Abstract
Chronic obstructive pulmonary disease (COPD) is caused by a complex interaction of environmental exposures, most commonly cigarette smoke, and genetic factors. Chronic cigarette smoke exposure in the mouse is a commonly used animal model of COPD. We aimed to expand our knowledge about the variable susceptibility of inbred strains to this model and test for genetic variants associated with this trait. To that end, we sought to measure differential susceptibility to cigarette smoke–induced emphysema in the mouse, identify genetic loci associated with this quantitative trait, and find homologous human genes associated with COPD. Alveolar chord length (CL) in 34 inbred strains of mice was measured after 6 months of exposure to cigarette smoke. After testing for association, we connected a murine candidate locus to a published meta-analysis of moderate-to-severe COPD. We identified deleterious mutations in a candidate gene in silico and measured gene expression in extreme strains. A/J was the most susceptible strain in our survey (Δ CL 7.0 ± 2.2 μm) and CBA/J was the least susceptible (Δ CL −0.3 ± 1.2 μm). By integrating mouse and human genome-wide scans, we identified the candidate gene Abi3bp. CBA/J mice harbor predicted deleterious variants in Abi3bp, and expression of the gene differs significantly between CBA/J and A/J mice. This is the first report of susceptibility to cigarette smoke–induced emphysema in 34 inbred strains of mice, and Abi3bp is identified as a potential contributor to this phenotype.
Keywords: emphysema, COPD, animal model, Abi3bp
Clinical Relevance
Cigarette smoke–induced emphysema in the mouse is a model of human emphysema that has contributed to our knowledge of the molecular mechanisms of chronic obstructive pulmonary disease. Although it was previously shown that inbred strains of mice have variable susceptibility to chronic cigarette smoke, this was only demonstrated in a limited number of strains. We show that mice of 34 inbred strains have a variable susceptibility to cigarette smoke–induced emphysema. We tested for genetic association with this trait and integrated the findings with the results of a meta-analysis of human chronic obstructive pulmonary disease genome-wide association studies. Using this approach, we identified the gene Abi3bp as a candidate gene contributing to emphysema susceptibility.
Chronic obstructive pulmonary disease (COPD) is defined as a progressive and irreversible airflow limitation, and is a major cause of worldwide morbidity and mortality (1). The disease is caused by a complex interaction of environmental exposures, most commonly cigarette smoke, and genetic factors (2). One of the major pathological processes that contribute to this disease is pulmonary emphysema (destruction of the distal airspaces), which results in a loss of elastic recoil in the lung and air trapping (3).
Studying emphysema in humans involves several challenges, notably the difficulty of accurately quantifying the extent of emphysema in the lung and accounting for factors such as body size and degree of inflation, as well as heterogeneous environmental effects. An alternative is to use mouse models of pulmonary emphysema in which environmental exposure is tightly controlled and the extent of emphysema can be directly measured in the lung. Mice have been shown to develop emphysema after chronic cigarette smoke exposure, and this approach has been successfully employed to model human disease (4). Studies using this model have greatly contributed to our understanding of the mechanisms of COPD (5). However, although variable susceptibility to cigarette smoke–induced emphysema has been demonstrated in five inbred strains of mice, such variability in other commonly used inbred strains has not been previously reported, nor have the genetic contributions to that variability been studied (6).
In humans, genome-wide association studies (GWASs) have been conducted to identify susceptibility variants associated with COPD (2). Although the majority of these studies tested for association with phenotypes such as COPD affection and spirometric traits, several recent studies have identified susceptibility loci associated with radiologic measures of emphysema, as well as specific patterns of emphysema (7–9). These studies demonstrated that some loci associated with emphysema are shared with COPD more broadly, whereas others, such as an association near the gene BICD1, appear to be uniquely associated with emphysema (8). To date, no genome-wide assessments using cross-strain comparisons of mouse models of COPD have been conducted, although such studies have effectively identified candidate genes in other lung diseases, including asthma and pulmonary hypertension (10, 11).
In this study, we confirm that mice have variable susceptibility to cigarette smoke–induced emphysema as measured by alveolar chord length (CL), and show that this trait is continuous when measured in 34 inbred strains. By testing for association of this quantitative trait across the genome and integrating our findings with a human genome-wide scan of COPD, we identify a candidate gene for future study.
Materials and Methods
Animal Experiments
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee of the University of Pittsburgh School of Medicine. Animals were housed in a pathogen-free barrier facility that maintains a 12-hour light/dark cycle in Plexiglas cages (one to four mice per cage) with free access to autoclaved water and irradiated pellet food. Animal health, weight, and overall behavior were monitored throughout the experiments.
Female mice of 34 inbred strains (129S1/SvImJ, A/J, AKR/J, BALB/cByJ, BALB/cJ, BPN/3J, BTBRT+tf/J, BUB/BnJ, C3H/HeJ,C57BL/10J, C57BL/6J, C57BLKS/J, C57BR/cdJ, C57L/J, CBA/J, CE/J, DBA/1J, DBA/2J, FVB/NJ, I/LnJ, KK/HlJ, LG/J, LP/J, MRL/MpJ, NOD/ShiLtJ, NON/ShiLtJ, NZO/HlLtJ, NZW/LacJ, P/J, PL/J, RIIIS/J, SJL/J, SM/J, and SWR/J) were exposed to 6 months of either room air (NS, n = 4–7/group) or cigarette smoke (SM, 4 cigarettes/d, 5 d/wk, n = 3–7/group) beginning at 10 weeks of age, as previously described (12). Kentucky Reference Cigarettes (1R5F) were obtained from the Tobacco and Health Research Institute of the University of Kentucky (Lexington, KY). After smoke exposure, the mice were killed and tracheostomized, and the lungs were removed and inflated with 10% buffered formalin to a constant pressure of 25 cm water for 10 minutes. The lungs were fixed for 24 hours in formalin and then embedded in paraffin. Serial midsagittal sections were obtained and stained with modified Gill’s stain. Using Scion Image software (version 4.0.2; Scion Corp., Frederick, MD), the mean alveolar CL was calculated on 10 randomly selected 200× fields per slide. Airway and vascular structures were masked from the analysis and the images were manually thresholded. The CL was determined in both horizontal and vertical planes, allowing for the calculation of alveolar airspace areas. Finally, the quantitative phenotype that was used in genetic association testing (SMCL; see further below) was determined by the following equation: log(SM − NS). This log-transformed value was used to ensure phenotype normality. Measurement of body weights and tissue elastance is described in more detail in the online supplement.
Genetic Association Testing
Testing for association between SMCL and 4 million single nucleotide polymorphisms (SNPs) published by the National Institute of Environmental Health Sciences was performed using the genome-wide efficient mixed model algorithm (GEMMA), which partially corrects for the complex population structure of the inbred strains in this survey (13, 14). These SNPs were aligned to NCBI37, and all genomic locations reported in this study reflect that unless otherwise noted. To account for nonvariant sites and sites in complete linkage disequilibrium in all 34 strains, we employed simpleM to determine the effective number of tests, which we then used to calculate Bonferroni-corrected significance thresholds in this study (15, 16).
Identification of Genomic Regions Enriched for Nominal Associations
A candidate region was defined as all SNPs yielding nominal associations (P < 10−3) that were within 1 Mb (Mb = 1 million base pairs) of another nominal SNP. The number of nominal associations in each region was recorded and 1 Mb was added in each direction to the location of the boundary SNPs to account for the significant linkage disequilibrium that can occur between inbred mouse strains (17).
Identification of Nominal Associations in Human GWAS Data
A region in the human genome that is homologous to the candidate region with the most significant association identified in the mouse strain survey was identified using mouse-human homology maps publicly available from the National Center for Biotechnology Information (18). Human homologs to all genes in the candidate region were identified. The human homologous region was identified as continuous blocks of these genes and separating noncoding regions.
Searching for Nominal SNPs in a Meta-analysis of Human COPD
The human homologous region was searched for nominal associations (P < 10−4) in a previously published meta-analysis comparing smokers with moderate-to-severe COPD and normal spirometry from the COPDGene, ECLIPSE, NETT/NAS, and Norway GenKOLS studies (19). Nominal associations were recorded and the closest genes to these associations were identified as genes of interest. Manhattan plots of searched regions were generated using LocusZoom (version 1.1; http://locuszoom.sph.umich.edu/locuszoom/) (20).
Identification of Candidate Genes
All variants that resulted in a change in the amino acid sequence of the encoded protein (missense variants, stop-gained variants, and frameshift variants) predicted to be deleterious by the ‘Sorting Tolerant From Intolerant’ (SIFT) algorithm (SIFT < 0.05) were identified in Ensembl (release 84, dbSNP142) in each mouse homolog of the genes of interest in this study (21, 22). The genotype of each of these coding sequence variants in CBA/J and A/J mice was downloaded from Ensembl and the genotype of any variant that differed between these two extreme strains was subsequently downloaded for all available strains included in this study.
mRNA Expression
Lung tissue was homogenized in Trizol solution (Thermo Fisher, Grand Island, NY) and total RNA was isolated according to the manufacturer’s instructions. RNA was quantified and used for reverse transcription with reverse transcriptase (Applied Biosystems, Grand Island, NY). Total cDNA was used for real-time PCR using primer and probe sets specific for the target genes (Applied Biosystems, Grand Island, NY). Relative fold change was calculated by comparing abundance normalized to GAPDH between samples using the ΔΔCT method.
Statistics
Two-tailed t tests were used to compare CLs after cigarette smoking or room air. Error bars, unless otherwise noted, represent the standard error of the mean (SEM). Statistics were calculated with R (http://cran.r-project.org).
Results
Susceptibility to Cigarette Smoke–Induced Emphysema Varies Continuously in Inbred Mouse Strains
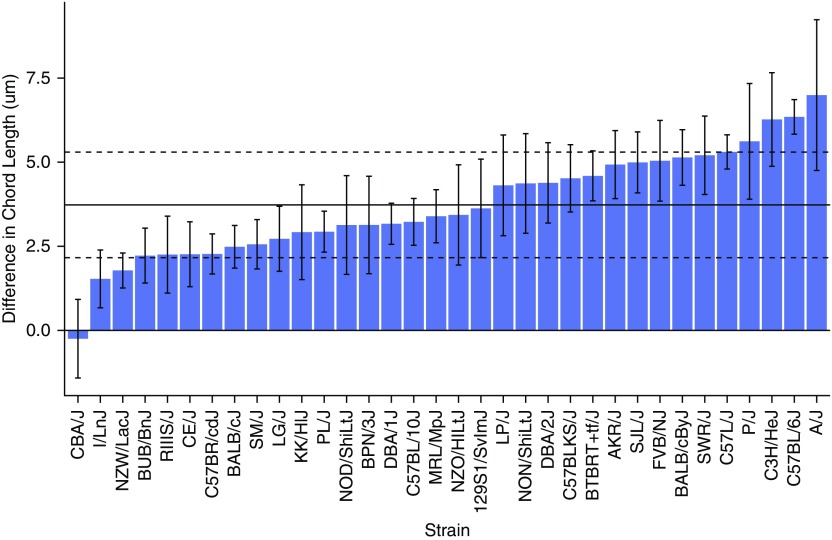
We measured the CL in 34 inbred mouse strains after 6 months of exposure to cigarette smoke (SM) or room air (no smoke exposure, NS) (Table 1). There was significant variability in the response to SM compared with NS (Table 1; Figure 1). This trait was continuous across the 34 inbred strains, with CBA/J mice having essentially no response to cigarette smoke (Δ CL −0.3 ± 1.2 μm) and A/J mice being the most susceptible to cigarette smoke (Δ CL 7.0 ± 2.2 μm) (Figure 1; Table 1). The mean change in CL across all strains was 3.7 ± 1.6 μm, and mice of the 129S1/SvImJ strain most closely reflected this mean. Although the majority of the strains demonstrated a significant change in CL after cigarette smoke exposure, seven strains (BPN/3J, CBA/J, ILn/J, KK/HlJ, NOD/ShiLtJ, NZO/HILtJ, and RIIIS/J) did not (Table 1). This trait did not always directly correlate with changes in body weight (Table E2 in the online supplement) or with other measures of cigarette smoke damage, such as tissue elastance (Table E3).
Table 1.
Variable Susceptibility to Chronic Cigarette Smoke Exposure as Measured by Alveolar Chord Length Is Seen in 34 Inbred Strains of Mice
| NS |
SM |
|||||||
|---|---|---|---|---|---|---|---|---|
| n | Mean CL | SEM | n | Mean CL | SEM | P value* | Δ CL# | |
| 129S1/SvImJ | 4 | 28.55 | 0.48 | 5 | 32.18 | 0.67 | 4.58E-02 | 3.63 |
| A/J | 5 | 33.78 | 0.74 | 6 | 40.77 | 2.11 | 1.83E-02 | 6.99 |
| AKR/J | 7 | 27.98 | 0.31 | 7 | 32.91 | 0.41 | 3.82E-04 | 4.93 |
| BALB/cByJ | 5 | 30.79 | 0.77 | 6 | 35.93 | 0.92 | 2.12E-04 | 5.14 |
| BALB/cJ | 5 | 30.16 | 0.79 | 6 | 32.64 | 1.15 | 4.10E-03 | 2.48 |
| BPN/3J | 4 | 27.49 | 1.00 | 5 | 30.62 | 1.07 | 6.13E-02 | 3.13 |
| BTBRT+tf/J | 5 | 31.00 | 0.83 | 6 | 35.59 | 0.57 | 9.79E-05 | 4.59 |
| BUB/BnJ | 4 | 30.79 | 0.91 | 5 | 33.01 | 0.73 | 3.28E-02 | 2.22 |
| C3H/HeJ | 5 | 40.49 | 0.41 | 6 | 46.76 | 0.48 | 1.95E-03 | 6.27 |
| C57BL/10J | 5 | 28.27 | 0.49 | 6 | 31.50 | 0.50 | 1.34E-03 | 3.23 |
| C57BL/6J | 5 | 29.67 | 0.79 | 6 | 36.02 | 0.90 | 8.21E-07 | 6.35 |
| C57BLKS/J | 5 | 34.05 | 0.32 | 6 | 38.57 | 0.85 | 1.27E-03 | 4.52 |
| C57BR/cdJ | 5 | 29.74 | 0.32 | 6 | 32.02 | 0.41 | 5.64E-03 | 2.27 |
| C57L/J | 5 | 32.29 | 0.56 | 6 | 37.60 | 0.25 | 2.35E-06 | 5.30 |
| CBA/J | 4 | 30.52 | 0.48 | 4 | 30.27 | 0.38 | 8.39E-01 | −0.25 |
| CE/J | 5 | 30.67 | 0.80 | 6 | 32.94 | 0.60 | 4.09E-02 | 2.26 |
| DBA/1J | 5 | 28.75 | 1.04 | 6 | 31.92 | 0.53 | 5.23E-04 | 3.17 |
| DBA/2J | 5 | 28.92 | 0.59 | 6 | 33.31 | 0.44 | 5.96E-03 | 4.38 |
| FVB/NJ | 5 | 28.73 | 0.69 | 6 | 33.77 | 0.28 | 2.63E-03 | 5.04 |
| I/LnJ | 5 | 33.12 | 1.28 | 6 | 34.65 | 0.78 | 1.13E-01 | 1.53 |
| KK/HlJ | 4 | 31.19 | 1.17 | 3 | 34.10 | 0.90 | 1.22E-01 | 2.92 |
| LG/J | 5 | 33.92 | 1.24 | 5 | 36.64 | 0.68 | 2.25E-02 | 2.72 |
| LP/J | 5 | 30.72 | 0.39 | 6 | 35.03 | 0.33 | 1.52E-02 | 4.31 |
| MRL/MpJ | 5 | 31.19 | 0.36 | 4 | 34.59 | 0.89 | 3.91E-03 | 3.39 |
| NOD/ShiLtJ | 4 | 27.80 | 0.36 | 5 | 30.93 | 1.09 | 6.80E-02 | 3.13 |
| NON/ShiLtJ | 5 | 40.03 | 0.59 | 6 | 44.40 | 0.62 | 1.49E-02 | 4.37 |
| NZO/HILtJ | 5 | 32.88 | 0.57 | 6 | 36.31 | 0.55 | 5.97E-02 | 3.43 |
| NZW/LacJ | 5 | 30.79 | 0.68 | 6 | 32.57 | 1.58 | 9.09E-03 | 1.78 |
| P/J | 5 | 29.23 | 0.76 | 6 | 34.85 | 0.59 | 1.41E-02 | 5.62 |
| PL/J | 5 | 30.02 | 1.13 | 6 | 32.96 | 0.93 | 6.19E-04 | 2.93 |
| RIIIS/J | 5 | 27.82 | 0.65 | 6 | 30.07 | 1.34 | 1.04E-01 | 2.25 |
| SJL/J | 5 | 29.50 | 0.31 | 6 | 34.50 | 0.51 | 6.58E-04 | 4.99 |
| SM/J | 5 | 32.11 | 0.55 | 4 | 34.67 | 0.60 | 1.27E-02 | 2.56 |
| SWR/J | 4 | 31.48 | 1.18 | 5 | 36.68 | 0.84 | 2.02E-03 | 5.20 |
Definition of abbreviations: CL, chord length; NS, no smoke exposure; SM, exposure to 6 months of cigarette smoke.
P values are the result of two-tailed t tests between SM and NS groups by strain.
Difference between mean SM CL and mean NS CL.
Figure 1.
Inbred strains of mice have variable susceptibility to cigarette smoke–induced emphysema. Shown is the absolute difference in chord length between mice exposed to cigarette smoke and those exposed to room air, with SEM. The solid horizontal line represents the mean difference in all strains and the dashed horizontal lines represent the mean of all strains ± SD.
The Genetic Contribution to Cigarette Smoke–Induced Emphysema Is Complex in the Mouse
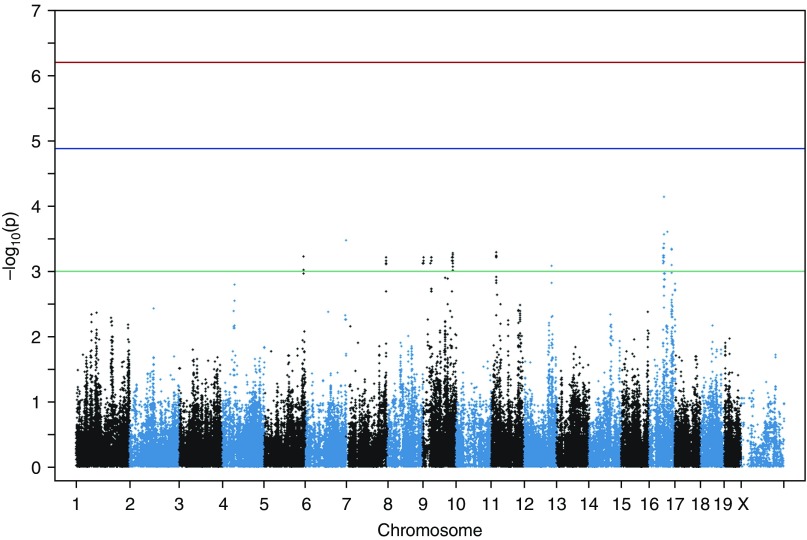
We tested for association of SMCL with known genotypes of 34 inbred strains of mice using GEMMA (13). No association with a single variant reached genome-wide (P < 6.3 × 10−7) or genome-wide suggestive (P < 1. 3 × 10−5) significance after correction for the 79,492 effective tests tested in this study. We determined the effective number of tests using simpleM for the specific strains in our panel (Figure 2) (15). The most significant association we identified was with rs46114044 located at chr16:54194946 (P = 7.21 × 10−5; Table E1). The top 50 most significant associations can be seen in Table E1. To account for the significant linkage disequilibrium in the mouse, we defined a 4.3 Mb region surrounding this association (see Materials and Methods) at chr16:54194946–58510508 as our candidate region to integrate with human GWAS results.
Figure 2.
Manhattan plot of associations with the log-transformed difference in chord length between mice exposed to cigarette smoke and mice exposed to room air. Each dot represents a single-nucleotide polymorphism (SNP), with its genomic location represented on the x-axis and the strength of association on the y-axis. The top threshold (red) represents genome-wide significance after Bonferroni correction of the effective number of tests, the middle threshold (blue) represents suggestive significance, and the bottom threshold (green) is drawn at P = 10−3 to demonstrate nominally associated SNPs that were used to identify candidate regions.
Identification of Candidate Associations from a Human GWAS
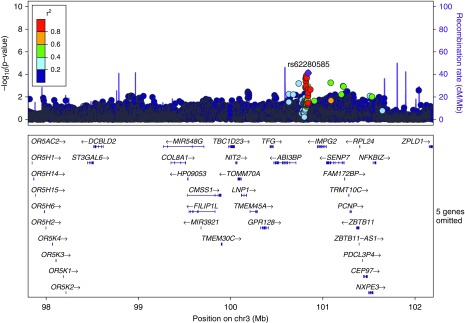
We mapped the candidate region from the mouse to its homologous region at 3q (chr3:97,806,017–102,198,685) in the human genome using a gene-based approach. We then searched the results of a previously published GWAS meta-analysis testing for genetic association with moderate-to-severe COPD in these human genomic regions, and identified any nominal associations yielding P < 1 × 10−4 (19). Only one association met this threshold, located at 3q12.2. The most significantly associated variant at this locus was rs62280585 (P = 7.72 × 10−5), which is located between the genes IMPG2 and ABI3BP (Figure 3).
Figure 3.
Manhattan plot of a human genomic region on chromosome 3 (chr3) syntenic to the mouse candidate region. A region at 3q12 in the human genome, homologous to a candidate region on chromosome 16 in the mouse genome, contains a single nominal association with COPD. The Manhattan plot shows the location on chromosome 3 and gene annotations on the x-axis and strength of association on the y-axis. Linkage disequilibrium with the most significant SNP estimated from the CEU population (Utah residents with Northern and Western European ancestry) genotyped in the 1,000 Genomes Project is illustrated by different-colored SNPs. The plot was created with LocusZoom (version 1.1).
Abi3bp Contains Nonsynonymous Mutations and Is Differentially Expressed Between Extreme Strains
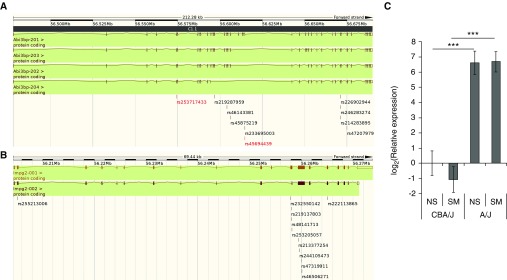
We attempted to identify coding nonsynonymous SNPs (cnSNPs) in the murine homologs of ABI3BP and IMPG2 to identify potentially causal variants. We compared the most susceptible strain, A/J, with the most resistant strain, CBA/J, and determined whether either strain held the alternate allele of a cnSNP predicted to be deleterious by SIFT. Two nonsynonymous mutations in Abi3bp, rs253717433 and rs45694439, are predicted to be deleterious, and the alternate allele of each of these variants occurs in CBA/J mice but not in A/J mice (Figure 4A). Of note, one of these variants, rs253717433, occurs in exon 5 of Abi3bp, which is shared among all known transcripts of the protein. The only other strain in this study harboring this allele is SM/J, which showed a small but significant response to cigarette smoke (ΔLm 2.6 ± 0.7 μm; Table 1). The alternate allele of rs45694439 is harbored by several susceptible strains, but this mutation only occurs in one of the four known transcripts of this gene (Figure 4A; Table E1). Neither of the extreme strains harbors the alternate allele of any of the nonsynonymous mutations in Impg2 with a predicted deleterious effect (Figure 4B).
Figure 4.
In silico and molecular evidence for Abi3bp. Nonsynonymous variants predicted to be deleterious by the ‘Sorting Tolerant from Intolerant’ (SIFT) algorithm in (A) Abi3bp and (B) Impg2 on mouse chromosome 3. A/J mice harbor the reference allele for all labeled variants, and CBA/J mice harbor the alternate allele of those labeled in red. (C) Relative gene expression of Abi3bp in A/J and CBA/J mice exposed to cigarette smoke (SM, n = 5/strain) or room air (NS, n = 5/strain) compared with CBA/J NS. RNA was isolated from whole lungs after 6 months of exposure to room air (NS) or cigarette smoke (SM). P values were determined by two-tailed t test of between-group comparisons (***P < 0.001).
Finally, we looked at the RNA expression of Abi3bp in the two extreme strains used in this study to identify potential functional effects. There was a significant difference in the levels of expression of this gene, with an ∼100-fold greater level of expression in A/J mice, which are susceptible to cigarette smoke–induced emphysema than in CBA/J mice, which are resistant (Figure 4C).
Discussion
In this study, we measured alveolar CL, a surrogate for airspace size and an important measure of emphysema, after chronic cigarette smoke exposure in 34 inbred strains of mice. This study adds to our understanding of the variable susceptibility to cigarette smoke–induced emphysema in mice, which was originally described in five inbred strains (NZWLac/J, C57BL6/J, A/J, SJ/L, and AKR/J) (6). By measuring this trait in 29 additional strains, we found that these mice exhibit a continuous variation in the response to chronic cigarette smoke exposure, ranging from showing essentially no response (CBA/J) to being extremely susceptible (A/J). Importantly, we identified several common inbred strains, in addition to the previously reported NZWLac/J, that are extremely resistant to cigarette smoke and show no significant change in CL after chronic smoke exposure. At the same time, we found that commonly used strains were comparatively more susceptible to cigarette smoke–induced emphysema than was previously reported; for example, C57BL/6J mice were the second most susceptible strain in our study.
As this was the first time that cigarette smoke–induced emphysema was measured in a large panel of inbred mice, it offered the opportunity to test for associations between the trait and previously genotyped variations among these strains. This inbred-strain–association approach has been used to detect association with a number of traits in mouse models of human disease and physiology (23, 24). However, due to the high interrelatedness of inbred strains, its statistical power has sometimes been limited for complex traits without supporting genetic data from an alternate approach, such as linkage analysis (14, 25). Although other approaches are available that may have greater power to achieve this goal, such as the Hybrid Mouse Diversity Panel, they also require phenotyping in many more strains of mice (26). Since the cigarette smoke–induced model of emphysema in mice is both time and labor intensive, we conducted a classic inbred-strain study hoping to improve our ability to detect true positives by combining our data with human GWAS results, an approach previously used in asthma (10).
Although the genetic evidence provided by human GWAS results offers improved resolution and additional confidence in the findings of an inbred-strain GWAS, murine genome-wide scans may also inform our interpretation of the results of future human COPD GWASs. Significant and replicated variants in human studies explain only a small portion of the heritability of COPD and other complex diseases (27, 28). Although some of this “missing” heritability is likely to be explained by rare and structural variation, it has been demonstrated that for some traits, a significant portion can be explained by a common variation failing to reach genome-wide significance due to lack of power or phenotypic heterogeneity (29). Genetic evidence from homologous loci in both mice and humans can thus be helpful for identifying associations that fail to meet significance thresholds but may still represent susceptibility loci.
By integrating our mouse and human genome-wide scans in this way, we identified Abi3bp as a candidate gene in emphysema in a relatively large (4.3 Mb) region that we originally identified in our murine inbred-strain association study. ABI family member 3 binding protein (ABI3BP, also known as TARSH and eratin) is an extracellular matrix–binding protein that was first identified in a yeast two-hybrid screen as binding to c-Abl, and appears to be predominantly expressed in the lung (30, 31). Of note, a recent study combining genetic association testing and expression quantitative trait loci studies in asthma identified ABI3BP as a candidate gene (32). Several studies have characterized ABI3BP as a putative tumor suppressor. Lung tumors exhibit marked reductions in expression and differential splicing of the gene product compared with healthy tissue (33, 34). Knockdown of Abi3bp by short hairpin RNA in mouse embryonic fibroblasts was shown to result in genomic instability, with a greater percentage of cells exhibiting multicentrosomes as well as increased colony formation compared with controls (35). Conversely, overexpression of ABI3BP reduced cellular proliferation in thyroid tumor cell lines via a p21-dependent senescence pathway (36). The gene appears to play a role in cellular proliferation in nontumor settings as well: mesenchymal and cardiac progenitor cells lacking ABI3BP demonstrate significant increases in proliferation and differentiation (37, 38). Additionally, a genome-wide copy-number variation study identified a significant association of ABI3BP with the chronic osteochondropathy Kashin-Beck disease (39). That study further demonstrated increased gene expression in the cartilage of patients, raising the possibility that growth restrictions in chondrocytes lead to the disrepair and loss of function observed in these patients (39). Taken together, these data suggest a role for ABI3BP in regulating cellular proliferation across a wide spectrum of tissues and disease states. In our study, we showed that CBA/J mice, which are highly resistant to cigarette smoke–induced emphysema, carry predicted nonsynonymous deleterious mutations in Abi3bp and have significantly lower expression of the Abi3bp transcript. It is intriguing to consider that a functional decrease of this gene, which controls the growth and differentiation of stem and tumor cells, may also protect against cigarette smoke–induced emphysema, potentially by promoting the growth and survival of the lung epithelium. Future studies using Abi3bp knockout mice will be necessary to delineate this relationship more clearly.
This study has limitations. Although we measured the response to cigarette smoke–induced emphysema in 34 inbred strains, we were still unable to detect any regions that met strict multiple testing correction thresholds. As mentioned above, this is due partially to the genetic complexity of the trait and partially to the power of inbred-strain surveys; a larger survey might be able to overcome this limitation. Since we did not identify any regions with genome-wide significance, we chose to use only the most significant association to identify candidate genes. Although this was useful for identifying Abi3bp, it remains possible that other nominal associations could be of interest with additional supporting evidence. In addition, although the use of a mouse model of human disease offers a unique opportunity to compare regions of association between organisms, it also is limited by the similarity of the model to the disease. We chose to use the alveolar CL, which has been used as a correlate for emphysema in many translational studies, but we recognize that this phenotype does not always directly mimic emphysema with airflow obstruction in humans. This is underlined by the fact that not all strains that developed significant changes in CL had similar changes in tissue elastance, another trait that has been used to measure response to cigarette smoke in the mouse (Table E3).
We have demonstrated that susceptibility to cigarette smoke–induced emphysema is a variable trait in 34 inbred strains of mice. Although we were not able to identify any single variants associated with this phenotype at a genome-wide level, we integrated our mouse genome-wide scan with the results of a human GWAS and identified a novel candidate gene, Abi3bp. As the absence of Abi3bp has been shown to increase cellular growth in both tumor and stem cells, its potential role in pulmonary emphysema is intriguing and requires more investigation.
Acknowledgments
Acknowledgments
The authors thank Chris Burton, Andy Metz, and Stephen Scott for their technical assistance with animal experiments.
COPDGene Investigators—Core Units
Administrative Center: James D. Crapo, M.D. (PI); Edwin K. Silverman, M.D., Ph.D. (PI); Barry J. Make, M.D.; Elizabeth A. Regan, M.D., Ph.D.
Genetic Analysis Center: Terri Beaty, Ph.D.; Ferdouse Begum, Ph.D.; Robert Busch, M.D.; Peter J. Castaldi, M.D., M.Sc.; Michael Cho, M.D.; Dawn L. DeMeo, M.D., M.P.H.; Adel R. Boueiz, M.D.; Marilyn G. Foreman, M.D., M.S.; Eitan Halper-Stromberg; Nadia N. Hansel, M.D., M.P.H.; Megan E. Hardin, M.D.; Craig P. Hersh, M.D., M.P.H.; Jacqueline Hetmanski, M.S., M.P.H.; Brian D. Hobbs, M.D.; John E. Hokanson, M.P.H., Ph.D.; Nan Laird, Ph.D.; Christoph Lange, Ph.D.; Sharon M. Lutz, Ph.D.; Merry-Lynn McDonald, Ph.D.; Margaret M. Parker, Ph.D.; Dandi Qiao, Ph.D.; Elizabeth A. Regan, M.D., Ph.D.; Stephanie Santorico, Ph.D.; Edwin K. Silverman, M.D., Ph.D.; Emily S. Wan, M.D.; Sungho Won.
Imaging Center: Mustafa Al Qaisi, M.D.; Harvey O. Coxson, Ph.D.; Teresa Gray; MeiLan K. Han, M.D., M.S.; Eric A. Hoffman, Ph.D.; Stephen Humphries, Ph.D.; Francine L. Jacobson, M.D., M.P.H.; Philip F. Judy, Ph.D.; Ella A. Kazerooni, M.D.; Alex Kluiber; David A. Lynch, M.B.; John D. Newell, Jr., M.D.; Elizabeth A. Regan, M.D., Ph.D.; James C. Ross, Ph.D.; Raul San Jose Estepar, Ph.D.; Joyce Schroeder, M.D.; Jered Sieren; Douglas Stinson; Berend C. Stoel, Ph.D.; Juerg Tschirren, Ph.D.; Edwin Van Beek, M.D., Ph.D.; Bram van Ginneken, Ph.D.; Eva van Rikxoort, Ph.D.; George Washko, M.D.; Carla G. Wilson, M.S.
PFT QA Center, Salt Lake City, UT: Robert Jensen, Ph.D.
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Ph.D.; Jim Crooks, Ph.D.; Camille Moore, Ph.D.; Matt Strand, Ph.D.; Carla G. Wilson, M.S.
Epidemiology Core, University of Colorado Anschutz Medical Campus, Aurora, CO: John E. Hokanson, M.P.H., Ph.D.; John Hughes, Ph.D.; Gregory Kinney, M.P.H., Ph.D.; Sharon M. Lutz, Ph.D.; Katherine Pratte, M.S.P.H.; Kendra A. Young, Ph.D.
COPDGene Investigators—Clinical Centers
Ann Arbor VA: Jeffrey L. Curtis, M.D.; Carlos H. Martinez, M.D., M.P.H.; Perry G. Pernicano, M.D.
Baylor College of Medicine, Houston, TX: Nicola Hanania, M.D., M.S.; Philip Alapat, M.D.; Mustafa Atik, M.D.; Venkata Bandi, M.D.; Aladin Boriek, Ph.D.; Kalpatha Guntupalli, M.D.; Elizabeth Guy, M.D.; Arun Nachiappan, M.D.; Amit Parulekar, M.D.
Brigham and Women’s Hospital, Boston, MA: Dawn L. DeMeo, M.D., M.P.H.; Craig Hersh, M.D., M.P.H.; Francine L. Jacobson, M.D., M.P.H.; George Washko, M.D.
Columbia University, New York, NY: R. Graham Barr, M.D., Dr.P.H.; John Austin, M.D.; Belinda D’Souza, M.D.; Gregory D.N. Pearson, M.D.; Anna Rozenshtein, M.D., M.P.H., FACR; Byron Thomashow, M.D.
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., M.D.; H. Page McAdams, M.D.; Lacey Washington, M.D.
HealthPartners Research Institute, Minneapolis, MN: Charlene McEvoy, M.D., M.P.H.; Joseph Tashjian, M.D.
Johns Hopkins University, Baltimore, MD: Robert Wise, M.D.; Robert Brown, M.D.; Nadia N. Hansel, M.D., M.P.H.; Karen Horton, M.D.; Allison Lambert, M.D., MHS; Nirupama Putcha, M.D., M.H.S.
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Torrance, CA: Richard Casaburi, Ph.D., M.D.; Alessandra Adami, Ph.D.; Matthew Budoff, M.D.; Hans Fischer, M.D.; Janos Porszasz, M.D., Ph.D.; Harry Rossiter, Ph.D.; William Stringer, M.D.
Michael E. DeBakey VAMC, Houston, TX: Amir Sharafkhaneh, M.D., Ph.D.; Charlie Lan, D.O.
Minneapolis VA: Christine Wendt, M.D.; Brian Bell, M.D.
Morehouse School of Medicine, Atlanta, GA: Marilyn G. Foreman, M.D., M.S.; Eugene Berkowitz, M.D., Ph.D.; Gloria Westney, M.D., M.S.
National Jewish Health, Denver, CO: Russell Bowler, M.D., Ph.D.; David A. Lynch, M.B.
Reliant Medical Group, Worcester, MA: Richard Rosiello, M.D.; David Pace, M.D.
Temple University, Philadelphia, PA: Gerard Criner, M.D.; David Ciccolella, M.D.; Francis Cordova, M.D.; Chandra Dass, M.D.; Gilbert D’Alonzo, D.O.; Parag Desai, M.D.; Michael Jacobs, Pharm.D.; Steven Kelsen, M.D., Ph.D.; Victor Kim, M.D.; A. James Mamary, M.D.; Nathaniel Marchetti, D.O.; Aditi Satti, M.D.; Kartik Shenoy, M.D.; Robert M. Steiner, M.D.; Alex Swift, M.D.; Irene Swift, M.D.; Maria Elena Vega-Sanchez, M.D.
University of Alabama, Birmingham, AL: Mark Dransfield, M.D.; William Bailey, M.D.; Surya Bhatt, M.D.; Anand Iyer, M.D.; Hrudaya Nath, M.D.; J. Michael Wells, M.D.
University of California, San Diego, CA: Joe Ramsdell, M.D.; Paul Friedman, M.D.; Xavier Soler, M.D., Ph.D.; Andrew Yen, M.D.
University of Iowa, Iowa City, IA: Alejandro P. Comellas, M.D.; John Newell, Jr., M.D.; Brad Thompson, M.D.
University of Michigan, Ann Arbor, MI: MeiLan K. Han, M.D., M.S.; Ella Kazerooni, M.D.; Carlos H. Martinez, M.D., M.P.H.
University of Minnesota, Minneapolis, MN: Joanne Billings, M.D.; Abbie Begnaud, M.D.; Tadashi Allen, M.D.
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, M.D.; Jessica Bon, M.D.; Divay Chandra, M.D., M.Sc.; Carl Fuhrman, M.D.; Joel Weissfeld, M.D., M.P.H.
University of Texas Health Science Center at San Antonio, San Antonio, TX: Antonio Anzueto, M.D.; Sandra Adams, M.D.; Diego Maselli-Caceres, M.D.; Mario E. Ruiz, M.D.
ECLIPSE Investigators—Bulgaria: Y. Ivanov, Pleven; K. Kostov, Sofia. Canada: J. Bourbeau, Montreal; M. Fitzgerald, Vancouver, BC; P. Hernandez, Halifax, NS; K. Killian, Hamilton, ON; R. Levy, Vancouver, BC; F. Maltais, Montreal; D. O'Donnell, Kingston, ON. Czech Republic: J. Krepelka, Prague. Denmark: J. Vestbo, Hvidovre. The Netherlands: E. Wouters, Horn-Maastricht. New Zealand: D. Quinn, Wellington. Norway: P. Bakke, Bergen. Slovenia: M. Kosnik, Golnik. Spain: A. Agusti, J. Sauleda, P. de Mallorca. Ukraine: Y. Feschenko, V. Gavrisyuk, L. Yashina, Kiev; N. Monogarova, Donetsk. UK: P. Calverley, Liverpool; D. Lomas, Cambridge; W. MacNee, Edinburgh; D. Singh, Manchester; J. Wedzicha, London. United States: A. Anzueto, San Antonio, TX; S. Braman, Providence, RI; R. Casaburi, Torrance CA; B. Celli, Boston; G. Giessel, Richmond, VA; M. Gotfried, Phoenix, AZ; G. Greenwald, Rancho Mirage, CA; N. Hanania, Houston; D. Mahler, Lebanon, NH; B. Make, Denver; S. Rennard, Omaha, NE; C. Rochester, New Haven, CT; P. Scanlon, Rochester, MN; D. Schuller, Omaha, NE; F. Sciurba, Pittsburgh; A. Sharafkhaneh, Houston; T. Siler, St. Charles, MO; E. Silverman, Boston; A. Wanner, Miami; R. Wise, Baltimore; R. ZuWallack, Hartford, CT.
ECLIPSE Steering Committee: H. Coxson (Canada), C. Crim (GlaxoSmithKline, USA), L. Edwards (GlaxoSmithKline, USA), D. Lomas (UK), W. MacNee (UK), E. Silverman (USA), R. Tal Singer (Co-chair, GlaxoSmithKline, USA), J. Vestbo (Co-chair, Denmark), J. Yates (GlaxoSmithKline, USA).
ECLIPSE Scientific Committee: A. Agusti (Spain), P. Calverley (UK), B. Celli (USA), C. Crim (GlaxoSmithKline, USA), B. Miller (GlaxoSmithKline, USA), W. MacNee (Chair, UK), S. Rennard (USA), R. Tal-Singer (GlaxoSmithKline, USA), E. Wouters (The Netherlands), J. Yates (GlaxoSmithKline, USA).
The ECLIPSE study (NCT00292552; GSK code SCO104960) was funded by GlaxoSmithKline.
The National Emphysema Treatment Trial was supported by NHLBI N01HR76101, N01HR76102, N01HR76103, N01HR76104, N01HR76105, N01HR76106, N01HR76107, N01HR76108, N01HR76109, N01HR76110, N01HR76111, N01HR76112, N01HR76113, N01HR76114, N01HR76115, N01HR76116, N01HR76118, and N01HR76119; the Centers for Medicare and Medicaid Services; and the Agency for Healthcare Research and Quality.
The Normative Aging Study is supported by the Cooperative Studies Program/ERIC of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center (MAVERIC). D.S. is supported by a VA Research Career Scientist award.
The Norway GenKOLS study (Genetics of Chronic Obstructive Lung Disease, GSK code RES11080) was funded by GlaxoSmithKline.
The COPDGene project is supported by the COPD Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens, and Sunovion.
Footnotes
This work was supported by grants 5R01HL107883-04 (S.D.S.), 5T32HL094295 (N.J.K.), R01HL089897 (J.D.C.), and R01HL089856 (E.K.S.) from the National Heart, Lung, and Blood Institute, National Institutes of Health; grant YFAC142013 from the Flight Attendant Medical Research Institute (A.D.G.); and the U.S. Department of Veterans Affairs.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health.
Author Contributions: Study design: all authors; data collection: J.E.R., A.D.G., A.S.L., Y.C., Y.Z., A.B., and S.D.S.; data quality control and analysis: J.E.R., A.D.G., A.S.L., Y.C., and Y.Z.; manuscript writing: J.E.R.; manuscript revision: all authors.
This article has an online data supplement, which is accessible from this issue's table of content online at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2016-0220OC on April 25, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.National Heart, Lung, and Blood Institute. Bethesda, MD: National Institutes of Health; 2012. Morbidity and mortality: 2012 chart book on cardiovascular, lung and blood diseases. [updated 2013 May; accessed 2016 Jun 21]. Available from: https://www.nhlbi.nih.gov/research/reports/2012-mortality-chart-book. [Google Scholar]
- 2.Berndt A, Leme AS, Shapiro SD. Emerging genetics of COPD. EMBO Mol Med. 2012;4:1144–1155. doi: 10.1002/emmm.201100627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macnee W. Pathogenesis of chronic obstructive pulmonary disease. Clin Chest Med. 2007;28:479–513, v. (v.). doi: 10.1016/j.ccm.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 4.Mahadeva R, Shapiro SD. Chronic obstructive pulmonary disease * 3: Experimental animal models of pulmonary emphysema. Thorax. 2002;57:908–914. doi: 10.1136/thorax.57.10.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hautamaki RD, Kobayashi DK, Senior RM, Shapiro SD. Requirement for macrophage elastase for cigarette smoke-induced emphysema in mice. Science. 1997;277:2002–2004. doi: 10.1126/science.277.5334.2002. [DOI] [PubMed] [Google Scholar]
- 6.Guerassimov A, Hoshino Y, Takubo Y, Turcotte A, Yamamoto M, Ghezzo H, Triantafillopoulos A, Whittaker K, Hoidal JR, Cosio MG. The development of emphysema in cigarette smoke-exposed mice is strain dependent. Am J Respir Crit Care Med. 2004;170:974–980. doi: 10.1164/rccm.200309-1270OC. [DOI] [PubMed] [Google Scholar]
- 7.Castaldi PJ, Cho MH, San José Estépar R, McDonald ML, Laird N, Beaty TH, Washko G, Crapo JD, Silverman EK COPDGene Investigators. Genome-wide association identifies regulatory Loci associated with distinct local histogram emphysema patterns. Am J Respir Crit Care Med. 2014;190:399–409. doi: 10.1164/rccm.201403-0569OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong X, Cho MH, Anderson W, Coxson HO, Muller N, Washko G, Hoffman EA, Bakke P, Gulsvik A, Lomas DA, et al. ECLIPSE Study NETT Investigators. Genome-wide association study identifies BICD1 as a susceptibility gene for emphysema. Am J Respir Crit Care Med. 2011;183:43–49. doi: 10.1164/rccm.201004-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, Budoff M, Austin JH, Washko GR, Carr JJ, et al. Genome-wide study of percent emphysema on computed tomography in the general population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Himes BE, Sheppard K, Berndt A, Leme AS, Myers RA, Gignoux CR, Levin AM, Gauderman WJ, Yang JJ, Mathias RA, et al. Integration of mouse and human genome-wide association data identifies KCNIP4 as an asthma gene. PLoS One. 2013;8:e56179. doi: 10.1371/journal.pone.0056179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kelly NJ, Radder JE, Baust JJ, Burton CL, Lai YC, Potoka KC, Agostini BA, Wood JP, Bachman TN, Vanderpool RR, et al. Mouse genome-wide association study of preclinical group II pulmonary hypertension identifies epidermal growth factor receptor. Am J Respir Cell Mol Biol. 2017;56:488–496. doi: 10.1165/rcmb.2016-0176OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shapiro SD, Goldstein NM, Houghton AM, Kobayashi DK, Kelley D, Belaaouaj A. Neutrophil elastase contributes to cigarette smoke-induced emphysema in mice. Am J Pathol. 2003;163:2329–2335. doi: 10.1016/S0002-9440(10)63589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou X, Stephens M. Genome-wide efficient mixed-model analysis for association studies. Nat Genet. 2012;44:821–824. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirby A, Kang HM, Wade CM, Cotsapas C, Kostem E, Han B, Furlotte N, Kang EY, Rivas M, Bogue MA, et al. Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics. 2010;185:1081–1095. doi: 10.1534/genetics.110.115014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34:100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao X. Multiple testing corrections for imputed SNPs. Genet Epidemiol. 2011;35:154–158. doi: 10.1002/gepi.20563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurie CC, Nickerson DA, Anderson AD, Weir BS, Livingston RJ, Dean MD, Smith KL, Schadt EE, Nachman MW. Linkage disequilibrium in wild mice. PLoS Genet. 2007;3:e144. doi: 10.1371/journal.pgen.0030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.National Center for Biotechnology Information. Homology maps. Rockville, MD: 2014 [accessed 2014 Sep 1]. Available from: http://www.ncbi.nlm.nih.gov/projects/homology/maps/
- 19.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, Demeo DL, Sylvia JS, Ziniti J, Laird NM, et al. NETT Genetics, ICGN, ECLIPSE and COPDGene Investigators. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. Lancet Respir Med. 2014;2:214–225. doi: 10.1016/S2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flicek P, Amode MR, Barrell D, Beal K, Billis K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fitzgerald S, et al. Ensembl 2014. Nucleic Acids Res. 2014;42:D749–D755. doi: 10.1093/nar/gkt1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 23.Lewis SR, Ahmed S, Dym C, Khaimova E, Kest B, Bodnar RJ. Inbred mouse strain survey of sucrose intake. Physiol Behav. 2005;85:546–556. doi: 10.1016/j.physbeh.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 24.Leme AS, Berndt A, Williams LK, Tsaih SW, Szatkiewicz JP, Verdugo R, Paigen B, Shapiro SD. A survey of airway responsiveness in 36 inbred mouse strains facilitates gene mapping studies and identification of quantitative trait loci. Mol Genet Genomics. 2010;283:317–326. doi: 10.1007/s00438-010-0515-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manenti G, Galvan A, Pettinicchio A, Trincucci G, Spada E, Zolin A, Milani S, Gonzalez-Neira A, Dragani TA. Mouse genome-wide association mapping needs linkage analysis to avoid false-positive Loci. PLoS Genet. 2009;5:e1000331. doi: 10.1371/journal.pgen.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flint J, Eskin E. Genome-wide association studies in mice. Nat Rev Genet. 2012;13:807–817. doi: 10.1038/nrg3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.So HC, Gui AH, Cherny SS, Sham PC. Evaluating the heritability explained by known susceptibility variants: a survey of ten complex diseases. Genet Epidemiol. 2011;35:310–317. doi: 10.1002/gepi.20579. [DOI] [PubMed] [Google Scholar]
- 28.Zhou JJ, Cho MH, Castaldi PJ, Hersh CP, Silverman EK, Laird NM. Heritability of chronic obstructive pulmonary disease and related phenotypes in smokers. Am J Respir Crit Care Med. 2013;188:941–947. doi: 10.1164/rccm.201302-0263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsuda S, Iriyama C, Yokozaki S, Ichigotani Y, Shirafuji N, Yamaki K, Hayakawa T, Hamaguchi M. Cloning and sequencing of a novel human gene that encodes a putative target protein of Nesh-SH3. J Hum Genet. 2001;46:483–486. doi: 10.1007/s100380170049. [DOI] [PubMed] [Google Scholar]
- 31.Uekawa N, Terauchi K, Nishikimi A, Shimada J, Maruyama M. Expression of TARSH gene in MEFs senescence and its potential implication in human lung cancer. Biochem Biophys Res Commun. 2005;329:1031–1038. doi: 10.1016/j.bbrc.2005.02.068. [DOI] [PubMed] [Google Scholar]
- 32.Nieuwenhuis MA, Siedlinski M, van den Berge M, Granell R, Li X, Niens M, van der Vlies P, Altmüller J, Nürnberg P, Kerkhof M, et al. Combining genomewide association study and lung eQTL analysis provides evidence for novel genes associated with asthma. Allergy. 2016;71:1712–1720. doi: 10.1111/all.12990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terauchi K, Shimada J, Uekawa N, Yaoi T, Maruyama M, Fushiki S. Cancer-associated loss of TARSH gene expression in human primary lung cancer. J Cancer Res Clin Oncol. 2006;132:28–34. doi: 10.1007/s00432-005-0032-1. [DOI] [PubMed] [Google Scholar]
- 34.Guimarães GS, Latini FR, Camacho CP, Maciel RM, Dias-Neto E, Cerutti JM. Identification of candidates for tumor-specific alternative splicing in the thyroid. Genes Chromosomes Cancer. 2006;45:540–553. doi: 10.1002/gcc.20316. [DOI] [PubMed] [Google Scholar]
- 35.Wakoh T, Uekawa N, Terauchi K, Sugimoto M, Ishigami A, Shimada J, Maruyama M. Implication of p53-dependent cellular senescence related gene, TARSH in tumor suppression. Biochem Biophys Res Commun. 2009;380:807–812. doi: 10.1016/j.bbrc.2009.01.171. [DOI] [PubMed] [Google Scholar]
- 36.Latini FR, Hemerly JP, Oler G, Riggins GJ, Cerutti JM. Re-expression of ABI3-binding protein suppresses thyroid tumor growth by promoting senescence and inhibiting invasion. Endocr Relat Cancer. 2008;15:787–799. doi: 10.1677/ERC-08-0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodgkinson CP, Naidoo V, Patti KG, Gomez JA, Schmeckpeper J, Zhang Z, Davis B, Pratt RE, Mirotsou M, Dzau VJ. Abi3bp is a multifunctional autocrine/paracrine factor that regulates mesenchymal stem cell biology. Stem Cells. 2013;31:1669–1682. doi: 10.1002/stem.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hodgkinson CP, Gomez JA, Payne AJ, Zhang L, Wang X, Dal-Pra S, Pratt RE, Dzau VJ. Abi3bp regulates cardiac progenitor cell proliferation and differentiation. Circ Res. 2014;115:1007–1016. doi: 10.1161/CIRCRESAHA.115.304216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang F, Guo X, Zhang Y, Wen Y, Wang W, Wang S, Yang T, Shen H, Chen X, Tian Q, et al. Genome-wide copy number variation study and gene expression analysis identify ABI3BP as a susceptibility gene for Kashin-Beck disease. Hum Genet. 2014;133:793–799. doi: 10.1007/s00439-014-1418-4. [DOI] [PubMed] [Google Scholar]