ABSTRACT
Human leishmaniases are widespread diseases with different clinical forms caused by about 20 species within the Leishmania genus. Leishmania species identification is relevant for therapeutic management and prognosis, especially for cutaneous and mucocutaneous forms. Several methods are available to identify Leishmania species from culture, but they have not been standardized for the majority of the currently described species, with the exception of multilocus enzyme electrophoresis. Moreover, these techniques are expensive, time-consuming, and not available in all laboratories. Within the last decade, mass spectrometry (MS) has been adapted for the identification of microorganisms, including Leishmania. However, no commercial reference mass-spectral database is available. In this study, a reference mass-spectral library (MSL) for Leishmania isolates, accessible through a free Web-based application (mass-spectral identification [MSI]), was constructed and tested. It includes mass-spectral data for 33 different Leishmania species, including species that infect humans, animals, and phlebotomine vectors. Four laboratories on two continents evaluated the performance of MSI using 268 samples, 231 of which were Leishmania strains. All Leishmania strains, but one, were correctly identified at least to the complex level. A risk of species misidentification within the Leishmania donovani, L. guyanensis, and L. braziliensis complexes was observed, as previously reported for other techniques. The tested application was reliable, with identification results being comparable to those obtained with reference methods but with a more favorable cost-efficiency ratio. This free online identification system relies on a scalable database and can be implemented directly in users' computers.
KEYWORDS: Leishmania, MALDI-TOF, database, identification online
INTRODUCTION
Leishmaniasis is caused by protozoan parasites that belong to the genus Leishmania and are transmitted by the bite of female phlebotomine sand flies. Leishmaniasis is a widespread disease, with 350 million people at risk and 12 million new cases per year in more than 98 countries (1). Currently, 53 Leishmania species have been described and are included in the two main phylogenetic lineages, euleishmania and paraleishmania. Among them, 31 species are involved in mammalian parasitism, and 20 species are associated with human leishmaniasis (2). Different clinical forms are commonly observed, including cutaneous, mucocutaneous, and visceral forms. Cutaneous leishmaniasis (CL) affects 1.5 million patients per year and displays a large spectrum of clinical forms, from small self-resolving papules to destructive mucosal lesions. The clinical presentation of CL depends on the Leishmania species, but the lesion characteristics and the probable place of contamination are not specific enough to determine the species involved (3). Nevertheless, the patient's travel history is a very important piece of information and could be considered a quality control for diagnostic results. Infections by two species of the Leishmania subgenus (Leishmania major and L. mexicana) show frequent spontaneous cures within a few months (3). On the other hand, the two main species of the Viannia subgenus (Leishmania braziliensis and L. panamensis-L. guyanensis) are associated with a 1 to 15% risk of delayed mucosal metastasis (4). Therefore, Leishmania species identification is especially relevant in CL and mucocutaneous leishmaniasis for therapeutic management (5). Moreover, Leishmania species identification remains fundamental for understanding complex epidemiological cycles and for setting up disease control measures. Several methods for Leishmania identification to the species level are available (reviewed in reference 6). Molecular methods allow Leishmania typing directly from clinical samples (e.g., skin biopsy specimens) or from parasite cultures (7). Multilocus enzyme electrophoresis (MLEE), which is based on parasite culture isolation, is considered by the World Health Organization to be the gold standard for parasite typing and is the only technique that has been evaluated for almost all currently identified Leishmania species (8). Nevertheless, this method is cumbersome, time-consuming, and used in only a few laboratories worldwide. Alternatively, PCR-based methods, such as restriction fragment length polymorphism (RFLP) or amplicon sequence analysis, have been proposed as methods to identify Leishmania species. Although they are simpler than MLEE, they remain time-consuming and are not available everywhere.
During the last decade, mass spectrometry (MS) has been adapted to microorganism identification. Nowadays, matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MS is integrated into the workflow of many routine laboratories, thus greatly simplifying pathogen identification and improving patient care (reviewed in reference 9). A few studies support the proof-of-concept for Leishmania species identification with this method (10–12). However, those studies were done by using in-house databases that are not available to other people working on leishmaniasis. As no reference mass-spectral library (MSL) for Leishmania is commercially available, the aim of this study was to construct and test the reliability of a reference MSL using samples from an international collection of Leishmania isolates in Montpellier, France (Centre de Ressources Biologiques des Leishmania [CRB-Leish]). This library is available through a free online mass-spectral identification (MSI) application.
(This study was presented in part as a poster at the 6th World Congress on Leishmaniasis, Toledo, Spain, 16 to 20 May 2017 [13].)
RESULTS
Construction and testing of the mass-spectral reference database (MSL).
The Leishmania MSL was built by using the mass spectra of 121 well-characterized Leishmania strains from the CRB-Leish collection (Table 1). The MSL was implemented in a specific Web-based application designed for mass-spectral identification (MSI application; https://biological-mass-spectrometry-identification.com/msi/) and then tested with an independent panel of 268 samples (231 Leishmania isolates and 37 outgroup controls) from four different laboratories (see Table 2 for a complete description of these samples). In each laboratory, the MS spectra of the allotted samples (four replicates for each sample) from the panel were used for sample identification with the MSI application and the reference MSL. For each submitted spectrum, the application gave a similarity score that includes three subscores (identification to the species level [score A], identification to the complex level [score B], and identification in a different complex [score C]). Each subscore ranged from 0 to 100 (100 indicates a perfect match between the compared spectra) (Fig. 1). For the 37 outgroup controls (Herpetomonas, Crithidia, Endotrypanum, bacteria, and fungi), no match with Leishmania was obtained, and the similarity score was systematically <17 (data not shown). The MSI application results were then compared with those obtained with the reference identification method used in that laboratory (Table 2). Overall, there was good concordance between methods; however, weakness in species differentiation, particularly within the L. braziliensis, L. guyanensis, and L. donovani complexes, was observed (Table 3). Specifically, only 6 (32%) of the 20 L. braziliensis isolates were well classified. However, among the 31 misidentified samples, only 1 was a wrong identification at the complex level and corresponded to 1 L. braziliensis isolate that was identified as L. guyanensis (Table 3).
TABLE 1.
Isolates used for mass-spectral library constructiona
| Section | Subgenus | Complex | Taxon (no. of isolates) | Isolate(s) (WHO code) used for MSL construction |
|---|---|---|---|---|
| Euleishmania | L. enriettii complex | L. enriettii | L. enriettii (2) | MCAV/BR/45/L88, MCAV/BR/95/CUR2 |
| L. martiniquensis (3) | MHOM/GP/2008/LEM5748, MHOM/MQ/92/MAR1, MHOM/MQ/94/LEM2870CL | |||
| L. siamensis (1) | MHOM/TH/2010/TR | |||
| Leishmania | L. donovani | L. archibaldi (8) | MHOM/ET/72/GEBRE1, MCAN/SD/98/LEM3556, MCAN/SD/99/LEM3796, MHOM/TR/2001/EP59, MHOM/SD/97/LEM3463, MHOM/SD/97/LEM3429, MHOM/SD/2002/MW116, MHOM/SD/2002/MW101 | |
| L. donovani (6) | MHOM/IN/00/DEVI, MHOM/IN/80/DD8, MHOM/IN/96/THAK57, MHOM/IN/99/8998, MHOM/TR/2012/LPS55, MHOM/LK/2010/OVN3 | |||
| L. infantum (7) | MHOM/EG/87/RTC2, MCAN/FR/87/RM1, MHOM/DZ/82/LIPA59, MHOM/TN/80/IPT1, MHOM/FR/78/LEM75, MHOM/DZ/83/LIPA96, MHOM/FR/88/LEM1265 | |||
| L. major | L. arabica (5) | MPSA/SA/83/JISH220, MCAN/SA/84/MD84, MPSA/SA/84/JISH238, MPSA/SA/84/JISH231, MPSA/SA/83/JISH224 | ||
| L. gerbilli (3) | MRHO/CN/60/GERBILLI, MRHO/SU/87/E11, MRHO/SU/88/KD/88984 | |||
| L. major (7) | MHOM/IL/80/FRIEDLIN2, MHOM/IL/2008/LRCL1350, MHOM/MA/81/LEM265, MHOM/MA/2003/LEM4685, MRHO/SU/59/Pstrain, MHOM/TN/2001/LEM4286, MHOM/SU/73/5ASKH | |||
| L. turanica (6) | MMEL/SU/79/MEL, MHOM/SU/64/VL, MRHO/SU/65/VL, MRHO/SU/74/95A, MRHO/SU/95/T9562R, MRHO/SU/91/T/91011L TM | |||
| L. mexicana | L. amazonensis (4) | MHOM/BR/72/M1845, MHOM/BR/73/M2269, MHOM/GF/2002/LAV003, MHOM/VE/2004/OHN/15 | ||
| L. aristidesi (1) | MORY/PA/69/GML3 | |||
| L. garnhami (1) | MHOM/VE/76/JAP78 | |||
| L. mexicana (4) | MHOM/BZ/82/BEL21, MNYC/BZ/62/M379, MHOM/VE/2004/CAP04, MHOM/HN/2002/LLM/1162 | |||
| L. pifanoi (1) | MHOM/VE/57/LL1 | |||
| L. tropica | L. aethiopica (6) | MHOM/ET/72/L100, MHOM/ET/91/KASSAYE, MHOM/ET/2004/LPN241, MHOM/JO/2006/LSL103, MHOM/ET/83/10383, MHOM/ET/89/LEM151 | ||
| L. killicki (4) | MHOM/DZ/2011/LIPA283, MHOM/TN/80/LEM163, MHOM/TN/80/LEM180, MHOM/TN/86/LEM904/CL | |||
| L. tropica (5) | MHOM/SU/74/K27, MHOM/GR/80/GRL35, MHOM/MA/95/LEM3015, MHOM/MA/2008/W38, MRAT/IQ/72/ADHANIS1 | |||
| Viannia | L. braziliensis | L. braziliensis (4) | MHOM/BR/75/M2904, MHOM/BR/82/LTB12JULY82, MHOM/GF/2003/LAV008, MHOM/BR/75/M2903 | |
| L. peruviana (5) | MHOM/00/2007/TIM24, MHOM/PE/84/LC26, MHOM/PE/84/UN56, MHOM/PE/84/L13A, MHOM/PE/84/L9 | |||
| L. guyanensis | L. guyanensis (5) | MHOM/GF/98/LEM3657, MHOM/GF/2003/LEM4570, MHOM/GF/2004/LAV016, MHOM/GF/2005/LPC410, MHOM/GF/79/LEM85 | ||
| L. panamensis (6) | MHOM/CR/2004/TIM13, MHOM/CR/2007/TIM23, MHOM/00/2004/LEI27, MHOM/EC/90/TERESA, MHOM/CO/89/UA403, MHOM/CR/2001/LCB29A | |||
| L. shawi (1) | MCEB/BR/84/M8408 | |||
| L. lainsoni | L. lainsoni (6) | IUBI/BR/2000/M12025, MCUN/BR/85/M9342, MHOM/BR/81/M6426, MHOM/PE/2013/LEM6459, MHOM/EC/93/489, MHOM/GF/2002/LEM4449 | ||
| L. naiffi (7) | MDAS/BR/79/M5533, MHOM/GF/LCB41, MHOM/GF/2006/LEM5108, MHOM/GF/2005/LEM5109, MHOM/GF/95/LBC1096, MHOM/00/94/CRE58, MHOM/00/99/TIM1 | |||
| Sauroleishmania | Sauroleishmania | L. adleri (1) | RLAT/KE/54/HU1433 | |
| L. gymnodactyli (1) | RGYM/SY/64/LV247 | |||
| L. hoogstraali (1) | RLIZ/SD/2000/LV31 | |||
| L. tarentolae (3) | RTAR/DZ/39/TARVI, RTAR/FR/77/LEM112, RTAR/FR/78/LEM126 | |||
| Paraleishmania | L. herreri | L. colombiensis (2) | IGOM/PA/85/E58234, IHAR/CA/86/CL500 | |
| L. deanei (2) | MCOE/BR/74/M2674, MCOE/BR/75/M2808 | |||
| L. equatoriensis (2) | MCHO/EC/82/LSP1.2, MCHO/EC/82/LSP1 | |||
| L. hertigi (1) | MCOE/PA/65/C8 |
The total number of isolates was 121.
TABLE 2.
List of Leishmania species tested by the 4 centers and the gold-standard method useda
| Panel description | No. of isolates tested (method) |
||||
|---|---|---|---|---|---|
| Montpellier | Cayenne | Barcelona | Marseille | Total | |
| L. adleri | 1 (MLEE) | 1 | |||
| L. aethiopica | 2 (MLEE) | 2 | |||
| L. amazonensis | 1 (RFLP) | 1 (hsp70 and rpoIILS) | 2 | ||
| L. archibaldi | 1 (MLEE) | 1 | |||
| L. braziliensis | 6 (MLEE), 2 (rpoIILS) | 4 (rpoIILS) | 7 (hsp70 and rpoIILS) | 1 (MLEE) | 20 |
| L. deanei | 1 (MLEE) | 1 | |||
| L. donovani | 3 (MLEE) | 4 (hsp70 and rpoIILS) | 7 | ||
| L. enriettii | 1 (MLEE) | 1 | |||
| L. gerbilli | 1 (MLEE) | 1 | |||
| L. guyanensis | 3 (MLEE), 51 (rpoIILS) | 15 (rpoIILS), 1 (RFLP) | 1 (hsp70 and rpoIILS) | 1 (MLEE) | 72 |
| L. gymnodactyli | 1 (rpoIILS) | 1 | |||
| L. infantum | 1 (MLEE) | 26 (hsp70 and rpoIILS) | 47 (MLEE) | 74 | |
| L. killicki | 3 (MLEE) | 3 | |||
| L. lainsoni | 3 (MLEE), 1 (rpoIILS) | 4 | |||
| L. major | 3 (MLEE), 3 (rpoIILS) | 4 (hsp70 and rpoIILS) | 7 (MLEE) | 17 | |
| L. mexicana | 4 (MLEE) | 1 (hsp70 and rpoIILS) | 5 | ||
| L. naiffi | 1 (rpoIIILS) | 1 (RFLP) | 2 | ||
| L. panamensis | 1 (hsp70 and rpoIILS) | 1 (MLEE) | 2 | ||
| L. peruviana | 4 (MLEE) | 1 (MLEE) | 5 | ||
| L. pifanoi | 1 (MLEE) | 1 (MLEE) | 2 | ||
| L. tropica | 2 (MLEE), 1 (rpoIILS) | 2 (hsp70 and rpoIILS) | 1 (MLEE) | 6 | |
| L. turanica | 1 (MLEE) | 1 (MLEE) | 2 | ||
| Trypanosoma brucei | 2 | 2 | |||
| Herpetomonas | 2 | 2 | |||
| Crithidia | 2 | 2 | |||
| Endotrypanum | 1 | 1 | |||
| Bacteria | 10 (16S rRNA) | 10 | |||
| Fungi | 20 (ITS2) | 20 | |||
| Total | 135 | 22 | 47 | 64 | 268 |
ITS2, internal transcribed spacer 2.
FIG 1.
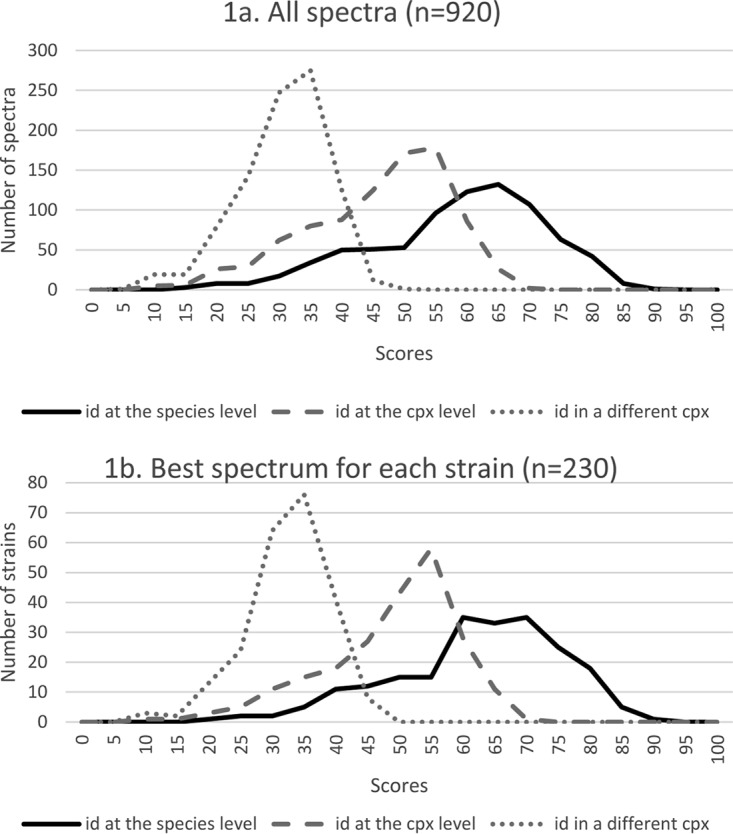
Distribution of the three similarity subscores for all spectra (four replicates for each isolate) (a) and for the spectrum with the best score for each isolate (b). id, identification; cpx, complex. The validation panel included 231 Leishmania isolates, but only 230 were identified by the MSI application.
TABLE 3.
List of the 31 misidentifications by MALDI-TOF MS and MS scores obtained
| Organism identified by reference method (no. of isolates) | Organism identified by MALDI-TOF MS | MS score(s) |
|---|---|---|
| L. braziliensis (13) | L. peruviana | 32.45, 35.17, 36.45, 38.42, 38.71, 43.49, 47.94, 48.44, 54.44, 56.24, 59.96, 65.8, 66.63 |
| L. braziliensis (1) | L. guyanensis | 36.48 |
| L. peruviana (1) | L. braziliensis | 63.83 |
| L. guyanensis (7) | L. panamensis | 41.08, 41.66, 41.95, 42.78, 43.14, 51.73, 60.21 |
| L. panamensis (1) | L. guyanensis | 55.8 |
| L. mexicana (1) | L. pifanoi | 69.57 |
| L. pifanoi (1) | L. mexicana | 55.79 |
| L. deanei (1) | L. hertigi | 43.84 |
| L. donovani (2) | L. archibaldi | 45.04, 53.57 |
| L. infantum (3) | L. archibaldi | 40.49, 47.22, 52.78 |
Score threshold assessment.
Analysis of the three similarity subscores for each sample (Fig. 1) relative to the results obtained with the reference methods indicated that most scores corresponding to a correct identification to the species or complex level were >35. However, the distinction between correct species identification and correct complex identification was more difficult to determine, because there was a clear overlap of the two score distributions (Fig. 1). Therefore, 15 different similarity score thresholds were tested, and positive predictive values (PPVs) (i.e., the proportion of true-positive results) were calculated for each threshold by taking into account only the highest similarity score among the four replicates for each sample (Table 4). The PPV for complex identification was always above 0.99, regardless of the considered threshold. Conversely, it was more difficult to distinguish between species belonging to the same complex, as shown by the PPV results at the species level. The number of misidentifications, particularly at the species level, decreased with higher threshold scores, whereas the number of unidentified strains (with similarity scores below the threshold) increased (Table 4). For instance, with a threshold of 40, all samples were correctly identified to the complex level (PPV = 1), but 13 isolates remained unidentified. These samples corresponded to five correct species identifications, seven correct complex identifications, and one wrong identification.
TABLE 4.
Identification capacities with the various tested score thresholds
| Score threshold | No. of isolates identified correctly to the taxon level | No. of isolates identified correctly to the complex or species level | No. of isolates with incorrect identification | No. of isolates for which the score threshold was not reached | PPV |
|
|---|---|---|---|---|---|---|
| Taxon level | Complex level | |||||
| 20 | 200 | 0 | 1 | 0 | 0.86 | 0.99 |
| 25 | 198 | 30 | 1 | 2 | 0.86 | 0.99 |
| 30 | 198 | 30 | 1 | 2 | 0.86 | 0.99 |
| 35 | 197 | 29 | 1 | 4 | 0.86 | 0.99 |
| 40 | 193 | 25 | 0 | 13 | 0.89 | 1 |
| 45 | 184 | 17 | 0 | 30 | 0.91 | 1 |
| 50 | 171 | 13 | 0 | 47 | 0.92 | 1 |
| 55 | 159 | 9 | 0 | 63 | 0.94 | 1 |
| 60 | 138 | 5 | 0 | 88 | 0.96 | 1 |
| 65 | 100 | 3 | 0 | 128 | 0.97 | 1 |
| 70 | 63 | 0 | 0 | 168 | 1 | 1 |
| 75 | 34 | 0 | 0 | 197 | 1 | 1 |
| 80 | 13 | 0 | 0 | 218 | 1 | 1 |
| 85 | 2 | 0 | 0 | 229 | 1 | 1 |
| 90 | 0 | 0 | 0 | 231 | ||
Culture conditions and testing of parasite concentrations.
To identify the best conditions for MALDI-TOF analysis, three strains from the CRB-Leish collection were used for comparing different culture media, parasite concentrations, and culture durations. Species identification for all three strains was obtained with all tested culture media (RPMI, SDM-79, and Schneider media). The best identification scores were obtained when MALDI-TOF analysis was performed with cultures of 3 × 106 Leishmania parasites/ml in the exponential phase of culture growth, regardless of the time required to obtain this concentration.
MS data acquisition.
In terms of identification performance, no difference was observed when mass spectra were acquired by using the Microflex or Autoflex instrument (Bruker Daltonics).
Online accessibility.
The four participating centers tested the online accessibility of the MSI application. The delay between the query submission and the Excel report acquisition for inputs containing up to 96 spectra was <1 min for all centers. Performances were equivalent with the Mac and Windows operating systems using any of the most used Web browsers (Google Chrome, Mozilla Firefox, Safari, and Internet Explorer).
DISCUSSION
The purpose of this study was to create and test the reliability of a database for Leishmania identification by MALDI-TOF MS using a free Web-based application to meet researchers' and public health needs. The database included spectra from 10 Leishmania complexes representing 33 species, thus covering the main diversity of this genus. Access to the MSI application requires an account and acceptance of a policy agreement. The account is password protected, and access codes are delivered for free after a discussion of the nature of the project, with the aim of ensuring that users are using the application for public health or scientific purposes.
Our results show that neither the medium used nor the time spent to reach the appropriate promastigote concentration significantly influenced the quality of spectra and, thus, the identification. The important point is to obtain a promastigote concentration of 3 × 106 parasites/ml from an exponential-phase culture. Once the sample is prepared, at least four replicates per strain are required, as previously recommended by Cassagne et al. (14). As already observed in many studies, the interpretation of results depends on the chosen score threshold (15, 16). In this study, the lower threshold considered was 20 because our experience with other organisms, such as fungi, indicates that nonspecific identifications are frequent below this threshold (17). With this threshold, 230 of the 231 Leishmania strains were correctly identified at least to the complex level. By increasing the threshold up to 40, the frequency of misidentification at the species level was greatly reduced; however, the number of samples that did not reach the threshold increased, despite these samples being correctly identified in most cases. In real life, the threshold choice can be influenced by many factors, and each user will have to weigh the advantages and disadvantages of each threshold, depending on the required level of accuracy of identification. Nevertheless, the MSI application allows obtaining reliable Leishmania identifications by MALDI-TOF MS in most cases. Moreover, as already seen with other databases, its reliability should increase progressively with the addition of new references (18).
Considering the complexes frequently involved in CL, we observed that L. major and L. tropica identifications were 100% correct. Moreover, our identification system performed well in differentiating L. killicki from L. tropica strains, although it was recently proposed that they could be considered synonymous (19). Conversely, many more incorrect species identifications were obtained for the L. braziliensis complex, which is composed of L. braziliensis and L. peruviana. According to the literature and our experience, very few differences are observed between these species using MLEE (20) or molecular methods (20–23). As the epidemiological data, clinical presentations, and treatments of CL caused by these two species are quite similar, identification to the complex level remains adequate. When considering a threshold value below 40, one L. braziliensis isolate was misidentified by the MSI application as L. guyanensis, a species belonging to the same subgenus but to a different complex. It is known that the L. braziliensis complex is very polymorphic, with a high recombination level, and is closely related to the L. guyanensis complex (24). Only one spectrum (although it obtained the highest similarity score) pointed to an identification of L. guyanensis (score of 36.48), while the other three replicates identified L. braziliensis (scores of 31.22 and 31.46) and L. panamensis (score of 31.98). Thus, in some cases, a general view of the results of the four scores could provide a better approach. For the L. guyanensis complex, which includes the species L. guyanensis, L. panamensis, and L. shawi, 100% and 90.4% of the identifications were correct at the complex and species levels, respectively. As for L. braziliensis and L. peruviana, L. shawi and L. panamensis are considered to be two clusters inside the L. guyanensis complex rather than two different species (21, 24–26).
As CL is a heterogeneous entity, and no therapeutic option is currently effective for all clinical forms, the species involved and the geographic area of contamination guide treatment decisions. Indeed, metastatic extension to the mucosa occurs in 1 to 3% of patients for some species. Fortunately, complex identification is sufficient to guide treatment management (5, 27, 28). Therefore, MALDI-TOF MS is adequate for the first-line identification of strains involved in CL or mucocutaneous leishmaniasis.
Some misidentifications also occurred within the L. donovani complex, which currently includes three species: L. donovani, L. infantum, and L. archibaldi. In our study, it was possible to identify 96% of L. infantum, 71% of L. donovani, and all L. archibaldi strains to the species level. All misidentified L. donovani isolates were erroneously identified as L. archibaldi. According to MLEE, the species L. archibaldi is characterized by a single enzyme, and some molecular studies suggest that the L. donovani complex is more likely to be a continuum in which L. archibaldi stands between the two other species (29). Therefore, some authors attribute the category of subspecies to L. infantum and L. donovani (30), and it is widely accepted that L. archibaldi should not be considered a real species (31–34). Anyway, there is no impact on therapeutic management because this complex is responsible for visceral forms of leishmaniasis, for which treatment does not depend on the species but on the therapeutic agents available in that country.
Although work is in progress to improve the sensitivity of the MSI application for the L. braziliensis, L. guyanensis, and L. donovani complexes, the clinical context and epidemiological data also need to be considered to confirm Leishmania identification. Therefore, the medical biologist should remain completely responsible for the diagnostic procedure.
Overall, the MSI application proved to be a solid identification tool, leading to identification results comparable to those obtained with reference techniques but faster. As shown by the large experience acquired with other microorganisms, the addition of new references will improve current performances; therefore, this identification tool will be further developed with the user community's help. The database could be regularly enriched and updated by users, by adding spectra representing new or rare species. Sharing access to a database, as we propose, should facilitate the implementation of studies requiring the identification of various organisms, by making available a common identification tool. Moreover, it provides an opportunity to implement collaborative research between groups working on related topics. Beyond the specific problem of identifying Leishmania, the approach presented here has also been applied to other organisms such as fungi (35), and we hope that similar initiatives will be implemented to build other identification systems, based on the collective construction of databases by scientific teams.
MATERIALS AND METHODS
Sample preparation. (i) Leishmania cultures.
Leishmania cultures were grown in RPMI (Sigma) and Schneider medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum or in SDM-79 medium at 25°C for 4 to 7 days.
(ii) Sample preparation for MALDI-TOF analysis.
Five milliliters of a promastigote culture containing about 3 × 106 parasites/ml in the exponential phase was harvested by centrifugation (10 min at 3,000 × g) and washed three times with 10 ml of 0.9% NaCl. Promastigote pellets were resuspended in one drop (approximately 20 μl) of 0.9% NaCl. One microliter of the solution was deposited onto the steal target and, once dried, covered with 1 μl of an α-cyano-4-hydroxycinnamic acid (HCCA) matrix (Sigma-Aldrich, Lyon, France). Ten replicates were prepared for every isolate included in the MSL, and four replicates were performed for each isolate in the panel to be tested.
(iii) Mass spectrum acquisition.
Mass spectra were acquired with a Microflex LT instrument (Bruker Daltonics) (three centers, Marseille, Montpellier, and Cayenne, France) or an Autoflex II TOF-TOF instrument (Bruker Daltonics) (one center, Barcelona, Spain) with the default acquisition parameters recommended by Bruker, as described previously by Cassagne et al. (11).
Identification system. (i) Reference database.
A total of 121 Leishmania isolates (Table 1) to be included in the MSL were selected from the CRB-Leish collection. All strains were previously identified by using MLEE as described previously by Rioux et al. (36), and for 50 isolates, species identification was also confirmed by multilocus sequence typing (29). Raw mass spectra were obtained with the Microflex LT instrument (Bruker Daltonics) in Montpellier and used to construct the database. The assignment of each isolate to a group/complex/species was based on the taxonomic classification proposed recently by Akhoundi et al. (2).
(ii) Identification system.
The MSL was implemented in a Web-based application designed for microbial mass spectrum identification (MSI application).
For identification, the raw mass spectra of a sample were compressed into a .zip file and submitted to the Web application. As soon as they were uploaded, raw spectra were subjected to several mathematical operations to eliminate background noise, smooth intensity values, and identify relevant peaks. A series of peak values was automatically built and compared with the peak values of all the reference spectra included in the database. The similarity between the spectrum to be identified and each reference spectrum was rated on a scale ranging from 0 to 100 (100 indicates a perfect match between the compared spectra). To establish the similarity level, the identification system relies on original algorithms, independently from the FlexAnalysis and MaldiBioTyper software provided by Bruker Daltonics. Many samples can be analyzed in one request provided that the number of spectra remains below a few hundred. Results can be exported as Excel files with a similarity score value for each spectrum. Reports may be stored in the user's account.
Implementation of the identification system. (i) Tested panel.
The panel of isolates used to test the online MSI application and the MSL database included 268 strains: 231 Leishmania isolates and 37 outgroup controls (Table 2). The panel was constituted by strains from the four laboratories that participated in the study (four subpanels): (i) 47 isolates were obtained in Barcelona (Secció de Parasitologia, Facultat de Farmàcia i Ciències de l'Alimentació, Universitat de Barcelona) and analyzed at the Hospital de la Santa Creu i Sant Pau (Barcelona, Spain); (ii) 22 isolates were obtained prospectively in French Guyana (Laboratoire de Parasitologie-Mycologie, Centre Hospitalier Universitaire de Cayenne); (iii) 64 strains, 31 originating from Maghreb countries (12 from the Institut Pasteur of Tunis, 19 from the Institut Pasteur of Alger) and 33 isolated in Marseille, were analyzed in Marseille, France; and (iv) 98 strains were selected from CRB-Leish in Montpellier, France, to increase the diversity of species and countries of origin of the strains. For each subpanel, various reference identification methods were used as gold standards, depending on the technique available in the laboratory, including MLEE (36), RFLP, sequencing of the heat shock protein 70 gene (hsp70) (22), or sequencing of the rpoIILS gene (2).
(ii) MSI performance and score threshold assessments.
Isolates from the panel were identified by using the MSI application and the MSL. The analysis report for each spectrum indicates (i) the species with the highest similarity score (this can be considered the isolate identification result) (score A); (ii) the best score obtained for a different species of the same complex, according to the taxonomy reported by Akhoundi et al. (2) (score B); and (iii) the best score obtained for a species in a different complex (score C). The identification results were compared to the results obtained with the gold-standard/reference method for each isolate. The threshold was established in two steps. First, the three scores for each spectrum, when available, were plotted to visualize their overlap (Fig. 1). Next, the performance of the identification system was tested with different thresholds, from 20 to 90, with increments of 5 units for each cutoff (Table 4). Threshold values below 20 were not considered because our previous experience with fungi (17) showed that misidentifications might occur at values below this threshold.
As suggested previously by Normand et al. (17), four replicates for each isolate were deposited, but only the replicate with the highest score was selected, and the identification corresponds to the one obtained for this replicate.
(iii) Testing of culture conditions (medium and duration) and parasite concentrations.
To identify the best culture conditions and parasite concentration for MS identification, three strains from CRB-Leish were selected: L. donovani MHOM/IN/2003/LEM4537, L. major MHOM/TN/93/LPN89, and L. guyanensis MHOM/GF/2004/LBC45. Three different media were tested: SDM-79, RPMI, and Schneider medium supplemented with 10% heat-inactivated fetal calf serum. Isolates were cultured at 25°C. Testing (in triplicate) was done on days 4, 6, 8, and 10 of culture by using five concentrations (from 102 to 106 parasites/ml).
(iv) Online accessibility.
The four centers tested MSI application accessibility and capacities for Mac and PC systems (Windows 7 and Windows 10) with several Web browsers (Google Chrome, Mozilla Firefox, Safari, and Internet Explorer).
REFERENCES
- 1.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M, WHO Leishmaniasis Control Team. 2012. Leishmaniasis worldwide and global estimates of its incidence. PLoS One 7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akhoundi M, Kuhls K, Cannet A, Votýpka J, Marty P, Delaunay P, Sereno D. 2016. A historical overview of the classification, evolution, and dispersion of Leishmania parasites and sandflies. PLoS Negl Trop Dis 10:e0004349. doi: 10.1371/journal.pntd.0004349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morizot G, Kendjo E, Mouri O, Thellier M, Pérignon A, Foulet F, Cordoliani F, Bourrat E, Laffitte E, Alcaraz I, Bodak N, Ravel C, Vray M, Grogl M, Mazier D, Caumes E, Lachaud L, Buffet PA, Cutaneous Leishmaniasis French Study Group. 2013. Travelers with cutaneous leishmaniasis cured without systemic therapy. Clin Infect Dis 57:370–380. doi: 10.1093/cid/cit269. [DOI] [PubMed] [Google Scholar]
- 4.Blum J, Lockwood DNJ, Visser L, Harms G, Bailey MS, Caumes E, Clerinx J, van Thiel PPAM, Morizot G, Hatz C, Buffet P. 2012. Local or systemic treatment for New World cutaneous leishmaniasis? Re-evaluating the evidence for the risk of mucosal leishmaniasis. Int Health 4:153–163. doi: 10.1016/j.inhe.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 5.Buffet PA, Rosenthal É, Gangneux J-P, Lightburne E, Couppié P, Morizot G, Lachaud L, Marty P, Dedet J-P, Société de Pathologie Exotique. 2011. Therapy of leishmaniasis in France: consensus on proposed guidelines. Presse Med 40:173–184. (In French.) doi: 10.1016/j.lpm.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 6.Van der Auwera G, Dujardin J-C. 2015. Species typing in dermal leishmaniasis. Clin Microbiol Rev 28:265–294. doi: 10.1128/CMR.00104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akhoundi M, Downing T, Votýpka J, Kuhls K, Lukeš J, Cannet A, Ravel C, Marty P, Delaunay P, Kasbari M, Granouillac B, Gradoni L, Sereno D. 31 January 2017. Leishmania infections: molecular targets and diagnosis. Mol Aspects Med doi: 10.1016/j.mam.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. 2010. Control of the leishmaniases. World Health Organization technical report series 949. World Health Organization, Geneva, Switzerland. [PubMed] [Google Scholar]
- 9.Clark AE, Kaleta EJ, Arora A, Wolk DM. 2013. Matrix-assisted laser desorption ionization–time of flight mass spectrometry: a fundamental shift in the routine practice of clinical microbiology. Clin Microbiol Rev 26:547–603. doi: 10.1128/CMR.00072-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mouri O, Morizot G, Van der Auwera G, Ravel C, Passet M, Chartrel N, Joly I, Thellier M, Jauréguiberry S, Caumes E, Mazier D, Marinach-Patrice C, Buffet P. 2014. Easy identification of Leishmania species by mass spectrometry. PLoS Negl Trop Dis 8:e2841. doi: 10.1371/journal.pntd.0002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassagne C, Pratlong F, Jeddi F, Benikhlef R, Aoun K, Normand A-C, Faraut F, Bastien P, Piarroux R. 2014. Identification of Leishmania at the species level with matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect 20:551–557. doi: 10.1111/1469-0691.12387. [DOI] [PubMed] [Google Scholar]
- 12.Culha G, Akyar I, Yildiz Zeyrek F, Kurt Ö, Gündüz C, Özensoy Töz S, Östan I, Cavus I, Gülkan B, Kocagöz T, Özbel Y, Özbilgin A. 2014. Leishmaniasis in Turkey: determination of Leishmania species by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS). Iran J Parasitol 9:239–248. [PMC free article] [PubMed] [Google Scholar]
- 13.Lachaud L, Fernández-Arévalo A, Cécile Normand A, Lami P, Nabet C, Donnadieu JL, Piarroux M, Djenad F, Cassagne C, Ravel C, Tebar S, Llovet T, Blanchet D, Demar M, Harrat Z, Aoun K, Bastien P, Muñoz C, Gállego M, Piarroux R. 2017. Abstr 6th World Congr Leishmaniasis, poster C1235. [Google Scholar]
- 14.Cassagne C, Normand A-C, Bonzon L, L'Ollivier C, Gautier M, Jeddi F, Ranque S, Piarroux R. 2016. Routine identification and mixed species detection in 6,192 clinical yeast isolates. Med Mycol 54:256–265. doi: 10.1093/mmy/myv095. [DOI] [PubMed] [Google Scholar]
- 15.Fernandes Santos A, Cayô R, Schandert L, Gales AC. 2013. Evaluation of MALDI-TOF MS in the microbiology laboratory. J Bras Patol Med Lab 49:191–197. doi: 10.1590/S1676-24442013000300006. [DOI] [Google Scholar]
- 16.Biswas S, Rolain J-M. 2013. Use of MALDI-TOF mass spectrometry for identification of bacteria that are difficult to culture. J Microbiol Methods 92:14–24. doi: 10.1016/j.mimet.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 17.Normand A-C, Cassagne C, Gautier M, Becker P, Ranque S, Hendrickx M, Piarroux R. 2017. Decision criteria for MALDI-TOF MS-based identification of filamentous fungi using commercial and in-house reference databases. BMC Microbiol 17:25. doi: 10.1186/s12866-017-0937-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Normand A-C, Cassagne C, Ranque S, L'Ollivier C, Fourquet P, Roesems S, Hendrickx M, Piarroux R. 2013. Assessment of various parameters to improve MALDI-TOF MS reference spectra libraries constructed for the routine identification of filamentous fungi. BMC Microbiol 13:76. doi: 10.1186/1471-2180-13-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaara D, Ravel C, Bañuls A-L, Haouas N, Lami P, Talignani L, El Baidouri F, Jaouadi K, Harrat Z, Dedet J-P, Babba H, Pratlong F. 2015. Evolutionary history of Leishmania killicki (synonymous Leishmania tropica) and taxonomic implications. Parasit Vectors 8:198. doi: 10.1186/s13071-015-0821-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bañuls AL, Dujardin JC, Guerrini F, De Doncker S, Jacquet D, Arevalo J, Noël S, Le Ray D, Tibayrenc M. 2000. Is Leishmania (Viannia) peruviana a distinct species? A MLEE/RAPD evolutionary genetics answer. J Eukaryot Microbiol 47:197–207. doi: 10.1111/j.1550-7408.2000.tb00039.x. [DOI] [PubMed] [Google Scholar]
- 21.Bañuls A-L, Hide M, Tibayrenc M. 2002. Evolutionary genetics and molecular diagnosis of Leishmania species. Trans R Soc Trop Med Hyg 96(Suppl 1):S9–S13. doi: 10.1016/S0035-9203(02)90045-3. [DOI] [PubMed] [Google Scholar]
- 22.Van der Auwera G, Ravel C, Verweij JJ, Bart A, Schönian G, Felger I. 2014. Evaluation of four single-locus markers for Leishmania species discrimination by sequencing. J Clin Microbiol 52:1098–1104. doi: 10.1128/JCM.02936-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Odiwuor S, Veland N, Maes I, Arévalo J, Dujardin J-C, Van der Auwera G. 2012. Evolution of the Leishmania braziliensis species complex from amplified fragment length polymorphisms, and clinical implications. Infect Genet Evol 12:1994–2002. doi: 10.1016/j.meegid.2012.03.028. [DOI] [PubMed] [Google Scholar]
- 24.Boité MC, Mauricio IL, Miles MA, Cupolillo E. 2012. New insights on taxonomy, phylogeny and population genetics of Leishmania (Viannia) parasites based on multilocus sequence analysis. PLoS Negl Trop Dis 6:e1888. doi: 10.1371/journal.pntd.0001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bañuls AL, Jonquieres R, Guerrini F, Le Pont F, Barrera C, Espinel I, Guderian R, Echeverria R, Tibayrenc M. 1999. Genetic analysis of Leishmania parasites in Ecuador: are Leishmania (Viannia) panamensis and Leishmania (Vianna) guyanensis distinct taxa? Am J Trop Med Hyg 61:838–845. doi: 10.4269/ajtmh.1999.61.838. [DOI] [PubMed] [Google Scholar]
- 26.Fraga J, Montalvo AM, De Doncker S, Dujardin J-C, Van der Auwera G. 2010. Phylogeny of Leishmania species based on the heat-shock protein 70 gene. Infect Genet Evol 10:238–245. doi: 10.1016/j.meegid.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 27.Blum J, Buffet P, Visser L, Harms G, Bailey MS, Caumes E, Clerinx J, van Thiel PPAM, Morizot G, Hatz C, Dorlo TPC, Lockwood DNJ. 2014. LeishMan recommendations for treatment of cutaneous and mucosal leishmaniasis in travelers, 2014. J Travel Med 21:116–129. doi: 10.1111/jtm.12089. [DOI] [PubMed] [Google Scholar]
- 28.Aronson N, Herwaldt BL, Libman M, Pearson R, Lopez-Velez R, Weina P, Carvalho EM, Ephros M, Jeronimo S, Magill A. 2016. Diagnosis and treatment of leishmaniasis: clinical practice guidelines by the Infectious Diseases Society of America (IDSA) and the American Society of Tropical Medicine and Hygiene (ASTMH). Clin Infect Dis 63:e202–e264. doi: 10.1093/cid/ciw670. [DOI] [PubMed] [Google Scholar]
- 29.El Baidouri F, Diancourt L, Berry V, Chevenet F, Pratlong F, Marty P, Ravel C. 2013. Genetic structure and evolution of the Leishmania genus in Africa and Eurasia: what does MLSA tell us. PLoS Negl Trop Dis 7:e2255. doi: 10.1371/journal.pntd.0002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraga J, Montalvo AM, Van der Auwera G, Maes I, Dujardin J-C, Requena JM. 2013. Evolution and species discrimination according to the Leishmania heat-shock protein 20 gene. Infect Genet Evol 18:229–237. doi: 10.1016/j.meegid.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 31.Lukes J, Mauricio IL, Schönian G, Dujardin J-C, Soteriadou K, Dedet J-P, Kuhls K, Tintaya KWQ, Jirků M, Chocholová E, Haralambous C, Pratlong F, Oborník M, Horák A, Ayala FJ, Miles MA. 2007. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc Natl Acad Sci U S A 104:9375–9380. doi: 10.1073/pnas.0703678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhls K, Mauricio IL, Pratlong F, Presber W, Schönian G. 2005. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect 7:1224–1234. doi: 10.1016/j.micinf.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Jamjoom MB, Ashford RW, Bates PA, Chance ML, Kemp SJ, Watts PC, Noyes HA. 2004. Leishmania donovani is the only cause of visceral leishmaniasis in East Africa; previous descriptions of L. infantum and “L. archibaldi” from this region are a consequence of convergent evolution in the isoenzyme data. Parasitology 129:399–409. doi: 10.1017/S0031182004005955. [DOI] [PubMed] [Google Scholar]
- 34.Mauricio IL, Stothard JR, Miles MA. 2004. Leishmania donovani complex: genotyping with the ribosomal internal transcribed spacer and the mini-exon. Parasitology 128:263–267. doi: 10.1017/S0031182003004578. [DOI] [PubMed] [Google Scholar]
- 35.Normand AC, Becker P, Gabriel F, Cassagne C, Accoceberry I, Gari-Toussaint M, Hasseine L, De Geyter D, Pierard D, Surmont I, Djenad F, Donnadieu JL, Piarroux M, Ranque S, Hendrickx M, Piarroux R. 21 June 2017. Online identification of fungi using MALDI-TOF mass-spectrometry: validation of a new Web application. J Clin Microbiol doi: 10.1128/JCM.00263-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rioux JA, Lanotte G, Serres E, Pratlong F, Bastien P, Perieres J. 1990. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann Parasitol Hum Comp 65:111–125. doi: 10.1051/parasite/1990653111. [DOI] [PubMed] [Google Scholar]


