Abstract
Two populations of alveolar macrophages (AMs) coexist in the inflamed lung: resident AMs that arise during embryogenesis, and recruited AMs that originate postnatally from circulating monocytes. The objective of this study was to determine whether origin or environment dictates the transcriptional, metabolic, and functional programming of these two ontologically distinct populations over the time course of acute inflammation. RNA sequencing demonstrated marked transcriptional differences between resident and recruited AMs affecting three main areas: proliferation, inflammatory signaling, and metabolism. Functional assays and metabolomic studies confirmed these differences and demonstrated that resident AMs proliferate locally and are governed by increased tricarboxylic acid cycle and amino acid metabolism. Conversely, recruited AMs produce inflammatory cytokines in association with increased glycolytic and arginine metabolism. Collectively, the data show that even though they coexist in the same environment, inflammatory macrophage subsets have distinct immunometabolic programs and perform specialized functions during inflammation that are associated with their cellular origin.
Keywords: macrophage programming, macrophage metabolism, acute lung injury
Clinical Relevance
In this study, the programming of resident alveolar macrophages and bone-marrow–derived, recruited alveolar macrophages are compared in an in vivo model of lung inflammation. RNA sequencing demonstrates unique signatures between cell types throughout the course of lung inflammation, with distinct differences in genes involved in proliferation, metabolism, and inflammatory cytokine production. Functional studies, including metabolic profiling, confirm these differences. This study builds upon a growing body of literature regarding the determinants of macrophage programming during inflammation, and it conclusively demonstrates that macrophages within the inflamed alveolus represent heterogeneous populations, each with dynamic and distinct functions.
Alveolar macrophages (AMs) have dual roles in the initiation and resolution of lung inflammation. They initiate innate immune responses by producing inflammatory cytokines that recruit neutrophils and other leukocytes, and regulate the resolution of inflammation and repair through clearance of cellular debris, interaction with the alveolar epithelium, and production of growth factors (1–5). To perform these diverse and seemingly contradictory functions, AMs undergo dynamic, multidimensional, stimulus-specific reprogramming (6, 7). The factors that regulate this reprogramming are not fully understood; however, it is clear that changes in metabolic flux and signaling through metabolic intermediates play critical roles (8–11). To date, the complex transcriptional and metabolic repertoires of AMs have been studied as a homogeneous group, at single time points, often in tissue culture or after application of stimulatory molecules ex vivo. The time-resolved transcriptional and metabolic changes that occur in individual populations of AMs during the course of inflammation in vivo have not been explored.
AMs lavaged from inflamed lungs are often studied as a homogeneous group. However, the inflammatory macrophage pool is comprised of two distinct populations. Resident AMs populate the lung during embryogenesis and self-renew throughout life, with little or no contribution from circulating monocytes (12, 13). In contrast, recruited AMs derive from circulating monocytes that traffic to sites of inflammation, where they mature into macrophages. In mice, intratracheal administration of inflammatory stimuli such as lipopolysaccharide (LPS) or influenza A leads to rapid accumulation of recruited AMs in the alveolar space. As inflammation resolves, these cells undergo apoptosis and are removed from the airspaces, leaving only a small number of recruited cells once alveolar integrity has been reestablished (14). Accordingly, the conserved process of monocyte recruitment and subsequent removal from the airspaces suggests that the AM subtypes have unique and necessary functions. Although recruited AMs can become transcriptionally and functionally indistinguishable from resident AMs after prolonged time in a shared microenvironment (15), their functional, transcriptional, and metabolic profiles have not been comprehensively assessed during inflammation.
The primary objective of this study was to compare global differences in the immunometabolic programming of resident and recruited AMs in a time-limited murine model of lung inflammation. We first performed RNA sequencing (RNAseq) of resident and recruited AMs from initiation through resolution of LPS-induced lung inflammation, and then assessed the metabolic profiles of AM subsets at the peak of inflammation. We validated our findings by assessing several key functions of resident and recruited AMs within the inflamed airspace. Our results indicate that although resident and recruited AMs exist in a shared environment, their cellular origin is the major determinant of how they are programmed during an acute inflammatory response. Major areas of difference include cell proliferation, inflammatory cytokine production, and metabolism.
Materials and Methods
Mice
This study was approved by and performed in accordance with the ethical standards of the Institutional Animal Care and Use Committee at National Jewish Health, Denver, Colorado. All of the mice used for RNAseq were C57B/6 males, 10–12 weeks of age, from The Jackson Laboratory (Bar Harbor, ME). Validation experiments were performed with mice bred in-house, with the exception of CCR2 knockout and CCR2 reporter mice (The Jackson Laboratory), and ranged from 8 to 14 weeks in age. Experiments with wild-type mice were initially performed in C57B/6 males, and male and female noninduced conditional transgenic mice on a C57B/6 background were used for validation.
Acute Lung Injury Model
LPS (20 μg; Escherichia coli O55:B5; List Biological Laboratories, Campbell, CA) in 50 μl of phosphate-buffered saline (PBS) was instilled directly into the tracheas of mice sedated with isoflurane (Baxter, Deerfield, IL) using a modified feeding needle.
Macrophage Isolation
Bronchoalveolar lavage (BAL) was performed with 10 serial lavages of 1 ml PBS containing 5 mM EDTA. BAL cells were washed twice in PBS. Lavages from five animals at each time point were pooled, except for Day 3, when 10 animals were used for each replicate. The staining protocol can be found in the online supplement. Fluorescence-activated cell sorting (FACS) was performed as described previously (15) using a Moflo XDP-100 with Summit v5.1 software (Beckman Coulter, Dako, Glostrup, Denmark) or BD FACSAria Cell Sorting System with BD FACSDiva software (BD Biosciences, Franklin Lakes, NJ) on unfixed specimens. Each sort was performed in triplicate.
RNA Isolation, Complementary DNA Generation, Microarray, and RNAseq Data Analysis
Isolated total RNA was processed for next-generation sequencing (NGS) library construction as developed in the NJH Genomics Facility for analysis on the Ion Proton NGS platform (Life Technologies, Carlsbad, CA). All data sets have been deposited in the National Center for Biotechnology Information/Gene Expression Omnibus under accession number GSE94749. See the online supplement for a detailed explanation of the analyses.
Flow Cytometry
Macrophages were harvested from BAL as described above. Cells were analyzed on an LSR II flow cytometer (BD Biosciences). Data were analyzed with FlowJo software v9.8.1 (Tree Star, Ashland, OR).
Proliferation Assays
Mice were injected intraperitoneally with 150 µl (2 mg) of bromodeoxyuridine (BrdU) solution (BD Biosciences) 24 h before harvest. Macrophages were harvested and BrdU- or Ki67-positive cells were quantified by flow cytometry. See the online supplement for additional details.
Microscopy
Sorted cells were examined after Wright-Giemsa staining of cytospin specimens (Fischer Diagnostics, Middletown, VA) and examined by light microscopy using a 100× objective on an Olympus BX51 microscope (Olympus Corporation, Tokyo, Japan).
Metabolic Profiling
AMs were isolated on Day 3 after LPS treatment as described above, except that the samples underwent neutrophil depletion before processing. For details regarding the methods used for metabolite extraction and ultra-high-pressure liquid chromatography/mass spectrometry (UHPLC/MS) analysis, see the online supplement.
Cytokine Analysis
For ex vivo experiments, cytokines were measured using a MULTI-SPOT Proinflammatory Panel 1 V-PLEX kit (catalog No. K15048D; Mesoscale Diagnostics, LLC, Rockville, MD) according to the manufacturer’s protocol. See the online supplement for further details.
Statistics
Significant differences in cytokine levels were determined using Student’s t test and proliferation levels were determined by one-way ANOVA with Fisher’s least significant difference test using GrapPad Prism version 6 (GraphPad Software, Inc., La Jolla, CA).
Results
Resident and Recruited AMs Exhibit Distinct Gene Expression Profiles throughout Lung Inflammation
To characterize resident and recruited AMs during acute and resolving lung inflammation, we used the LPS model of acute lung injury (16, 17). As previously described, peak neutrophil inflammation occurred 3 days after intratracheal LPS instillation (14, 18). Although recruited AMs were present at this time, their numbers peaked later (Day 6) and then slowly declined until epithelial integrity was fully restored at Day 12 (Figure 1A) (14). By examining AMs during homeostasis (referred to as Day 0) and 3, 6, 9, and 12 days after LPS administration, we aimed to capture the changes in programming between resident and recruited AMs at peak inflammation, resolution of inflammation, and subsequent return to the resting state.
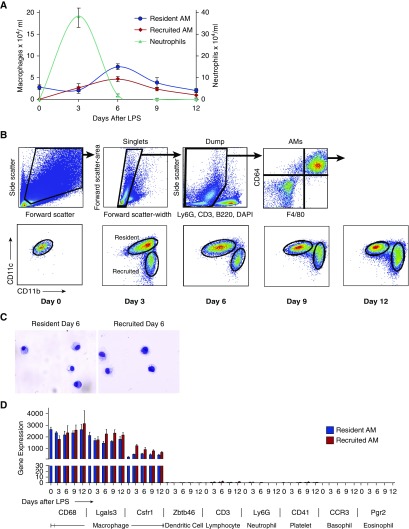
Figure 1.
Sort strategy and validation of RNA sequencing (RNAseq) data. Mice were treated with intratracheal LPS and macrophages were isolated from bronchoalveolar lavage (BAL). (A) Leukocyte counts from BAL after LPS-induced lung injury. Error bars represent SEM, n = 4–7 animals per time point. (B) Forward scatter and side scatter were used to exclude lymphocytes and monocytes (smaller and with lower side scatter than macrophages), and doublets were excluded using the pulse width. Dead and contaminating cells were further eliminated using DAPI, CD3, NK1.1, B220, and Ly6G. Alveolar macrophages (AMs) were further identified by high expression of CD64 and F4/80. AMs were then divided into resident (CD11chigh, CD11blow) or recruited (CD11clow, CD11bhigh) populations. (C) Sorted resident (left) and recruited (right) AMs were examined using light microscopy to ensure purity. (D) Gene transcript profiles of resident (blue bars) and recruited (red bars) AMs were evaluated for genes highly expressed by macrophages (CD68, Lgals3, and Csfr1) and for genes expressed by potential contaminating cells, including lymphocytes (CD3), neutrophils (Ly6G), eosinophils (Prg2), basophils (CCR3), dendritic cells (Zbtb46), and platelets (CD41).
At each time point, we separated mature macrophages using FACS. The gating strategies were designed to eliminate neutrophils, lymphocytes, and nonviable cells. Monocytes were also excluded (see Figures E1A and E1B in the online supplement) and mature macrophages were stringently selected. From the macrophage gate, we distinguished resident and recruited macrophages by their expression of CD11c and CD11b, a strategy we previously validated using lung-shielded bone-marrow chimeras and fluorescent PKH dye labeling (14). In line with our previous work, we observed that during late stages of the inflammatory response, the boundaries between resident and recruited AM populations became less distinct. Accordingly, cells between gates were excluded from analysis because their origins could not be confidently determined (Figure 1B).
Although our previous studies clearly demonstrated that recruited macrophages originated from bone marrow, they did not specifically demonstrate that the macrophages derived from monocytes recruited along a chemokine gradient (14). We therefore sought to determine whether the recruited macrophages identified by our gating strategies were truly derived from monocytes that were recruited to the alveolus rather than from monocytes that simply leaked into injured airspaces. Because previous studies have shown the importance of CCL2 in driving classical monocytes (CCR2+ Ly6G+ CX3CR1lo monocytes) (3), we investigated the CCL2/CCR2 axis. As a first step, we confirmed the presence of CCL2 in the BAL during peak accumulation of the recruited AM pool. We next demonstrated that recruited macrophages express high levels of CCR2, and as a final step confirmed that CCR2-deficient mice had decreased numbers of recruited AMs in their lungs (Figure E1C). Taken as a whole, these data suggest that recruited AMs are truly recruited to the lungs, and that the CCL2/CCR2 axis plays a role.
To validate our sorting strategy, we first examined the morphology of isolated populations using light microscopy (Figure 1C). Contamination by other cell types was negligible at all time points. We next demonstrated that both resident and recruited AMs expressed high levels of commonly recognized macrophage markers, and that transcripts expressed by potentially contaminating cell types, including neutrophils, lymphocytes, eosinophils, basophils, dendritic cells, and platelets, were expressed at insignificant levels (Figure 1D). As a final step to validate the sorting strategy and identify cell-surface markers unique to each AM population, we assessed the gene and protein expression of several surface markers (Figure E2). Notably, Siglec-F, CD206, and CD169 were present at high levels on resident AMs, whereas Ly6C was highly expressed on recruited AMs.
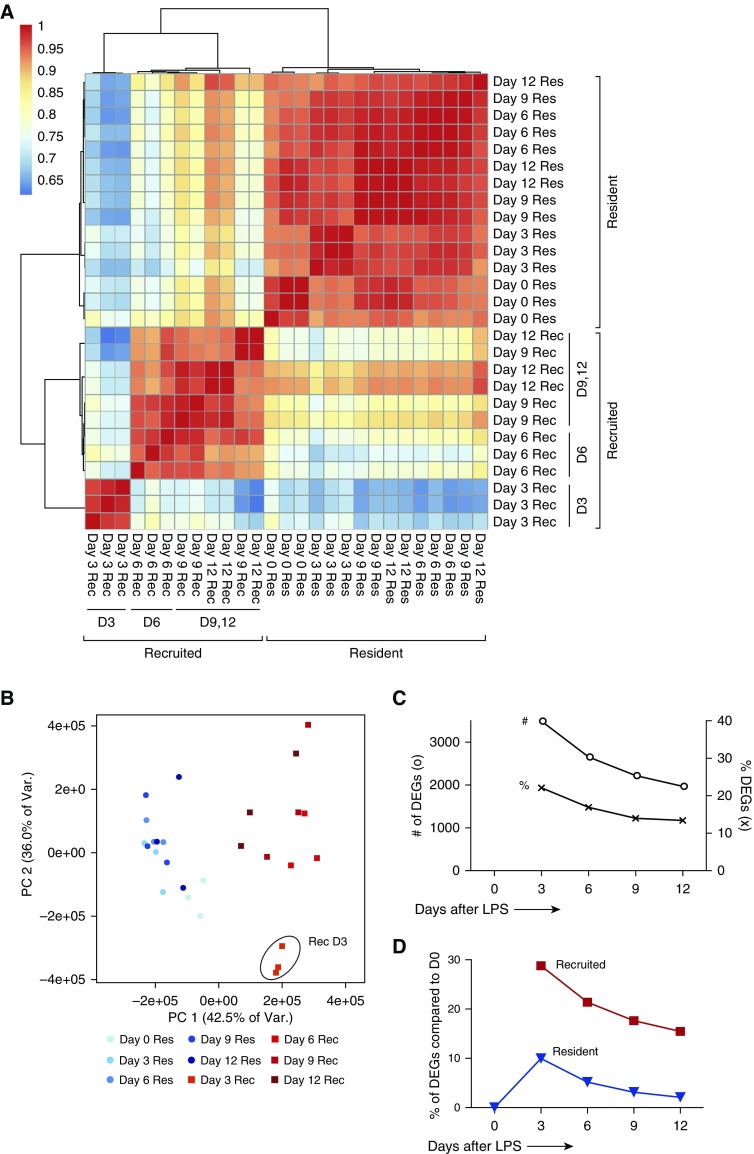
We began our transcriptome analyses by comparing the global similarity in expression among all samples in an unsupervised manner (Figures 2A and 2B). Hierarchical clustering and principal component analysis, using > 17,000 unique transcripts (each detected in all three replicates of at least one time point or AM type) demonstrated that recruited AMs at Day 3 were the most distinct population, followed by recruited AMs from resolution time points. Intriguingly, resident AMs clustered together across all time points. After 9 and 12 days, resident and recruited AMs began to share more transcript expression but remained markedly dissimilar. We next performed a pairwise analysis of gene expression between resident and recruited AMs at each time point. Several thousand mRNA transcripts differentiated resident and recruited populations throughout the course of lung inflammation (Figure 2C). At the peak of neutrophilic inflammation (Day 3), more than 3,000 genes, or 22% of the genes analyzed, were differentially expressed between resident and recruited AMs by a factor of at least two, and more than 13% remained differentially expressed after Day 12. Our methods also allowed us to examine the change in gene expression within each AM subset over time. We performed pairwise comparisons between resident AMs during homeostasis (Day 0) and each other sample to gauge the longitudinal changes in programming (Figure 2D). At peak inflammation, only 10% of the transcripts were differentially expressed in resident AMs compared with naive AMs, and only 2% were differentially expressed by Day 12. In comparison, recruited AMs demonstrated striking transcriptional changes over time, with more than 28% of recruited AM transcripts affected on Day 3, and more than 15% of transcripts showing sustained differential expression at Day 12. Overall, these results demonstrate substantially distinct gene-expression profiles between resident and recruited AMs at all points examined. The most dynamic changes occurred in the recruited population, with only a slight convergence of programming toward that of preinflammatory, naive AMs.
Figure 2.
Global analysis of resident and recruited AM transcriptomes after LPS treatment. (A) Individual replicates of resident (Res, blue) and recruited (Rec, red) AM transcriptomes were clustered based on pairwise Pearson correlation coefficients using the Ward method of the hclust function in R version 3.1.0. (B) Principal component (PC) analysis of Res and Rec samples at each time point. Day 3 recruited samples are circled. (C) Pairwise comparisons of resident and recruited AM transcriptomes at each time point expressed as the percentage of the genes that differed between cell types (x), and the number of differentially expressed genes (o) between cell types. Qualifying genes had at least a 2-fold change and an adjusted P value ≤ 0.1. (D) Pairwise comparison of resident AMs (blue triangle) or recruited AMs (red square) with naive resident AMs (same criteria as in C). # of DEGs, number of differentially expressed genes; % of DEGs, percent differentially expressed genes.
Recruited and Resident AMs Regulate Distinct Cellular Pathways
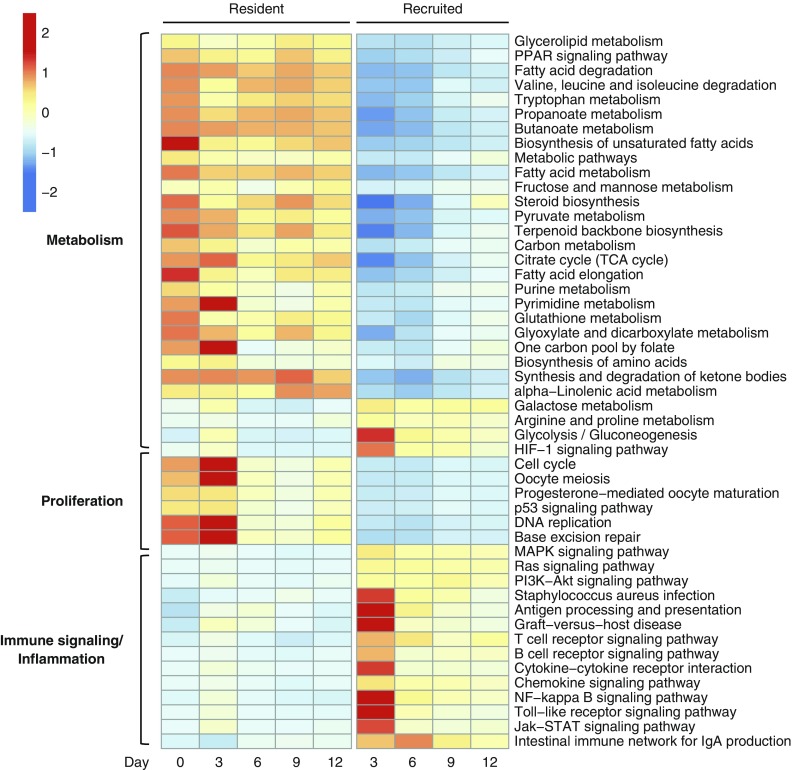
To ascribe functional roles to the transcriptional profiles of AM subsets, we performed a pathway analysis of differentially expressed genes at each time point using the Kyoto Encyclopedia of Genes and Gene Products (KEGG) database. More than 150 unique pathways were represented at one or more time points (Table E1; Figure E3). The greatest number of enriched pathways was noted at Day 3. Investigation of the pathways revealed several prominent themes: recruited AMs demonstrated enrichment in pathways involved in immune signaling, inflammation, and glycolytic and arginine metabolism, whereas resident macrophages had enrichment in pathways related to proliferation, the tricarboxylic acid (TCA) cycle, amino acids, and fatty acid metabolism (Figure 3).
Figure 3.
Pathway analysis of RNAseq. Selected KEGG pathways demonstrate enrichment of differentially expressed genes (compared with naive resident AMs). The median expression profile (transcripts per million [TPM]) of all differentially expressed genes in the pathway is scaled relative to the row mean. Differentially expressed genes are defined as those with at least a 2-fold change in either direction and adjusted P value ≤ 0.1.
Resident AMs, but Not Recruited AMs, Proliferate during Lung Inflammation
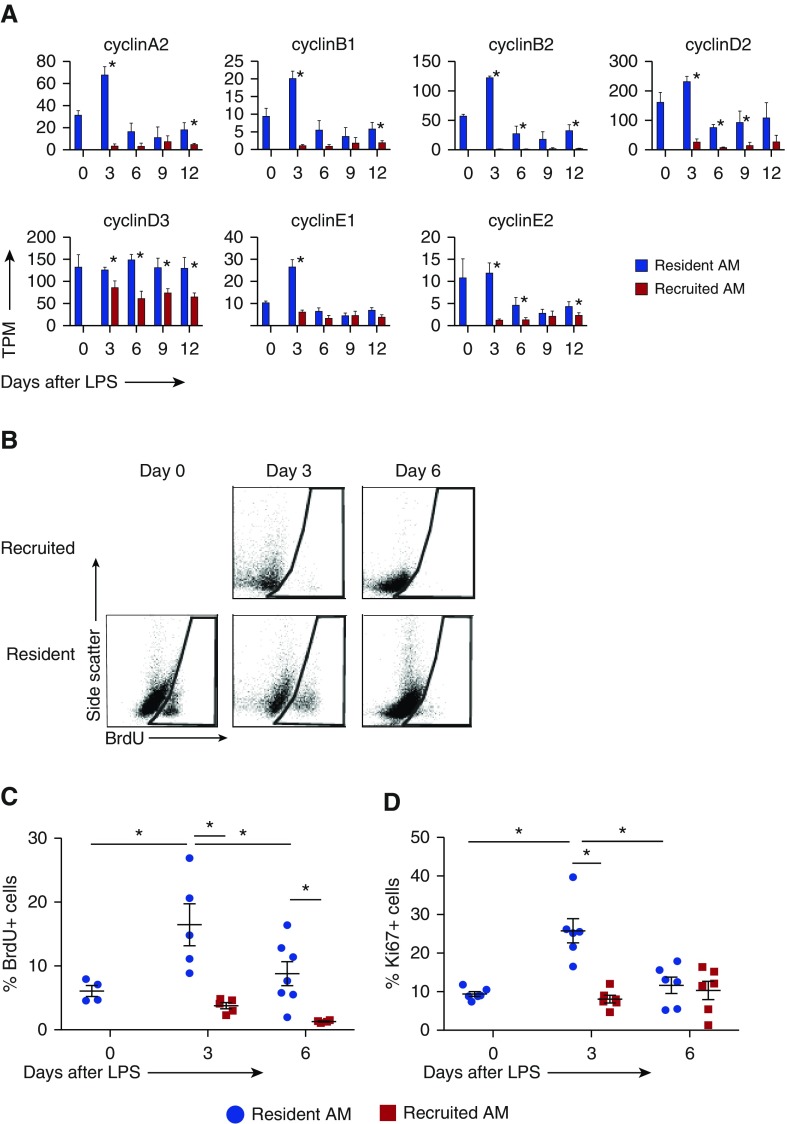
To investigate the differences in cellular proliferation as suggested by enrichment of cell-cycle and DNA-replication pathways, we examined the expression levels of classical cyclins. These were significantly increased in resident AMs, particularly at Day 3 (Figure 4A). Expression of cyclin-dependent kinases was also substantially increased in resident compared with recruited AMs (Figure E4).
Figure 4.
Proliferation of resident versus recruited AMs. (A) Expression levels of classical cyclins from RNAseq data, shown as TPM. Error bars represent SD between replicates. (B–D) Proliferation of resident and recruited AMs measured by BrdU incorporation and Ki67 staining via flow cytometry. BrdU was injected 24 h before harvest. (B) Representative flow plots of BrdU-positive cells. Positive gates were drawn based on fluorescence minus one controls at each time point. (C and D) Percent BrdU-positive and Ki67-positive resident (blue circle) versus recruited (red square) AMs in BAL. Error bars represent SD. n = 4–7 animals per time point. *P < 0.05.
The proportion of proliferating cells in each AM population was determined by BrdU incorporation and Ki67 staining (Figures 4B–4D). To avoid capturing replication of recruited cells before they entered the alveolar space, BrdU was administered intraperitoneally 24 hours before harvest. Resident AMs exhibited low-level BrdU incorporation and Ki67 staining during homeostasis that increased at peak inflammation (Day 3) and returned to baseline levels by Day 6. Conversely, recruited AMs proliferated at very low levels throughout inflammation. These results demonstrate a marked difference in proliferative potential between resident and recruited AMs, with high levels of DNA replication in resident AMs and minimal replication in recruited AMs.
Recruited and Resident AMs Express Distinct Inflammatory Mediators
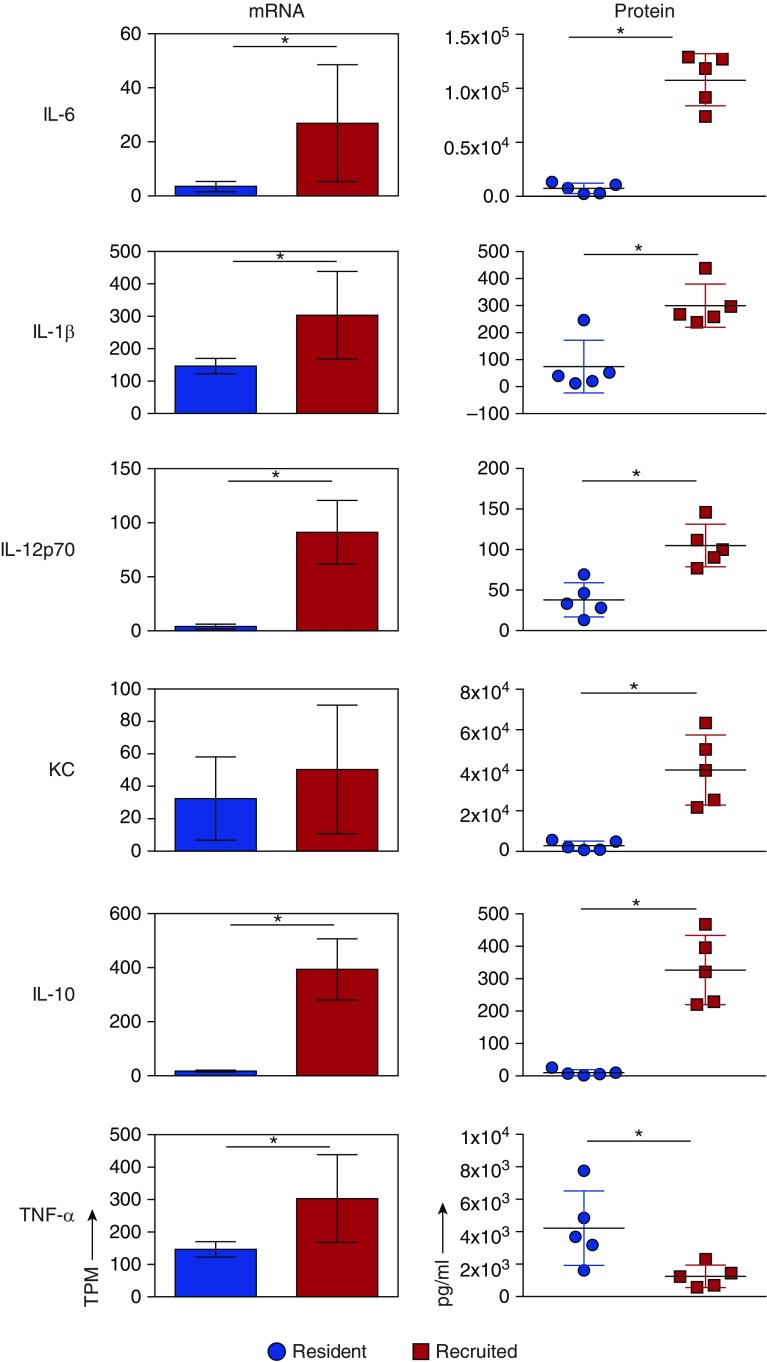
Given that pathways for inflammation and immune signaling were enriched in recruited AMs, we examined the mRNA expression levels of inflammatory cytokines thought to be important in lung injury (Figure 5, left panel), and complemented this approach with ex vivo cytokine production assays using AMs isolated on Day 3 after LPS administration (Figure 5, right panel). Recruited AMs produced higher levels of IL-1β, IL-6, IL-12p70, keratinocyte chemokine (KC), and IL-10 compared with resident cells. Of note, although resident AMs showed lower TNF-α gene expression, they produced more TNF-α in culture. These results highlight the unique inflammatory signaling that occurs between the AM subgroups, and suggest that both populations contribute to inflammation at its peak. To determine whether these findings fit within the context of classical (M1) versus alternative (M2) macrophage polarization, we compared the gene transcripts of commonly recognized activation markers between resident and recruited AMs after LPS administration and naive resident AMs (Figure E5). The resulting data do not support a distinct pro- or anti-inflammatory pattern of programming in either resident or recruited AMs. Rather, they suggest that both cell types contribute to inflammation and repair over the course of LPS-induced lung injury (Figure E5).
Figure 5.
Inflammatory cytokine profiles of resident and recruited AMs. Resident and recruited AMs were isolated 3 days after LPS treatment. Right panel: cytokine concentrations (in pg/ml) were measured in conditioned media after 24 h of culture. Left panel: gene expression of proinflammatory cytokines using freshly isolated macrophages. mRNA levels are expressed as TPM. IL-12α mRNA levels are shown. There was no significant difference in IL-12β transcript levels between resident and recruited AMs. n = 3 per group, mean and SD shown. KC, chemokine ligand 1. Error bars represent SD. Significance testing was done using Student’s t test for protein levels and the Wald test in DESeq2 (adjusted for multiple testing) for RNAseq. *P < 0.05.
Resident and Recruited Macrophages Have Distinct Metabolic Profiles during Inflammation
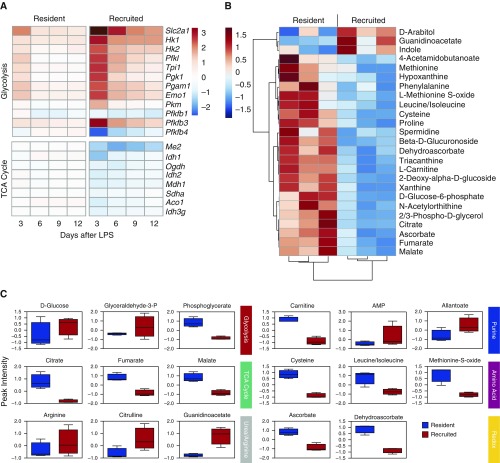
To confirm the differences in metabolic programming suggested by the pathway analysis, we first compared the gene-expression profiles of key enzymes involved in carbohydrate metabolism (Figure 6A). Compared with naive resident AMs, recruited AMs had increased expression of enzymes involved in glycolysis and decreased expression of TCA-cycle enzymes. Conversely, transcription of TCA-cycle enzymes remained stable in resident AMs and expression of glycolytic enzymes persisted at low levels.
Figure 6.
Metabolic profiles of resident and recruited AMs. (A) Base-2 log fold-change expression of transcripts involved in glycolytic and TCA-cycle metabolism in resident and recruited AMs after LPS treatment relative to naive AMs is shown as a heat map. (B) Hierarchical clustering of the top 25 metabolites differentially detected in resident and recruited AMs at Day 3 (the relative abundance is shown). (C) Peak intensity of select metabolites from major metabolic pathways in resident (blue) and recruited (red) AMs at Day 3. The box represents the 25th–75th percentile of peak intensity (atomic units), n = 3. The horizontal line represents the median value. Error bars represent minimum and maximum values. TCA, tricarboxylic acid.
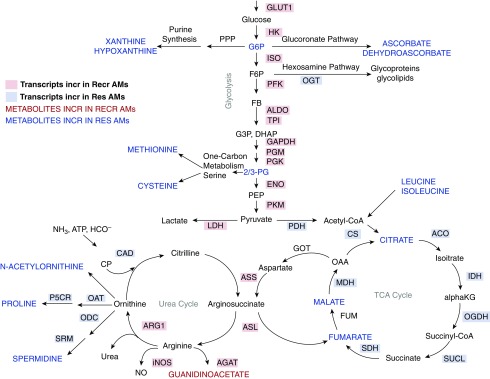
We next performed unbiased, whole-cell metabolomic profiling of resident and recruited AMs isolated from LPS-treated mice at Day 3 using UHPLC/MS (Table E2). As predicted by the RNAseq data, TCA-cycle intermediates, including citrate, fumarate, and malate, were present at significantly greater levels in resident AMs compared with recruited AMs. In addition, there were significant increases in metabolites involved in amino acid metabolism and glutathione homeostasis in the resident AMs. Although there were trends toward increased glucose and glyceraldehyde-3-phosphate in the recruited AMs, no statistical difference in glycolytic intermediates was measured. However, recruited AMs demonstrated dramatic increases in arginine metabolism (Figures 6B and 6C). Together with RNAseq, these data confirm that AMs of different origins display distinct metabolic landscapes in inflamed alveolar spaces (Figure 7).
Figure 7.
Integration of RNAseq and metabolomics data. Major metabolic pathways are illustrated. Gene transcripts and metabolite levels were compared between Res and Recr AMs at Day 3 after LPS treatment. Transcripts differentially upregulated (at least a 2-fold change and adjusted P value ≤ 0.1) in resident AMs are highlighted in blue, and those increased in recruited AMs are highlighted in red. Metabolites detected at higher levels in resident AMs are in blue font and those elevated in recruited AMs are in red font. 2/3-PG, 2/3-phosophoglycerate; ACO, aconitase; AGAT, arginine:glycine amidinotransferase; ALDO, aldolase; αKG, α-ketoglutarate; ARG1, arginase 1; ASL, arginosuccinate lyase; ASS, arginosuccinate synthase; CAD, carbamoyl phosphate synthetase; CP, carbamoyl phosphate; CS, citrate synthase; DHAP, dihydroxyacetone phosphate; ENO, enolase; F6P, fructose 6-phosphate; FB, fructose 1,6-bisphosphate; FUM, fumarase; G3P, glyceraldehyde 3-phosphate; G6P, glucose 6-phosphate; GLUT1, solute carrier family 2, facilitated glucose transporter 1; GOT, aspartate aminotransferase; HK, hexokinase; IDH, isocitrate dehydrogenase; iNOS, inducible nitric oxide synthase; ISO, phosphoglucose isomerase; LDH, lactate dehydrogenase; MDH, malate dehydrogenase; NO, nitric oxide; OAA, oxaloacetic acid; OAT, ornithine aminotransferase; ODC, ornithine decarboxylase; OGDH, α-ketoglutarate dehydrogenase; OGT, O-linked N-acetylglucosamine transferase; P5CR, pyrroline-5-carboxylate reductase; PDH, pyruvate dehydrogenase; PEP, phosphoenolpyruvate; PFK, phosphofructokinase; PGM, phosphoglycerate mutase; PGK, phosphoglycerate kinase; PKM, pyruvate kinase; PPP, pentose phosphate shunt; SDH, succinate dehydrogenase; SRM, spermine synthase; SUCL, succinate-CoA ligase; TPI, triose phosphate isomerase.
Discussion
Inflammatory AMs are often studied as a homogeneous population. However, our data clearly demonstrate that resident and recruited AMs have dramatically different transcriptional and metabolic signatures, despite the fact that they coexist in the same environment. To our knowledge, this study is the first to integrate metabolic and transcriptional data to comprehensively examine the programming of embryonic-derived versus postnatal-derived macrophages during an acute inflammatory process. These data provide evidence that unique cellular functions and metabolic profiles in inflammatory AM subsets are dictated by cellular origin.
Taken as a whole, our findings suggest that the metabolic programming and functional status of resident and recruited AMs are quite different. Accordingly, we propose that the AM subsets serve different roles during acute inflammation and its resolution, and suggest a framework by which divisions of labor and time-resolved differences in resident and recruited AMs functions within the inflamed airspace can be conceptualized. To begin, we postulate that the primary role of resident AMs is to maintain homeostasis within the alveolus. Hence, in < 3 days after initiation of the inflammatory cascade, proinflammatory mediator production by resident AMs has already begun to turn off and the cells begin to express gene profiles and metabolites consistent with replenishment of the AM pool and maintenance of alveolar integrity. Conversely, our data show that recruited AMs augment acute inflammation through production of inflammatory mediators and metabolic reprogramming associated with cytotoxic and antimicrobial functions. After inflammation resolves and homeostasis begins to be restored, the recruited AMs undergo apoptosis, leaving the resident AMs to serve once again as homeostatic regulators and airspace sentinels.
Our finding that resident AMs proliferate after LPS-induced lung injury is in keeping with the well-characterized self-renewal of tissue macrophages during homeostasis (12, 19–22) and transient proliferation of tissue resident macrophages in certain inflammatory settings (21, 23–26). Notably, up to 20% of resident AMs incorporated BrdU at the peak of inflammation. Whether these cells represent a highly proliferative subset of AMs (such as a progenitor cell pool) or all resident AMs have a proliferative capacity is unknown. Notably, proliferation of recruited AMs within the airspaces appeared negligible in our model. However, they most likely maintain an ability to proliferate in the proper setting. For example, when resident AMs are ablated by clodronate or radiation, recruited AMs can fill and maintain the niche (15).
Previous studies by our group demonstrated that recruited AMs undergo apoptosis and are removed from the airspace during resolution of inflammation (14, 18). In the study presented here, 4′,6-diamidino-2-phenylindole was used to exclude nonviable cells during cell sorting. Because this nuclear dye does not detect cells in early stages of programmed cell death (up to 20% of recruited AMs during Days 6–12) (14), it is possible that some early apoptotic cells were isolated during our sorting. Whether this has bearing on our transcriptional data is unclear, but we think it is unlikely given that mRNA is degraded almost immediately after apoptosis is initiated (27, 28).
Our study was not designed to differentiate whether the same recruited AMs existed throughout the full course of inflammation or rapid cell turnover with ongoing immigration of recruited AMs was responsible for maintaining their numbers. However, time-dependent downregulation of classical monocyte markers (i.e., Ly6C) with concomitant upregulation of “mature” macrophage markers (i.e., CD206) suggests the former. Furthermore, studies by our group using labeled blood monocytes suggest that these cells arrive in bulk during early inflammation and are steadily removed from the airspaces during resolution (unpublished data). Intriguingly, enhanced macrophage proliferation has been associated with anti-inflammatory programming, raising the possibility that local proliferation of resident tissue macrophages plays a role in anti-inflammatory signaling (23). Additional investigations into the kinetics and regulation of cell turnover and the effect of macrophage proliferation on inflammation will be important in furthering our understanding of inflammatory lung diseases.
Our findings of divergent metabolic and immune signaling profiles between resident and recruited AMs complement an emerging literature elucidating the link between metabolism and immune regulation in macrophages (8, 10, 29–33). At first glance, it appears that recruited AMs recapitulate classical proinflammatory programs described for in vitro stimulated bone-marrow–derived or elicited peritoneal macrophages, including increased glycolytic enzyme transcription, decreased mitochondrial function, induction of nitric oxide synthase (Nos2), and inflammatory cytokine production (10, 31, 32). However, we demonstrate several notable differences between recruited AMs found in the acutely inflamed lung and previously described “proinflammatory” modules derived from cultured macrophages. First, in addition to transcripts associated with typical inflammatory macrophages, recruited AMs simultaneously expressed high levels of transcripts associated with “alternative activation,” such as Arg1 and Ym1 (Figure E5) (34). Furthermore, metabolic profiling of recruited AMs demonstrated a dichotomous array of arginine metabolites. Metabolism of arginine has long been recognized as a determinant of macrophage programming. In this context, arginine can be metabolized through Nos2 to generate nitric oxide, which is critical for antimicrobial and inflammatory functions, or it can be hydrolyzed by arginase to ornithine and urea, whose downstream products are important for wound healing, cell growth, and differentiation (31, 35). At the peak of inflammation, recruited AMs in our study produced high levels of both Arg1 and Nos2, and contained metabolites from both pathways. We postulate that the simultaneous expression of Nos2 and Arg1 in recruited AMs represents a pivotal inflammatory checkpoint in limiting nitric oxide–induced inflammation, and further implicates the recruited AMs in both pro- and anti-inflammatory functions within the alveolar space (36). Whether simultaneous expression of these enzymes occurs in a single cell or subpopulations of recruited AMs antagonize each other will be determined through the use of single-cell studies.
In contrast to recruited AMs, resident AMs isolated from the inflamed lung exhibited programming more consistent with that described for “alternatively activated” macrophages (31, 32). Our findings show that resident AMs have increased cellular proliferation, enhanced expression of enzymes important in hexosamine and N-glycosylation pathways, and production of TCA-cycle intermediates and amino acids—all of which have been described in anti-inflammatory macrophages (23, 31, 32). Although transcription of glycolytic enzymes and inflammatory mediators was less pronounced in resident cells than in recruited cells, glycolytic intermediates and the inflammatory cytokine TNF-α were detected at significant levels, suggesting that resident AMs also contribute to the acute inflammatory milieu, and in fact likely contribute to its initiation. Intriguingly, pathway analysis suggested increased processing of glutathione in resident AMs, and metabolic profiling detected higher levels of the antioxidants ascorbate and dehydroascorbate during inflammation compared with recruited AMs. Although the exact role of redox metabolism in macrophage polarization is unclear (31, 37, 38), future investigations into the overall effect of these different intracellular redox programs on cell function may inform our understanding of the intricate relationship between redox metabolism and macrophage activation.
Gene expression of glycolytic enzymes was significantly higher in recruited AMs than in resident AMs; however, concentrations of glycolytic metabolites were similar between the two populations. Several potential mechanisms may explain this finding. First, if recruited AMs use glycolysis as the major source of cellular ATP during peak inflammation, the rapid flux through this pathway may result in rapid conversion of intermediates and thus low detection using our methods. Alternatively, the single time point profiled may not adequately resolve the kinetic effects of changing glycolytic enzyme levels, including the recently described nonenzymatic feedback of enzymes on glycolytic rates and inflammation (33, 39). Future metabolomic studies using labeled glucose will be necessary to clarify the rate of glycolytic flux and fate of glucose-derived carbons during inflammation.
The goal of this study was to explore the differences in AM programming throughout the course of lung inflammation. Despite a modest amount of variability between replicate samples due to variation in the inflammatory response to LPS between animals, the reproducibility of the data was high, especially at early time points. We chose Day 3 as the initial point of analysis because at earlier time points, recruited-AM numbers were exceedingly low and isolation was technically challenging due to the overwhelming presence of neutrophils. Our study employed a sterile, self-limited model of LPS-induced lung inflammation, a well-characterized mimic of the most common inflammatory lung disease, bacterial pneumonia. Whether our observations about AM programming will extend to other models of inflammation is unknown. However, our findings are supported by data from allergen-induced models that demonstrate differential immunomodulatory effects of macrophage subsets on disease progression (25). Although we characterized the inflammatory and metabolic profiles of resident and recruited AMs at the point of greatest dissimilarity, Day 3, the time-resolved changes and convergence of immunometabolic programming during resolution of lung injury are the focus of ongoing studies.
It is important to note that in this study we identified macrophage populations using previously validated and well-accepted cell-surface markers. However, it is possible that additional subpopulations of AMs exist. For example, AMs that adhere tightly to the alveolar epithelium and are not removed by simple lavage would have evaded our analysis. Along similar lines, the methods employed in this study did not allow for reliable separation of macrophages by compartment (i.e., alveolar space versus tissue interstitium). Accordingly, the programming of interstitial macrophages was not assessed. The roles played by these macrophages in inflammation and tissue repair remain unclear and represent a target for future studies.
In summary, the integrated transcriptional and metabolic characterization of resident and recruited AMs in this study demonstrates for the first time that macrophages of different origins are uniquely programmed within the shared inflammatory alveolar environment. Our study demonstrates that resident AMs proliferate locally, contain a metabolic profile consistent with TCA cycle and amino acid synthesis, and at the time points studied contribute little to inflammatory cytokine production. Recruited AMs, however, appear to be primed for inflammatory signaling with high levels of arginine metabolism and cytokine production. These results suggest that AM subtypes may be differentially targeted in inflammatory lung disease, and provide an impetus for considering cell origin when studying AMs.
Footnotes
This work was supported by National Institutes of Health grants HL109517, HL34303, AI110408, and F32HL131397. The Cancer Center Flow Cytometry Shared Resource at the University of Colorado–Anschutz Medical Campus is supported by National Institutes of Health grant P30CA046934.
Author Contributions: K.J.M. performed research, analyzed data, and wrote the manuscript. L.B. and M.P.M. performed research and contributed to the manuscript. S.M.T. and A.L.M. contributed to the manuscript. T.D. and S.M.L. performed the bioinformatics and contributed to the manuscript. T.E.F. and B.P.O’C. designed research. A.D’A. and J.A.R. performed research, analyzed data, and contributed to the manuscript. D.L.B contributed to the conceptual design of the research and to the manuscript. C.V.J. contributed to the manuscript. W.J.J. designed research, analyzed data, and wrote the manuscripxt.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2017-0061OC on April 19, 2017
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Cakarova L, Marsh LM, Wilhelm J, Mayer K, Grimminger F, Seeger W, Lohmeyer J, Herold S. Macrophage tumor necrosis factor-alpha induces epithelial expression of granulocyte-macrophage colony-stimulating factor: impact on alveolar epithelial repair. Am J Respir Crit Care Med. 2009;180:521–532. doi: 10.1164/rccm.200812-1837OC. [DOI] [PubMed] [Google Scholar]
- 2.Herold S, Tabar TS, Janssen H, Hoegner K, Cabanski M, Lewe-Schlosser P, Albrecht J, Driever F, Vadasz I, Seeger W, et al. Exudate macrophages attenuate lung injury by the release of IL-1 receptor antagonist in gram-negative pneumonia. Am J Respir Crit Care Med. 2011;183:1380–1390. doi: 10.1164/rccm.201009-1431OC. [DOI] [PubMed] [Google Scholar]
- 3.Maus UA, Waelsch K, Kuziel WA, Delbeck T, Mack M, Blackwell TS, Christman JW, Schlöndorff D, Seeger W, Lohmeyer J. Monocytes are potent facilitators of alveolar neutrophil emigration during lung inflammation: role of the CCL2-CCR2 axis. J Immunol. 2003;170:3273–3278. doi: 10.4049/jimmunol.170.6.3273. [DOI] [PubMed] [Google Scholar]
- 4.Narasaraju T, Ng HH, Phoon MC, Chow VT. MCP-1 antibody treatment enhances damage and impedes repair of the alveolar epithelium in influenza pneumonitis. Am J Respir Cell Mol Biol. 2010;42:732–743. doi: 10.1165/rcmb.2008-0423OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huynh ML, Fadok VA, Henson PM. Phosphatidylserine-dependent ingestion of apoptotic cells promotes TGF-beta1 secretion and the resolution of inflammation. J Clin Invest. 2002;109:41–50. doi: 10.1172/JCI11638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xue J, Schmidt SV, Sander J, Draffehn A, Krebs W, Quester I, De Nardo D, Gohel TD, Emde M, Schmidleithner L, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lampropoulou V, Sergushichev A, Bambouskova M, Nair S, Vincent EE, Loginicheva E, Cervantes-Barragan L, Ma X, Huang SC, Griss T, et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan Z, Xie N, Banerjee S, Cui H, Fu M, Thannickal VJ, Liu G. The monocarboxylate transporter 4 is required for glycolytic reprogramming and inflammatory response in macrophages. J Biol Chem. 2015;290:46–55. doi: 10.1074/jbc.M114.603589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tannahill GM, Curtis AM, Adamik J, Palsson-McDermott EM, McGettrick AF, Goel G, Frezza C, Bernard NJ, Kelly B, Foley NH, et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Través PG, de Atauri P, Marín S, Pimentel-Santillana M, Rodríguez-Prados JC, Marín de Mas I, Selivanov VA, Martín-Sanz P, Boscá L, Cascante M. Relevance of the MEK/ERK signaling pathway in the metabolism of activated macrophages: a metabolomic approach. J Immunol. 2012;188:1402–1410. doi: 10.4049/jimmunol.1101781. [DOI] [PubMed] [Google Scholar]
- 12.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, Becker CD, See P, Price J, Lucas D, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yona S, Kim KW, Wolf Y, Mildner A, Varol D, Breker M, Strauss-Ayali D, Viukov S, Guilliams M, Misharin A, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38:79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen WJ, Barthel L, Muldrow A, Oberley-Deegan RE, Kearns MT, Jakubzick C, Henson PM. Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. Am J Respir Crit Care Med. 2011;184:547–560. doi: 10.1164/rccm.201011-1891OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibbings SL, Goyal R, Desch AN, Leach SM, Prabagar M, Atif SM, Bratton DL, Janssen W, Jakubzick CV. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood. 2015;126:1357–1366. doi: 10.1182/blood-2015-01-624809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthay MA, Ware LB, Zimmerman GA. The acute respiratory distress syndrome. J Clin Invest. 2012;122:2731–2740. doi: 10.1172/JCI60331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matute-Bello G, Downey G, Moore BB, Groshong SD, Matthay MA, Slutsky AS, Kuebler WM Acute Lung Injury in Animals Study Group. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol. 2011;44:725–738. doi: 10.1165/rcmb.2009-0210ST. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kearns MT, Barthel L, Bednarek JM, Yunt ZX, Henson PM, Janssen WJ. Fas ligand-expressing lymphocytes enhance alveolar macrophage apoptosis in the resolution of acute pulmonary inflammation. Am J Physiol Lung Cell Mol Physiol. 2014;307:L62–L70. doi: 10.1152/ajplung.00273.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soucie EL, Weng Z, Geirsdóttir L, Molawi K, Maurizio J, Fenouil R, Mossadegh-Keller N, Gimenez G, VanHille L, Beniazza M, et al. Lineage-specific enhancers activate self-renewal genes in macrophages and embryonic stem cells. Science. 2016;351:aad5510. doi: 10.1126/science.aad5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosas M, Davies LC, Giles PJ, Liao CT, Kharfan B, Stone TC, O’Donnell VB, Fraser DJ, Jones SA, Taylor PR. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davies LC, Rosas M, Smith PJ, Fraser DJ, Jones SA, Taylor PR. A quantifiable proliferative burst of tissue macrophages restores homeostatic macrophage populations after acute inflammation. Eur J Immunol. 2011;41:2155–2164. doi: 10.1002/eji.201141817. [DOI] [PubMed] [Google Scholar]
- 22.Tarling JD, Lin HS, Hsu S. Self-renewal of pulmonary alveolar macrophages: evidence from radiation chimera studies. J Leukoc Biol. 1987;42:443–446. [PubMed] [Google Scholar]
- 23.Jenkins SJ, Ruckerl D, Cook PC, Jones LH, Finkelman FD, van Rooijen N, MacDonald AS, Allen JE. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gundra UM, Girgis NM, Ruckerl D, Jenkins S, Ward LN, Kurtz ZD, Wiens KE, Tang MS, Basu-Roy U, Mansukhani A, et al. Alternatively activated macrophages derived from monocytes and tissue macrophages are phenotypically and functionally distinct. Blood. 2014;123:e110–e122. doi: 10.1182/blood-2013-08-520619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zasłona Z, Przybranowski S, Wilke C, van Rooijen N, Teitz-Tennenbaum S, Osterholzer JJ, Wilkinson JE, Moore BB, Peters-Golden M. Resident alveolar macrophages suppress, whereas recruited monocytes promote, allergic lung inflammation in murine models of asthma. J Immunol. 2014;193:4245–4253. doi: 10.4049/jimmunol.1400580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zigmond E, Samia-Grinberg S, Pasmanik-Chor M, Brazowski E, Shibolet O, Halpern Z, Varol C. Infiltrating monocyte-derived macrophages and resident kupffer cells display different ontogeny and functions in acute liver injury. J Immunol. 2014;193:344–353. doi: 10.4049/jimmunol.1400574. [DOI] [PubMed] [Google Scholar]
- 27.Del Prete MJ, Robles MS, Guáo A, Martínez-A C, Izquierdo M, Garcia-Sanz JA. Degradation of cellular mRNA is a general early apoptosis-induced event. FASEB J. 2002;16:2003–2005. doi: 10.1096/fj.02-0392fje. [DOI] [PubMed] [Google Scholar]
- 28.Thomas MP, Liu X, Whangbo J, McCrossan G, Sanborn KB, Basar E, Walch M, Lieberman J. Apoptosis triggers specific, rapid, and global mrna decay with 3′ uridylated intermediates degraded by DIS3L2. Cell Reports. 2015;11:1079–1089. doi: 10.1016/j.celrep.2015.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang L, Xie M, Yang M, Yu Y, Zhu S, Hou W, Kang R, Lotze MT, Billiar TR, Wang H, et al. PKM2 regulates the Warburg effect and promotes HMGB1 release in sepsis. Nat Commun. 2014;5:4436. doi: 10.1038/ncomms5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, Lu Y, Martinez J, Bi Y, Lian G, Wang T, Milasta S, Wang J, Yang M, Liu G, et al. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1α-dependent. Proc Natl Acad Sci USA. 2016;113:1564–1569. doi: 10.1073/pnas.1518000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jha AK, Huang SC, Sergushichev A, Lampropoulou V, Ivanova Y, Loginicheva E, Chmielewski K, Stewart KM, Ashall J, Everts B, et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 2015;42:419–430. doi: 10.1016/j.immuni.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Prados JC, Través PG, Cuenca J, Rico D, Aragonés J, Martín-Sanz P, Cascante M, Boscá L. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 33.Palsson-McDermott EM, Curtis AM, Goel G, Lauterbach MA, Sheedy FJ, Gleeson LE, van den Bosch MW, Quinn SR, Domingo-Fernandez R, Johnston DG, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the Warburg effect in LPS-activated macrophages Cell Metab 20152165–80.[Published erratum appears in Cell Metab 21:347.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stables MJ, Shah S, Camon EB, Lovering RC, Newson J, Bystrom J, Farrow S, Gilroy DW. Transcriptomic analyses of murine resolution-phase macrophages. Blood. 2011;118:e192–e208. doi: 10.1182/blood-2011-04-345330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rath M, Müller I, Kropf P, Closs EI, Munder M. Metabolism via arginase or nitric oxide synthase: two competing arginine pathways in macrophages. Front Immunol. 2014;5:532. doi: 10.3389/fimmu.2014.00532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redente EF, Higgins DM, Dwyer-Nield LD, Orme IM, Gonzalez-Juarrero M, Malkinson AM. Differential polarization of alveolar macrophages and bone marrow-derived monocytes following chemically and pathogen-induced chronic lung inflammation. J Leukoc Biol. 2010;88:159–168. doi: 10.1189/jlb.0609378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haschemi A, Kosma P, Gille L, Evans CR, Burant CF, Starkl P, Knapp B, Haas R, Schmid JA, Jandl C, et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.He C, Carter AB. The metabolic prospective and redox regulation of macrophage polarization. J Clin Cell Immunol. 2015;6:pii:371. doi: 10.4172/2155-9899.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolf AJ, Reyes CN, Liang W, Becker C, Shimada K, Wheeler ML, Cho HC, Popescu NI, Coggeshall KM, Arditi M, et al. Hexokinase is an innate immune receptor for the detection of bacterial peptidoglycan. Cell. 2016;166:624–636. doi: 10.1016/j.cell.2016.05.076. [DOI] [PMC free article] [PubMed] [Google Scholar]