Abstract
Background/Aim:
Recently, AT-rich interactive domain-containing 1A protein (ARID1A) has been identified as a novel tumor suppressor gene in gastric cancer (GC) and colorectal cancer (CRC). However, the clinicopathologic value of ARID1A mutation or protein level in GC and CRC patients is controversial. Hence, we conducted a meta-analysis on the relationship between ARID1A aberrations and clinicopathologic parameters in GC and CRC.
Materials and Methods:
Relevant published studies were selected from PubMed and EMBASE. The effect sizes of ARID1A mutation or level on the patient's clinicopathologic parameters were calculated by prevalence rate or odds ratio (OR) or hazard ratio (HR), respectively. The effect sizes were combined using a random-effects model.
Results:
The frequency of ARID1A mutation and loss of ARID1A protein expression in GC patients was 17% and 27%, respectively. The loss of ARID1A protein expression of GC patients was significantly associated with advanced tumor depth (OR = 1.8, P = 0.004), lymph node metastasis (OR = 1.4, P = 0.001), and unfavorable adjusted overall survival (HR = 1.5, P < 0.001). ARID1A mutation of GC was significantly associated with microsatellite instability (MSI) (OR = 24.5, P < 0.001) and EBV infection (OR = 2.6, P = 0.001). The frequency of ARID1A mutation and ARID1A protein expression loss in CRC patients was approximately 12–13%. Interestingly, the loss of ARID1A protein expression in CRC patients was significantly associated with poorly differentiated grade (OR = 4.0, P < 0.001) and advanced tumor depth (OR = 1.8, P = 0.012).
Conclusion:
Our meta-analysis revealed that ARID1A alterations may be involved in the carcinogenesis of GC by EBV infection and MSI. The loss of ARID1A protein expression may be a marker of poor prognosis in GC and CRC patients.
Keywords: ARID1A, colorectal cancer, gastric cancer, meta-analysis
INTRODUCTION
Recent studies of gastric cancer (GC) and colorectal cancer (CRC) patients using next-generation sequencing have highlighted the new tumor suppressor gene, AT-rich interactive domain-containing 1A protein (ARID1A).[1,2,3,4,5,6,7] The prevalence of ARID1A mutation in GC and CRC ranged from 8–31% and 6–39%, respectively.[1,2,3,4,5,6,7,8,9,10,11,12,13,14] ARID1A encodes a large nuclear protein, which forms Switch/Sucrose nonfermentable chromatin remodeling complex and plays a critical regulatory role in cellular processes including development, differentiation, proliferation, and DNA repair.[15]
The majority of ARID1A mutations are mostly insertion/deletion and nonsense mutations, which lead to protein truncation and consequent rapid degradation.[1,16] Therefore, loss of ARID1A protein expression is highly correlated with ARID1A mutation.[1,16] Several reports have indicated the loss of ARID1A protein expression in GC and CRC.[1,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32] However, clinicopathologic significances of ARID1A mutation or protein expression loss in GC and CRC patients remains unclear.[1,2,3,4,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32] Therefore, we conducted a meta-analysis to clarify the clinicopathologic characteristics of ARID1A mutation or protein expression loss in GC and CRC patients.
MATERIAL AND METHODS
Data collection and selection criteria for meta-analysis
The search was conducted according to the PRISMA (Preferred Reporting Items for Systemic Reviews and Meta-Analyses) guidelines.[33] We searched PubMed (http://www.ncbi.nlm.nih.gov/pubmed) and EMBASE (www.embase.com) using the keywords: [(ARID1A) and (stomach cancer or gastric cancer) or (colorectal cancer or colon cancer or rectal cancer]. In addition, we manually explored the reference lists of identified articles. Duplicate data or overlapping articles were excluded by examining the authors' names and affiliations. When multiple articles were published by the same authors or group, the most informative or recent article was selected. Original articles reporting cases of ARID1A mutation or protein expression level in GC and CRC published before October 2016 were included. We excluded review articles without original data, conference abstracts, case reports, cell line studies, and articles lacking clinicopathologic data for meta-analysis. Geographic or language restrictions were not applied. Study quality was independently scored by two reviewers according to the Newcastle-Ottawa Scale,[34] which is frequently used for case-control studies with a maximum case-control score of 9. The selection process of the articles is shown in Figure 1.
Figure 1.

The flowchart of article selection for meta-analysis
Data pooling and statistics
A meta-analysis was performed as previously described.[35] Briefly, effect sizes for each study were calculated by prevalence rate or odds ratio (OR) or hazard ratio (HR), and the corresponding 95% confidence interval (CI) using the Mantel-Haenszel method or the Cohen method. The prevalence rate or OR or HR was combined using a random-effects model (DerSimonian-Laird method). Statistical heterogeneity among studies was evaluated using the Cochrane Q test and I2 statistics. The I2 statistic refers to the percentage of variation across studies due to heterogeneity rather than chance and does not inherently depend on the number of studies considered [I2 = 100% × (Q − df)/Q]. We clarified the cutoff of I2 statistics for assignment of low (<25%), moderate (25–50%), and high (>50%) heterogeneities. If I2 value was >25%, subgroup analysis was conducted. Sensitivity analyses were performed for influence of each study on the pooled prevalence rate, OR, or HR by serially omitting an individual study and pooling the remaining studies. The Publication bias was determined by funnel plots and Egger's tests for the degree of asymmetry. Publication bias was assumed as present if P value was <0.1. The pooled analysis was performed using Comprehensive Meta-analysis Software version 2.0 (Biostat, Englewood, NJ, USA).
RESULTS
Gastric cancer
ARID1A protein level
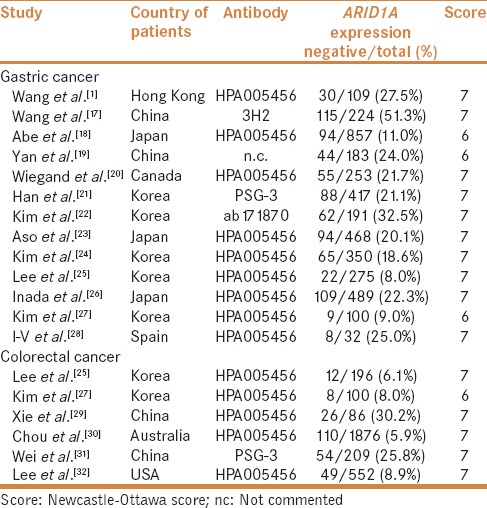
Thirteen studies reported the frequency of ARID1A protein level among 3948 cases of GC in 285 Caucasian and 3663 Asian patients [Table 1].[1,17,18,19,20,21,22,23,24,25,26,27,28] Pooled analysis indicated the loss of ARID1A protein expression in 26.7% of GC patients (95% CI: 14.8–43.1). Moreover, loss of ARID1A protein expression did not differ according to ethnicity (P = 0.839).
Table 1.
Characteristics of individual studies of ARID1A expression loss included in the meta-analysis
Ten studies described ARID1A protein level in GC patients according to gender.[1,17,18,19,20,21,22,23,24,26] ARID1A protein expression loss was observed in 522 of 2462 male and 234 of 1079 female GC patients. ARID1A protein level showed no association with gender (OR = 0.982, 95% CI: 0.797–1.209; P = 0.862, Q = 11.767, I2 = 23.515).
Nine studies reported ARID1A protein level according to the histologic subtype of GC.[1,18,19,20,21,22,23,24,25] ARID1A protein expression loss was found in 285 of 1625 intestinal type and 219 of 1234 diffuse type GC patients. ARID1A protein expression loss was not associated with the histologic subtype of GC (OR = 0.825, 95% CI: 0.671–1.014; P = 0.068, Q = 7.413, I2 = 0.000).
Five studies presented ARID1A protein level according to the tumor depth.[18,20,21,23,24] ARID1A protein expression loss was observed in 71 of 637 early gastric cancer (EGC) and 324 of 1708 advanced gastric cancer (AGC) patients. The loss of ARID1A protein expression was significantly associated with AGC (OR = 1.813, 95% CI: 1.203–2.733; P = 0.004, Q = 8.030, I2 = 50.186). Subgroup analysis revealed that the association between tumor depth and ARID1A protein level differed according to ethnicity [Table 2].
Table 2.
Subgroup analysis

Ten studies presented ARID1A protein level according to the lymph node metastasis.[1,17,18,19,20,22,23,24,25,26] ARID1A protein expression loss was observed in 474 of 2085 cases with lymph node metastasis and 215 of 1275 cases without lymph node metastasis. The loss of ARID1A protein expression was significantly associated with lymph node metastasis (OR = 1.432, 95% CI: 1.158–1.772; P = 0.001, Q = 10.979, I2 = 18.026) [Figure 2].
Figure 2.
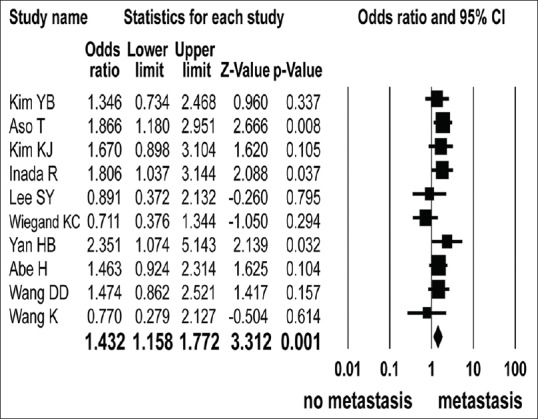
Odd ratios with corresponding 95% confidence intervals of individual studies and pooled data for the association between negative expression of ARID1A protein in gastric cancer patients and lymph node metastasis. Forest plot demonstrates the effect sizes and 95% CIs for each study and overall
Five studies described ARID1A protein level according to the clinical stage.[1,21,22,25,26] ARID1A protein expression loss was observed in 175 of 742 of the stage III, IV and 136 of 720 of stage I, II GC patients. There was no significant association between stage and ARID1A protein expression loss (OR = 1.195, 95% CI: 0.861–1.658; P = 0.287, Q = 5.626, I2 = 28.901). Subgroup analysis revealed that the association between stage and ARID1A protein level was not different according to the antibody type [Table 2].
Five[1,17,19,24,26] and four[17,20,24,26] studies presented the univariate unadjusted and multivariate adjusted survival outcomes of GC patients according to the ARID1A protein level. The number of patients in each study ranged from 109 to 489, for a total of 1355 and 1316 patients, respectively. The estimated unadjusted and adjusted HRs ranged from 0.511–1.981 and 1.36–1.663, respectively. The prognostic variables used in the multivariate survival model were patient's sex, histologic type, and clinical stage. The loss of ARID1A protein expression was significantly associated with unfavorable adjusted overall HR (HR = 1.508, 95% CI: 1.249–1.820; P < 0.001, Q = 0.834, I2 = 0.000) [Figure 3], but not unadjusted overall HR (HR = 1.388, 95% CI: 0.937–2.055; P = 0.102, Q = 16.449, I2 = 75.683). Subgroup analysis revealed that the kind of antibody used did not influence the relationship between ARID1A protein expression loss and unadjusted overall HR [Table 2].
Figure 3.
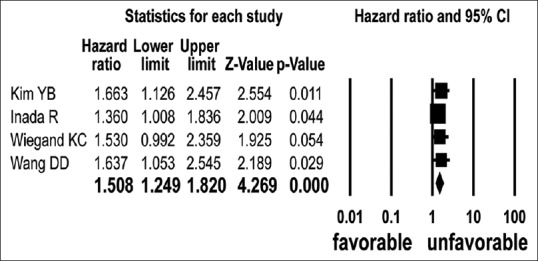
Hazard ratios and pooled data for overall survival according to ARID1A expression in multivariate analysis
Six studies reported ARID1A protein level according to Epstein-Barr virus (EBV) infection[1,18,20,21,23,24]. ARID1A protein expression loss was detected in 71 of 189 EBV-associated gastric cancer (EBVaGC) cases and 353 of 2258 non-EBVaGC cases. The loss of ARID1A protein expression was significantly associated with EBVaGC (OR = 3.351, 95% CI: 2.156–5.210; P < 0.001, Q = 8.301, I2 = 39.796) [Figure 4]. Subgroup analysis revealed that the association between EBVaGC and ARID1A protein expression loss was not different according to the ethnicity and antibody type [Table 2].
Figure 4.
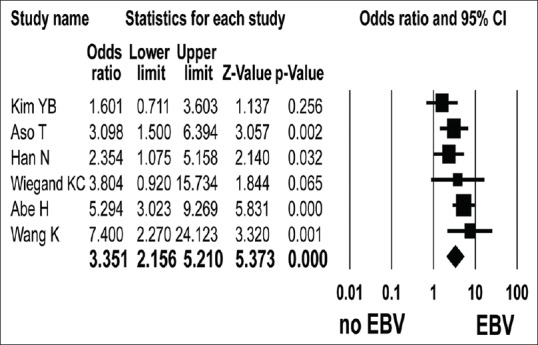
Forest plot of odds ratios with corresponding 95% confidence intervals for the association between negative expression of ARID1A protein and EBV associated gastric cancer
ARID1A mutation
A total of 10 studies reported the frequency of ARID1A mutation among 1036 GC patients, of which 551 were Caucasian and 485 were Asian [Table 3].[1,2,3,4,5,8,9,10,11,12] Pooled analysis indicated ARID1A mutation in 16.8% of patients with GC (95% CI: 11.7–23.7). ARID1A mutation did not differ according to ethnicity (P = 0.453).
Table 3.
Characteristics of individual studies of ARID1A mutation included in the meta-analysis
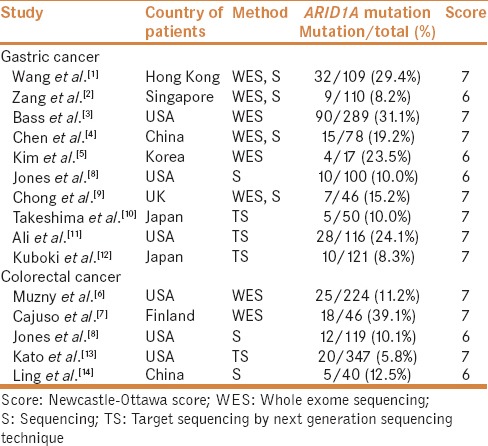
Four studies described ARID1A mutation in 325 cases with III or IV stage and 246 cases with I or II stage.[1,2,3,4] ARID1A mutation was observed in 70 of 325 stage III and IV cases and 69 of 246 stage I and II cases. The stage was not significantly associated with ARID1A mutation (OR = 0.782, 95% CI: 0.526–1.164; P = 0.226, Q = 1.309, I2 = 0.000).
Four studies reported ARID1A mutation according to the microsatellite instability (MSI).[1,2,3,5] ARID1A mutation was found in 80 of 103 MSI GC cases and 55 of 422 stable MSI GC cases. ARID1A mutation was significantly associated with MSI (OR = 24.495, 95% CI: 13.633–44.012; P < 0.001, Q = 0.503, I2 = 0.000). Three studies presented ARID1A mutation in EBVaGC.[1,3,4] ARID1A mutation was detected in 30 of 77 EBVaGC cases and 107 of 399 non-EBVaGC cases. ARID1A mutation was significantly associated with EBVaGC (OR = 2.572, 95% CI: 1.445–4.577; P = 0.001, Q = 0.530, I2 = 0.000).
Colorectal cancer
Six studies reported the frequency of ARID1A protein level in 3019 CRC patients [Table 1].[25,27,29,30,31,32] Pooled analysis indicated the loss of ARID1A protein expression in 11.7% of CRC patients (95% CI: 6.1–21.4). Five studies presented the prevalence of ARID1A mutation in 776 CRC patients [Table 3].[6,7,8,13,14] Pooled analysis indicated ARID1A mutation in 13.0% of patients with CRC (95% CI: 6.4–24.6). ARID1A mutation and protein level did not differ with ethnicity (P = 0.958 and P = 0.119, respectively).
Five studies focused on ARID1A protein level in CRC by histologic grade.[25,29,30,31,32] ARID1A protein expression loss was found in 86 of 415 poorly differentiated and 139 of 2099 well to moderately differentiated CRC patients. The loss of ARID1A protein expression was significantly associated with poorly differentiated CRC (OR = 3.952, 95% CI: 2.206–7.081; P < 0.001, Q = 9.440, I2 = 57.627) [Figure 5].
Figure 5.
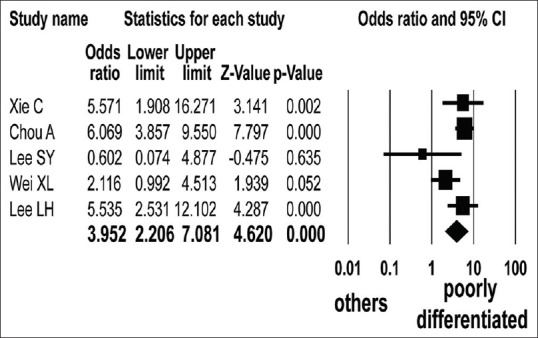
Forest plot of odds ratios with corresponding 95% confidence intervals for the association between negative expression of ARID1A protein and histologic grade of colorectal cancer
Four studies described ARID1A protein level in CRC patients according to the tumor depth.[25,29,31,32] ARID1A protein expression loss was observed in 24 of 269 cases with T1,2 and 117 of 773 cases with T3,4 CRC patients. The loss of ARID1A protein expression was significantly associated with advanced tumor depth (OR = 1.849, 95% CI: 1.146–2.984; P = 0.012, Q = 0.793, I2 = 0.000).
Four studies reported ARID1A protein level in CRC patients according to the stage.[25,29,30,31] The ARID1A protein expression loss was observed in 89 of 1118 cases with stage I, II and 112 of 1231 cases with stage III, IV CRC patients. Stage showed no significant association with ARID1A protein level (OR = 1.139, 95% CI: 0.837–1.550; P = 0.409, Q = 1.779, I2 = 0.000).
Publication bias and sensitivity analysis
Funnel plots and the Egger's regression tests of pooled result of association between clinical stage of GC and ARID1A protein level indicated the possibility of publication bias. However, other pooled analyses showed no evidence of publication bias (data not shown).
Sensitivity analyses revealed that individual study affected the results of histologic subtype, tumor depth, stage, and unadjusted HR according to ARID1A protein level in GC. The study of Bass et al. influenced the result of EBV association.[3] However, other studies did not affect the OR or HR with the 95% CIs.
DISCUSSION
This pooled analysis using 1036 and 3948 GC patients confirmed that ARID1A mutation and ARID1A protein expression loss were detected in 17% and 27% cases, respectively. The loss of ARID1A protein expression in GC patients was significantly associated with advanced tumor depth, lymph node metastasis, and unfavorable overall survival. This meta-analysis confirmed that ARID1A mutation or protein expression loss was significantly associated with MSI and EBV infection of GC patients. In addition, the frequency of ARID1A mutation or loss of ARID1A protein expression occurred in 12–13% of CRC patients. The loss of ARID1A protein expression was significantly associated with poorly differentiated histologic grade CRC and advanced tumor depth.
To our knowledge, this is the first meta-analysis investigating the relationship between ARID1A mutation or protein expression loss and clinicopathologic parameters of GC and CRC patients. The role of ARIDA1 aberration in individual GC and CRC patient is currently unclear due to the small sample size and heterogeneous patient population. Wang et al. claimed that ARIDA1 alterations were associated with better prognosis in GC.[1] In contrast, some studies showed that loss of ARID1A protein expression was associated with poor prognostic factors.[17,21,22,26] The previous published data about the associations between ARID1A alteration and clinicopathologic parameters of CRC had not been consistent. The results of pooled analysis in our study indicated that the loss of ARID1A protein expression may be a marker of poor prognosis in individual GC and CRC patient.
The association between ARIDA1 aberration and MSI is a very important issue. This meta-analysis revealed that ARID1A mutation causes dramatic increases in MSI GC patients. Thus, ARID1A may be a driver gene targeted by MSI pathway. Due to deficiency of data, we were unable to study the association between ARIDA1 aberration and MSI of CRC.
Our meta-analysis indicated that ARID1A mutation or loss of its protein expression in GC was associated with EBVaGC. The ARID1A mutation or protein expression loss showed approximately three-fold increase in EBVaGC compared with non-EBVaGC patients. Histologically, EBVaGC is characterized by dense lymphocytic infiltration within or surrounding GC.[36] Interestingly, the lymphoid infiltration is one of the histologic features of GC with MSI.[37] The association between ARID1A mutation or its protein expression loss and subtype with lymphocytic infiltration is suggestive that ARID1A aberration may be related to the immune surveillance of this GC subtype.
Our study had a few limitations. First, the immunohistochemical results of ARID1A protein level vary with the kind of antibody used, tissue fixation time, and data interpretation. Second, the data of ARID1A alteration according to clinicopathologic parameters were insufficient. Lastly, we roughly classified patients into Caucasian and Asian groups, which could lead to discordance between the current result and original data.
Our meta-analysis indicated that GC or CRC with ARID1A alteration might be a marker of poor prognosis. The ARID1A alteration of GC may result from different epigenetic factors.
Financial support and sponsorship
This work was supported by Korea University Grant (K1710601) and Mid-career Researcher Program through National Research Foundation of Korea (NRF) grant (grant number: 2016 R1A2B4012030) funded by the Ministry of Education, Science and Technology.
Conflicts of interest
There are no conflicts of interest.
Acknowledgments
Young-Sik Kim and Hoiseon Jeong contributed equally to this work.
REFERENCES
- 1.Wang K, Kan J, Yuen ST, Shi ST, Chu KM, Law S, et al. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat Genet. 2011;43:1219–23. doi: 10.1038/ng.982. [DOI] [PubMed] [Google Scholar]
- 2.Zang ZJ, Cutcutache I, Poon SL, Zhang SL, McPherson JR, Tao J, et al. Exome sequencing of gastric adenocarcinoma identifies recurrent somatic mutations in cell adhesion and chromatin remodeling genes. Nat Genet. 2012;44:570–4. doi: 10.1038/ng.2246. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K, Yang D, Li X, Sun B, Song F, Cao W, et al. Mutational landscape of gastric adenocarcinoma in Chinese: Implications for prognosis and therapy. Proc Natl Acad Sci USA. 2015;112:1107–12. doi: 10.1073/pnas.1422640112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TM, Jung SH, Kim MS, Baek IP, Park SW, Lee SH, et al. The mutational burdens and evolutionary ages of early gastric cancers are comparable to those of advanced gastric cancers. J Pathol. 2014;234:365–74. doi: 10.1002/path.4401. [DOI] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cajuso T, Hanninen UA, Kondelin J, Gylfe AE, Tanskanen T, Katainen R, et al. Exome sequencing reveals frequent inactivating mutations in ARID1A, ARID1B, ARID2 and ARID4A in microsatellite unstable colorectal cancer. Int J Cancer. 2014;135:611–23. doi: 10.1002/ijc.28705. [DOI] [PubMed] [Google Scholar]
- 8.Jones S, Li M, Parsons DW, Zhang X, Wesseling J, Kristel P, et al. Somatic mutations in the chromatin remodeling gene ARID1A occur in several tumor types. Hum Mutat. 2012;33:100–3. doi: 10.1002/humu.21633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chong IY, Cunningham D, Barber LJ, Campbell J, Chen L, Kozarewa I, et al. The genomic landscape of oesophagogastric junctional adenocarcinoma. J Pathol. 2013;231:301–10. doi: 10.1002/path.4247. [DOI] [PubMed] [Google Scholar]
- 10.Takeshima H, Niwa T, Takahashi T, Wakabayashi M, Yamashita S, Ando T, et al. Frequent involvement of chromatin remodeler alterations in gastric field cancerization. Cancer Lett. 2015;357:328–38. doi: 10.1016/j.canlet.2014.11.038. [DOI] [PubMed] [Google Scholar]
- 11.Ali SM, Sanford EM, Klempner SJ, Rubinson DA, Wang K, Palma NA, et al. Prospective comprehensive genomic profiling of advanced gastric carcinoma cases reveals frequent clinically relevant genomic alterations and new routes for targeted therapies. Oncologist. 2015;20:499–507. doi: 10.1634/theoncologist.2014-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuboki Y, Yamashita S, Niwa T, Ushijima T, Nagatsuma A, Kuwata T, et al. Comprehensive analyses using next-generation sequencing and immunohistochemistry enable precise treatment in advanced gastric cancer. Ann Oncol. 2016;27:127–33. doi: 10.1093/annonc/mdv508. [DOI] [PubMed] [Google Scholar]
- 13.Kato S, Schwaederle M, Daniels GA, Piccioni D, Kesari S, Bazhenova L, et al. Cyclin-dependent kinase pathway aberrations in diverse malignancies: Clinical and molecular characteristics. Cell Cycle. 2015;14:1252–9. doi: 10.1080/15384101.2015.1014149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ling C, Wang L, Wang Z, Xu L, Sun L, Yang H, et al. A pathway-centric survey of somatic mutations in Chinese patients with colorectal carcinomas. PLoS One. 2015;10:e0116753. doi: 10.1371/journal.pone.0116753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–68. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 16.Bosse T, ter Haar NT, Seeber LM, v Diest PJ, Hes FJ, Vasen HF, et al. Loss of ARID1A expression and its relationship with PI3K-Akt pathway alterations, TP53 and microsatellite instability in endometrial cancer. Mod Pathol. 2013;26:1525–35. doi: 10.1038/modpathol.2013.96. [DOI] [PubMed] [Google Scholar]
- 17.Wang DD, Chen YB, Pan K, Wang W, Chen SP, Chen JG, et al. Decreased expression of the ARID1A gene is associated with poor prognosis in primary gastric cancer. PLoS One. 2012;7:e40364. doi: 10.1371/journal.pone.0040364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abe H, Maeda D, Hino R, Otake Y, Isogai M, Ushiku AS, et al. ARID1A expression loss in gastric cancer: Pathway-dependent roles with and without Epstein-Barr virus infection and microsatellite instability. Virchows Arch. 2012;461:367–77. doi: 10.1007/s00428-012-1303-2. [DOI] [PubMed] [Google Scholar]
- 19.Yan HB, Wang XF, Zhang Q, Tang ZQ, Jiang YH, Fan HZ, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis. 2014;35:867–76. doi: 10.1093/carcin/bgt398. [DOI] [PubMed] [Google Scholar]
- 20.Wiegand KC, Sy K, Kalloger SE, Li-Chang H, Woods R, Kumar A, et al. ARID1A/BAF250a as a prognostic marker for gastric carcinoma: A study of 2 cohorts. Hum Pathol. 2014;45:1258–68. doi: 10.1016/j.humpath.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 21.Han N, Kim MA, Lee HS, Kim WH. Loss of ARID1A expression is related to gastric cancer progression, Epstein-Barr virus infection, and mismatch repair deficiency. Appl Immunohistochem Mol Morphol. 2016;24:320–5. doi: 10.1097/PAI.0000000000000199. [DOI] [PubMed] [Google Scholar]
- 22.Kim KJ, Jung HY, Oh MH, Cho H, Lee JH, Lee HJ, et al. Loss of ARID1A expression in gastric cancer: Correlation with mismatch repair deficiency and clinicopathologic features. J Gastric Cancer. 2015;15:201–8. doi: 10.5230/jgc.2015.15.3.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aso T, Uozaki H, Morita S, Kumagai A, Watanabe M. Loss of ARID1A, ARID1B, and ARID2 expression during progression of gastric cancer. Anticancer Res. 2015;35:6819–27. [PubMed] [Google Scholar]
- 24.Kim YB, Ham IH, Hur H, Lee D. Various ARID1A expression patterns and their clinical significance in gastric cancers. Hum Pathol. 2016;49:61–70. doi: 10.1016/j.humpath.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 25.Lee SY, Kim DW, Lee HS, Ihn MH, Oh HK, Park DJ, et al. Loss of AT-rich interactive domain 1A expression in gastrointestinal malignancies. Oncology. 2015;88:234–40. doi: 10.1159/000369140. [DOI] [PubMed] [Google Scholar]
- 26.Inada R, Sekine S, Taniguchi H, Tsuda H, Katai H, Fujiwara T, et al. ARID1A expression in gastric adenocarcinoma: Clinicopathological significance and correlation with DNA mismatch repair status. World J Gastroenterol. 2015;21:2159–68. doi: 10.3748/wjg.v21.i7.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim MS, Je EM, Yoo NJ, Lee SH. Loss of ARID1A expression is uncommon in gastric, colorectal, and prostate cancers. APMIS. 2012;120:1020–2. doi: 10.1111/j.1600-0463.2012.02930.x. [DOI] [PubMed] [Google Scholar]
- 28.Ibarrola-Villava M, Llorca-Cardenosa MJ, Tarazona N, Mongort C, Fleitas T, Perez-Fidalgo JA, et al. Deregulation of ARID1A, CDH1, cMET and PIK3CA and target-related microRNA expression in gastric cancer. Oncotarget. 2015;6:26935–45. doi: 10.18632/oncotarget.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie C, Fu L, Han Y, Li Q, Wang E. Decreased ARID1A expression facilitates cell proliferation and inhibits 5-fluorouracil-induced apoptosis in colorectal carcinoma. Tumour Biol. 2014;35:7921–7. doi: 10.1007/s13277-014-2074-y. [DOI] [PubMed] [Google Scholar]
- 30.Chou A, Toon CW, Clarkson A, Sioson L, Houang M, Watson N, et al. Loss of ARID1A expression in colorectal carcinoma is strongly associated with mismatch repair deficiency. Hum Pathol. 2014;45:1697–703. doi: 10.1016/j.humpath.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 31.Wei XL, Wang DS, Xi SY, Wu WJ, Chen DL, Zeng ZL, et al. Clinicopathologic and prognostic relevance of ARID1A protein loss in colorectal cancer. World J Gastroenterol. 2014;20:18404–12. doi: 10.3748/wjg.v20.i48.18404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee LH, Sadot E, Ivelja S, Vakiani E, Hechtman JF, Sevinsky CJ, et al. ARID1A expression in early stage colorectal adenocarcinoma: An exploration of its prognostic significance. Hum Pathol. 2016;53:97–104. doi: 10.1016/j.humpath.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wells GA, Shea B, O'Connell D, Peterson J, Welch V, Losos M, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomized studies in meta-analyses. Available from: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp .
- 35.Lee JH, Kim Y, Choi JW, Kim YS. The association between papillary thyroid carcinoma and histologically proven Hashimoto's thyroiditis: A meta-analysis. Eur J Endocrinol. 2013;168:343–9. doi: 10.1530/EJE-12-0903. [DOI] [PubMed] [Google Scholar]
- 36.Shinozaki-Ushiku A, Kunita A, Fukayama M. Update on Epstein-Barr virus and gastric cancer (review) Int J Oncol. 2015;46:1421–34. doi: 10.3892/ijo.2015.2856. [DOI] [PubMed] [Google Scholar]
- 37.dos Santos NR, Seruca R, Constancia M, Seixas M, Sobrinho-Simoes M. Microsatellite instability at multiple loci in gastric carcinoma: Clinicopathologic implications and prognosis. Gastroenterology. 1996;110:38–44. doi: 10.1053/gast.1996.v110.pm8536886. [DOI] [PubMed] [Google Scholar]


