Abstract
FUT2 is a gene for a fucosyltransferase that encodes expression of ABO blood group antigens found on gastrointestinal mucosa and secretions. We hypothesized that the fecal microbiomes of healthy subjects, with blood group antigens A, B, and O, have differing compositions. We analyzed 33 fecal and blood specimens from healthy subjects for FUT2 genotype, and the fecal microbiome was determined by 454 pyrosequencing. Our data show that being a blood group secretor is associated with less diversity at higher orders of taxonomy; and the presence of blood group A antigens in the secretor subjects are associated with an expansion families of bacteria within the gut. Furthermore, our study confirms the previous findings that secretors and nonsecretors have differing bacterial taxa. This extends the previous findings by demonstrating that the impact of being a nonsecretor is higher than that of individual blood group antigens. Additionally, we demonstrate that both secretor status and blood group antigen expression especially affect the Lachnospiraceae family of bacteria within the gut microbiome, with lower abundances noted in nonsecretors and higher abundances in secretors of various blood groups. We further note specific differences in blood group A-secretors demonstrating that the genus Blautia is lower in the group A-secretors compared with the non-A-secretors and that this reduction is accompanied by higher abundances of members of the Rikenellaceae, Peptostreptococcaceae, Clostridiales, and Turicibacter. This study offers a first insight into the relationship between the fecal microbiome and blood group antigens in secretors.
Keywords: fucosyltransferase- FUT2, blood group antigens, bacteria, gastrointestinal microbiome, gastrointestinal secretions
humans harbor a large number of bacterial and microbial species on and inside their body, and there is considerable variation in the resident microbiome composition from one person to another (31). The major factors that affect variability in microbiome composition between individuals have largely been unexplained and can be important to understanding both health and disease related processes ranging from skin diseases to oral, vaginal, and gastrointestinal (GI) health. In fact, the hygiene hypothesis and its supporting data suggest that sterility in the environment can lead to increased frequency of autoimmune diseases such as asthma, allergies, eczema, and inflammatory bowel disease (22). Data to date also implicate gut microbiota composition in the development of cancer, infectious illnesses (i.e., Clostridium difficile colitis), cardiovascular disease, and various other diseases (19, 29, 37). Therefore, understanding the reasons for the variation in microbiome composition in humans has the potential to improve our ability to manipulate the microbiome for therapeutic gains in health maintenance and disease management (1, 20).
Current data also suggest that microbiome composition in the human gut is partly due to genetic and partly due to environmental factors. Additionally, the interpersonal variation in bacterial composition is actually higher than temporal variation due to external factors, such as dietary changes (5, 9, 22). The human gut mostly harbors four main bacterial phyla: Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria. The majority of the interpersonal variation in the gut bacterial microbiome occurs at the lower orders of taxonomy (4). Little is known about the various genetic determinants of gut microbiota composition.
One such gene implicated in playing a role in gut microbiota composition is fucosyltransferase-2 (FUT2). FUT2 encodes α-1, 2-fucosyltransferase for the expression of ABH blood group antigens on mucosal surfaces (8, 9). Polymorphisms within the FUT2 gene have been relatively well described (8, 34) and determine the ability to secrete blood group antigens into gastrointestinal secretions. Individuals who are nonsecretors of blood group antigens have homozygous noncoding variants in FUT2 and do not have ABH antigens expressed on the mucosal secretions and surfaces (13). Such single nucleotide polymorphisms (SNP) that lead to a nonsecretor trait are found in 20% of the European and African populations (13). The most common of these is the rs601338 (W143X, G428A) polymorphism, noted in Western populations (33). The nonsecretor homozygous genotype in FUT2 has been linked to disease, has been found to be associated with protection from Norovirus gene group-2 (GG2) infections and Helicobacter pylori gastritis (in which the bacterial organisms attach to the gastric epithelium via H and Lewis b antigens) (8, 13), and has also been related to the gut microbiome changes in patients with inflammatory bowel disease (20).
Despite this known association between FUT2 and the gut microbiota and microbiota-related illnesses, there is little to no data about the relationship of overall gut bacterial microbiome composition and common blood group antigens (A, B, O) in secretors specifically (15). Furthermore, enzymes that degrade the common blood group antigens have been known to exist in the feces since the 1930s (23–25), and studies thereafter have confirmed their bacterial origin and specificity for particular blood groups (11).
In this study, we aimed to examine the associations between gut bacterial microbiome composition and ABO blood group antigens in healthy individuals who are secretors of these antigens based on FUT2 polymorphisms. We hypothesized that there would be significant differences between those who are secretors of blood group antigens and those who are not. We also hypothesized that there would be differences between secretors of the A, B, and O blood group antigens and gut microbiota composition.
MATERIALS AND METHODS
Subjects and samples.
Healthy subjects (n = 33) were recruited via flyers and research advertisements at Rush University Medical Center (RUMC) as part of two ongoing studies (ORA: 07121103; ORA: L04092807). All of these healthy subjects gave written informed consent to the use of their samples (i.e., blood and feces) and data and were also part of an RUMC Institutional Review Board-approved GI repository. The collection procedures were the same for all subjects. Subjects were instructed to keep the fecal samples in a freezer at home and to bring them within 24 h of defecation to the outpatient clinic on ice. Blood and fecal samples were both stored at −80°C until the time of analysis. All subjects completed RUMC structured demographic, lifestyle, and medical history questionnaires. All questionnaire packets were labeled with a sequential patient number to maintain patient confidentiality and serve as the patient identifier for the study. All work for this study was carried out in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) for experiments involving humans and Uniform Requirements for manuscripts submitted to biomedical journals published by the International Committee of Medical Journal Editors.
Inclusion criteria of the healthy subjects included: a normal medical history, complete blood count, and comprehensive metabolic profile. Exclusion criteria for the subjects included: 1) any colonic diseases (i.e., inflammatory bowel disease, irritable bowel syndrome) including primary gastrointestinal malignancies and 2) the use of antibiotics and immunosuppressants within 3 mo before sample collection. The characteristics of the subjects (n = 33) are shown in Table 1.
Table 1.
Demographic characteristics of study subjects
| All Subjects | ||||
|---|---|---|---|---|
| Subject Characteristics (n = 33) | A (n = 10) | O (n = 10) | B (n = 10) | AB (n = 3) |
| Age, yr | 45.5 ± 16.0 | 40.0 ± 11.5 | 48.6 ± 10.0 | 65 ± 12.5 |
| Sex, men/women | 5/5 | 5/5 | 4/6 | 3/0 |
| Race | ||||
| White (Caucasian) | 8 (80%) | 8 (80%) | 4 (40%) | 2 (67%) |
| Black (African American) | 2 (20%) | 2 (20%) | 6 (60%) | 1 (33%) |
| Ethnicity | ||||
| Non-Hispanic/Latino | 8 (80%) | 9 (90%) | 10 (100%) | 3 (100%) |
| Hispanic/Latino | 2 (20%) | 1 (10%) | 0 (0%) | 0 (0%) |
| Secretor Subjects | ||||
| Secretors (n = 27) | A (n = 8) | O (n = 9) | B (n = 8) | AB (n = 2) |
|---|---|---|---|---|
| Age, yr | 45.8 ± 18.0 | 47.8 ± 11.4 | 47.5 ± 10.6 | 72.0 ± 4.24 |
| Sex, men/women | 4/4 | 5/4 | 3/5 | 2/0 |
| Race | ||||
| White (Caucasian) | 7 (88%) | 8 (89%) | 3 (37.5%) | 2 (100%) |
| Black (African American) | 1 (12%) | 1 (11%) | 5 (62.5%) | 0 (0%) |
| Ethnicity | ||||
| Non-Hispanic/Latino | 6 (75%) | 8 (89%) | 8 (100%) | 2 (100%) |
| Hispanic/Latino | 2 (25%) | 1 (11%) | 0 (0%) | 0 (0%) |
| Nonsecretor Subjects | ||||
| Nonsecretors (n = 6) | all (n = 6) |
| Age, yr | 51.0 ± 7.6 |
| Sex, men/women | 3/3 |
| Race | |
| White (Caucasian) | 2 (33%) |
| Black (African American) | 4 (67%) |
| Ethnicity | |
| Non-Hispanic/Latino | 6 (100%) |
| Hispanic/Latino | 0 (0%) |
n, Number of subjects; % = Percent; means ± SD.
Blood typing.
Blood group antigens were determined by reverse blood typing kits with Reagent Red Blood Cells (Bio-Rad Laboratories, Hercules, CA) and determined by back-typing, using the serum samples of the subjects, as is typically done in our blood bank. The sera were mixed with commercial red blood cell reagents (Bio-Rad) and placed in the centrifuge at room temperature at 3,500 rpm for 15 s. The samples were each assessed for coagulation to determine presence of antibodies to the A and B antigens according to standard blood bank procedures. Based on the antibodies present in the samples and whether coagulation was present, blood type was determined for each subject.
16S rDNA sequencing of fecal samples.
The fecal microbiome was sequenced using Roche 454 pyrosequencing in (n = 33) healthy subjects. DNA was extracted with a commercially available kit, FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH) using the manufacturer’s recommended protocol. The adequacy of the amount of extracted DNA from samples was verified with fluorometric quantitation (Qubit, Life Technologies, Grand Island, NY), and samples with inadequate amounts of template DNA were not sequenced. Forward primer 28F: 5′ GAGTTTGATCNTGGCTCAG 3′ and reverse primer 519R: 5′ GTNTTACNGCGGCKGCTG 3′ were used to pyrosequence the 16S rDNA on a 454 GS FLX platform, with barcoding, using titanium kits at Research and Testing Laboratories (Lubbock, TX) for the analysis of the bacterial 16S rRNA phylotypes (28). Python scripts in the Quantitative Insights Into Microbial Ecology (QIIME) software pipeline (VirtualBox versions 1.5, 1.6, and 1.7) and other custom scripts were used to process the sequencing files at Research and Testing Laboratories and Rush University (3, 12, 27, 36). The sequence outputs were filtered for low-quality sequences (defined as any sequences that are <200 bps or >1,000 bps; sequences with any nucleotide mismatches to either the barcode or primers; sequences with homopolymer runs >6; sequences with an average quality score of <25; sequences with ambiguous bases >6; and sequences truncated at the reverse primer). Sequences were denoised with USEARCH (6), and chimera were checked with UCHIME (7) and Chimera Slayer (10). Operational taxonomic units (OTUs) were picked with uclust (6) at a 97% similarity threshold, and representative sequences were generated. Sequences were aligned with PyNAST (2), and taxonomy assignment was performed in QIIME 1.6VB against the QIIME 1.6 version of Greengenes database (16, 35) using the RDP classifier (35) at a 80% bootstrap value threshold. An approximately-maximum-likelihood phylogenetic tree was created using FastTree v2.1.3 (18). Multivariate reduction analyses using principal coordinates with a Unifrac metric was done to determine the global microbiome composition based on OTUs using QIIME VB 1.6 distances. Analysis of similarities (ANOSIM) implementation in QIIME VB 1.7 was used to perform a randomization test of significance of pseudo F values, with 999 randomizations for each model, on rarified sequence data and was used to statistically assess differences in beta diversity. Linear discriminant analysis effect size (LEFSE) was calculated (26), and graphs of the data were generated using its open source Galaxy implementation at the Huttenhower laboratory website (https://huttenhower.sph.harvard.edu/galaxy/). Bacterial taxa that were not present in at least 10 of the samples (i.e., taxa that were present in only a handful of subjects) were not reported as differentially abundant, to prevent erroneous results stemming from absence of taxa rather than their presence. SPSS (v.19.0.0; Chicago, IL) was used to analyze clinical metadata. In SPSS, t-tests or ANOVA was used to analyze differences for parametric data satisfying test assumptions; Kruskal-Wallis, Mann-Whitney, or median tests were used to analyze nonparametric data; χ2 or Fisher exact tests were used to detect differences in proportions between groups, as appropriate. Microsoft Excel and PowerPoint were also used to generate plots of bacterial taxa.
FUT2 genotyping.
The subjects were genotyped for FUT2 using a standard PCR-Sanger sequencing protocol: Two primers were designed spanning an 806 bp region around the SNP site (rsID# 601338) and were used for amplification. The primers (CCTTTCTCCTTTCCCATG and AAATCTTTGGCAGGTGAG) were synthesized as standard oligonucleotides by Integrated DNA Technologies (IDT, Coralville, IA). PCR reactions were established using DreamTaq Green 2× PCR mastermix in total volumes of 25 μl per reaction. PCR conditions were 95°C for 5 min, followed by 35 cycles of 95°C for 30 s, 62°C for 30 s, and 72°C for 2 min. A final step of 72°C for 5 min was performed. PCR reactions were purified using a 0.6X AMPure XP cleanup (Agencourt, Beckman Coulter). Sequencing was performed on an ABI 3730xl Sequencer (Applied Biosystems, Foster City, CA). Primer design, amplification, and sequencing were performed at the DNA Services Core Facility at the University of Illinois at Chicago.
RESULTS AND DISCUSSION
Subject demographics.
Demographic information about the healthy subjects (n = 33) is shown in Table 1. Six subjects were nonsecretors of blood group antigens by FUT2 genotyping, and 27 were secretors (with 22 of those being heterozygote secretors and 5 being homozygote secretors). There were no statistically significant differences between age, sex, race and ethnicity, and FUT2 genotype in those with a blood group A, B, O, and AB (P > 0.05, all). There were no statistically significant demographic differences in age, sex, race and ethnicity, and FUT2 genotype between secretors and nonsecretors of blood group antigens (P > 0.05, all). Among the secretors, there were more African American subjects in those with blood group B (P = 0.045).
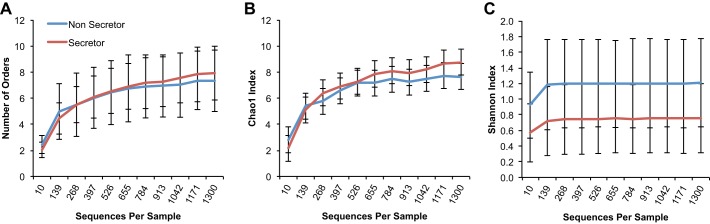
Nonsecretors have a trend for higher alpha diversity (i.e., within sample diversity) compared with secretors at the phylum, class and order level, but this difference is not detectable at the lower levels of taxonomy such as the family, genus, and OTU level.
At the OTU level, we noted no statistically significant differences between secretors and nonsecretors in alpha diversity measures, such as richness of the number of OTUs, Chao1 richness, Shannon index, Phylogenetic Diversity Whole Tree (PD-WT) index (data not shown). However, secretors had numerically higher means for richness, Chao1, and Shannon indexes, similar to what has been reported in prior publications (34). At higher levels of taxonomy, however, instead of secretors being more diverse, we noted a trend for a higher diversity in the nonsecretors starting at the order level in the Shannon diversity index (P = 0.069) compared with the secretors (Fig. 1). This trend continued to show a higher diversity in the nonsecretors at the class level (P = 0.076) (data not shown) and at the phylum level (P = 0.069) (data not shown) in the Shannon index, suggesting that the nonsecretor status may affect microbiota composition at higher levels of taxonomy, differently. The observed number of orders and the Chao1 richness at the phylum (data not shown), class (data not shown), and order levels were not different (Fig. 1); suggesting the trend for the higher diversity noted in the nonsecretors is related to the changes in relative abundance of various bacterial taxa, rather than increases in the number of bacterial taxa (i.e., their presence). This finding suggests that having the secretor gene may select for certain bacterial phyla, classes, and orders, and particular taxa within the latter, and then expand into numerous OTUs, whereas nonsecretors are less selective at higher orders of taxonomy and allow for multiple phyla, classes, and orders that compete to determine microbiome composition.
Fig. 1.
Shannon index trends higher in nonsecretors vs. secretors at the order level. y-Axis shows alpha diversity as assessed by the following indexes: Richness (i.e., total number of orders) (P > 0.05) (A), Chao1 index (P > 0.05) (B), and Shannon index (P = 0.069) (C). x-Axis depicts sequences per sample. Statistical comparisons were made at the maximum rarified depth.
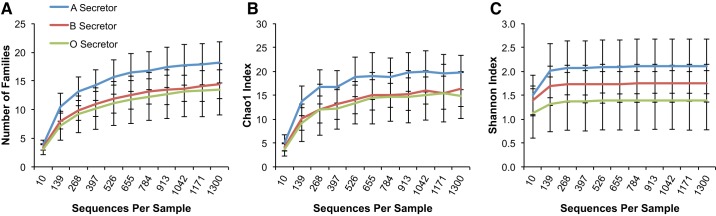
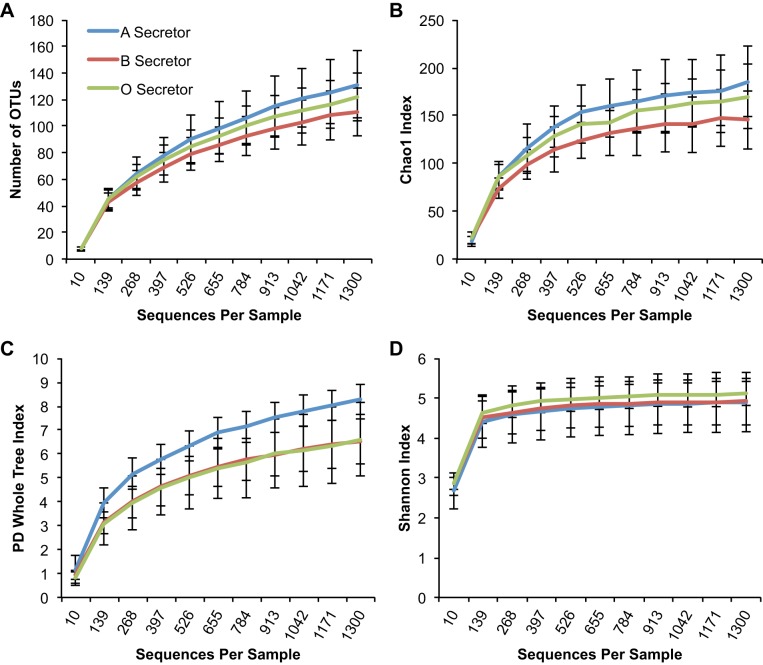
Blood group A-secretors are phylogenetically more diverse than other blood group secretors at the OTU level.
When the effect of blood groups A, B, and O was examined in the secretor subset of subjects, A-secretors had an increased number of families (P = 0.029) and Chao1 richness at the family level (P = 0.044) compared with other secretors carrying blood group antigens B and O (Fig. 2). Furthermore, A-secretors had a higher PD-WT index (P = 0.041) [Fig. 3, showing an increased phylogenetic diversity within this group, compared with non-A-secretors (i.e., B-and O-secretors)]. Interestingly, the two individuals who were AB-secretors appeared to have a very similar PD-WT index to A-secretors, and the PD-WT index was numerically higher than that of B- and O-secretors. These findings suggest that the presence of blood group A antigens in the gut mucosal surface in secretor subjects is associated with an expansion of families of bacteria, and blood group A secretion may confer an environment that is more accommodating to a greater number of bacterial families.
Fig. 2.
Alpha diversity is higher in A-secretors vs. non-A-secretors at the family level. Among the secretor samples, blood group A-secretors have a significant increase in both the number of families and Chao1 index compared with blood group B- and O-secretors. y-Axis shows alpha diversity as assessed by the following indexes: Richness (i.e., total number of families) (P = 0.029) (A), Chao1 index (P = 0.044) (B), and Shannon index (P > 0.05) (C). x-Axis depicts sequences per sample. Statistical comparisons were made at the maximum rarified depth.
Fig. 3.
Phylogenetic diversity is higher in A-secretors vs. non-A-secretors at the Operational taxonomic unit (OTU) level. Among the secretor samples, blood group A-secretors have a significant increase in the Phylogenetic Diversity (PD)-Whole Tree index compared with blood group B- and O-secretors. y-Axis shows alpha diversity as assessed by the following indexes: Richness (i.e., total number of OTUs) (P > 0.05) (A), Chao1 index (P > 0.05) (B), PD-Whole Tree Index (P = 0.041) (C), Shannon index (P > 0.05) (D). x-Axis depicts sequences per sample. Statistical comparisons were made at the maximum rarified depth.
Secretors and nonsecretors have a differing bacterial composition (i.e., differ in beta diversity).
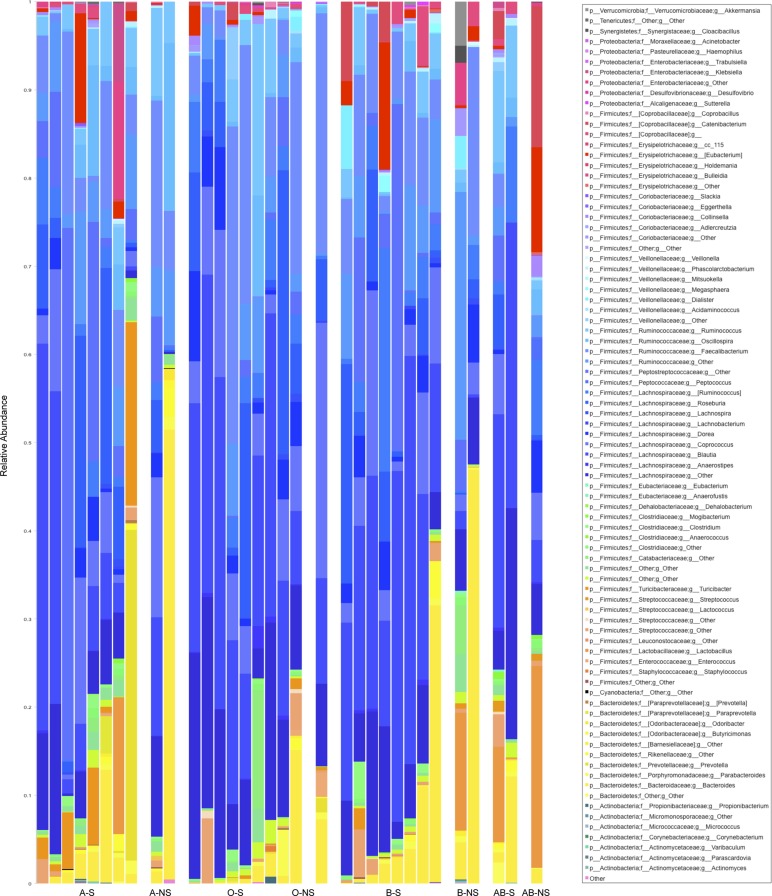
The stacked histogram of bacterial phyla and genera taxa present in our samples are shown in Fig. 4. We calculated the unweighted and weighted Unifrac distances between all pairs of samples and compared secretors vs. nonsecretors with ANOSIM. When all samples were analyzed, the microbiota compositions in the secretors and the nonsecretors were significantly different according to both the weighted and unweighted Unifrac metric (ANOSIM: R = 0.35, P = 0.034 and R = 0.34, P = 0.022, respectively). When we also took into account the blood groups in addition to the secretor status in all samples, we found that the differences in bacterial composition remained significant by the weighted Unifrac metric (ANOSIM: R = 0.14, P = 0.05), and the unweighted Unifrac metric (ANOSIM: R = 0.17, P = 0.022), albeit at lower R values. This finding implies that the differences noted in bacterial composition are more strongly associated with secretor status than blood group. Among the secretors, we did not find any differences between heterozygote secretors vs. homozygote secretors with either the unweighted Unifrac or the weighted Unifrac metric (data not shown).
Fig. 4.
Stacked histogram of bacterial genera by secretor status and blood group. Each column in the stacked histogram represents 1 sample and has different color bars within it, and these bars are proportional to the percent relative bacterial abundance of each genera within the sample, summing up to 100%. y-Axis represents the relative abundance (i.e., percent abundance) of each genus. The order of samples is organized along the x-axis by blood group and secretor status: A-S (A-secretors); A-NS (A-nonsecretors); O-S (O-secretors); O-NS (O-nonsecretors); B-S (B-secretors); B-NS (B-nonsecretors); AB-S (AB-secretors); AB-NS (AB-nonsecretors) going from left to right of the graph. Total number of subjects is 33.
Blood group A-secretors have a differing dominant bacterial composition than secretors of other blood group antigens by Unifrac analysis.
Because we found that secretor status affected bacterial composition significantly, we then did a second analysis, in which we eliminated nonsecretors and examined only the secretor subjects to determine the effect of individual blood groups on bacterial composition. The elimination of the nonsecretors was done to make sure the differences by the presence of a specific blood group antigen were not masked by the relatively large effect of being a nonsecretor. When we compared the three blood groups A, B, O among the secretors only, we noted no significant differences in overall bacterial composition using the unweighted and weighted Unifrac metrics and the ANOSIM test (R = 0.03, P = 0.25; R = 0.02 P = 0.24, respectively). However, when we examined boxplots of Unifrac distances in the secretors between those within blood group A, those within blood group B, and those within blood group O, there were notable statistically significant differences (Fig. 5, A and B), suggesting that samples from A-secretors are farther from each other and that A-secretors harbor bacterial compositional differences from non-A-secretors.
Fig. 5.

Boxplots of Unifrac distances between of secretors of the same blood group. A: unweighted Unifrac distances in the y-axis. B: weighted Unifrac distances in the y-axis. The distribution of distances between each pair of A-secretors (i.e., blood group A to blood group A secretor distances) is shown at left along the x-axis. The distribution of distances between each pair of B-secretors (i.e., blood group B to blood group B secretor distances) is shown in the mid part of the graph along the x-axis. The distribution of distances between each pair of O-secretors (i.e., blood group O to blood group O secretor distances) is shown right along the x-axis. Horizontal black lines reflect median values. Significant P values are shown only, by blue horizontal lines, and represent Student t-test results, adjusted for multiple comparisons using a Bonferroni correction.
To further investigate this latter finding, we then looked at the bacterial composition in A-secretors against all other secretors (i.e., the group B- and O-secretors) using weighted Unifrac distances: There was a significant difference in overall bacterial composition (ANOSIM: R = 0.27, P = 0.023). On the contrary, the unweighted Unifrac distances did not show a difference in composition of blood group A-secretors to other blood group secretors (ANOSIM: R = 0.07, P = 0.253). These findings also held true when combining blood group A with those two secretor subjects who had blood group AB and comparing them against all other secretor subjects (data not shown). Weighted Unifrac takes into account bacterial composition, as well as abundance, as opposed to unweighted Unifrac, which treats each bacterial OTU as equal and is more comparable with a presence or absence type of analysis. As such, finding significant differences with the weighted Unifrac analysis but not finding such differences with unweighted Unifrac analysis suggests that there is an effect of blood group A secretion on the dominant (i.e., more abundant) bacterial taxa rather than all taxa (including those that are rare).
Lastly, we also looked at the bacterial composition in B-secretors against all other secretors (i.e., the group A- and O-secretors) using the unweighted Unifrac distances because unweighted Unifrac distances were significantly shorter among B-secretors compared with O-secretors and A-secretors (Fig. 5A). However, using the ANOSIM test, we noted no differences in overall bacterial composition in the B-secretors against non-B-secretors.
Bacterial taxa associated with secretors and with various blood groups.
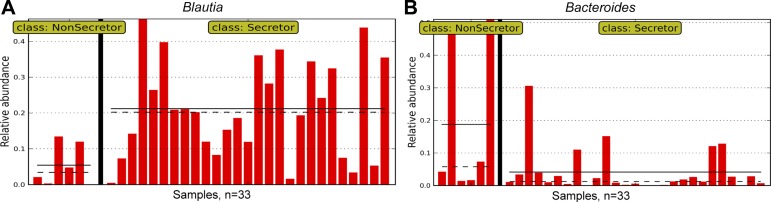
To determine which bacterial taxa were specifically associated with the compositional changes noted, we conducted an LEFSE analysis that examines for large differences in bacterial composition and is adjusted for multiple comparisons. When LEFSE analysis was done comparing secretors to the nonsecretors, at the genus level, the abundance of Blautia (within the Lachnospiraceae family) appeared to be higher in the secretors with a linear discriminant analysis (LDA) score (log 10) of 4.895 (Fig. 6A), whereas the abundance of the Bacteroides genus (within the Bacteroidaceae family) appeared lower with an LDA score (log10) of −4.95 (Fig. 6B). These data confirm the above beta diversity findings that certain microbiota could be preferentially selected to reside and grow in the gastrointestinal tract based on the presence of the secretor phenotype.
Fig. 6.
Linear discriminant analysis effect size (LEFSE) analysis indicates genera associated with secretor status. The genus Blautia is more abundant in the secretors compared with nonsecretors; and the genus Bacteroides is more abundant in nonsecretors compared with secretors. Each column represents the relative abundance in a single sample. y-Axis represents the relative abundance (i.e., percent abundance) value. A: the relative abundance of Blautia in all samples. B: the relative abundance of Bacteroides in all samples. The nonsecretor subjects are shown to the left of the graph, and the secretor subjects are shown to the right. The continuous and dotted black horizontal lines represent mean and median abundance values, respectively.
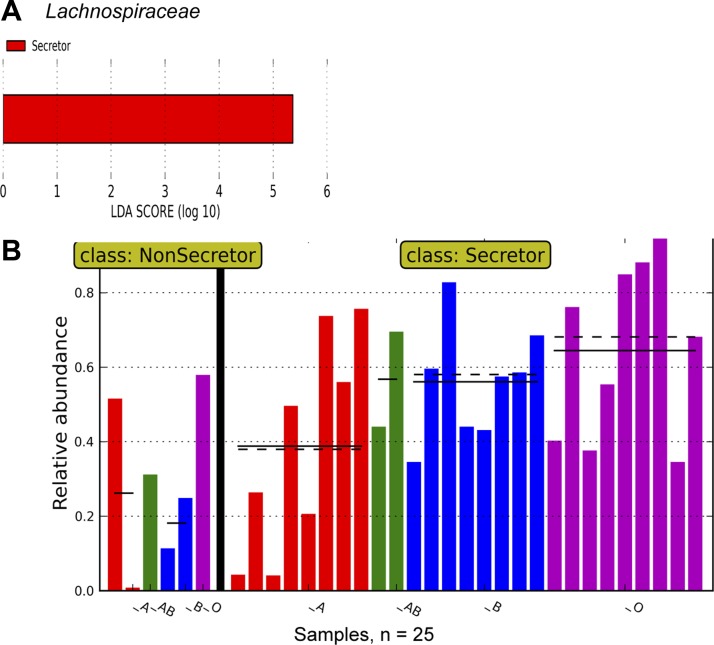
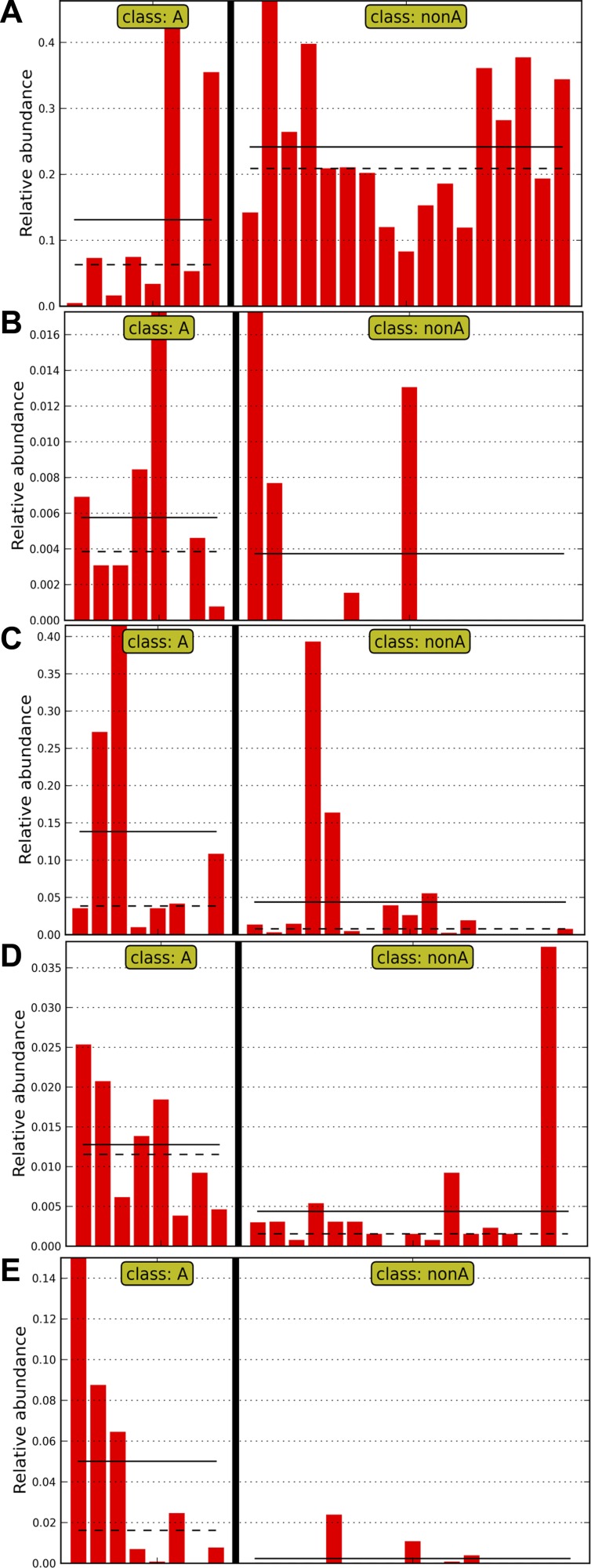
When we performed a nested analysis, comparing secretors to nonsecretors first and then comparing individual blood groups, at the family level, there was a significant increase in Lachnospiraceae in secretors with an LDA score (log 10) >5 (Fig. 7, A and B). Interestingly, there also seemed to be a gradation of bacterial abundance in Lachnospiraceae, with the lowest relative abundance in blood group A-secretors and the highest relative abundance in blood group O-secretors (Fig. 7B). When we examined which Lachnospiraceae were affected, it was notable that Blautia was lower in the group A-secretors compared with the non-A-secretors, with an LDA score (log 10) of −4.80 (Fig. 8A). This reduction was accompanied by a rise in other taxa in the A-secretors: There were increases in unclassified bacteria within the Rikenellaceae (LDA score (log10) = 3.66) (Fig. 8B) and Peptostreptococcaceae families (LDA score (log10) = 4.60) (Fig. 8C); in unclassified bacteria within the Clostridiales order (LDA score (log10) = 3.93) (Fig. 8D); and Turicibacter (LDA score (log10) = 4.44) (Fig. 8E) in the A-secretors compared with the non-A-secretors.
Fig. 7.
LEFSE analysis by secretor status and blood group indicates a rise in Lachnospiraceae in secretors and variation in Lachnospiraceae abundance by blood group among the secretors. A: the results of LEFSE analysis when both secretor status (primary grouping) and blood group (subgrouping) are taken into account in the analysis, and the linear discriminant analysis (LDA) score (log 10) in secretors vs nonsecretors is shown, depicting an increased relative abundance of Lachnospiraceae. B: the relative abundance of Lachnospiraceae in all samples. Each column represents the relative abundance in a single sample. y-Axis represents the relative abundance (i.e., percent abundance) value. The nonsecretor subjects are shown to the left of the graph, and the secretor subjects are shown to the right. Samples from individuals with blood group A are colored red, those with blood group AB are colored green, those with blood group B are colored blue, and those with blood group O are colored magenta. Lachnospiraceae are least abundant in A-secretors with an increasing abundance in B-secretors and the highest abundance in O-secretors. The continuous and dotted black horizontal lines represent mean and median abundance values, respectively.
Fig. 8.
LEFSE analysis indicates bacterial taxa associated with A-secretors vs. non-A-secretors (i.e., B-secretors and O-secretors). In all of the panels, each column represents the relative abundance in a single sample, and the y-axis represents the relative abundance (i.e., percent abundance) value. Samples from subjects who are blood group A-secretors are shown to the left of the graph, and those from non-A-secretors (i.e., B-secretors and O-secretors) are shown to the right. The continuous and dotted black horizontal lines in each panel represent mean and median abundance values, respectively. A: within the family Lachnospiraceae, the genus Blautia is more abundant in the non-A-secretor samples compared with A-secretor samples. Conversely, genera within the Rikenellaceae family shown in B, genera within the Peptostreptococcaceae family shown in C, genera within the Clostridiales order shown in D, and Turicibacter shown in E are more abundant in A-secretor samples.
Conclusions
This study offers a first insight, using sequencing technology, into the relationship between fecal microbiome composition and ABO blood group antigens. Our data show that being a blood group secretor is associated with less diversity at higher orders of taxonomy, and the presence of blood group A antigens in the gut mucosa in secretor subjects is associated with an expansion of families of bacteria within the gut. Furthermore, our study confirms the previous findings that secretors and nonsecretors harbor differing bacterial taxa (15, 20, 32–34) and extends the previous findings by demonstrating that the impact of being a nonsecretor on gut bacterial microbiome composition is higher than that of an individual ABO blood group (15, 33, 34). Additionally, we demonstrate that both secretor status and ABO blood group antigen expression especially affect the Lachnospiraceae family of bacteria within the gut microbiome with lowest abundances noted in nonsecretors and increasing abundances in secretors, in the order of blood group A to B to O. Moreover, we note further specific bacterial taxa differences in blood group A-secretors. These findings suggest that part of the interindividual variation in the GI tract microbiome could be related to the expression of blood group antigens and secretor status.
To date, there has been only a single published study that has examined the association between A, B, O histo-antigens and gut microbiota composition (15). In this prior study, %G+C nucleotides were analyzed in DNA from fecal samples coming from (n = 11–17) secretor subjects per blood group from blood group A, B, O, and AB. Subjects were primarily female (≥85%). A higher %G+C was noted in samples from blood group A-secretors. A denaturing gradient gel electrophoresis (DGGE) was also done in this latter study, and the microbiota fingerprints in subjects having the B antigen (i.e., B- and AB-secretors) seemed different than A- and O-secretors. The changes in %G+C content in A-secretors in this study mirror our observations and certainly suggest the presence of microbial taxa rich in these nucleotides. Interestingly, one can also note that Lachnospiraceae low in A-secretors are a member of the Firmicutes phylum, which is traditionally low in G+C content. The exact taxa responsible for %G+C change were not detectable in the prior study probably due to the low resolution of the DGGE technique, which was a state of the art bacterial profiling in the past. The high-throughput sequence technique employed in our study is superior in its resolution to DGGE for detecting relative abundance differences, and this is likely the reason we are able to note compositional differences that were not detected previously.
The mechanism involved in the microbial associations and blood groups observed in our study may be related to the chemical structure of the various blood group antigens: In the case of O-secretors, only the H-antigen composed of a chain of β-D-galactose, β-D-N-acetylglucosamine, β-D-galactose, and 2-linked, α-L-fucose is present. For the secretion of the A-antigen, a glycosyltransferase bonds an α-N-acetylgalactosamine residue to the D-galactose end of the H antigen. For the secretion of the B-antigen, a different glycosyltransferase bonds α-D-galactose to the D-galactose end of the H-antigen. It is certainly conceptually plausible that certain bacterial taxa may have a selective advantage by producing enzymes that specifically target and cleave these different residues on the host’s secretions. For example, most Lachnospiraceae are known to possess β-galactosidases (30), and therefore it may not be surprising to note them more abundantly in B- and O-secretors since the chemical structure of both blood groups B and O antigens contain D-galactose residues in the terminal part of the chain. Furthermore, it has also been well described that certain bacterial strains can have cell surface transporters allowing for preferential utilization of N-acetylgalactosamine in some bacteria (14), which may confer a selective advantage to colonizing A-secretors. Unfortunately, structural differences in bacterial utilization and transport pathways have been characterized only for a small fraction of the intestinal bacteria in general, preventing any definitive conclusions about individual bacterial families observed in our study. Furthermore, such studies may be difficult to undertake owing to the challenges in growing multiple members of the anaerobic bacteria that may allow bacteria the ability to adapt and change their carbohydrate utilization, depending on environmental conditions.
The strengths of this study include using high-throughput technologies to examine both known and unknown bacterial taxa, examination of not only secretor status but also the impact of blood groups, and the analyses of the results at various levels of taxonomy to deduce patterns. Of additional consideration is the fact that our samples were collected in an attempted match by age, sex, and race across blood types studied here, to decrease confounding as much as possible.
Our limitations include the fact that we have used fecal samples, which carry advantages and disadvantages: While fecal samples are thought to grossly mirror members of the GI community on the mucosal surface, especially in the healthy state, it is plausible that mucosa-associated bacteria (such as those obtained by biopsies of the colon) could be affected by FUT2 expression to a larger extent than those found in fecal samples. Other possibilities also include that mucosal microbes are those bacteria that are long-term commensal agents selected by the host, while microbes within the fecal stream in the lumen are more transient bacteria (17). In addition to the effects of secretions at the mucosal surface, luminal secretions such as saliva and gastric and intestinal juices could contain variable levels of secretion of blood group antigens, affecting microbiome composition in a given individual. These types of samples were not examined in our study. Another limitation of our study is the sample size. While our sample size is comparable to other published studies mentioned above, our findings should be confirmed with a bigger sample size to increase the reliability and validity of results. Thus, our current findings provide the rationale for undertaking a bigger study. Furthermore, it would be highly valuable to examine microbiome composition in those individuals who are AB-secretors, to gain further insights into the relationship between blood group antigens and gut microbiome composition. Such a study would require a large number of subjects to be screened, when one considers that only 4% of the population has both blood group antigens A and B (21). Lastly, our study has not factored in the influence of environmental variables in microbiome composition such as diet in humans in conjunction with FUT2 expression, which should be explored in future work.
In conclusion, this study takes us one step further toward understanding the interindividual variations in the gut microbiome. Our findings not only suggest an impact of being able to secrete blood group antigens into GI secretions but also suggest that blood group antigens themselves could help shape gut bacterial composition in health. Going forward, studies examining gut microbiome composition in humans should take into account secretor status and blood group. Further understanding the relationship between blood group antigens and gut bacterial colonization could enable individualized and/or targeted (hence potentially more effective) modulation of the microbiome with various interventions such as customized pre- or probiotics or fecal transplantation for therapeutic gains both for preserving health and for disease treatment.
GRANTS
This study was partially supported with funds through National Institutes of Health Grants DK-071838 and CA-204808 to E. A. Mutlu.
DISCLOSURES
Research and Testing Laboratory, Lubbock, TX, and DNA Services core facility at the University of Illinois at Chicago, Chicago, IL, were paid for the work performed.
AUTHOR CONTRIBUTIONS
A.G. and P.A.E. performed experiments; A.G. and E.A.M. interpreted results of experiments; A.G., P.A.E., and R.S. prepared figures; A.G. drafted manuscript; A.G., P.A.E., R.S., and E.A.M. edited and revised manuscript; E.A.M. conceived and designed research; E.A.M. analyzed data; E.A.M. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank all of the subjects participating in this study. The authors thank Dr. Yan Sun for performing the pyrosequencing experiments and Dr. Lars Koenig for initial bioinformatics analyses at Research and Testing Laboratory, Lubbock, TX. The authors thank Dr. Stefan Green and the staff at DNA Services Core Facility at the University of Illinois at Chicago, Chicago, IL, for the FUT2 genotyping work. The authors also thank Patricia Hano and Dr. Bruce McLeod at the Rush University Blood Bank who provided supplies for blood typing and taught and supervised A. Gampa in the technical aspects of the blood typing assay. The authors thank Sue L. Mikolaitis for assistance in locating subject data and samples from the tissue bank.
REFERENCES
- 1.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, Antolín M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, Foerstner KU, Friss C, van de Guchte M, Guedon E, Haimet F, Huber W, van Hylckama-Vlieg J, Jamet A, Juste C, Kaci G, Knol J, Lakhdari O, Layec S, Le Roux K, Maguin E, Mérieux A, Melo Minardi R, M’rini C, Muller J, Oozeer R, Parkhill J, Renault P, Rescigno M, Sanchez N, Sunagawa S, Torrejon A, Turner K, Vandemeulebrouck G, Varela E, Winogradsky Y, Zeller G, Weissenbach J, Ehrlich SD, Bork P; MetaHIT Consortium . Enterotypes of the human gut microbiome. Nature 473: 174–180, 2011. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26: 266–267, 2010. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336, 2010. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 326: 1694–1697, 2009. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science 308: 1635–1638, 2005. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461, 2010. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 7.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200, 2011. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrer-Admetlla A, Sikora M, Laayouni H, Esteve A, Roubinet F, Blancher A, Calafell F, Bertranpetit J, Casals F. A natural history of FUT2 polymorphism in humans. Mol Biol Evol 26: 1993–2003, 2009. doi: 10.1093/molbev/msp108. [DOI] [PubMed] [Google Scholar]
- 9.Gaskins HR, Croix JA, Nakamura N, Nava GM. Impact of the intestinal microbiota on the development of mucosal defense. Clin Infect Dis 46, Suppl 2: S80–S86, 2008. doi: 10.1086/523336. [DOI] [PubMed] [Google Scholar]
- 10.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Petrosino JF, Knight R, Birren BW; Human Microbiome Consortium . Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504, 2011. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoskins LC, Boulding ET. Degradation of blood group antigens in human colon ecosystems. I. In vitro production of ABH blood group-degrading enzymes by enteric bacteria. J Clin Invest 57: 63–73, 1976. doi: 10.1172/JCI108270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishak HD, Plowes R, Sen R, Kellner K, Meyer E, Estrada DA, Dowd SE, Mueller UG. Bacterial diversity in Solenopsis invicta and Solenopsis geminata ant colonies characterized by 16S amplicon 454 pyrosequencing. Microb Ecol 61: 821–831, 2011. doi: 10.1007/s00248-010-9793-4. [DOI] [PubMed] [Google Scholar]
- 13.Kelly RJ, Rouquier S, Giorgi D, Lennon GG, Lowe JB. Sequence and expression of a candidate for the human Secretor blood group alpha(1,2)fucosyltransferase gene (FUT2). Homozygosity for an enzyme-inactivating nonsense mutation commonly correlates with the non-secretor phenotype. J Biol Chem 270: 4640–4649, 1995. doi: 10.1074/jbc.270.9.4640. [DOI] [PubMed] [Google Scholar]
- 14.Leyn SA, Gao F, Yang C, Rodionov DA. N-acetylgalactosamine utilization pathway and regulon in proteobacteria: genomic reconstruction and experimental characterization in Shewanella. J Biol Chem 287: 28047–28056, 2012. doi: 10.1074/jbc.M112.382333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mäkivuokko H, Lahtinen SJ, Wacklin P, Tuovinen E, Tenkanen H, Nikkilä J, Björklund M, Aranko K, Ouwehand AC, Mättö J. Association between the ABO blood group and the human intestinal microbiota composition. BMC Microbiol 12: 94, 2012. doi: 10.1186/1471-2180-12-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, Andersen GL, Knight R, Hugenholtz P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–618, 2012. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nava GM, Stappenbeck TS. Diversity of the autochthonous colonic microbiota. Gut Microbes 2: 99–104, 2011. doi: 10.4161/gmic.2.2.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One 5: e9490, 2010. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raffals LE, Chang EB. Navigating the microbial basis of inflammatory bowel diseases: seeing the light at the end of the tunnel. Gut Liver 10: 502–508, 2016. doi: 10.5009/gnl15491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rausch P, Rehman A, Künzel S, Häsler R, Ott SJ, Schreiber S, Rosenstiel P, Franke A, Baines JF. Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (Secretor) genotype. Proc Natl Acad Sci USA 108: 19030–19035, 2011. doi: 10.1073/pnas.1106408108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reid ME, Lomas-Francis C, Olsson ML. The Blood Group Antigen Factsbook. London: Academic, 2012. [Google Scholar]
- 22.Rook GA. Hygiene and other early childhood influences on the subsequent function of the immune system. Dig Dis 29: 144–153, 2011. doi: 10.1159/000323877. [DOI] [PubMed] [Google Scholar]
- 23.Schiff F, Akune M. Blutgruppen und Physiologie. Munch Med Wochenschr 78: 657–660, 1931. [Google Scholar]
- 24.Schiff F, Weiler G. Fermente und Blutgruppen. I. Biochem Z 235: 454–465, 1931. [Google Scholar]
- 25.Schiff F, Weiler G. Fermente und Blutgruppen. II. Biochem Z 239: 489–492, 1931. [Google Scholar]
- 26.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol 12: R60, 2011. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen R, Ishak HD, Estrada D, Dowd SE, Hong E, Mueller UG. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc Natl Acad Sci USA 106: 17805–17810, 2009. doi: 10.1073/pnas.0904827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith DM, Snow DE, Rees E, Zischkau AM, Hanson JD, Wolcott RD, Sun Y, White J, Kumar S, Dowd SE. Evaluation of the bacterial diversity of pressure ulcers using bTEFAP pyrosequencing. BMC Med Genomics 3: 41, 2010. doi: 10.1186/1755-8794-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonnenburg JL, Bäckhed F. Diet-microbiota interactions as moderators of human metabolism. Nature 535: 56–64, 2016. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stackebrandt E. The Family Lachnospiraceae. In: The Prokaryotes, edited by Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E. New York: Springer, 2014, p. 197–201. [Google Scholar]
- 31.Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS, Giglio MG, Hallsworth-Pepin K, Lobos EA, Madupu R, Magrini V, Martin JC, Mitreva M, Muzny DM, Sodergren EJ, Versalovic J, Wollam AM, Worley KC, Wortman JR, Young SK, Zeng Q, Aagaard KM, Abolude OO, Allen-Vercoe E, Alm EJ, Alvarado L, Andersen GL, Anderson S, Appelbaum E, Arachchi HM, Armitage G, Arze CA, Ayvaz T, Baker CC, Begg L, Belachew T, Bhonagiri V, Bihan M, Blaser MJ, Bloom T, Bonazzi V, Paul Brooks J, Buck GA, Buhay CJ, Busam DA, Campbell JL, Canon SR, Cantarel BL, Chain PSG, Chen I-MA, Chen L, Chhibba S, Chu K, Ciulla DM, Clemente JC, Clifton SW, Conlan S, Crabtree J, Cutting MA, Davidovics NJ, Davis CC, DeSantis TZ, Deal C, Delehaunty KD, Dewhirst FE, Deych E, Ding Y, Dooling DJ, Dugan SP, Michael Dunne W, Scott Durkin A, Edgar RC, Erlich RL, Farmer CN, Farrell RM, Faust K, Feldgarden M, Felix VM, Fisher S, Fodor AA, Forney LJ, Foster L, Di Francesco V, Friedman J, Friedrich DC, Fronick CC, Fulton LL, Gao H, Garcia N, Giannoukos G, Giblin C, Giovanni MY, Goldberg JM, Goll J, Gonzalez A, Griggs A, Gujja S, Kinder Haake S, Haas BJ, Hamilton HA, Harris EL, Hepburn TA, Herter B, Hoffmann DE, Holder ME, Howarth C, Huang KH, Huse SM, Izard J, Jansson JK, Jiang H, Jordan C, Joshi V, Katancik JA, Keitel WA, Kelley ST, Kells C, King NB, Knights D, Kong HH, Koren O, Koren S, Kota KC, Kovar CL, Kyrpides NC, La Rosa PS, Lee SL, Lemon KP, Lennon N, Lewis CM, Lewis L, Ley RE, Li K, Liolios K, Liu B, Liu Y, Lo C-C, Lozupone CA, Dwayne Lunsford R, Madden T, Mahurkar AA, Mannon PJ, Mardis ER, Markowitz VM, Mavromatis K, McCorrison JM, McDonald D, McEwen J, McGuire AL, McInnes P, Mehta T, Mihindukulasuriya KA, Miller JR, Minx PJ, Newsham I, Nusbaum C, O’Laughlin M, Orvis J, Pagani I, Palaniappan K, Patel SM, Pearson M, Peterson J, Podar M, Pohl C, Pollard KS, Pop M, Priest ME, Proctor LM, Qin X, Raes J, Ravel J, Reid JG, Rho M, Rhodes R, Riehle KP, Rivera MC, Rodriguez-Mueller B, Rogers Y-H, Ross MC, Russ C, Sanka RK, Sankar P, Fah Sathirapongsasuti J, Schloss JA, Schloss PD, Schmidt TM, Scholz M, Schriml L, Schubert AM, Segata N, Segre JA, Shannon WD, Sharp RR, Sharpton TJ, Shenoy N, Sheth NU, Simone GA, Singh I, Smillie CS, Sobel JD, Sommer DD, Spicer P, Sutton GG, Sykes SM, Tabbaa DG, Thiagarajan M, Tomlinson CM, Torralba M, Treangen TJ, Truty RM, Vishnivetskaya TA, Walker J, Wang L, Wang Z, Ward DV, Warren W, Watson MA, Wellington C, Wetterstrand KA, White JR, Wilczek-Boney K, Wu YQ, Wylie KM, Wylie T, Yandava C, Ye L, Ye Y, Yooseph S, Youmans BP, Zhang L, Zhou Y, Zhu Y, Zoloth L, Zucker JD, Birren BW, Gibbs RA, Highlander SK, Methé BA, Nelson KE, Petrosino JF, Weinstock GM, Wilson RK, White O; Human Microbiome Project Consortium . Structure, function and diversity of the healthy human microbiome. Nature 486: 207–214, 2012. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong M, McHardy I, Ruegger P, Goudarzi M, Kashyap PC, Haritunians T, Li X, Graeber TG, Schwager E, Huttenhower C, Fornace AJ Jr, Sonnenburg JL, McGovern DP, Borneman J, Braun J. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn’s disease risk polymorphism. ISME J 8: 2193–2206, 2014. doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wacklin P, Mäkivuokko H, Alakulppi N, Nikkilä J, Tenkanen H, Räbinä J, Partanen J, Aranko K, Mättö J. Secretor genotype (FUT2 gene) is strongly associated with the composition of Bifidobacteria in the human intestine. PLoS One 6: e20113, 2011. doi: 10.1371/journal.pone.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wacklin P, Tuimala J, Nikkilä J, Sebastian Tims, Mäkivuokko H, Alakulppi N, Laine P, Rajilic-Stojanovic M, Paulin L, de Vos WM, Mättö J. Faecal microbiota composition in adults is associated with the FUT2 gene determining the secretor status. PLoS One 9: e94863, 2014. doi: 10.1371/journal.pone.0094863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Werner JJ, Koren O, Hugenholtz P, DeSantis TZ, Walters WA, Caporaso JG, Angenent LT, Knight R, Ley RE. Impact of training sets on classification of high-throughput bacterial 16s rRNA gene surveys. ISME J 6: 94–103, 2012. doi: 10.1038/ismej.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolcott RD, Gontcharova V, Sun Y, Dowd SE. Evaluation of the bacterial diversity among and within individual venous leg ulcers using bacterial tag-encoded FLX and titanium amplicon pyrosequencing and metagenomic approaches. BMC Microbiol 9: 226, 2009. doi: 10.1186/1471-2180-9-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zackular JP, Baxter NT, Chen GY, Schloss PD. Manipulation of the gut microbiota reveals role in colon tumorigenesis. mSphere 1: e00001–e00015, 2015. doi: 10.1128/mSphere.00001-15. [DOI] [PMC free article] [PubMed] [Google Scholar]