Abstract
Nearly all mungbean cultivars are completely susceptible to seed bruchids (Callosobruchus chinensis and Callosobruchus maculatus). Breeding bruchid-resistant mungbean is a major goal in mungbean breeding programs. Recently, we demonstrated in mungbean (Vigna radiata) accession V2802 that VrPGIP2, which encodes a polygalacturonase inhibiting protein (PGIP), is the Br locus responsible for resistance to C. chinensis and C. maculatus. In this study, mapping in mungbean accession V2709 using a BC11F2 population of 355 individuals revealed that a single major quantitative trait locus, which controlled resistance to both C. chinensis and C. maculatus, was located in a 237.35 Kb region of mungbean chromosome 5 that contained eight annotated genes, including VrPGIP1 (LOC106760236) and VrPGIP2 (LOC106760237). VrPGIP1 and VrPGIP2 are located next to each other and are only 27.56 Kb apart. Sequencing VrPGIP1 and VrPGIP2 in “V2709” revealed new alleles for both VrPGIP1 and VrPGIP2, named VrPGIP1-1 and VrPGIP2-2, respectively. VrPGIP2-2 has one single nucleotide polymorphism (SNP) at position 554 of wild type VrPGIP2. This SNP is a guanine to cystine substitution and causes a proline to arginine change at residue 185 in the VrPGIP2 of “V2709”. VrPGIP1-1 has 43 SNPs compared with wild type and “V2802”, and 20 cause amino acid changes in VrPGIP1. One change is threonine to proline at residue 185 in VrPGIP1, which is the same as in VrPGIP2. Sequence alignments of VrPGIP2 and VrPGIP1 from “V2709” with common bean (Phaseolus vulgaris) PGIP2 revealed that residue 185 in VrPGIP2 and VrPGIP1 contributes to the secondary structures of proteins that affect interactions between PGIP and polygalacturonase, and that some amino acid changes in VrPGIP1 also affect interactions between PGIP and polygalacturonase. Thus, tightly linked VrPGIP1 and VrPGIP2 are the likely genes at the Br locus that confer bruchid resistance in mungbean “V2709”.
Keywords: bruchid, seed weevil, polygalacturonase inhibitor, PGIP, Callosobruchus, insect resistance, mungbean
Introduction
Mungbean [Vigna radiata (L.) Wilczek] is an important legume crop of Asia. It is cultivated on ~6 million hectares, mainly in Asia (Somta et al., 2007). The crop is popularly grown for seeds after rice or wheat because of its early maturity (60–75 day), drought tolerance and symbiotic relationship with rhizobia, which fix atmospheric nitrogen to the soil. Seeds of mungbean contain high levels of proteins (20–25%) and carbohydrate (60–75%), and thus, they are an important human dietary staple (Somta et al., 2007).
Bruchids or seed weevils (Coleoptera: Bruchidae) are serious pests that damage legume seeds after harvest. Bruchids infest seeds in the field and after harvest, and the latter can results in total loss of a seed lot within 3–4 months (Srinives et al., 2007). The bruchid-damaged seeds are inedible and cannot be used for agricultural and commercial purposes. Azuki bean weevil (Callosobruchus chinensis L.) and cowpea weevil (Callosobruchus muculatus F.) are the major bruchid species that feed on seeds of mungbean and other food legumes, including cowpea (Vigna unguiculata L.), black gram [Vigna mungo (L.) Hepper], Bambara groundnut [Vigna subterranea (L.) Verdc.], azuki bean [Vigna angularis (Willd.) Ohwi and Ohashi], soybean [Glycine max (L.) Merr.], chickpea (Cicer arietinum L.) and pigeon pea [Cajanus cajan (L.) Millsp.]. Although C. chinensis and C. maculatus originated in Asia and Africa, respectively, they are currently present in almost every continent owing to international seed/grain trading. Generally, bruchids are controlled by chemical fumigation and dusting. However, these chemicals are not only harmful to humans and the environment, but also increase production costs. Additionally, chemical control is not practical for small-landholder farmers. Thus, a major goal in mungbean breeding programs is to develop bruchid-resistant cultivar(s) (Somta et al., 2007; Srinives et al., 2007).
Several bruchid-resistant mungbean germplasms have been identified. Fujii and Miyazaki (1987) reported that wild mungbean (V. radiata var. sublobata) accession TC1966 is completely resistant to C. chinensis and C. maculatus. Talekar and Lin (1992) found that cultivated mungbean accessions V2709 and V2802 are highly resistant to C. chinensis. Somta et al. (2008) demonstrated that V2709 and V2802 are also highly resistant to C. maculatus, while the cultivated mungbean accessions V1128 and V2817 are completely resistant to both bruchids.
Kitamura et al. (1988) showed that resistance to C. chinensis in “TC1966” is controlled by a single dominant gene, Br. Somta et al. (2007) demonstrated that resistance to C. chinensis and C. maculatus in “V2709” and “V2802” is controlled by a single dominant gene with some modifiers. Kaga and Ishimoto (1998) fine mapped the Br gene in “TC1966” to a region of 0.7 cM between restriction fragment length polymorphism (RFLP) markers Bng110 and Bng143. The Br gene is only 0.2 cM away from the Bng143. Chotechung et al. (2011) found that expressed sequence tag-simple sequence repeat (EST-SSR) marker DMB-SSR158 (DMB158 in the original report) co-segregated perfectly with the Br gene in “V2802”. Later, Chotechung et al. (2016) narrowed the genome region of the Br gene to a 38-kb segment on chromosome 5 and found that a gene, VrPGIP2, encoding polylacturonase-inhibiting protein (PGIP; also known as polylacturonase inhibitor) was probably responsible for the bruchid resistance. They also showed that “V2802”, “V1128”, “V2817”, and “TC1966” had the same VrPGIP2 allele.
For “V2709”, the mode of bruchid-resistance inheritance is the same as for “V2808” and “TC1966” (Somta et al., 2007). However, the EST-SSR marker DMB-SSR158, which is located on the VrPGIP2 sequence, did not reveal a polymorphism between “V2709” and bruchid-susceptible mungbean “Kamphaeng Saen 1” (KPS1; Chotechung et al., 2011), which was the same susceptible parent used by Chotechung et al. (2016). This suggested that the gene or allele for bruchid resistance in “V2709” is different from that of “V2802”.
In this study, we report the identification of new alleles for bruchid resistance in mungbean accession V2709. The objectives of this study were to (i) fine map the Br locus in “V2709”, (ii) identify candidate gene(s) for the Br locus in “V2709” and (iii) identify mutation(s) responsible for the resistance.
Materials and methods
Plant population and DNA extraction
A BC11F2 mapping population was developed from “KPS1” and “V2709” (Supplementary Figure S1). “KPS1” is susceptible to C. chinensis and C. maculatus, while “V2709” is resistant to both bruchids (Somta et al., 2008). In total, 355 BC11F2 plants together with “KPS1” and “V2709” were planted in an experimental field of Kasetsart University, Kamphaeng Saen Campus, Nakhon Pathom, Thailand from February to April 2013. Seeds from each BC11F2 plant were harvested for bruchid-resistance evaluation. Genomic DNA from all of the mungbean plants was extracted from young leaf tissues using a modified cetyl trimethylammonium bromide (CTAB) method (Lodhi et al., 1994).
Evaluation for bruchid resistance
Cultures of C. chinensis and C. maculatus were reared on “KPS1” seeds. The resistance evaluation was carried out as per Somta et al. (2007) with minor modifications. In brief, 30 to 40 intact seeds from each BC11F2 plant were placed into a transparent plastic box. Twenty pairs (males and females) of 1- to 3-day-old bruchids were introduced into the box, kept for 7 day for egg laying, and then removed from the box. Parental seeds were tested and replicated five times. The seeds were maintained at 28°C and 70% relative humidity. Then, 60 day after insect introduction, the numbers of damaged seeds were counted and the percentages calculated.
Segregation analysis
The monogenic inheritance of bruchid resistance in “V2709” (Somta et al., 2007; Chotechung et al., 2011) was confirmed. BC11F2 plants were classified into two classes; plants with 0–80%-damaged seeds were classified as resistant, which included the homozygous resistant genotype (highly resistant, with 0–20%-damaged seeds) and heterozygous resistant genotype (moderately resistant, with 21–80%-damaged seeds), while plants with 81–100%-damaged seeds were classified as the homozygous susceptible genotype (Somta et al., 2007). A chi-square (χ2) test was conducted to determine a 3 (resistance):1 (susceptible) goodness of fit using software R-program 2.0.10 (R Development Core Team, 2012).
DNA marker analysis
In total, 25 DNA markers were developed based on the chromosome 5 sequence of mungbean (Supplementary Table S1). These markers, together with 52 SSR and sequence-tagged site (STS) markers located on mungbean chromosome 5 associated with the bruchid resistance reported by Chotechung et al. (2011, 2016), Hong et al. (2015), and Liu et al. (2016), were used to detect polymorphisms between “KPS1” and “V2709” (Supplementary Table S1). PCR amplification, gel electrophoresis and DNA band visualization were carried out as per Somta et al. (2009). SSR markers showing polymorphisms between “KPS1” and “V2709” were used to genotype the BC11F2 plants.
Quantitative trait locus (QTL) analysis
A genetic linkage map was constructed using software QTL IciMapping 4.0 (Meng et al., 2015). Markers were grouped by a log of odds (LOD) value of 3.0. The recombination frequency between markers was converted into genetic map distances (centimorgan; cM) using the Kosambi mapping function (Kosambi, 1944).
The QTL for bruchid resistance was mapped using the inclusive composite interval mapping (ICIM) method (Li et al., 2007) by QTL IciMapping 4.0. The significant LOD threshold for the QTL was determined by a 5,000 permutation test at P = 0.01. ICIM was performed at 1-cM steps, and the P-value for entering variables (PIN) was 0.001.
Sequencing of VrPGIP1 and VrPGIP2 in “V2709”
VrPGIP2 in “V2709” was amplified and sequenced following the procedures described by Chotechung et al. (2016). VrPGIP1 was amplified and sequenced in “KPS1” and “V2802” using the primer pair Br_03940_F1 (5′-CGACTAGCACCGGAAATTA-3′)/Br_03940_R13 (5′-CTCAACTTGGTTAATGATGCTTA-3′), and in “V2709” using the primer pair Br_03940_F1 (5′-CGACTAGCACCGGAAATTA-3′)/Br_03940_R14 (5′- ACACTGCACACGTGTCAAA−3′). PCR amplification and sequencing protocols were the same as those for VrPGIP2. NCBI nucleotide accession numbers for VrPGIP1 and VrPGIP2 are MF398961 and MF398954, respectively, for “KPS1”, MF496138 and MF398959, respectively, for “V2709”, and MF398960 and MF398955, respectively, for “V2802”.
The sequences of VrPGIP1 and VrPGIP2 from “KPS1”, “V2709”, and “V2802” were aligned using ClustalW (Larkin et al., 2007). The predicted protein sequences of both genes in these accessions were also aligned to detect protein sequence variation. In addition, the amino acids of VrPGIP1 and VrPGIP2 were aligned with PvPGIP2 (P58822.1), which is the only PGIP that has a known crystal structure (Di Matteo et al., 2003), to determine their primary protein structures.
Phylogenetic analysis of PGIPs
VrPGIP1 and VrPGIP2 sequences and PGIP sequences of chickpea (C. arietinum L.) [CaPGIP1 (XP_004504732.1) and CaPGIP2 (XP_012572409.1)], common bean (Phaseolus vulgaris L.) [PvPGIP1 (CAI11357.1), PvPGIP2 (P58822.1), PvPGIP3 (CAI11359.1) and PvPGIP4 (CAI11360.1)], soybean (G. max Merr.) [GmPGIP1 (CAI99392.1), GmPGIP2 (CAI99393.1), GmPGIP3 (CAI99394.1), GmPGIP4 (CAI99395.1), GmPGIP5 (XP_003524769.1) and GmPGIP7 (XP_003531070.1)] and Medicago truncatula [MtPGIP1 (XP_013457276.1), MtPGIP2, (XP_013457278.1) and MtPGIP3 (XP_013457280.1)] were subjected to phylogenic analysis using software Phylogeny.fr (Dereeper et al., 2008; http://www.phylogeny.fr).
Results
Bruchid resistance in the BC11F2 population
Seeds of “V2709” were free from damage by C. chinensis and C. maculatus, while seeds of “KPS1” were all damaged by both bruchid species. In the BC11F2 population, the level of seed's damaged ranged from 0 to 100%, with means of 44.19% for C. chinensis and 44.60% for C. maculatus. There was a very high correlation (r = 0.97, d.f. = 317, P < 0.0001) between the percentages of damaged seeds caused by the two bruchid species, suggesting that the resistance to C. chinensis and to C. maculatus in “V2709” is controlled by the same gene(s). The frequency distribution of the percentage of damaged seeds was nearly discontinuous and bimodal (Figure 1); however, there was a clear continuous distribution for the resistance group (1–50%-damaged seeds).
Figure 1.
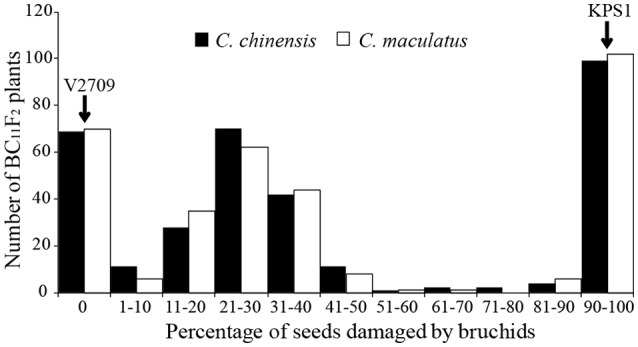
Frequency distribution of the percentage of seeds damaged by Callosobruchus chinensis and Callosobruchus maculatus in the mungbean BC11F2 population derived from “KPS1” and “V2709” as recipient and donor parents, respectively.
The segregation of plants in the BC11F2 population that showed resistance and susceptibility to C. chinensis and C. maculatus did not fit the 3 (resistant):1 (susceptible) ratio (χ2 = 5.24, P = 0.0221 and χ2 = 9.36, P = 0.0022, respectively). This result contrasted previous findings reported by Somta et al. (2007) and Chotechung et al. (2011) that a single dominant gene, Br, controls bruchid resistance in “V2709”. This contradiction may result from our BC11F2 population being a composite population derived from six BC11F2 families each having between 41 and 75 plants. Families with small number of plants may cause the distortion of Br locus-associated resistance in the composite population. In addition, modifying factors for the resistance presented in “V2709” (Somta et al., 2007) may also cause the distortion in resistance segregation.
QTL mapping for bruchid resistance in “V2709”
In total, 77 DNA markers located near the Br locus on chromosome 5 of mungbean were screened for polymorphisms between “V2709” and “KPS1”, and 19 polymorphic markers were selected to construct a linkage map with a length of 84.12 cM.
A single major QTL for C. chinensis (qBrc5.1) and a single major QTL for C. maculatus (qBrm5.1) were detected by ICIM (Table 1 and Supplementary Figure S2). qBrc5.1 and qBrm5.1 were both located at 34.0 cM, between markers VrID1 and Vr-SSR017. These QTLs accounted for 95.58 and 96.30% of the total variation of seeds damage caused by C. chinensis and C. maculatus, respectively. The additive and dominant effects of qBrc5.1 were 49.07 and 45.00%, respectively, while those of qBrm5.1 were 48.72 and 44.17%, respectively. At both QTLs, allele(s) from “V2709” decreased the seed damage caused by bruchids. Because qBrc5.1 and qBrm5.1 were at the same position and conferred similar genetic effects for the resistance, we considered them as the same locus and named it “qBr5.1”.
Table 1.
QTL conferring resistance to bruchids (Callosobruchus chinensis and Callosobruchus maculatus) identified by inclusive composite interval mapping in the BC11F2 mungbean population [KPS1 × (KPS1 × V2709)].
| Bruchid speceis | QTL name | Position (cM) | LOD score | Interval markers | 1-LOD support interval (cM) | PVEa (%) | Additive effect | Dominance effect |
|---|---|---|---|---|---|---|---|---|
| C. chinensis | qRcc5.1 | 34.0 | 252.17 | VrID1–VrBr-SSR017 | 33.5–34.5 | 93.34 | 49.07 | 45.00 |
| C. maculatus | qRcm5.1 | 34.0 | 263.16 | VrID1–VrBr-SSR017 | 33.5–34.5 | 93.84 | 48.72 | 44.17 |
The resistance was determined based on the percentage of seeds damaged by bruchids.
Phenotypic variance explained by the QTL.
The genomic confidence region of the qBr5.1 was between markers VRID5 and VrBr-SSR037, covering ~237.35 Kb (Vr05: 5,410,272–5,647,621), in which there were eight annotated protein-encoding genes, including VrPGIP1 (LOC106760236) and VrPGIP2 (LOC106760237), based on the annotation by NCBI (Figure 2 and Supplementary Table S2).
Figure 2.
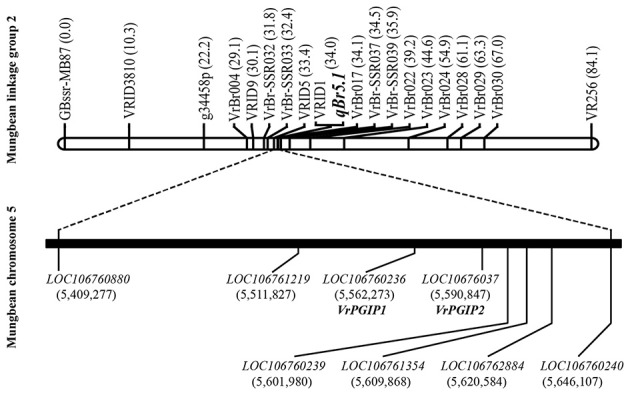
Location of the QTL qBr5.1 controlling resistance to Callosobruchus chinensis and Callosobruchus maculatus in mungbean accession V2709. The QTL analysis was performed using BC11F2 population [KPS1 × (KPS1 × V2709)]. The 1-LOD confidence region that corresponds to chromosome 5 of the mungbean reference genome is shown. NCBI's annotated genes in the confidence region of the qBr5.1 are also shown. Numbers in parenthesis after each marker indicate positions in centiMorgan (cM) units. Numbers in parenthesis under each gene indicate positions on mungbean chromosome 5.
New alleles of VrPGIP1 and VrPGIP2 in “V2709”
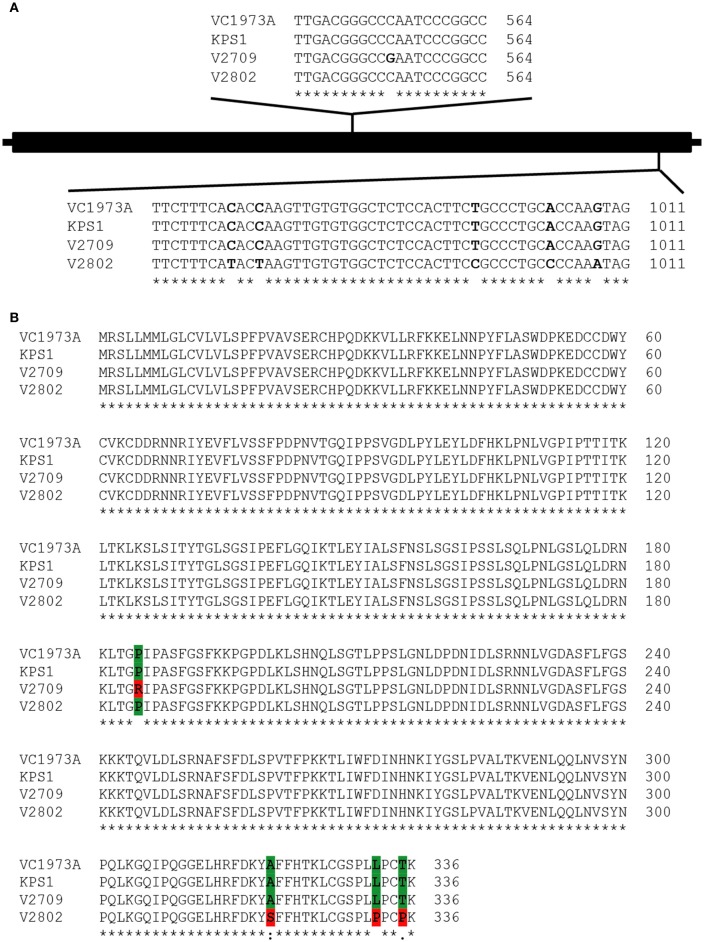
VrPGIP2 is an intronless gene with 1,011 nucleotides, and it translates into a 336-amino acid protein (Chotechung et al., 2016). It is the same gene as LOC106760237 annotated by NCBI. A sequence alignment of VrPGIP2 genes from “V2802” and KPS1 showed seven SNPs (at nucleotide positions 573, 958, 969, 972, 995, 1,003 and 1,008) between the two mungbean accessions. The SNPs at positions 958, 995 and 1,003 caused amino acid changes in VrPGIP2 at positions 320, 332, and 335 in “V2802” (Chotechung et al., 2016). In this study, the sequencing of VrPGIP2 in “V2709” revealed that VrPGIP2 was 1,011 nucleotides, which was the same as in “V2802” and “KPS1”. A sequence alignment of VrPGIP2 from “V2709” and “KPS1” revealed a single SNP at the 554 position (Figure 3A). The nucleotide at this position was cytosine in “KPS1” and guanine in “V2709”. This SNP caused an amino acid change at position 185(from arginine to proline) of the VrPGIP2 protein in “V2709” (Figure 3B). There were no polymorphisms between “V2802” and “KPS1” at this position (Figure 3A). We designated the VrPGIP2 allele in “V2802” as VrPGIP2-1 and that in “V2709” as VrPGIP2-2. We also aligned VrPGIP2 of “V2709” with PvPGIP2. The alignment showed that both VrPGIP2 and PvPGIP2 have 10 leucine-rich regions, and the amino acid change in VrPGIP2 of “V2709” is in leucine-rich region 5. The residue 185 of VrPGIP2, which is polymorphic between “V2709” and “KPS1” is equivalent to residue 193 of PvPGIP2 (Figure 6A).
Figure 3.
Single nucleotide polymorphisms in the VrPGIP2 (LOC106760237) sequences from mungbean accession VC1973A (reference sequence), KPS1, V2709, and V2802. (A) “VC1973A” and “KPS1” are susceptible to bruchids, while “V2802” and “V2709” are resistant to bruchids. Polymorphic sites are presented in bold color. (B)The alignment of the protein sequences encoded by VrPGIP2 in these mungbean accessions is also shown. Polymorphic sites are colored.
VrPGIP1 was annotated by Kang et al. (2014), and it had three exons with an open reading frame of 1,302 nucleotides that encodes a 433-amino acid protein (Chotechung et al., 2016). However, the current mungbean genome annotation by NCBI showed that VrPGIP1 (LOC106760236) is intronless, has 1,011 nucleotides and encodes a 336-amino acid protein, which is the same as VrPGIP2. We conducted Sanger sequencing of this gene in “KPS1”, “V2709”, and “V2802”, and successfully obtained the ORF sequences for “KPS1” and “V2802” but we only obtained a continuous 960-bp sequence (including start codon) for “V2709”. We obtained the full sequence of VrPGIP1 from our in-house whole genome sequence (WGS) data of “V2709”. However, 100% of the partial VrPGIP1 sequence of “V2709” obtained from the Sanger sequencing was identical to the full VrPGIP1 sequence generated from the WGS (data not shown). This indicated the high accuracy of the VrPGIP1 sequence in “V2709” generated by the WGS. A sequence alignment of VrPGIP2 genes from “V2709”, “V2802”, “KPS1” and the reference sequence (“VC1973A”) showed that the sequences from “KPS1”, “VC1973A”, and “V2802” were identical and that there were 43 SNPs between “V2709” and “KPS1” (Figure 4). Among those SNPs, 20 caused amino acid changes in “V2709”, when compared with “KPS1” (Figure 5). Interestingly, of those amino acid changes, the change at position 185 (from threonine to proline) was similar to the change in VrPGIP2. We named the VrPGIP1 allele in “V2709” as VrPGIP1-1.
Figure 4.
Alignment of the coding sequences of VrPGIP1 (LOC106760236) from the accession VC1973A (reference sequence), KPS1, V2709, and V2802. “VC1973A” and “KPS1” are susceptible to bruchids, while “V2802” and “V2709” are resistant to bruchids. Polymorphic sites are colored.
Figure 5.
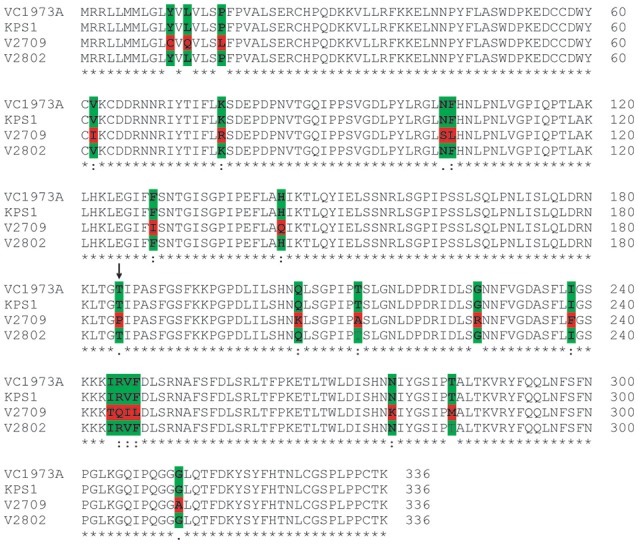
Alignment of the protein sequences encoded by VrPGIP1 (LOC106760236) from the accession VC1973A (reference sequence), KPS1, V2709, and V2802. “VC1973A” and “KPS1” are susceptible to bruchids, while “V2802” and “V2709” are resistant to bruchids. Polymorphic sites are present in bold color. Arrow indicates position of important amino acid for formation of secondary structure of the VrPGIP protein and PG recognition.
Phylogeny of VrPGIP1 and VrPGIP2
A phylogenetic analysis was conducted to determine relationships of VrPGIP1 and VrPGIP2 with PGIPs from common bean, soybean, chickpea and M. truncatula. The phylogenetic tree revealed that VrPGIP1 and VrPGIP2 were closely related to each other but distantly related to the PGIPs from the other legumes (Supplementary Figure S3). Among the other legumes, VrPGIP1 and VrPGIP2 showed the closest relationship with MtPGIP3 of M. truncatula.
Discussion
Although the wild mungbean accession TC1966, which carries the Br locus that confers resistance to C. chinensis and C. maculatus, has been reported since 1987 (Fujii and Miyazaki, 1987), only a few cultivars possessing the Br locus have been released to farmers (Lee et al., 2000; Yao et al., 2015). This is mainly due to skepticism about the safety of the Br locus. The chemical responsible for bruchid resistance at the Br locus is not known, and toxicity studies of the bruchid-resistant mungbeans carrying this locus are very limited (Miura et al., 1996; Yao et al., 2015). Kitamura et al. (1990) reported that the chemical involved in bruchid resistance was heat-stable and possibly a polysaccharide. Cyclo-peptide alkaloids, called Vignatic acid A and Vignatic acid B, isolated from a bruchid-resistant breeding line were detrimental to C. chenensis (Sugawara et al., 1996). A cysteine-rich protein designated VrCRP/VrD1 (defensin) identified from a bruchid-resistant breeding line inhibited the development of C. chenensis larva (Chen et al., 2002). However, gene mapping (Kaga and Ishimoto, 1998) and linkage analyses (Isemura et al., 2012) revealed that the loci controlling these chemicals are not in the Br locus.
Chotechung et al. (2016) demonstrated that VrPGIP2, which encodes a PGIP, is the candidate gene for the Br locus in mungbean “V2802”, although a role for VrPGIP1, which also encodes a PGIP and is located next to VrPGIP2, could not be eliminated. VrPGIP2 in mungbean may confer bruchid resistance by inhibiting the digestive polygalacturonases (PGs) of bruchids that degrade pectin in the seeds, producing an energy source for the bruchids and helping other digestive enzymes to gain access to their substrates in bruchid food (Chotechung et al., 2016). In this study, we located a single major QTL for bruchid resistance with a high genetic effect (phenotypic variance explained (PVE) > 95%) on chromosome 5 using mungbean accession “V2709”. The genomic confidence region of the QTL (VrID5 to VrBr-SSR037) covered ~237.35 Kb (Vr05: 5,410,272–5,647,621) with eight protein-encoding genes, including VrPGIP1 and VrPGIP2 (Figure 1 and Supplementary Table S2).
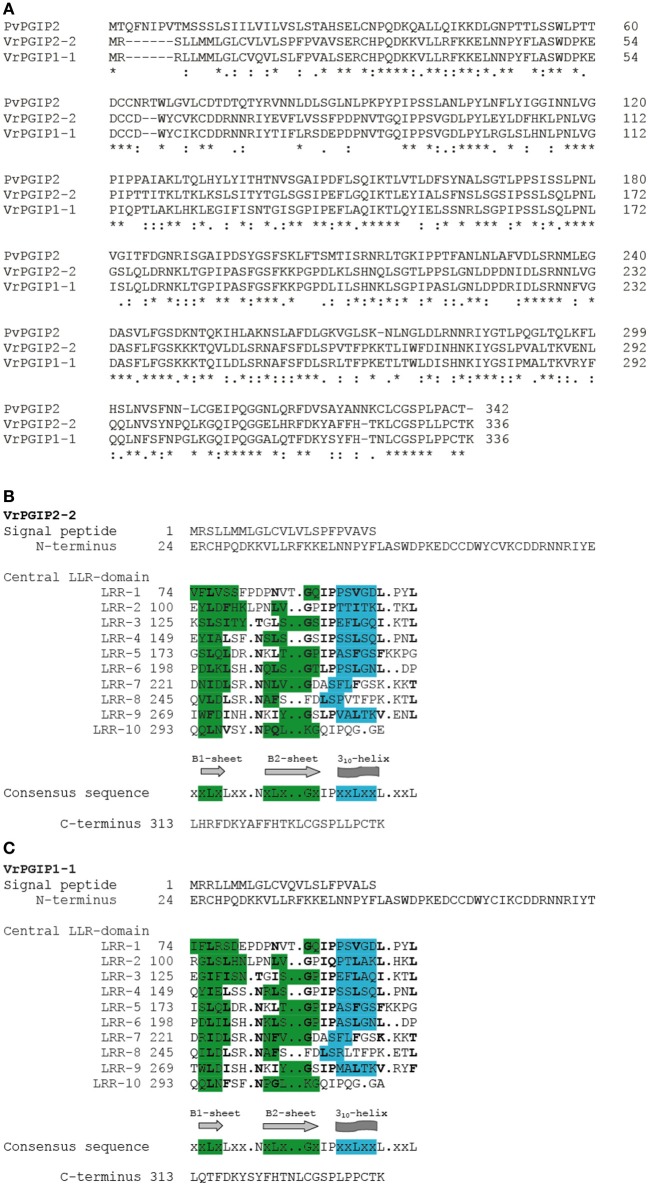
We sequenced VrPGIP1 and VrPGIP2 in mungbean “V2709” and found new alleles, VrPGIP1-1 and VrPGIP2-2, respectively (Figures 3A, 4). The VrPGIP2 encoded by VrPGIP2-2 possessed an amino acid change at the position 185, as compared with the wild type protein (Figure 3B). Comparing the sequence of VrPGIP2 with PvPGIP2, which is the only PGIP that has a known crystal structure, we found that residue 185 of the VrPGIP2 primary sequence (Figure 6B) corresponds to residue 164 of the crystal structure of PvPGIP2 (Di Matteo et al., 2003). Residue 164 is a site that contributes to the formation of the secondary structure of the PvPGIP2 protein (Di Matteo et al., 2003), and it is also a putative positive selection site of PvPGIP2 that plays an important role in PG recognition (Casasoli et al., 2009). The VrPGIP1 encoded by VrPGIP1-1 showed many amino acid changes when compared with the wild type, and one change that occurred at position 185, which is the same position of the single amino acid change in VrPGIP2, resulted in the same amino acid at this position in VrPGIP1 and VrPGIP2 in “V2709” (Figures 5, 6C). PGs are important digestive enzymes found in the midgut of Callosobruchus maculatus (Pedra et al., 2003; Pauchet et al., 2010; Nogueira et al., 2012; Santana, 2013) and Phaedon cochleariae (mustard leaf beetle) (Kirsch et al., 2012), a coleopteran insect similar to the Callosobruchus species. A semi-purified PGIP from orange flavedo inhibited an endoPG purified from the larvae of sugarcane rootstalk borer weevil (Diaprepes abbreviates) (Doostdar et al., 1997), while PvPGIP3 and PvPGIP4 from the common bean strongly inhibited the PG activities of the mirid bugs Lygus rugulipennis and Adelphocoris lineolatus (D'Ovidio et al., 2004; Frati et al., 2006). Insect pectinase complexes, including PGs, are mainly found in Coleopteran and Hemipteran insects and are involved in plant penetration, the softening of plant material, and the digestion of young plant tissues and grains (Calderón-Cortés et al., 2012). Kirsch et al. (2016) showed that the rice weevil Sitophilus oryzae, a coleopteran insect that infests both cereal and legume seeds, uses PGs and pectin methylesterases synergistically to degrade pectin. If this was also the case in C. chinensis and C. maculatus, VrPGIP2-2 and also VrPGIP2-1 could confer resistance to these bruchids by inhibiting the ability of PG to degrade pectin, which would result in the retardation and arrest of insect growth and development.
Figure 6.
Alignment and putative secondary structures of VrPGIP2-2, VrPGIP1-1 and PvPGIP2 sequences. (A) Alignment of VrPGIP2-2, VrPGIP1-1 and PvPGIP2 sequences. PvPGIP2 is the only PGIP that secondary structure has been determined (Di Matteo et al., 2003). (B) Putative residues contributing to the secondary structure of VrPGIP2-2 and (C) of VrPGIP2-1 are based on PvPGIP2 and highlighted in green (Sheets B1 and Sheets B2) and blue (310-helix).
Hong et al. (2015) demonstrated that two closely linked QTLs (~7-cM apart) with similar effects (PVE for each QTL was ~45%) controlled C. chinesis resistance in “V2709” although a genetic study revealed the monogenic inheritance of the trait. This contradicts our finding that only one single major QTL with a PVE greater than 98% on mungbean chromosome 5 controlled bruchid resistance in “V2709”. It is difficult to compare the results between the two studies because only one marker (GBssr-MB87) is common and it is far from the QTLs in both studies. Although the RP marker that is closely linked to a QTL reported by Hong et al. (2015) showed a polymorphism between the parents of our mapping population, it produced weak DNA bands that were difficult to unambiguously score. Nonetheless, the probable location of RP on chromosome 5 is between 5,210,000 and 5,520,000 (Chotechung et al., 2016).
In addition to VrPGIP genes that show an association with bruchid resistance (this study and Chotechung et al., 2011), g39185 encoding a BURP domain-containing protein has been identified as a candidate gene for bruchid resistance in mungbean. g39185 was identified by comparing transcript and protein profiles between bruchid-susceptible and bruchid-resistant mungbean, which derived its resistance from “TC1966” (Lin et al., 2016). g39185 is at position 5,236,101 of mungbean chromosome 5 (Lin et al., 2016) which is ~326.7 and ~354.7 Kbp from VrPGIP1 and VrPGIP2, respectively, and is very near the possible location of the RP marker reported by Hong et al. (2015). BURP domain-containing proteins have only been found in plants. Although many BURP proteins have been isolated from plants, their expression patterns are diverse and some functions are still unknown. Generally, BURP domain-containing proteins play important roles in maintaining normal plant metabolism or development (Shao et al., 2011; Tang et al., 2014; Li et al., 2016). Based on the information reported by Lin et al. (2016), g39185 corresponds to Vradi05g03810 on the reference mungbean chromosome 5, and Vradi05g03810 is annotated as LOC106759697 in the GenBank database. LOC106759697 encodes a PG (https://www.ncbi.nlm.nih.gov/gene/?term$=$LOC106759697). Three forms of PGs were predicted from LOC106759697. A BLASTN algorithm-based search revealed that the gene was also most similar to a gene from azuki bean (e-value = 0.0, identity = 95%) that encodes a RESPONSIVE TO DEHYDRATION22 (RD22)-like protein of the BURP family. The RD22 protein is responsive to abiotic stresses, such as drought and salinity (Abe et al., 2003; Hanana et al., 2008). Based on the known and/or possible physiological function of PGIP and our results from gene sequencing and mapping in this study and in Chotechung et al. (2016), VrPGIP2 and/or VrPGIP1 is most likely the Br locus. Nonetheless, our results in this study indicated that the tightly linked genes VrPGIP1 and VrPGIP2 are likely the genes at the Br locus that confer bruchid resistance in the mungbean “V2709”. In addition, the different genetic bases of the bruchid resistance under the Br loci in “V2709” and “V2802” provides plant breeders the opportunity to develop new mungbean cultivar(s) with durable resistance to bruchids by pyramiding or rotating the VrPGIP1 and VrPGIP2 resistance genes.
Author contributions
AnK and AlK carried out DNA marker analysis and data analysis. PrS, AnK, TY, and PeS prepared plant materials and conducted insect resistance evaluation. JC, XY, and XC performed DNA sequencing, and analyzed sequencing results. PrS initiated and coordinated the study, and analyzed the results. AnK, JC, PrS, XC, and PeS wrote the manuscript. All authors approved the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to the Center for Agricultural Biotechnology, Kasetsart University, Joint Legume Research Center between Kasetsart University and Jiangsu Academy of Agricultural Sciences, and Institute of Industrial Crops, Jiangsu Academy of Agricultural Sciences/Jiangsu Key Laboratory for Horticultural Crop Genetic Improvement, for molecular lab facilities.
Footnotes
Funding. This work was financially supported by the National Natural Science Foundation of China (Grant no. 31501370). This work was partially supported by the Center for Advanced Studies for Agriculture and Food, Institute for Advanced Studies, Kasetsart University under the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission, Ministry of Education, Thailand.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01692/full#supplementary-material
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderón-Cortés N., Quesada M., Watanabe H., Cano-Camacho H., Oyama K. (2012). Endogenous plant cell wall digestion: a key mechanism in insect evolution. Annu. Rev. Ecol. Evol. Syst. 43, 45–71. 10.1146/annurev-ecolsys-110411-160312 [DOI] [Google Scholar]
- Casasoli M., Federici L., Spinelli F., Di Matteo A., Vella N., Scaloni F., et al. (2009). Integration of evolutionary and desolvation energy analysis identifies functional sites in a plant immunity protein. Proc. Natl. Acad. Sci. U.S.A. 106, 7666–7671. 10.1073/pnas.0812625106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. C., Lin C. Y., Kuan C. C., Sung H. Y., Chen C. S. (2002). A novel defensin encoded by a mungbean cDNA exhibits insecticidal activity against bruchid. J. Agric. Food Chem. 50, 7258–7263. 10.1021/jf020527q [DOI] [PubMed] [Google Scholar]
- Chotechung S., Chankaew S., Srinives P., Somta P. (2011). Identification of DNA markers associated with bruchid resistance in mungbean. Khon Khan Agric. J. 39, 221–226. [Google Scholar]
- Chotechung S., Somta P., Chen J., Yimram T., Chen X., Srinives P. (2016). A gene encoding a polygalacturonase-inhibiting protein (PGIP) is a candidate gene for bruchid (Coleoptera: bruchidae) resistance in mungbean (Vigna radiata). Theor. Appl. Genet. 129, 1673–1683. 10.1007/s00122-016-2731-1 [DOI] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Matteo A., Federici L., Mattei B., Salvi G., Johnson K. A., Savino C., et al. (2003). The crystal structure of polygalacturonase-inhibiting protein (PGIP)., a leucine-rich repeat protein involved in plant defense. Proc. Natl. Acad. Sci. U.S.A. 100, 10124–10128. 10.1073/pnas.1733690100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doostdar H., McCollum T. G., Mayer R. T. (1997). Purification and characterization of an endo-polygalacturonase from the gut of West Indies sugarcane rootstalk borer weevil (Diaprepes abbreviatus L.) larvae. Comp. Biochem. Phys. B 118, 861–867. 10.1016/S0305-0491(97)00285-X [DOI] [Google Scholar]
- D'Ovidio R., Raiola A., Capodicasa C., Devoto A., Pontiggia D., Roberti S., et al. (2004). Characterization of the complex locus of bean encoding polygalacturonase-inhibiting proteins reveals subfunctionalization for defense against fungi and insects. Plant Physiol. 135, 2424–2435. 10.1104/pp.104.044644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frati F., Galletti R., De Lorenzo G., Salerno G., Conti E. (2006). Activity of endo-polygalacturonases in mirid bugs (Heteroptera: Miridae) and their inhibition by plant cell wall proteins (PGIPs). Eur. J. Entomol. 103, 515–522. 10.14411/eje.2006.067 [DOI] [Google Scholar]
- Fujii K., Miyazaki S. (1987). Infestation resistance of wild legumes (Vigna sublobata) to azuki bean weevil., Callosobruchus chinensis (L.) (Coleoptera: Bruchidae) and its relationship with cytogenetic classification. Appl. Entmol. Zool. 22, 229–230. 10.1303/aez.22.229 [DOI] [Google Scholar]
- Hanana M., Deluc L., Fouquet R., Daldoul S., Léon C., Barrieu F., et al. (2008). Identification and characterization of “rd22” dehydration responsive gene in grapevine (Vitis vinifera L.). C. R. Biol. 331, 569–578. 10.1016/j.crvi.2008.05.002 [DOI] [PubMed] [Google Scholar]
- Hong M. G., Kim K. H., Ku J. H., Jeong J. K., Seo M. J., Park C. H., et al. (2015). Inheritance and quantitative trait loci analysis of resistance genes to bruchid and bean bug in mungbean (Vigna radiata L. Wilczek). Plant Breed. Biotechnol. 3, 39–46. 10.9787/PBB.2015.3.1.039 [DOI] [Google Scholar]
- Isemura T., Kaga A., Tabata S., Somta P., Srinives P., Shimizu T., et al. (2012). Construction of a genetic linkage map and genetic analysis of domestication related traits in mungbean (Vigna radiata). PLoS ONE 7:e41304. 10.1371/journal.pone.0041304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga A., Ishimoto M. (1998). Genetic localization of a bruchid resistance gene and its relationship to insecticidal cyclopeptide alkaloids., the vignatic acids in mungbean (V. radiata L.Wilczek). Mol. Gen. Genet. 258, 378–384. 10.1007/s004380050744 [DOI] [PubMed] [Google Scholar]
- Kang Y. J., Kim S., Kim M. Y., Lestari P., Kim K. H., Ha B. K., et al. (2014). Genome sequence of mungbean and insights into evolution within Vigna species. Nat. Commun. 5:5443. 10.1038/ncomms6443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch R., Heckel D. G., Pauchet Y. (2016). How the rice weevil breaks down the pectin network: enzymatic synergism and sub-functionalization. Insect Biochem. Mol. 71, 72–82. 10.1016/j.ibmb.2016.02.007 [DOI] [PubMed] [Google Scholar]
- Kirsch R., Wielsch N., Vogel H., Svatoš A., Heckel D. G., Pauchet Y. (2012). Combining proteomics and transcriptome sequencing to identify active plant-cell-wall-degrading enzymes in a leaf beetle. BMC Genomics 13:587. 10.1186/1471-2164-13-587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura K., Ishimoto M., Ishii S. (1990). Bruchid resistance factors in Phaseolus and Vigna legumes, in Bruchids and Legumes: Economics., Ecology and Coevolution, eds. Fujii K., Gatehouse A. M. R., Johnson C. D., Mitchel R., Yoshida T. (Dordrecht: Kluwer Academic Publishers; ), 229–240. [Google Scholar]
- Kitamura K., Ishimoto M., Sawa M. (1988). Inheritance of resistance to infestation with azuki bean weevil in Vigna sublobata and successful incorporation to V. radiata. Jap. J. Breed. 38, 459–464. 10.1270/jsbbs1951.38.459 [DOI] [Google Scholar]
- Kosambi D. (1944). The estimation of map distances from recombination values. Ann. Eugen. 12, 172–175. 10.1111/j.1469-1809.1943.tb02321.x [DOI] [Google Scholar]
- Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lee Y. H., Moon J. K., Park K. Y., Ku J. H., Yun H. T., Chung W. K., et al. (2000). A new mungbean cultivar with bruchid resistance., “Jangannogdu”. Korean J. Breed. 32, 296–297. 10.1603/EC14113 [DOI] [Google Scholar]
- Li H., Ye G., Wang J. (2007). A modified algorithm for the improvement of composite interval mapping. Genetics 175, 361–374. 10.1534/genetics.106.066811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Chen X., Chen Z., Cai R., Zhang H., Xiang Y. (2016). Identification and expression analysis of BURP domain-containing genes in Medicago truncatula. Front. Plant Sci. 7:485. 10.3389/fpls.2016.00485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W. J., Ko C. Y., Liu M. S., Kuo C. Y., Wu D. C., Chen C. Y., et al. (2016). Transcriptomic and proteomic research to explore bruchid-resistant genes in mungbean isogenic lines. J. Agric. Food Chem. 64, 6648–6658. 10.1021/acs.jafc.6b03015 [DOI] [PubMed] [Google Scholar]
- Liu M. S., Kuo T. C. Y., Ko C. Y., Wu D. C., Li K. Y., Lin W. J., et al. (2016). Genomic and transcriptomic comparison of nucleotide variations for insights into bruchid resistance of mungbean (Vigna radiata [L.] R. Wilczek). BMC Plant Biol. 16:46. 10.1186/s12870-016-0736-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodhi M. A., Ye G. N., Weeden N. F., Reisch B. I. (1994). A simple and efficient method for DNA extraction from grapevine cultivars and Vitis species. Plant Mol. Biol. Rep. 12, 6–13. 10.1007/BF02668658 [DOI] [Google Scholar]
- Meng L., Li H., Zhang L., Wang J. (2015). QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 3, 269–283. 10.1016/j.cj.2015.01.001 [DOI] [Google Scholar]
- Miura K., Ishimoto M., Yamanaka N., Miyazaki S., Hiramatsu M., Nakajima Y., et al. (1996). Effects of bruchid-resistant mungbean meal on growth and blood-biochemical values in mice. JIRCAS J. 3, 23–31. [Google Scholar]
- Nogueira F. C. S., Silva C. P., Alexandre D., Samuels R. I., Soares E. L., Aragão F. J. L., et al. (2012). Global proteome changes in larvae of Callosobruchus maculatus (Coleoptera: Chrysomelidae: Bruchinae) following ingestion of a cysteine proteinase inhibitor. Proteomics 12, 2704–2715. 10.1002/pmic.201200039 [DOI] [PubMed] [Google Scholar]
- Pauchet Y., Wilkinson P., Chauhan R., Ffrench-Constant R. H. (2010). Diversity of beetle genes encoding novel plant cell wall degrading enzymes. PLoS ONE 5:e15635. 10.1371/journal.pone.0015635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedra J. H. F., Brandt A., Westerman R., Lobo N., Li H. M., Romero-Severson J., et al. (2003). Transcriptome analysis of the cowpea weevil bruchid: identification of putative proteinases and alpha-amylases associated with food breakdown. Insect Mol. Biol. 12, 405–412. 10.1046/j.1365-2583.2003.00425.x [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2012). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; Available online at: https://www.r-project.org/. [Google Scholar]
- Santana M. C. (2013). Identificação de Poligalacturonases Expressas No Sistema Digestório de Callosobruchus Maculatus (Coleoptera: Chrysomelidae: Bruchinae). Thesis, Universidade Federal de Santa Catarina. [Google Scholar]
- Shao Y., Wei G., Wang L., Dong Q., Zhao Y., Chen B., et al. (2011). Genome-wide analysis of BURP domain-containing genes in Populus trichocarpa. J. Integr. Plant Biol. 53, 743–755. 10.1111/j.1744-7909.2011.01068.x [DOI] [PubMed] [Google Scholar]
- Somta C., Somta P., Tomooka N., Ooi P. A. C., Vaughan D. A., Srinives P. (2008). Characterization of new sources of mungbean (Vigna radiata (L) Wilczek) resistance to bruchids., Callosobruchus spp (Coleoptera: Bruchidae). J. Stored Prod. Res. 44, 316–321. 10.1016/j.jspr.2008.04.002 [DOI] [Google Scholar]
- Somta P., Ammaranan C., Ooi P. A. C., Srinives P. (2007). Inheritance of seed resistance to bruchids in cultivated mungbean (Vigna radiata (L) Wilzcek). Euphytica 155, 49–55. 10.1007/s10681-006-9299-9 [DOI] [Google Scholar]
- Somta P., Seehalak W., Srinives P. (2009). Development, characterization and cross-species amplifcation of mungbean (Vigna radiata) genic microsatellite markers. Conserv. Genet. 10, 1939–1943. 10.1007/s10592-009-9860-x [DOI] [Google Scholar]
- Somta P., Srinives P. (2007). Genome research in mungbean [Vigna radiata (L.) Wilczek] and blackgram [V. mungo (L.) Hepper]. ScienceAsia 33(Suppl. 1), 69–74. 10.2306/scienceasia1513-1874.2007.33(s1).069 [DOI] [Google Scholar]
- Srinives P., Somta P., Somta C. (2007). Genetics and breeding of resistance to bruchids (Callosobruchus spp) in Vigna crops: a review. NU. Sci. J. 4, 1–17. [Google Scholar]
- Sugawara F., Ishimoto M., Le-Van N., Koshino H., Uzawa J., Yoshida S., et al. (1996). Insecticidal peptide from mungbean: a resistant factor against infestation with azuki bean weevil. J. Agric. Food. Chem. 44, 3360–3364. 10.1021/jf960166c [DOI] [Google Scholar]
- Talekar N. S., Lin C. L. (1992). Characterization of Callosobruchus chinensis (Coleoptera: Bruchidae) resistance in mungbean. J. Econ. Entomol. 85, 1150–1153. 10.1093/jee/85.4.1150 [DOI] [Google Scholar]
- Tang Y., Cao Y., Qiu J., Gao Z., Ou Z., Wang Y., et al. (2014). Expression of a vacuole-localized BURP-domain protein from soybean (SALI3-2) enhances tolerance to cadmium and copper stresses. PLoS ONE 9:e98830. 10.1371/journal.pone.0098830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y., Cheng X., Ren G. (2015). A 90-day study of three bruchid-resistant mung bean cultivars in Sprague-Dawley rats. Food Chem. Toxicol. 76, 80–85. 10.1016/j.fct.2014.11.024 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.