2-bis(heptanoylthio) glycerol-3-phosphorylcholine (Cayman Chemical), and 0.5 mM 5,5′-dithio-bis-(2-nitrobenzoic acid (Cayman Chemical).
Bacterial growth
Bacteria were grown in broth culture, as previously described, or were grown on blood agar plates. P. gingivalis was inoculated into basal (peptone, trypticase, and yeast extract) medium supplemented with hemin and menadione (Sigma, St. Louis, MO) and BHI (22). Culture purity was verified by Gram stain, lack of growth in aerobic culture, and formation of uniform colonies when inoculated on BHI agar plates and grown under anaerobic conditions. The suspension cultures were incubated for 4 days in an anaerobic chamber flushed with N2 (80%), CO2 (10%), and H2 (10%) at 37°C and the bacteria were harvested by centrifugation (2,000 g for 2 h).
Lipid extraction, fractionation, and characterization
Lipids were extracted from lyophilized bacterial pellets using the method of Bligh and Dyer (23). Lyophilized bacterial samples (0.5 g) were dissolved in chloroform:methanol:water (1.33:2.67:1, v/v/v, 4 ml), as previously described (22). The mixture was vortexed at 15 min intervals for 2 h and the mixture was supplemented with 0.75 ml of chloroform and 0.75 ml of (2 N KCl + 0.5 N K2HPO4). The mixture was vortexed and centrifuged (2,000 g) at 20°C for 4 h. The lower organic phase was removed and dried under nitrogen.
Human brain samples from neurologically normal subjects (ages were not recorded) were obtained as fresh frozen samples (Rocky Mountain MS Center at the University of Colorado, University of Colorado Denver, Department of Neurology, Aurora, CO) and were stored frozen until processing. Fresh blood samples were collected from healthy adults using aseptic venipuncture. Carotid artery samples from otherwise healthy individuals between the ages of 16 and 36 (n = 6) were provided as fresh frozen samples from the National Disease Research Interchange, Philadelphia, PA. Carotid endarterectomy samples were surgically excised under sterile operating room conditions from older individuals, though the donors’ ages were not recorded. A portion of each endarterectomy sample was submitted for pathological evaluation and the remainder of the sample was stored frozen until tissue processing, as described below. Carotid artery, brain, and blood samples were extracted using the same Bligh and Dyer method. New glass test tubes were used for extracting lipids from human samples, as well as for storing lipid extracts. The dried lipid extracts were dissolved in chloroform and the total lipid weight was determined using a Cahn Electrobalance. For mass spectral analysis of lipid extracts from individual carotid artery samples, 1 mg of each lipid sample was aliquoted and a known amount of synthetic D9-Lipid 654 internal standard (24) (0.2 ng/sample, actual negative ion mass: m/z 662.5) was added to each artery sample. Individual human brain and serum samples were extracted using the same total lipid extraction procedure and the individual lipid samples were stored as dry pellets at −20°C until analysis.
For the determination of Lipid 654 levels in the combined endarterectomy samples, the dried lipid extracts of a dozen carotid artery endarterectomy samples (n = 12 endarterectomy samples) were reconstituted in neutral HPLC solvent (hexane:isopropanol:water, 6:8:0.75, v/v/v, 24 ml), vortexed, and combined (22). The combined endarterectomy lipid extract was centrifuged at 2,500 g for 10 min and the supernatant removed for HPLC analysis. Semipreparative HPLC fractionation was accomplished using a Shimadzu HPLC system equipped with dual pumps (LC-10ADvp), automated controller (SCL-10Avp), and in-line UV detector (SPD-10Avp). Lipids were fractionated using normal phase isocratic separation (Ascentis®Si, 25 cm × 10 mm, 5 μm; Supelco Analytical) with a solvent flow of 1.8 ml/min and 1 min fractions. HPLC solvent was composed of hexane:isopropanol:water (6:8:0.75, v/v/v). The effluent was monitored at 205 nm. For pooled lipid extracts of carotid endarterectomies, replicate fractionations were pooled and dried under nitrogen. The dried samples were reconstituted in HPLC solvent for mass spectrometric analysis, as described below. Fractions containing Lipid 654 were pooled and dried before additional HPLC fractionation.
Acidic HPLC fractionation of Lipid 654
Fractions containing Lipid 654 were further fractionated by eluting over the same HPLC column at 1.8 ml/min, but using HPLC solvent supplemented with 0.1% acetic acid. Recovery of Lipid 654 was verified by ESI-MS or MS/MS as described below.
Enzymatic hydrolysis of synthetic or P. gingivalis Lipid 654
Lipid 654 preparations were dissolved in CHCl3 and were aliquoted into 5 ml conical reaction vials and dried. The vials then received 1 ml of Tris buffer (10 mM, pH 7.4) with 150 mM NaCl and 10 mM CaCl2 (referred to as Tris buffer). The samples were sonicated for 20 s (3 watts). PP PLA2, HBV PLA2, or human Rs PLA2 enzyme preparations were added to the Lipid 654 suspensions and the samples incubated for the indicated times. Human RLp PLA2 hydrolysis of Lipid 654 was performed using Tris buffer containing 5 mM EDTA and no CaCl2. At the conclusion of enzyme incubations, the samples were acidified with glacial acetic acid (25 μl) and extracted with CHCl3 (1 ml × 3 extractions). The extracts were pooled and dried under nitrogen.
MS
Lipid samples in HPLC fractions from either semi-preparative neutral or acidic fractionations were dissolved in the neutral HPLC solvent described above and were injected over a normal phase column (Ascentis®Si, 3 cm × 2.1 mm, 5 μm; Supelco Analytical) interfaced with an API Qtrap 4000 instrument (Sciex). The lipid extracts from individual carotid arteries were first analyzed using a Shimadzu autosampler (SIL-10ADvp) with an intervening solvent wash between samples to minimize bacterial lipid carryover. Lipid samples of specific categories were analyzed as a group during the same analysis session. For the analysis of human and bacterial samples shown in Fig. 4, a manual injector was used for sample introduction. However, mass spectrometric analysis also included evaluation of 14 bacterial samples and eight serum samples using the autosampler introduction with intervening solvent washes between samples. Isocratic neutral HPLC solvent was delivered using a Shimadzu LC-10ADvp pump at a flow rate of 120 μl/min. Total ion chromatograms were acquired using negative ion mode and a mass range of 100–1,800 amu, and MS/MS acquisitions used parameters optimized for specific lipid products. Collision energies for negative ion products were typically between −30 and −55 V optimized for each precursor ion under investigation. Negative ion ESI was carried out at −4,500 V, with a declustering potential of −90 V, focusing potential of −350 V, and entrance potential of −10 V. MRM negative ion transitions for Lipid 654 were m/z 653.5-131.1, 653.5-306.2, 653.5-349.3, and 653.5-381.4 and ion transitions for Lipid 430 were m/z 430.2-140.9, 430.3-173.1, and 430.3-382.3. Lipid 430/Lipid 654 ratios were calculated using only the m/z 430.3-382.3 and 653.5-381.4 transitions. The synthetic D9-Lipid 663 internal standard was monitored using m/z 662.5-390.5, 662.5-358.4, and 662.5-315.3 transitions.
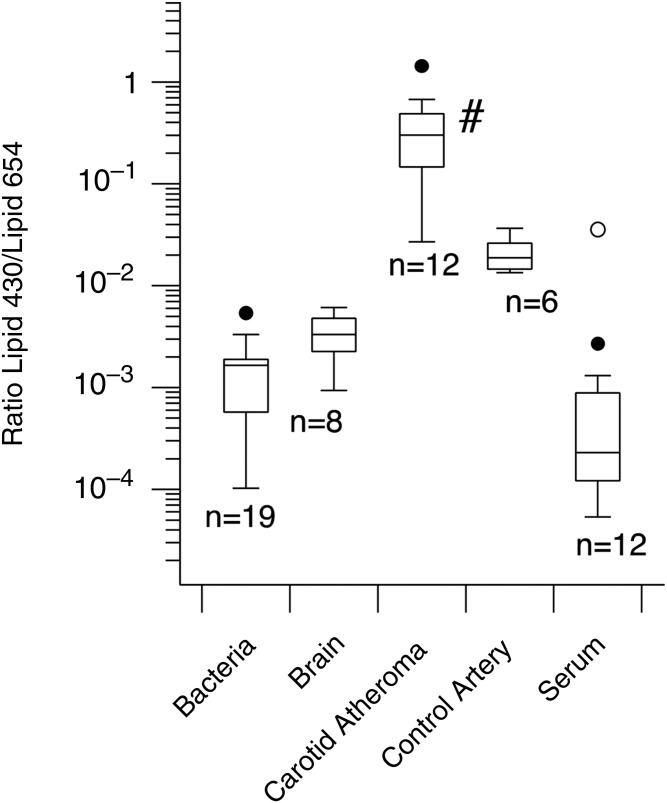
Fig. 4.
Lipid 430/Lipid 654 ratios for carotid endarterectomies, normal carotid artery samples, human brain samples, bacteria lipid extracts, and serum from healthy subjects. Lipid samples were extracted as described in the Materials and Methods and were analyzed using ESI-MRM. Lipid 430 and Lipid 654 were quantified using ESI-MRM and ratios were calculated from electronically integrated peaks. The results are depicted as Box plots and show the median ratios of Lipid 430/Lipid 654 for each category of samples (n = number of samples). Kruskal-Wallis ANOVA revealed significant differences between sample medians (P = 2.199 × 10−8). The median Lipid 430/Lipid 654 ratio for carotid endarterectomy lipid extracts (#) was significantly elevated over the median ratio in bacterial samples (P = 4.83 × 10−6), brain samples (P = 0.00021), control arteries (P = 0.00145), and serum samples (P = 0.00005) by Mann-Whitney U test.
Synthesis of Lipid 654 and Lipid 430
The (3R)- and (3S)-(15-methyl-3-((13-methyltetradecanoyl)oxy)hexadecanoyl)-glycyl-L-serine stereoisomers, abbreviated as L-serine-(R)- and -(S)-Lipid 654, were prepared according to a recent report. The details of the synthesis and verification of the enantio-enriched (R)- and (S)-Lipid 654 diastereomers can be found in (25). Details of the synthesis of diastereomeric Lipid 430 can also be found in (25).
Statistical analysis
Statistical testing of replicate samples revealed that results depicted in Figs. 3 and 4 were not normally distributed and were depicted as Box plots. Statistical testing included Kruskal-Wallis ANOVA with pairwise comparisons using Mann Whitney U tests. Results in Fig. 5 were normally distributed (depicted as the mean ± SD) and were analyzed by one factor ANOVA with pairwise comparisons using the Fisher LSD test.
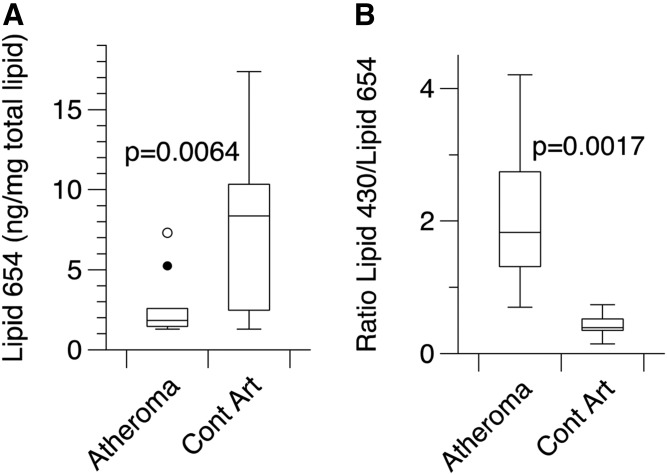
Fig. 3.
Recovery of Lipid 654 in lipid extracts of carotid endarterectomy samples or carotid artery samples from young individuals. Carotid endarterectomy samples (Atheromas, n = 10) were recovered at the time of surgical excision; a portion of each endarterectomy sample was removed for pathological evaluation and the remainder of the sample was frozen until processing. Arteries from young adults (Cont Art, n = 6) were recovered at necropsy and fresh frozen as processed at the National Disease Research Interchange. Frozen samples were thawed and minced into small pieces. The samples were extracted for 3 days in organic solvent according to the procedure of Bligh and Dyer and the recovered lipids were dried under a stream of nitrogen. The recovered lipids were dissolved in chloroform and 1 mg was aliquoted and was supplemented with D9-Lipid 654 internal standard (0.2 ng/mg total lipid). Samples were then dissolved in 500 μl of HPLC solvent and analyzed by MRM-MS, as described in the Materials and Methods. An autosampler was used for sample introduction to the mass spectrometer and each sample was followed by a solvent wash to ensure negligible lipid carryover between samples. Each P value was determined using the Mann-Whitney U test. A: Comparison of the Lipid 654 levels versus total lipid (in nanograms per milligram). B: Comparison of Lipid 430/Lipid 654 ratios in the same artery lipid extracts.
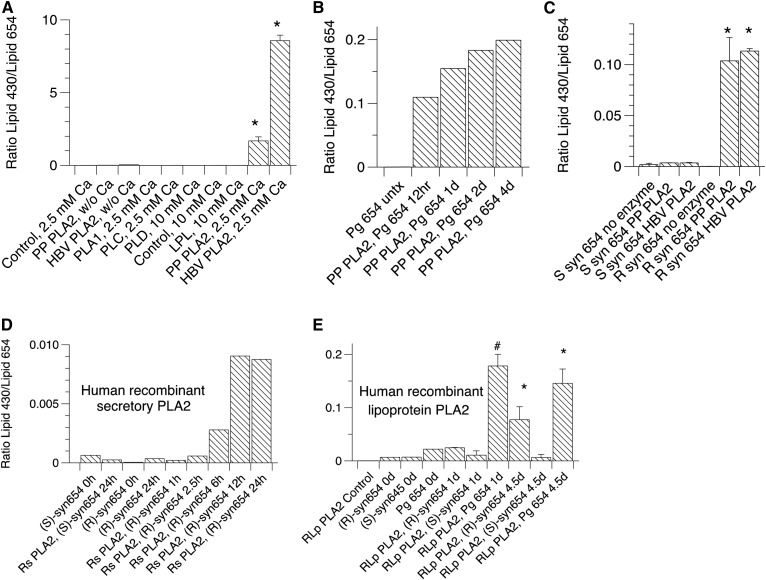
Fig. 5.
Hydrolysis of (R)- or (S)-synthetic or bacterial Lipid 654 by various PLA2 lipid preparations. A: P. gingivalis Lipid 654 was aliquoted (10 μg per vial) and sonicated for 30 s (3 watts) in 10 mM Tris (pH 7.4) with the indicated concentrations of CaCl2. For samples with no CaCl2 added, the medium was supplemented with 5 mM EDTA (pH 7.4). The samples were stirred for 4 days at room temperature and were acidified with acetic acid followed immediately by extraction with CHCl3 (3 × 1 ml extractions). The extracts were dried and analyzed for levels of Lipid 654 and Lipid 430 using ESI-MRM. PP PLA2, HBV PLA2, LPL, and PLA1, PLC, and PLD (100 U each) were used for these hydrolyses. Results are depicted as mean ± SD (n = 2). Significant differences were noted for PP PLA2 hydrolysis or HBV hydrolysis of P. gingivalis Lipid 654 (*) versus controls (by one factor ANOVA with Fisher LSD pairwise comparisons). For (B) through (E), either bacterial or synthetic Lipid 654 was aliquoted (5 μg per vial) and sonicated for 30 s (3 watts) in 10 mM Tris (pH 7.4) with 10 mM CaCl. The samples were incubated for 3 days at room temperature unless otherwise noted, and were extracted and analyzed as described above. B: Time course of P. gingivalis Lipid 654 hydrolysis by PP PLA2 (100 U per sample). C: Stereoselective hydrolysis of synthetic (R)-Lipid 654 over synthetic (S)-Lipid 654 by either PP PLA2 or HBV PLA2. Results are depicted as mean ± SD (n = 3). Significant differences were noted for PP PLA2 hydrolysis or HBV hydrolysis of (R)-synthetic Lipid 654 [(R)-syn654] (*) versus (S)-synthetic Lipid 654 [(S)-syn654] and controls (by one factor ANOVA with Fisher LSD pairwise comparisons). D: Time course of human Rs PLA2 hydrolysis of (R)-syn654. (S)-syn654 is shown for comparison. E: Time course of human RLp PLA2 hydrolysis of P. gingivalis Lipid 654, (R)-syn654, and (S)-syn654. Results are depicted as mean ± SD (n = 2). Significant differences were noted for RLp PLA2 hydrolysis of P. gingivalis Lipid 654 (Pg 654) or (R)-syn654 versus (S)-syn654 (*) or between Pg 654 versus (S)-syn654 (#) (by one factor ANOVA with Fisher LSD pairwise comparisons).
RESULTS
Recovery of Lipid 430 and Lipid 654 in common Bacteroidetes bacteria and carotid endarterectomy lipid extracts
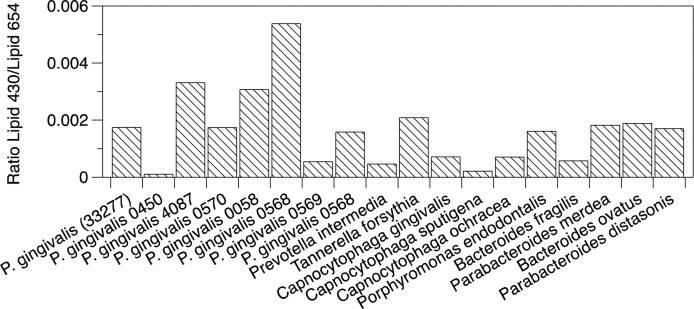
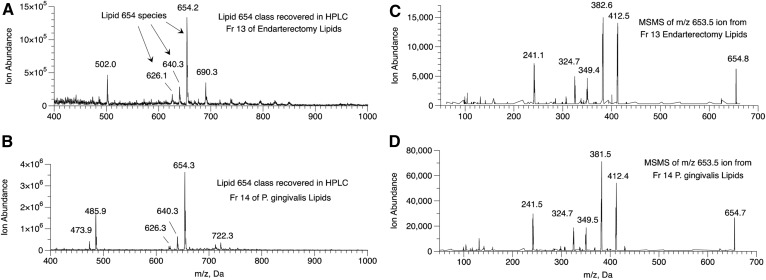
Total lipid extracts from fourteen oral Bacteroidetes bacterial species and four intestinal Bacteroidetes bacterial species were analyzed for Lipid 430 and Lipid 654 as described in the Materials and Methods. The Lipid 430/Lipid 654 ratios ranged from 0.0001 to 0.0055 (Fig. 1) and these ratios were averaged for all bacterial samples. Next, lipid extracts from carotid endarterectomies (n = 12) were pooled and fractionated first by normal phase HPLC, as previously described (26). The fractions containing Lipid 654 were combined and refractionated over the same column using HPLC solvent supplemented with 0.1% acetic acid (26). In order to minimize the likelihood of bacterial lipid carryover from columns previously used for bacterial lipid separations, a separate semipreparative HPLC column was used only for the fractionation of carotid artery lipids. Also, solvent wash runs were used to confirm the lack of carryover between runs. Lipid 654 was recovered primarily in one fraction (HPLC fraction 13) and Fig. 2 shows the MS spectrum indicating the characteristic three negative ions for the Lipid 654 class (Fig. 2A). Lipids of P. gingivalis were fractionated using the same method and the maximum abundance of the Lipid 654 lipid class was observed in HPLC fraction 14 (Fig. 2B). MS/MS analysis of the most abundant species of Lipid 654 (negative molecular ion of m/z 653.5) recovered in fraction 13 of pooled endarterectomy lipids revealed the spectrum shown in Fig. 2C. Figure 2D shows the MS/MS spectrum of the most abundant molecular ion of the Lipid 654 class recovered in HPLC fraction 14 of P. gingivalis lipids. This evidence demonstrates that lipid extracts of human carotid endarterectomies were contaminated with the Lipid 654 class.
Fig. 1.
Recovery of Lipid 430 and Lipid 654 in common Bacteroidetes bacteria. The listed bacteria were prepared either in broth culture or were streaked on blood agar plates. Bacteria grown in broth culture were harvested during the late log phase of growth. The intestinal species, Bacteroides fragilis, Bacteroides ovatus, Parabacteroides medea, and Parabacteroides distasonis, were grown on blood agar plates. Bacterial samples were collected by centrifugation or scraping colonies from blood agar plates. Total lipids were extracted and the samples were analyzed for Lipid 654 and Lipid 430 using ESI-MRM. Lipid 430/Lipid 654 ratios were calculated from electronically integrated peaks and are shown for oral and gastrointestinal Bacteroidetes.
Fig. 2.
Lipid 654 recovered in pooled carotid atheroma lipid extracts. A: Lipid extracts from 12 carotid endarterectomy samples were pooled and fractionated using first neutral HPLC solvent fractionation followed by pooling fractions containing Lipid 654 and refractionating with acidic HPLC solvent. Lipid 654 was recovered primarily in one HPLC fraction (HPLC fraction 13). ESI-MS of HPLC fraction 13 revealed the masses of the three characteristic species of the Lipid 654 class (m/z 654.2, 640.3, and 626.1). B: ESI-MS of HPLC fraction 14 lipids of P. gingivalis lipids demonstrating the characteristic species of Lipid 654. ESI-MS/MS of the m/z 653.5 molecular ion from HPLC fraction 13 of carotid endarterectomy lipids (C) and ESI-MS/MS spectrum of the m/z 653.5 molecular ion from HPLC fraction 14 of P. gingivalis lipids (D).
Quantifying Lipid 654 levels in lipid extracts of carotid artery samples
We next examined lipid extracts of individual carotid endarterectomy samples (n = 10) and the individual control and endarterectomy carotid artery lipid extracts were first analyzed using an autosampler introduction system with a solvent wash between individual artery lipid samples. Using this approach, all carotid artery lipid extracts were shown to contain detectable levels of Lipid 654. Each milligram aliquot of arterial lipid extract was spiked with D9-Lipid 654 internal standard (0.2 ng, negative ion mass: m/z 662.5) in order to quantify the actual level of Lipid 654 in these arterial lipid extracts. Figure 3A shows the median level of Lipid 654 per milligram in total lipid extracts of carotid endaraterectomy or control carotid artery samples and Fig. 3B shows the range of Lipid 430/Lipid 654 ratios measured in the same samples. The median level of Lipid 654 per milligram of total lipid was 4-fold less in carotid endarterectomy lipid samples compared with control artery samples. Figure 3B shows, on average, that the atheroma samples had a median Lipid 430/Lipid 654 ratio that was at least 10-fold higher than that in control carotid artery samples.
Recovery of Lipid 430 and Lipid 654 in bacterial, human brain, and serum samples
The median Lipid 430/Lipid 654 ratios for lipid extracts of carotid endarterectomy and control carotid artery samples were compared with bacterial, brain, and serum lipid extracts (Fig. 4). These lipid samples, including the carotid lipid extracts, were manually injected and all samples within a given category were analyzed as a group during a single session. The median Lipid 430/Lipid 654 ratio observed in bacterial lipid extracts was 180 times less than that observed in carotid endarterectomies, 11 times less than that observed in carotid artery samples from healthy young subjects, and 7 times greater than that observed in serum samples from healthy subjects. Lipid extracts from human brain samples revealed a median Lipid 430/Lipid 654 ratio approximately 90 times less than that observed in carotid endarterectomies. The differences between the medians for these sample categories are significant using the Kruskal-Wallace ANOVA (P = 2.199 × 10−8). The carotid artery samples injected by autosampler (Fig. 3) showed essentially the same relative decrease in the Lipid 430/Lipid 654 ratio as that observed with manually injected lipid extracts (Fig. 4). Reanalysis of most of the bacterial lipid extracts using an autosampler introduction with intervening solvent washes yielded a median Lipid 430/Lipid 654 ratio of 0.00112 (n = 14 bacterial lipid samples). Most of the serum samples were also reanalyzed using autosampler introduction and this revealed a median Lipid 430/Lipid 654 ratio of 0.0035 (n = 8).
Hydrolysis of Lipid 654 to Lipid 430 by lipase enzymes
Hydrolysis of Lipid 654 to Lipid 430 in diseased arteries will depend on the presence of host enzymes that can mediate deacylation of Lipid 654. Macrophage activation is associated with increased activity of PLA1, PLA2, PLC, and PLD. In addition, LPL has also been implicated in altering the course of atherosclerosis. The next goal of this investigation was to evaluate commercial preparations of these enzymes for their capacity to hydrolyze Lipid 654 (Fig. 5A). This survey revealed that only PLA2 preparations, including PP and HBV PLA2 enzyme preparations, hydrolyzed Lipid 654 to Lipid 430. These enzymes represent different isoforms of secretory PLA2 and, therefore, both require calcium for hydrolysis of phospholipids. Lipid 654 hydrolysis to Lipid 430 by either of these enzyme preparations required calcium as shown in Fig. 5A. However, PLA1, PLC, PLD, and LPL enzyme preparations in the presence of calcium did not promote hydrolysis of Lipid 654 to Lipid 430.
We next compared these secretory PLA2 preparations with human Rs and RLp PLA2 preparations for their capacity to hydrolyze Lipid 654. Because PLA2 enzymes are known to hydrolyze phospholipids containing only L-glycerol, we also evaluated whether these PLA2 preparations were enantioselective in hydrolyzing Lipid 654. Highly enriched bacterial (Fig. 2B) or synthetic Lipid 654 was aliquoted into vials, dried, and sonicated in Tris buffer, as described in the Materials and Methods. According to the time course of P. gingivalis Lipid 654 hydrolysis by PP PLA2, approximately 12 h was required for 50% hydrolysis of Lipid 654 (Fig. 5B). Figure 5C shows that only synthetic L-serine-(R)-Lipid 654, but not L-serine-(S)-Lipid 654, was hydrolyzed by either PP PLA2 or HBV PLA2. Treatment of Lipid 654 isolated from pooled endarterectomy samples (shown in Fig. 2A, B) for 3 days with either PP PLA2 or HBV PLA2 revealed a 9.655-fold or a 4.25-fold increase, respectively, in the Lipid 430/Lipid 654 ratios, indicating that at least a portion of the Lipid 654 isolated from human endarterectomies is comprised of (R)-Lipid 654. For Fig. 5D, L-serine-(R)- or -(S)-Lipid 654 (5 μg per vial or 7.65 mM) was sonicated in specified vials containing 1 ml Tris buffer and was supplemented with human Rs PLA2 (0.024 U per sample). For Fig. 5E, L-serine-(R)- or -(S)-Lipid 654 (5 μg per vial) were sonicated in vials containing 1 ml Tris buffer containing 5 mM EDTA and no calcium, followed by the addition of human RLp PLA2 (0.044 U added per sample). These samples were incubated at 37°C for the indicated times. Both recombinant human secretory and lipoprotein PLA2 preparations hydrolyzed Lipid 654 in a manner consistent with the known stereoselective hydrolysis of phospholipids containing only L-glycerol. Finally, this evidence shows that mammalian PLA2 enzymes are capable of hydrolyzing Lipid 654 to Lipid 430, providing a possible explanation for the observed increase in the Lipid 430 to Lipid 654 ratio in carotid atheromas.
DISCUSSION
Serine dipeptide lipids produced by oral and intestinal Bacteroidetes bacteria are consistently recovered in lipid extracts of carotid arteries, including atherosclerotic artery samples. The absolute amount of Lipid 654 as a percentage of the total arterial lipid was 4-fold lower in the carotid endarterectomies compared with the control carotid artery samples, which would argue that Lipid 654 is not important in atherogenesis. However, the lower level of Lipid 654 recovered in endarterectomy lipid extracts could be related to the substantial accumulation of cholesterol, triglycerides, and other atherogenic lipids in the carotid endarterectomies. By contrast, if atherogenic lipids and Lipid 654 accumulate coincidentally, the Lipid 654 levels would remain essentially unchanged as a percentage of the total lipid present. Furthermore, if atherogenic lipid accumulation causes a net decrease in the amount of Lipid 654 in lipid extracts of diseased arteries, it should also be reflected by a similar dilution of Lipid 430 in the same samples. Because the median Lipid 654 level in endarterectomy samples decreased while the median Lipid 430/Lipid 654 ratio increased when compared with control artery samples, this argues that Lipid 654 is deacylated to Lipid 430, resulting in a net loss of Lipid 654. This evidence suggested that conversion of Lipid 654 to Lipid 430 was increased in carotid endarterectomies when compared with carotid artery samples. The median Lipid 430/Lipid 654 ratio observed in Bacteroidetes bacteria is also inconsistent with the direct invasion of artery walls by live bacteria. Otherwise, the Lipid 430/Lipid 654 ratios would be similar between carotid artery and bacterial samples. Finally, the median Lipid 430/Lipid 654 ratio in carotid endarterectomy samples was significantly higher than that observed in human brain samples, despite the known vascularity of brain tissue. Therefore, the increased level of Lipid 430 relative to Lipid 654 observed in carotid endarterectomy samples is not reflected by similar changes in blood or brain tissues, suggesting a local conversion process in diseased arteries.
We previously reported that Lipid 654 mediates effects on osteoblasts through engagement of TLR2 (26). However, Lipid 430 is at least twice as potent on a weight basis in mediating cellular effects on bone cells when compared with Lipid 654. While purified bacterial Lipid 654 can mediate effects on cells, it is not clearly established that the actual metabolically active lipid constituent is in fact Lipid 654. The present report demonstrated that Lipid 430 can be produced from Lipid 654 by enzymatic treatment with PLA2. The present study also showed that the apparent Lipid 654 breakdown to Lipid 430 is most prominent in diseased artery samples, but the exact location of the lipid deposition and metabolism was not clear. Because commercially available PLA2 preparations require days of exposure to observe substantial hydrolysis of Lipid 654, it does not appear feasible to test the hydrolysis of Lipid 654 in artery tissue homogenates given the likelihood of bacterial growth and production of necrosis-related proteolytic enzymes. Because these bacterial serine lipids are amenable to mass spectrometric detection and quantification, multidimensional MALDI-TOF evaluation of serial sections of endarterectomy samples should clarify the location and relative levels of bacterial serine lipid deposits in the artery walls relative to the location of atheromas. This experimental approach is now feasible.
For the evaluation of enzymatic deacylation of Lipid 654 to Lipid 430, we examined the capacity of several common PL enzyme preparations (PLA1, PLA2, PLC, and PLD), as well as LPL, to hydrolyze Lipid 654 to Lipid 430. These PL enzyme classes were selected either because of their expression in activated macrophages or because of their association with atherosclerosis. LPL was included because of its association with atherosclerosis (27), though reports vary as to whether LPL is proatherogenic (28) or protective against atherosclerosis (29). We show here that recombinant PLA2 preparations, including human Rs PLA2 type V and human RLp PLA2, catalytically hydrolyze Lipid 654 to Lipid 430, despite the fact that these serine lipids are not phospholipids. Isoforms of secretory PLA2 previously identified in atherosclerotic lesions include PLA2 IIA, IID, IIE, III, V, and X (30). Of these isoforms, PLA2 isoform III (HBV PLA2) and human Rs PLA2 V were shown to hydrolyze Lipid 654. Though PP PLA2 is a type IB form of secretory PLA2 and has not been reported in atherosclerotic lesions, it also hydrolyzed Lipid 654 to Lipid 430. Secretory PLA2 is reported to be elevated in atherosclerotic arteries in conjunction with chronic inflammation associated with atheroma formation (31, 32). Lipoprotein PLA2 expression in artery walls and blood is associated with increased risk of atherosclerosis development and serious cardiac sequelae (33–35). While increased PLA2 activity is associated with increased arachidonic acid release and subsequent prostaglandin production, the evidence presented here suggests that increased PLA2 activity could account for the increased conversion of Lipid 654 to Lipid 430 in diseased arteries. Although it is not established whether Lipid 430 can activate macrophages, endothelial cells, or arterial smooth muscle cells, Lipid 430 is reported to activate human embryonic kidney cells expressing either human or mouse TLR2 (12) and will inhibit bone cell differentiation (26). Because the shift in the Lipid 430 to Lipid 654 ratio was most pronounced in carotid endarterectomies, our results suggest that Lipid 654 hydrolysis to Lipid 430 through PLA2 enzyme activity may promote atherogenesis through engagement of TLR2.
TLRs, in particular TLR2 and TLR4, are reported to be important in development of atherosclerosis (16, 18). TLR9 appears to play a lesser role in atherosclerosis promotion. Lipid 654 does not engage TLR4 (12). Lipid 654 exposure to artery walls is potentially important in atherogenesis for several reasons. First, increased TLR2 expression is observed in arterial endothelial cells in areas of nonlaminar flow of blood, such as artery curvatures, bifurcations, and other locations where atheromas are typically observed (19, 20). Second, genetically engineered mice deficient in LDL or ApoE receptor function demonstrate increased susceptibility to atherosclerosis that is reversed if mice are simultaneously rendered TLR2 deficient (18, 20). Finally, TLR2 engagement is reported to increase PLA2 expression in monocytes (36), macrophages (37), and synoviocytes (38). Though PLA2 enzymes have been implicated in promoting atherogenesis, recent reports indicate that pharmacological inhibition of PLA2 has little value in preventing serious cardiovascular events secondary to atherosclerosis. This is particularly true for darapladib that specifically inhibits lipoprotein PLA2 (39–41). A recent clinical trial was terminated because of an increased risk of cardiovascular events for subjects taking the secretory PLA2 inhibitor, varespladib (41). However, PLA2 inhibitor trials included relatively short-term treatment regimens. If PLA2-mediated hydrolysis of Lipid 654 to Lipid 430 is important in atherogenesis, inhibition of PLA2 would be required for extended periods to show benefit. Evidence presented here suggests that bacterial serine lipids accumulate in carotid arteries early in life before substantial atherogenesis is evident and the conversion to Lipid 430 is increased with atheroma formation, suggesting greater PLA2 activity in atheromas.
As shown here, Lipid 654 and related lipids are produced by many members of the phylum Bacteroidetes within the human microbiome and, in the case of the gastrointestinal tract, the biomass of Bacteroides species exposed to the host is substantial. Oral species in the phylum Bacteroidetes could also contribute to the accumulation of serine dipeptide lipids in arteries, but the total oral Bacteroidetes phylum biomass is considerably less than that of the gut. Normal body function is expected to lead to transient bacteremias from either oral or gut microbiomes, but passive transfer/uptake of bacterial lipids into the host cannot be excluded. However, secondary prevention of atherosclerosis using antibiotics has shown inconsistent results (42, 43). Dietary restrictions or modifications that minimize colonization of the oral cavity or intestinal tract with species of the phylum Bacteroidetes have not been reported. Given that serine dipeptide lipids of Bacteroidetes bacteria are recovered in readily detectable amounts in endarterectomies and are likely metabolized within mammalian tissues, it becomes important to establish whether these lipids promote atherogenesis in experimental animals. Furthermore, it is important to determine whether vascular and immune cells are capable of metabolizing these lipids and to characterize the biological properties of these lipids on macrophages and cells of the vascular wall. The fact that these lipids appear in lipid extracts of carotid arteries in relatively young individuals suggests that the process of bacterial lipid deposition is followed by subsequent metabolism later in life in association with the atherogenesis process. It is also not clear whether the cholesterol and oxidized lipid product accumulation in artery walls is mechanistically related to bacterial lipid deposition and/or engagement of TLR2. TLR2 is expressed in arteries in part because of the flow characteristics of blood, but this does not necessarily correlate with cell activation. Furthermore, expression of TLR2 in different cell types associated with atheroma formation could vary depending on the location of the deposited bacterial lipids. Given that bacterial serine dipeptide lipids are now established to contaminate human arteries, the question of how these lipids are transported and deposited in arteries becomes important. Future studies will address these possibilities.
Footnotes
Abbreviations:
- BHI
- brain heart infusion
- HBV
- honey bee venom
- MRM
- multiple reaction monitoring
- PL
- phospholipase
- PP
- porcine pancreatic
- RLp
- recombinant lipoprotein
- Rs
- recombinant secretory
- TLR
- Toll-like receptor
This work was supported by National Institutes of Health Grant DE021055 to F.C.N. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Salyers A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38: 293–313. [DOI] [PubMed] [Google Scholar]
- 2.Eckburg P. B., Bik E. M., Bernstein C. N., Purdom E., Dethlefsen L., Sargent M., Gill S. R., Nelson K. E., and Relman D. A.. 2005. Diversity of the human intestinal microbial flora. Science. 308: 1635–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.How K. Y., Song K. P., and Chan K. G.. 2016. Porphyromonas gingivalis: an overview of periodontopathic pathogen below the gum line. Front. Microbiol. 7: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qi M., Miyakawa H., and Kuramitsu H. K.. 2003. Porphyromonas gingivalis induces murine macrophage foam cell formation. Microb. Pathog. 35: 259–267. [DOI] [PubMed] [Google Scholar]
- 5.Gibson F. C. III, and Genco C. A.. 2007. Porphyromonas gingivalis mediated periodontal disease and atherosclerosis: disparate diseases with commonalities in pathogenesis through TLRs. Curr. Pharm. Des. 13: 3665–3675. [DOI] [PubMed] [Google Scholar]
- 6.Haraszthy V. I., Zambon J. J., Trevisan M., Zeid M., and Genco R. J.. 2000. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 71: 1554–1560. [DOI] [PubMed] [Google Scholar]
- 7.Stelzel M., Conrads G., Pankuweit S., Maisch B., Vogt S., Moosdorf R., and Flores-de-Jacoby L.. 2002. Detection of Porphyromonas gingivalis DNA in aortic tissue by PCR. J. Periodontol. 73: 868–870. [DOI] [PubMed] [Google Scholar]
- 8.Fiehn N. E., Larsen T., Christiansen N., Holmstrup P., and Schroeder T. V.. 2005. Identification of periodontal pathogens in atherosclerotic vessels. J. Periodontol. 76: 731–736. [DOI] [PubMed] [Google Scholar]
- 9.Armingohar Z., Jørgensen J. J., Kristoffersen A. K., Abesha-Belay E., and Olsen I.. 2014. Bacteria and bacterial DNA in atherosclerotic plaque and aneurysmal wall biopsies from patients with and without periodontitis. J. Oral Microbiol. 6: 23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorn B. R., Dunn W. A. Jr., and Progulske-Fox A.. 1999. Invasion of human coronary artery cells by periodontal pathogens. Infect. Immun. 67: 5792–5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deshpande R. G., Khan M. B., and Genco C. A.. 1998. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect. Immun. 66: 5337–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clark R. B., Cervantes J. L., Maciejewski M. W., Farrokhi V., Nemati R., Yao X., Anstadt E., Fujiwara M., Wright K. T., Riddle C., et al. 2013. Serine lipids of Porphyromonas gingivalis are human and mouse Toll-like receptor 2 ligands. Infect. Immun. 81: 3479–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomi K., Kawasaki K., Kawai Y., Shiozaki M., and Nishijima M.. 2002. Toll-like receptor 4-MD-2 complex mediates the signal transduction induced by flavolipin, an amino acid-containing lipid unique to Flavobacterium meningosepticum. J. Immunol. 168: 2939–2943. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki K., Gomi K., Kawai Y., Shiozaki M., and Nishijima M.. 2003. Molecular basis for lipopolysaccharide mimetic action of Taxol and flavolipin. J. Endotoxin Res. 9: 301–307. [DOI] [PubMed] [Google Scholar]
- 15.Farrokhi V., Nemati R., Nichols F. C., Yao X., Anstadt E., Fujiwara M., Grady J., Wakefield D., Castro W., Donaldson J., et al. 2013. Bacterial lipodipeptide, Lipid 654, is a microbiome-associated biomarker for multiple sclerosis. Clin. Transl. Immunology. 2: e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moghimpour Bijani F., Vallejo J. G., and Rezaei N.. 2012. Toll-like receptor signaling pathways in cardiovascular diseases: challenges and opportunities. Int. Rev. Immunol. 31: 379–395. [DOI] [PubMed] [Google Scholar]
- 17.Madan M., and Amar S.. 2008. Toll-like receptor-2 mediates diet and/or pathogen associated atherosclerosis: proteomic findings. PLoS One. 3: e3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mullick A. E., Tobias P. S., and Curtiss L. K.. 2005. Modulation of atherosclerosis in mice by Toll-like receptor 2. J. Clin. Invest. 115: 3149–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Curtiss L. K., and Tobias P. S.. 2009. Emerging role of Toll-like receptors in atherosclerosis. J. Lipid Res. 50 (Suppl.): S340–S345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtiss L. K., Black A. S., Bonnet D. J., and Tobias P. S.. 2012. Atherosclerosis induced by endogenous and exogenous Toll-like receptor (TLR)1 or TLR6 agonists. J. Lipid Res. 53: 2126–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laman J. D., Schoneveld A. H., Moll F. L., van Meurs M., and Pasterkamp G.. 2002. Significance of peptidoglycan, a proinflammatory bacterial antigen in atherosclerotic arteries and its association with vulnerable plaques. Am. J. Cardiol. 90: 119–123. [DOI] [PubMed] [Google Scholar]
- 22.Nichols F. C., Riep B., Mun J., Morton M. D., Bojarski M. T., Dewhirst F. E., and Smith M. B.. 2004. Structures and biological activity of phosphorylated dihydroceramides of Porphyromonas gingivalis. J. Lipid Res. 45: 2317–2330. [DOI] [PubMed] [Google Scholar]
- 23.Bligh E. G., and Dyer W. J.. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917. [DOI] [PubMed] [Google Scholar]
- 24.Dietz C., Clark R. B., Nichols F. C., and Smith M. B.. 2017. Convergent synthesis of a deuterium labeled serine dipeptide lipid for analysis of biological samples. J. Labelled Comp. Radiopharm. 60: 274–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dietz C., Hart T. K., Nemati R., Yao X., Nichols F. C., and Smith M. B.. 2016. Structural verification via convergent total synthesis of dipeptide-lipids isolated from Porphyromonas gingivalis. Tetrahedron. 72: 7557–7569. [Google Scholar]
- 26.Wang Y. H., Nemati R., Anstadt E., Liu Y., Son Y., Zhu Q., Yao X., Clark R. B., Rowe D. W., and Nichols F. C.. 2015. Serine dipeptide lipids of Porphyromonas gingivalis inhibit osteoblast differentiation: relationship to Toll-like receptor 2. Bone. 81: 654–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobayashi J., and Mabuchi H.. 2015. Lipoprotein lipase and atherosclerosis. Ann. Clin. Biochem. 52: 632–637. [DOI] [PubMed] [Google Scholar]
- 28.Pentikäinen M. O., Oksjoki R., Oörni K., and Kovanen P. T.. 2002. Lipoprotein lipase in the arterial wall: linking LDL to the arterial extracellular matrix and much more. Arterioscler. Thromb. Vasc. Biol. 22: 211–217. [DOI] [PubMed] [Google Scholar]
- 29.Tsutsumi K. 2003. Lipoprotein lipase and atherosclerosis. Curr. Vasc. Pharmacol. 1: 11–17. [DOI] [PubMed] [Google Scholar]
- 30.Rosenson R. S., and Hurt-Camejo E.. 2012. Phospholipase A2 enzymes and the risk of atherosclerosis. Eur. Heart J. 33: 2899–2909. [DOI] [PubMed] [Google Scholar]
- 31.Bobryshev Y. V., Crozier J. A., Lord R. S., Tran D., Jamal O. S., Parsson H. N., and Scott K. F.. 1996. Expression of secretory group II phospholipase A2 by CD1a positive cells-in human atherosclerotic plaques. Atherosclerosis. 127: 283–285. [DOI] [PubMed] [Google Scholar]
- 32.Menschikowski M., Hagelgans A., and Siegert G.. 2006. Secretory phospholipase A2 of group IIA: is it an offensive or a defensive player during atherosclerosis and other inflammatory diseases? Prostaglandins Other Lipid Mediat. 79: 1–33. [DOI] [PubMed] [Google Scholar]
- 33.Häkkinen T., Luoma J. S., Hiltunen M. O., Macphee C. H., Milliner K. J., Patel L., Rice S. Q., Tew D. G., Karkola K., and Ylä-Herttuala S.. 1999. Lipoprotein-associated phospholipase A(2), platelet-activating factor acetylhydrolase, is expressed by macrophages in human and rabbit atherosclerotic lesions. Arterioscler. Thromb. Vasc. Biol. 19: 2909–2917. [DOI] [PubMed] [Google Scholar]
- 34.Kolodgie F. D., Burke A. P., Skorija K. S., Ladich E., Kutys R., Makuria A. T., and Virmani R.. 2006. Lipoprotein-associated phospholipase A2 protein expression in the natural progression of human coronary atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 26: 2523–2529. [DOI] [PubMed] [Google Scholar]
- 35.Fenning R. S., Burgert M. E., Hamamdzic D., Peyster E. G., Mohler E. R., Kangovi S., Jucker B. M., Lenhard S. C., Macphee C. H., and Wilensky R. L.. 2015. Atherosclerotic plaque inflammation varies between vascular sites and correlates with response to inhibition of lipoprotein-associated phospholipase A2. J. Am. Heart Assoc. 4: e001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lindner S. C., Kohl U., Maier T. J., Steinhilber D., and Sorg B. L.. 2009. TLR2 ligands augment cPLA2alpha activity and lead to enhanced leukotriene release in human monocytes. J. Leukoc. Biol. 86: 389–399. [DOI] [PubMed] [Google Scholar]
- 37.Ruipérez V., Astudillo A. M., Balboa M. A., and Balsinde J.. 2009. Coordinate regulation of TLR-mediated arachidonic acid mobilization in macrophages by group IVA and group V phospholipase A2s. J. Immunol. 182: 3877–3883. [DOI] [PubMed] [Google Scholar]
- 38.Sommerfelt R. M., Feuerherm A. J., Skuland T., and Johansen B.. 2015. Cytosolic phospholipase A2 modulates TLR2 signaling in synoviocytes. PLoS One. 10: e0119088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White H. D., Held C., Stewart R., Tarka E., Brown R., Davies R. Y., Budaj A., Harrington R. A., Steg P. G., Ardissino D., et al. ; STABILITY Investigators. 2014. Darapladib for preventing ischemic events in stable coronary heart disease. N. Engl. J. Med. 370: 1702–1711. [DOI] [PubMed] [Google Scholar]
- 40.O’Donoghue M. L., Braunwald E., White H. D., Lukas M. A., Tarka E., Steg P. G., Hochman J. S., Bode C., Maggioni A. P., Im K., et al. ; SOLID-TIMI 52 Investigators. 2014. Effect of darapladib on major coronary events after an acute coronary syndrome: the SOLID-TIMI 52 randomized clinical trial. JAMA. 312: 1006–1015. [Erratum. 2014. JAMA. 312: 1473.] [DOI] [PubMed] [Google Scholar]
- 41.Kokotou M. G., Limnios D., Nikolaou A., Psarra A., and Kokotos G.. 2017. Inhibitors of phospholipase A2 and their therapeutic potential: an update on patents (2012–2016). Expert Opin. Ther. Pat. 27: 217–225. [DOI] [PubMed] [Google Scholar]
- 42.Muhlestein J. B. 2003. Antibiotic treatment of atherosclerosis. Curr. Opin. Lipidol. 14: 605–614. [DOI] [PubMed] [Google Scholar]
- 43.Vainas T., Stassen F. R., Schurink G. W., Tordoir J. H., Welten R. J., van den Akker L. H., Kurvers H. A., Bruggeman C. A., and Kitslaar P. J.. 2005. Secondary prevention of atherosclerosis through chlamydia pneumoniae eradication (SPACE Trial): a randomised clinical trial in patients with peripheral arterial disease. Eur. J. Vasc. Endovasc. Surg. 29: 403–411. [DOI] [PubMed] [Google Scholar]