ABSTRACT
Diagnosing chronic Chagas disease (CD) requires antibody–antigen detection methods, which are traditionally based on enzymatic assay techniques whose performance depend on the type and quality of antigen used. Previously, 4 recombinant chimeric proteins from the Instituto de Biologia Molecular do Paraná (IBMP-8.1 to 8.4) comprising immuno-dominant regions of diverse Trypanosoma cruzi antigens showed excellent diagnostic performance in enzyme-linked immunosorbent assays. Considering that next-generation platforms offer improved CD diagnostic accuracy with different T. cruzi-specific recombinant antigens, we assessed the performance of these chimeras in liquid microarrays (LMAs). The chimeric proteins were expressed in Escherichia coli and purified by chromatography. Sera from 653 chagasic and 680 healthy individuals were used to assess the performance of these chimeras in detecting specific anti-T. cruzi antibodies. Accuracies ranged from 98.1 to 99.3%, and diagnostic odds ratio values were 3,548 for IBMP-8.3, 4,826 for IBMP-8.1, 7,882 for IBMP-8.2, and 25,000 for IBMP-8.4. A separate sera bank (851 samples) was employed to assess cross-reactivity with other tropical diseases. Leishmania, a pathogen with high similarity to T. cruzi, showed cross-reactivity rates ranging from 0 to 2.17%. Inconclusive results were negligible (0 to 0.71%). Bland–Altman and Deming regression analysis based on 200 randomly selected CD-positive and negative samples demonstrated interchangeability with respect to CD diagnostic performance in both singleplex and multiplex assays. Our results suggested that these chimeras can potentially replace antigens currently used in commercially available assay kits. Moreover, the use of multiplex platforms, such as LMA assays employing 2 or more IBMP antigens, would abrogate the need for 2 different testing techniques when diagnosing CD.
KEYWORDS: human Chagas disease, Trypanosoma cruzi, chimeric antigens, liquid microarray, singleplex and multiplex assays
INTRODUCTION
Chagas disease (CD) is a life-threatening neglected tropical condition affecting approximately 5.7 million people in 21 Latin America countries, of which Brazil, Mexico, and Argentina are home to >60% of the estimated total number of infected individuals (1). Human migration has contributed to the worldwide distribution of infection, transforming this disease into a global health problem (2, 3). The vector-borne protozoan parasite Trypanosoma cruzi is the causative agent of CD. Transmission mainly occurs when contaminated urine/feces of hematophagous insects of the Triatominae family enters a bite site wound or mucosal membrane, by blood transfusions, and by the consumption of contaminated beverages or food (4).
Two distinct stages occur during the natural course of CD progression. Initially, an acute phase presents as a nonspecific oligosymptomatic febrile illness lasting for approximately 2 to 3 months with abundant parasitemia. A small number of cases are accompanied by myocarditis and other lethal complications. This parasite can only be observed by staining thick and thin blood smears during the initial phase. During the lifelong chronic stage, parasites remain hidden in target tissues, notably in the digestive system and cardiac muscles. This phase is initially characterized by an asymptomatic clinical course lasting 2 to 3 decades, after which approximately 10% and 20% of infected individuals develop digestive and heart complications, respectively (5). Due to low parasitemia and high levels of specific anti-T. cruzi antibodies, diagnosis in this chronic phase is traditionally performed by serological methods, including enzyme-linked immunosorbent assays (ELISAs), indirect immunofluorescence assays, and indirect hemagglutination inhibition assays (6). Because no standardized reference test is commercially available, the World Health Organization advises the use of two distinct techniques for CD diagnosis (7) and the Brazilian Health Ministry recommends 2 serological methods involving distinct antigen preparations, both of which must be performed concomitantly (6). Next-generation diagnostic platforms have improved the accuracy of CD diagnosis by using different T. cruzi-specific recombinant proteins in a variety of detection systems, such as chemiluminescence (8), surface plasmon resonance (9, 10), and bead-based technologies, including cytometry bead arrays (11) and liquid microarrays (LMAs) (12).
In countries where CD is endemic, the screening of blood donors for T. cruzi is mandatory to prevent CD transmission by blood transfusions. Accordingly, numerous tests must be performed on a daily basis in these areas. LMA is considered appropriate for detecting and quantifying multiple analytes in multiplex assays using relatively small sample volumes with high-throughput potential. Using this technique, it is possible to incorporate up to 500 color-coded fluorescent magnetic bead sets, each with 2 spectrally different fluorophore ratios, making each bead set distinguishable by its fluorescence emission when excited by a laser (13, 14). Because LMA technology permits the detection of many analytes simultaneously in each test sample, this method could potentially be singularly employed for CD diagnosis as a substitute for ELISAs and other traditional serological methods. These serological assays employ either fractionated lysates of T. cruzi at the epimastigote stage or recombinant proteins, which can produce inconclusive results or cross-reactivity with related diseases. Therefore, chimeric proteins have been proposed to improve the assay's accuracy to diagnose CD. Recently, a phase I study was performed with 4 chimeric proteins from the Instituto de Biologia Molecular do Paraná (IBMP-8.1, 8.2, 8.3, and 8.4) to detect specific anti-T. cruzi antibodies using both ELISA and LMA (15), demonstrating that each antigen accurately discriminated CD-positive from CD-negative samples. In addition, no significant differences were observed with respect to the diagnostic performances of the ELISA and LMA test methods. Data from a subsequent phase II study confirmed the high performance of these proteins in ELISAs (16). In the present study, we aimed to assess the diagnostic performance of the IBMP chimeras to diagnose CD using LMA in singleplex and multiplex formats.
RESULTS
LMA performance.
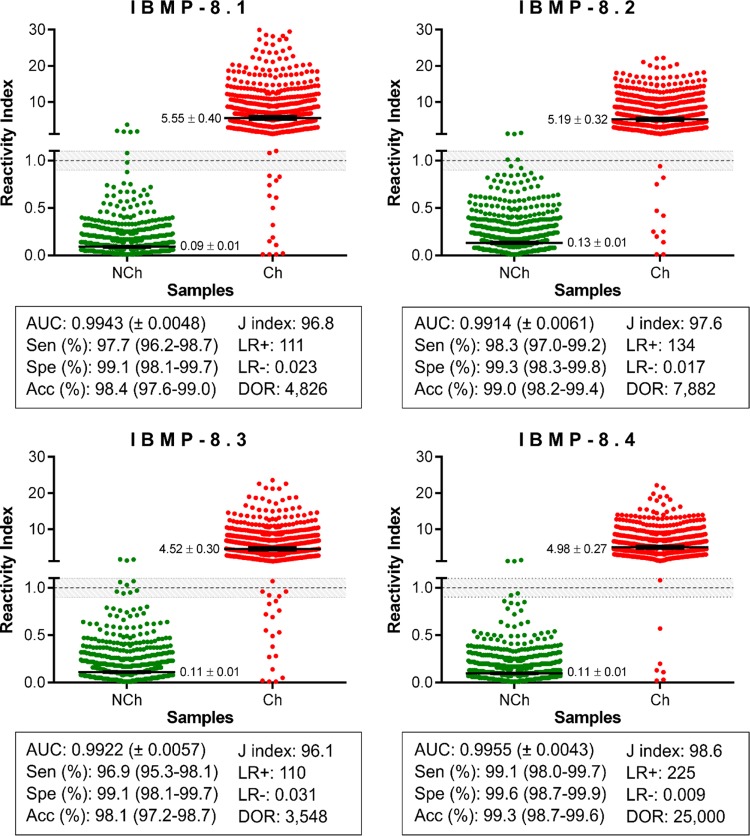
Using 1,333 sera from chagasic (Ch) and nonchagasic (NCh) individuals, the LMA performance and reactivity index (RI) distributions of all IBMP chimeras were assessed, as shown in Fig. 1. Area under the curve (AUC) values were >99%, revealing excellent overall diagnostic accuracy. IgG levels in Ch samples were variable, ranging from 4.52 for IBMP-8.3 and 4.98 for IBMP-8.4 to 5.19 for IBMP-8.2 and 5.55 for IBMP-8.1. Out of 653 Ch samples, IBMP-8.4-LMA showed 99.1% sensitivity with only 6 cases classified as false negatives; with these samples 4 were also classified as false negatives for all other antigens. Higher numbers of false negatives were observed for IBMP-8.1 (15 cases), IBMP-8.2 (11 cases), and IBMP-8.3 (20 cases), with corresponding sensitivity values of 97.7%, 98.3%, and 96.9%, respectively. Nevertheless, no statistically significant differences were detected with respect to IBMP protein sensitivity. Regarding the NCh samples, the IBMP chimeras showed specificity values >99.0%, and RI values ≤0.13 for all chimeras, with statistical differences observed only in relation to IBMP-8.2.
FIG 1.
Singleplex IBMP chimeric antigen assay of serum samples from chagasic (Ch) and nonchagasic (NCh) individuals. The cutoff value was established as reactivity index (RI) = 1.0, and the shadowed area represents the gray zone (RI = 1.0 ± 0.10). Geometric means (±95% CI) are represented by horizontal lines with corresponding results for each group. Acc, accuracy; AUC, area under the curve; DOR, diagnostic odds ratio; LR, likelihood ratio; Sen, sensitivity; Spe, specificity.
Relatively few Ch and NCh samples were considered inconclusive: 3 (0.23%) in the IBMP-8.1 assay, 5 (0.38%) in the IBMP-8.2 assay, 12 (0.90%) in the IBMP-8.3 assay, and 3 (0.23%) in the IBMP-8.4 assay. IBMP-8.4 was found to most accurately diagnose CD (99.3%), followed by IBMP-8.2 (99.0%), IBMP-8.1 (98.4%), and IBMP-8.3 (98.1%). The Youden index was the highest for IBMP-8.4 (98.6%), followed by IBMP-8.2 (97.6%), IBMP-8.1 (96.8%), and IBMP-8.3 (96.1%) proteins. The test performance was summarized by the diagnostic odds ratio (DOR) value, which reached 25,000 for IBMP-8.4, 7,882 for IBMP-8.2, 4,826 for IBMP-8.1, and 3,548 for IBMP-8.3.
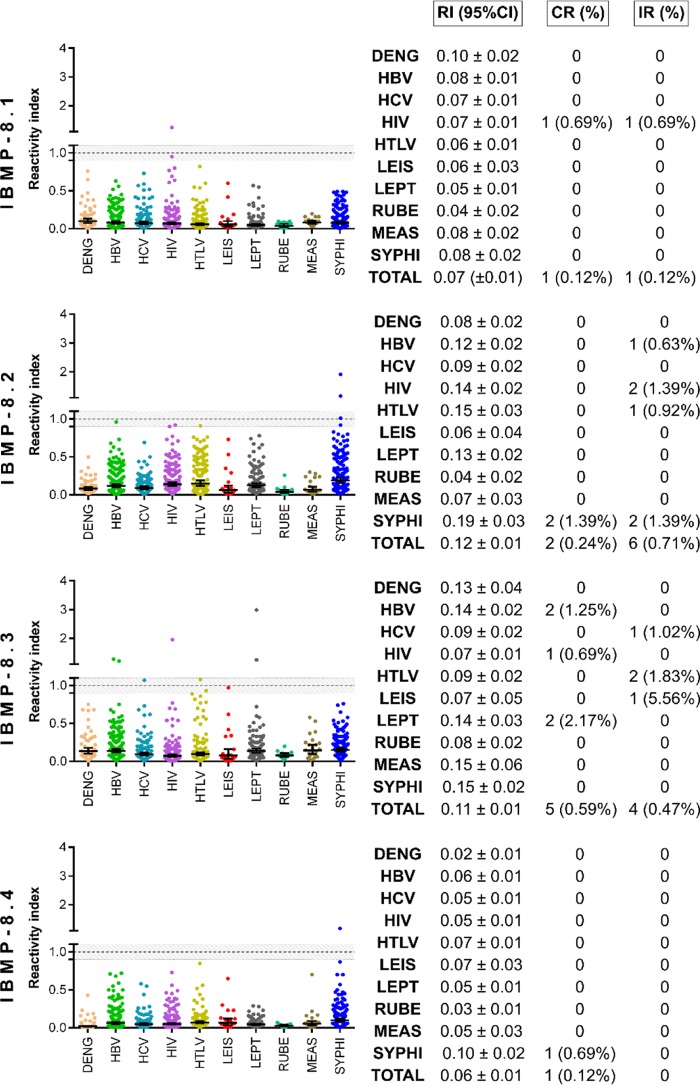
Potential cross-reactivity (RI ≥ 1.0) of the IBMP chimeras was assessed using serum samples from 851 individuals with unrelated diseases. As shown in Fig. 2, the incidence of cross-reactivity was negligible: 0.12% (1/851) for IBMP-8.1 and IBMP-8.4, 0.24% (2/851) for IBMP-8.2, and 0.59% (5/581) for IBMP-8.3. Moreover, a very low frequency of inconclusive results was observed: 0.12% (1/851) for IBMP-8.1, 0.71% (6/851) for IBMP-8.2, and 0.47% (4/851) for IBMP-8.3 (Fig. 2). Notably, we found no inconclusive results in relation to the IBMP-8.4 protein. Regarding the Leishmania spp. samples, none exhibited any cross-reactivity with the 4 IBMP chimeras, and only 1 showed an inconclusive result with respect to IBMP-8.3.
FIG 2.
Analysis of IBMP chimera cross-reactivity with sera from individuals with unrelated diseases. The cutoff value was established as reactivity index = 1.0, and the shadowed area represents the gray zone (RI = 1.0 ± 0.10). Geometric means (±95% CI) are represented by horizontal lines, with the corresponding results shown for each group. (CR, cross-reaction); DENG, Dengue; HBV, hepatitis B virus; HCV, hepatitis C virus; HIV, human immunodeficiency virus; HTLV, human T-cell lymphotropic virus; IR, inconclusive results; LEIS, leishmaniasis; LEPT, leptospirosis; MEAS, measles; RI, reactivity index; RUBE, rubella virus; SYPHI, syphilis.
Comparison of singleplex versus multiplex IBMP antigen performance.
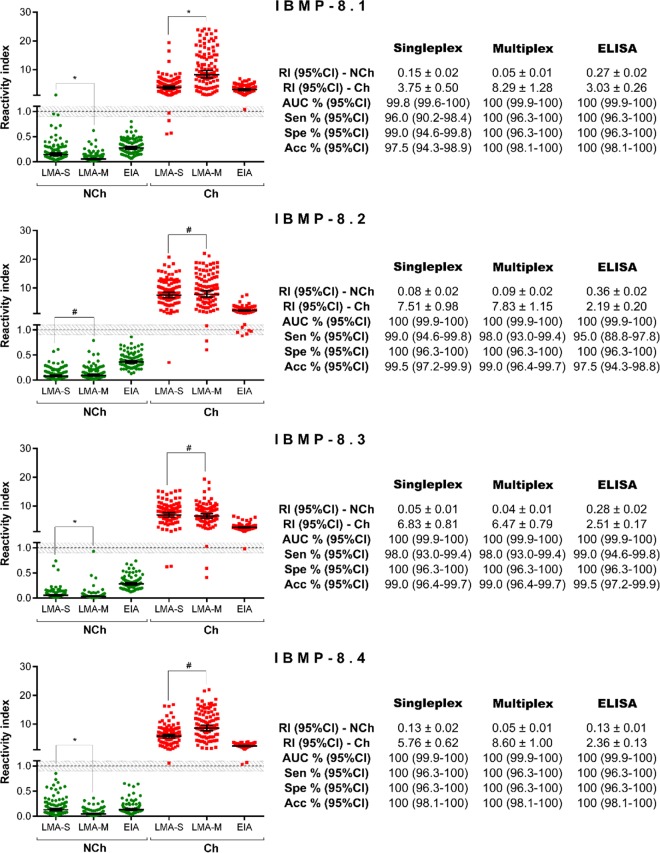
No significant differences were observed with respect to LMA performance when assaying 100 Ch and 100 NCh samples in singleplex or multiplex assays (Fig. 3). The AUCs were >99.7%. The level of agreement between the expected results ranged from 95.0% (κ = 0.950 [0.907 to 0.993]) for IBMP-8.1 to 99.0% (κ = 0.990 [0.970 to 1.01]) for IBMP-8.2, while the IBMP-8.3 and-8.4 chimeras showed 100% agreement. Despite the high level of agreement seen and the consistency in performance of the parameters evaluated, NCh samples yielded lower signals when assayed with IBMP-8.1, IBMP-8.3, and IBMP-8.4 in the multiplex assay. Regarding the Ch samples, differences in RI values were observed only in the samples assayed by the IBMP-8.1 chimera in multiplex assays. For comparison purposes, ELISA performances are also described in Fig. 3.
FIG 3.
Singleplex and multiplex IBMP chimeric antigen assays of serum samples from chagasic (Ch) and nonchagasic (NCh) individuals. The cutoff value was established as reactivity index = 1.0, and the shadowed area represents the gray zone (RI = 1.0 ± 0.10). Geometric means (±95% CI) are represented by horizontal lines, with the corresponding results shown for each group. Acc, accuracy; AUC, area under the curve; EIA, ELISA; LMA-M, multiplex liquid microarray; LMA-S, singleplex liquid microarray; RI, reactivity index; Sen, sensitivity; Spe, specificity.
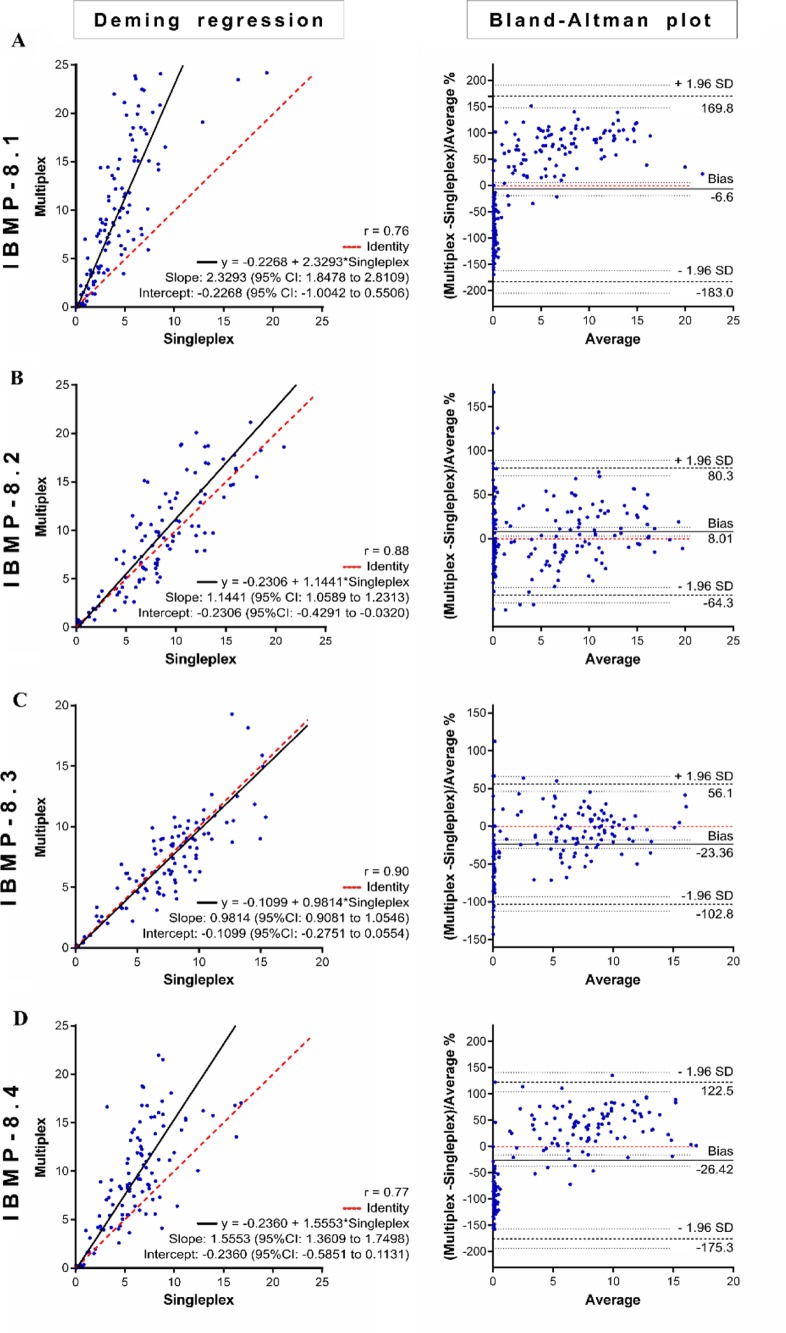
Figure 4 graphically illustrates the strength of agreement between the singleplex and multiplex data for each protein assayed by Deming regression fit analysis (left panels) and Bland–Altman plots (right panels). The IBMP-8.1 antigen multiplex aligned poorly with the singleplex method under Deming regression fit analysis, described by the equation y = −0.2268 + 2.3293x, with an intercept of −0.2268 (95% confidence interval [CI], −1.0042 to 0.5506), a slope of 2.3293 (95% CI, 1.8478 to 2.8109), and an R-squared value of 0.76 (Fig. 4A). The mean bias derived from the Bland–Altman difference plot was −6.6% (95% CI, −19.5 to 5.89%) with the limits of agreement (LoAs) ranging between −183.0% and 169.8%. Although Deming regression fit analysis indicated significant proportional bias, Bland–Altman analysis presented no significant bias with respect to the means, as the line of equality fell within the confidence interval. We observed that all data points fell within the LoAs, which is consistent with the expectation that only 5% would fall outside these limits. The IBMP-8.2 antigen (Fig. 4B) showed good agreement between the singleplex and multiplex assays, with an R-squared value of 0.88, an intercept of −0.2306 (95% CI, −0.4291 to −0.0320), and a slope of 1.1441 (95% CI, 1.0589 to 1.2313). The mean bias was 8.01% (95% CI, 2.9 to 13.12%) with LoA values ranging from −64.3% to 80.3%, which indicated statistical significance since the line of equality fell outside the CI. We observed 8 points (4.0%) outside the LoAs, which is consistent with the 5% expectation. Figure 4C shows a good fit between these 2 methods using IBMP-8.3, with an R-squared value of 0.90, an intercept of −0.1099 (95% CI, −0.2751 to 0.0554), and a slope of 0.9814 (95% CI, 0.9081 to 1.0546). The mean bias was −23.36% (95% CI, −28.98 to −17.74%) with LoA values ranging from −102.8% to 56.1%. Despite the absence of significance regarding the slope under Deming regression analysis, the mean bias derived from Bland–Altman analysis indicated that the multiplex results were up to −23.36% less than those obtained with the singleplex method. Nine points (4.5%) fell beyond the LoAs. For IBMP-8.4 (Fig. 4D), the correlation coefficient between the 2 singleplex and multiplex tests was 0.77. Deming regression analysis showed a slope of 1.5553 (95% CI, 1.3609 to 1.7498) and an intercept of −0.2360 (95% CI, −0.5851 to 0.1131), while the Bland–Altman plot showed a mean bias of −26.42% (95% CI, −36.95 to −15.89). The slope value is indicative of significant proportional bias, as evidenced by an up to 20% variation between the results obtained with the singleplex and multiplex techniques. Just 1 point (0.5%) fell outside the LoA.
FIG 4.
Deming regression fit (left) and Bland–Altman plots (right) comparing single- and multiplex methods of detecting anti-T. cruzi IgG, using the IBMP-8.1 (A), IBMP-8.2 (B), IBMP-8.3 (C), and IBMP-8.4 (D) chimeras.
DISCUSSION
The T. cruzi IBMP recombinant antigenic proteins employed herein have already been shown to be sensitive and specific for CD diagnosis when assessed by ELISA (16), although their performances using other approaches remains to be elucidated. A phase I study previously conducted by our group using ELISAs and LMAs showed high performance when these antigens were assayed using a small set of samples comprised of only 300 sera from CD-positive and CD-negative individuals (15). Here, we expanded the sample size to 1,333 sera and found AUC values higher than 99% for all 4 proteins. These data are in accordance with results from a phase II study, where these same proteins were tested by ELISA (16), thereby indicating the high discriminative power these antigens potentially possess with respect to other diagnostic platforms. Most importantly, these IBMP chimeric proteins provided much better AUC values than did T. cruzi cell lysates, single recombinant proteins, or other recombinant chimeric proteins commonly used in diagnostic kits (17, 18). In addition, differences greater than 4.40 were seen between the RI signals from the positive and negative samples for all proteins, providing further evidence of their high discriminatory capability. Moreover, the RI signals obtained from positive samples assayed by LMA were up to 56% stronger than those previously obtained by ELISA (16). Conversely, the average RI signals from negative samples were 32% lower by LMA. Finally, the total number of inconclusive results was very low, ranging from 0.23% to 0.90%, again reinforcing the optimal discriminatory power of these IBMP proteins combined with next-generation diagnostic platforms.
Performance assessments were carried out with the LMA assays to determine the diagnostic sensitivity, specificity, and accuracy for CD. Despite the fact that no differences were observed in sensitivity and specificity, the IBMP-8.4 protein produced more accurate results than IBMP-8.2. Nonetheless, this difference was almost negligible, considering that the 95% CI values practically overlapped. LMA assay performance was comparable to previously published data with ELISAs (16). With the exception of the IBMP-8.2 antigen, both testing methods offered similar performance. When evaluated by LMA, the IBMP-8.2 protein showed 99.0% accuracy, while it showed 96.6% accuracy by ELISA. According to a previous study, the lower value obtained by ELISA was probably due to the amino acid sequence of this protein, which impaired its recognition by specific anti-T. cruzi antibodies from CD-positive samples collected in distinct geographical regions (16). However, this discrepancy in accuracy may also be the result of characteristics inherent to each diagnostic platform used. Indeed, the median fluorescence intensity (MFI) of the detection antibody corresponds to an average of 100 bead readings; i.e., a single serum sample is analyzed 100 times per antigen versus just once in an ELISA reaction. This level of precision improves the limit of detection by LMA assays (13, 19). These performance results were corroborated by the J index and DOR. In addition to accuracy, the J index measures the effectiveness of a diagnostic marker by considering the sensitivity and specificity together as a single parameter, and we found that the J index value was >0.96 for all chimeras. The DOR is a global performance parameter that summarizes the diagnostic accuracy of a given testing method (20). It can vary from 0 to infinity, with higher values indicating improved discriminatory diagnostic testing. The DOR for IBMP-8.4 (25, 000) was greater than that obtained for IBMP-8.1 (4, 826), IBMP-8.2 (7, 882), and IBMP-8.3 (3, 548). These data agree with data from a previous study using ELISA that highlighted the IBMP-8.4 protein as the best antigen for diagnosing CD (16).
Considering the large number of sera from patients with unrelated diseases used to assess cross-reactivity, the small number of samples that cross-reacted was irrelevant. This was expected due to the low similarity between the IBMP sequences and those deposited in the NCBI database for other pathogens, including Leishmania spp. Furthermore, cross-reacting samples also presented a weak RI signal. Similarly, inconclusive results using this same panel were statistically irrelevant, particularly with respect to IBMP-8.1 and 8.4. These findings are consistent with previous results obtained when assessing cross-reactivity in ELISAs (16). As such, the authors are confident that all of these chimeric proteins can be safely employed in diagnostic platforms in areas with endemic CD, as well as other infectious diseases.
We also comparatively assayed 100 CD-positive and 100 CD-negative samples by the singleplex and multiplex LMA approaches. Both methods were highly efficient in distinguishing CD-positive and CD-negative samples. Regarding the CD-positive samples, a significant difference was seen in the RI signal intensity only with respect to the IBMP-8.1 protein, whereas, in the CD-negative samples, lower RI signals for IBMP-8.1, 8.3, and 8.4 proteins were observed in multiplex assays compared to those observed in singleplex assays. Despite these discrepancies, the performance parameters were identical for both methods. Deming regression analysis showed a substantial proportional bias for the IBMP-8.1, 8.2, and 8.4 proteins, suggesting that these methods are not in complete agreement throughout the measurement range involving CD-positive and CD-negative samples, as evidenced by the Bland–Altman plots, especially regarding IBMP-8.1 and IBMP-8.4. This finding indicates the highly linear nature of multiplex assays compared to the singleplex approach. Regression analysis also showed a systematic negative bias only with respect to the IBMP-8.2 protein, indicating that results obtained using this antigen under multiplex assays were slightly higher, by a constant amount, than those produced by the singleplex method. This was probably due to inadequate blanking, a miss-set 0 calibration point, or some other type of interference in the assay (21). Although this bias seems to indicate a substantial difference between the singleplex and multiplex LMA techniques, it does not affect diagnostic accuracy; i.e., both methods are sufficiently interchangeable for CD diagnosis. As multiplex assays inherently involve similar analysis time and serum volumes compared to singleplex methods, multiplex methods are crucial for assessing outbreaks involving the screening of large populations, as well as for routine testing at blood donation centers. Furthermore, multiplex approaches can be used to effectively screen for several diseases concomitantly, unlike traditional serological testing, in which only 1 condition is evaluated. Thus, multiplexing not only reduces costs, analysis time, and the serum volume required, but also enables the incorporation of multiple markers for infectious diseases (14, 22, 23), cancer, and other conditions (24–26). Although LMA-based technology offers several advantages, it nonetheless requires a significant laboratory infrastructure, a well-trained workforce, and substantial financial investment.
In conclusion, the results described herein indicate that these 4 T. cruzi IBMP recombinant antigenic proteins can be safely used for CD diagnosis in both LMA platforms evaluated, as well as in ELISA-based assays (16). Moreover, the accuracy of LMA was shown not to vary among these IBMP antigens, regardless of use of singleplex or multiplex techniques, suggesting that these chimeras can potentially replace those currently used in commercially available assay kits. Accordingly, a multiplex LMA assay employing 2 or more IBMP antigens would abrogate the need for using 2 different tests when diagnosing CD.
MATERIALS AND METHODS
Ethical considerations.
The Institutional Review Board (IRB) for Human Research at the Aggeu Magalhães Institute of the Oswaldo Cruz Foundation (Recife, Pernambuco, Brazil) provided ethical approval to conduct this study (CAEE: 15812213.8.0000.5190). To protect patient privacy, the IRB required that samples be coded to mask patient identification, thus eliminating the need for verbal or written consent.
Subjects and sample collection.
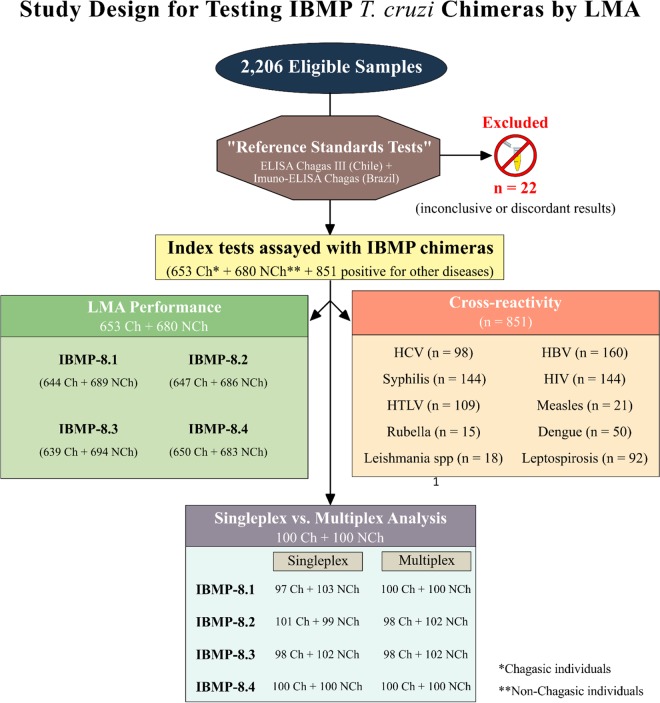
Previously collected human sera were provided by the biorepositories of the Hemope Foundation (Recife, Pernambuco), the Central Laboratory for Public Health-LACEN (Recife, Pernambuco), the Reference Laboratory for Chagas Disease (Fiocruz-Recife, Pernambuco), the Molecular Biology Institute of Paraná (IBMP-Paraná), and the Laboratory for Research on Chagas Disease (Federal University of Goiás-Goiás). Samples from 653 chagasic (Ch) and 680 nonchagasic (NCh) individuals were utilized to assess the performance of T. cruzi IBMP chimeras in diagnosing CD by LMA. This panel was composed of samples from Brazilian states where Chagas is endemic and nonendemic (Bahia-BA, Minas Gerais-MG, Goiás-GO, Pernambuco-PE, and Paraná-PR), as well as from Brazilian and international commercial suppliers (National Panel for Blood Screening Quality Control, Fiocruz, RJ, Brazil; Boston Biomedical Inc., Norwood, MA, USA; SeraCare Life Sciences Inc., Milford, MA, USA). Samples from individuals with dengue virus (n = 50), hepatitis B virus (n = 160), hepatitis C virus (n = 98), human immunodeficiency virus (n = 144), human T-cell lymphotropic virus (n = 109), leishmaniasis (n = 18), leptospirosis (n = 92), rubella virus (n = 15), measles (n = 21), and syphilis (n = 144) were used to assess cross-reactivity between the IBMP chimeras and proteins associated with unrelated diseases. Before LMA analysis, all sera were reevaluated using 2 commercial ELISAs, namely, the Imuno-ELISA Chagas test (Wama Diagnostica, São Paulo, Brazil; batch 14D061) and the ELISA Chagas III test (BIOSChile, Ingeniaría Genética S.A., Santiago, Chile; batch 1F130525) (27). Each sample was assigned a numeric code in the laboratory to ensure a blinded analysis.
Acquisition of recombinant chimeric proteins.
Immuno-dominant sequence selection, synthetic gene construction, and recombinant chimeric protein expression were performed, as previously described (15). Briefly, T. cruzi synthetic gene constructs were obtained from a commercial supplier (GenScript, Piscataway, NJ, USA) and subcloned into the pET28a expression vector (Novagen, Madison, WI, USA). Chimeric antigens were expressed as soluble proteins in Escherichia coli BL21-Star (DE3) cells grown in LB medium supplemented with 0.5 M isopropyl-β-d-1-thiogalactopyranoside (IPTG). Recombinant expression of the chimeras was checked by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (28). Chimeras were purified by both ion-exchange and liquid affinity chromatography. Concentrations were determined by performing a fluorimetric assay (Qubit 2.0, Invitrogen Technologies, Carlsbad, CA, USA).
IBMP antigen coupling to microsphere beads and in-house LMA procedures.
The IBMP antigen-coupling protocol employed herein was performed as previously described (15). Briefly, 2 × 106 microsphere beads were washed with activation buffer (100 mM sodium phosphate, pH 6.3) and chemically activated using 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride and N-hydroxysulfosuccinimide (Sigma, St. Louis, MO, USA), both diluted to 50 mg/ml of ultrapure water (18.2 MΩ). Activated beads were subsequently incubated with 200 μl of antigen diluted in coupling buffer at previously determined concentrations (15). These suspensions were incubated at 250 rpm under horizontal agitation for 2 h at 37°C. Next, the beads were washed 3 times with wash buffer (phosphate-buffered saline [PBS], containing 1% bovine serum albumin [BSA] and 0.05% Tween 20). The final bead suspensions were adjusted to a concentration of 50 × 103 microspheres/ml in wash buffer and stored overnight at 2 to 8°C in low-binding tubes. For LMA analysis, a previously prepared E. coli lysate (diluted to 2%) (29) was mixed with 50 μl of serum sample (diluted 1:200) and 50 μl of bead suspension, placed in a 96-well plate, and incubated under agitation for 15 min at 37°C. The beads were then washed twice. Phycoerythrin-conjugated, goat anti-human IgG (Moss Substrates, Pasadena, MA, USA), diluted 1:1,000, was added and the plates were incubated under agitation for 15 min at 37°C. The beads were then washed with sheath fluid and resuspended in 200 μl of the same solution. For the multiplex LMA assay, 2,500 beads of each set were mixed together in a final volume of 50 μl/well, following the assay protocol described above. The results were interpreted using a Luminex 200 BioAnalyzer (Luminex Corp. Austin, TX, USA) with xPONENT software (version 3.1.871.0). For bead identification, a minimum of 100 beads bearing a unique fluorescent signature was detected per region, measured in terms of the median fluorescence intensity (MFI) per sample in accordance with the manufacturer instructions.
Singleplex versus multiplex LMA.
A total of 100 Ch and 100 NCh samples were randomly selected to compare the performance and concordance among the IBMP chimeric antigens, either singleplexed (assayed individually using a single bead type) or multiplexed (each antigen assayed together with different bead types).
Data analysis.
Data were encoded and analyzed using GraphPad Prism 6 graphing software (San Diego, CA, USA). Descriptive statistics are presented as geometric means ± standard deviation (SD). The Shapiro–Wilk test was used to test data normality, and homogeneity of variance was verified using Levene's test. When these 2 assumptions were confirmed, Student's t test was used for sample comparisons; otherwise, the Wilcoxon signed-rank test was employed. All analyses were 2-tailed, and a P < 0.05 was considered significant. Cutoff point analysis was used to establish a maximum MFI to distinguish positive and negative samples. The threshold was set by determining the greatest area under the receiver operating characteristic (ROC) curve. Data are displayed via scatter plot and are presented in terms of the reactivity index (RI; i.e., ratio of the sample MFI to the cutoff MFI), with results ≥1.00 considered positive. RI values within 1.0% ± 10% were considered indeterminate and deemed as inconclusive (shown as a gray zone). LMA performance was evaluated using a dichotomous approach with respect to sensitivity, specificity, accuracy, Youden index (J), the likelihood ratio, and the diagnosis odds ratio (DOR) (30). Confidence intervals (CI) were calculated to assess the precision of these parameters, with a confidence level of 95%. Singleplex versus multiplex LMA results were compared using Cohen's kappa coefficient (κ), the Bland–Altman plot, and Deming regression analysis. The strength of agreement was interpreted as nearly perfect (0.81 < κ ≤ 1.0), substantial (0.61 < κ ≤ 0.80), moderate (0.41 < κ ≤ 0.60), fair (0.21 < κ ≤ 0.40), slight (0 < κ ≤ 0.20), or poor (κ ≤ 0) agreement (31). Bland–Altman plots with limits of agreement (LoAs) were generated to assess the variability and magnitude between the singleplex and multiplex assays (32). Deming regression was used to mathematically determine the agreement between the singleplex and multiplex techniques, as well as proportional bias (slope, 95% CI) and systematic bias (intercept, 95% CI). Deming regression analysis revealed a null hypothesis when the intercept and slope were 0 and 1, respectively. A checklist and flowchart (Fig. 5) are provided according to the Standards for Reporting of Diagnostic Accuracy studies (STARD) guidelines (33).
FIG 5.
STARD flowchart. Standards for Reporting of Diagnostic Accuracy studies (STARD) description of the study design.
ACKNOWLEDGMENTS
This work was supported by Conselho Nacional de Desenvolvimento Científico e Tecnológico-CNPq (grant 404242/2012-0), Fundação de Amparo à Ciência e Tecnologia do Estado de Pernambuco-FACEPE (grant APQ-1257-2.11/12), and RVE Finep (grant 01.13.0283.00-reference 0473/12).
Yara de Miranda Gomes, Wayner Vieira de Sousa, and Marco Aurélio Krieger are research fellows supported by CNPq proc. no. 304543/2012-8, 306222/2013-2, and 590032/2011-9, respectively.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare that no competing interests exist.
REFERENCES
- 1.World Health Organization. 2015. Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Wkly Epidemiol Rec 90:33–44. [PubMed] [Google Scholar]
- 2.Schmunis GA, Yadon ZE. 2010. Chagas disease: a Latin American health problem becoming a world health problem. Acta Trop 115:14–21. doi: 10.1016/j.actatropica.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Manne-Goehler J, Reich MR, Wirtz VJ. 2015. Access to care for Chagas disease in the United States: a health systems analysis. Am J Trop Med Hyg 93:108–113. doi: 10.4269/ajtmh.14-0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amato Neto V, Lopes M, Umezawa ES, Aveiro Ruocco M, Dias JC. 2000. Outras formas de transmissão do Trypanosoma cruzi. Rev Patol Trop 29:115–129. [Google Scholar]
- 5.Rassi A Jr, Rassi A, Marcondes de Rezende J. 2012. American trypanosomiasis (Chagas disease). Infect Dis Clin North Am 26:275–291. doi: 10.1016/j.idc.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 6.Ministério da Saúde Secretaria de Vigilância em Saúde. 2005. Brazilian consensus on Chagas disease. Rev Soc Bras Med Trop 38(Suppl 3):7–29. (In Portuguese.) doi: 10.1590/S0037-86822005000100002. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. 2007. WHO consultation on international biological reference preparations for Chagas diagnostic tests. World Health Organization, Geneva, Switzerland: http://www.who.int/bloodproducts/ref_materials/WHO_Report_1st_Chagas_BRP_consultation_7-2007_final.pdf. [Google Scholar]
- 8.Abras A, Gállego M, Llovet T, Tebar S, Herrero M, Berenguer P, Ballart C, Martí C, Muñoz C. 2016. Serological diagnosis of chronic Chagas disease: is it time for a change? J Clin Microbiol 54:1566–1572. doi: 10.1128/JCM.00142-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luz JG, Souto DE, Machado-Assis GF, de Lana M, Kubota LT, Luz RC, Damos FS, Martins HR. 2015. Development and evaluation of a SPR-based immunosensor for detection of anti-Trypanosoma cruzi antibodies in human serum. Sens Actuators B Chem 212:287–296. doi: 10.1016/j.snb.2015.01.135. [DOI] [Google Scholar]
- 10.Luz JG, Souto DE, Machado-Assis GF, de Lana M, Luz RC, Martins-Filho OA, Damos FS, Martins HR. 2016. Applicability of a novel immunoassay based on surface plasmon resonance for the diagnosis of Chagas disease. Clin Chim Acta 454:39–45. doi: 10.1016/j.cca.2015.12.025. [DOI] [PubMed] [Google Scholar]
- 11.Teixeira-Carvalho A, Campos FM, Geiger SM, Rocha RD, de Araújo FF, Vitelli-Avelar DM, Andrade MC, Araújo MS, Lemos EM, de Freitas Carneiro Proietti AB, Sabino EC, Caldas RG, Freitas CR, Campi-Azevedo AC, Elói-Santos SM, Martins-Filho OA. 2015. FC-TRIPLEX Chagas/Leish IgG1: a multiplexed flow cytometry method for differential serological diagnosis of Chagas disease and leishmaniasis. PLoS One 10:e0122938. doi: 10.1371/journal.pone.0122938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Foti L, Fonseca BP, Nascimento LD, Marques CF, da Silva ED, Duarte CA, Probst CM, Goldenberg S, Pinto AG, Krieger MA. 2009. Viability study of a multiplex diagnostic platform for Chagas disease. Mem Inst Oswaldo Cruz 104:136–141. doi: 10.1590/S0074-02762009000900019. [DOI] [PubMed] [Google Scholar]
- 13.Hoare R, Thompson KD, Herath T, Collet B, Bron JE, Adams A. 2016. Development, characterisation and application of monoclonal antibodies for the detection and quantification of infectious salmon anaemia virus in plasma samples using Luminex bead array technology. PLoS One 11:e0159155. doi: 10.1371/journal.pone.0159155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christopher-Hennings J, Araujo KP, Souza CJ, Fang Y, Lawson S, Nelson EA, Clement T, Dunn M, Lunney JK. 2013. Opportunities for bead-based multiplex assays in veterinary diagnostic laboratories. J Vet Diagn Invest 25:671–691. doi: 10.1177/1040638713507256. [DOI] [PubMed] [Google Scholar]
- 15.Santos FL, Celedon PA, Zanchin NI, Brasil Tde A, Foti L, Souza WV, Silva ED, Gomes YM, Krieger MA. 2016. Performance assessment of four chimeric Trypanosoma cruzi antigens based on antigen-antibody detection for diagnosis of chronic Chagas disease. PLoS One 11:e0161100. doi: 10.1371/journal.pone.0161100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santos FL, Celedon PA, Zanchin NI, Souza WV, da Silva ED, Foti L, Krieger MA, Gomes YM. 2017. Accuracy of chimeric proteins in the serological diagnosis of chronic Chagas disease—a phase II study. PLoS Negl Trop Dis 11:e0005433. doi: 10.1371/journal.pntd.0005433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia VS, Gonzalez VD, Marcipar IS, Gugliotta LM. 2014. Immunoagglutination test to diagnose Chagas disease: comparison of different latex-antigen complexes. Trop Med Int Health 19:1346–1354. doi: 10.1111/tmi.12379. [DOI] [PubMed] [Google Scholar]
- 18.Santamaría AL, De Rissio AM, Riarte A, Garavaglia PA, Bruballa AC, Rodríguez MA, Irazu LE, Ruiz AM, García GA. 2013. Use of an enzyme-linked immunosorbent assay that utilizes the Tc13Tul antigen of Trypanosoma cruzi to monitor patients after treatment with benznidazole. Diagn Microbiol Infect Dis 76:197–205. doi: 10.1016/j.diagmicrobio.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 19.Kuller L, Watanabe R, Anderson D, Grant R. 2005. Development of a whole-virus multiplex flow cytometric assay for antibody screening of a specific pathogen-free primate colony. Diagn Microbiol Infect Dis 53:185–193. doi: 10.1016/j.diagmicrobio.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 20.Glas AS, Lijmer JG, Prins MH, Bonsel GJ, Bossuyt PM. 2003. The diagnostic odds ratio: a single indicator of test performance. J Clin Epidemiol 56:1129–1135. doi: 10.1016/S0895-4356(03)00177-X. [DOI] [PubMed] [Google Scholar]
- 21.Westgard JO. 2008. How do you use statistics to estimate analytical errors? p137–152. In Westgard JO. (ed), Basic method validation, 3rd ed Westgard QC, Inc., Madison, WI. [Google Scholar]
- 22.Leva A, Eibach D, Krumkamp R, Käsmaier J, Rubbenstroth D, Adu-Sarkodie Y, May J, Tannich E, Panning M. 2016. Diagnostic performance of the Luminex xTAG gastrointestinal pathogens panel to detect rotavirus in Ghanaian children with and without diarrhoea. Virol J 13:132. doi: 10.1186/s12985-016-0588-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caboré RN, Piérard D, Huygen K. 2016. A Belgian serosurveillance/seroprevalence study of diphtheria, tetanus and pertussis using a Luminex xMAP technology-based pentaplex. Vaccines (Basel) 4:E16. doi: 10.3390/vaccines4020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Argiris A, Lee SC, Feinstein T, Thomas S, Branstetter BF 4th, Seethala R, Wang L, Gooding W, Grandis JR, Ferris RL. 2011. Serum biomarkers as potential predictors of antitumor activity of cetuximab-containing therapy for locally advanced head and neck cancer. Oral Oncol 47:961–966. doi: 10.1016/j.oraloncology.2011.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Djoba Siawaya JF, Roberts T, Babb C, Black G, Golakai HJ, Stanley K, Bapela NB, Hoal E, Parida S, van Helden P, Walzl G. 2008. An evaluation of commercial fluorescent bead-based luminex cytokine assays. PLoS One 3:e2535. doi: 10.1371/journal.pone.0002535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shadfan BH, Simmons AR, Simmons GW, Ho A, Wong J, Lu KH, Bast RC Jr, McDevitt JT. 2015. A multiplexable, microfluidic platform for the rapid quantitation of a biomarker panel for early ovarian cancer detection at the point-of-care. Cancer Prev Res (Phila) 8:37–48. doi: 10.1158/1940-6207.CAPR-14-0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santos FL, de Souza WV, Barros Mda S, Nakazawa M, Krieger MA, Gomes YM. 2016. Chronic Chagas disease diagnosis: A comparative performance of commercial enzyme immunoassay tests. Am J Trop Med Hyg 94:1034–1039. doi: 10.4269/ajtmh.15-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Crestani S, Leitolis A, Lima LF, Krieger MA, Foti L. 2016. Enhanced target-specific signal detection using an Escherichia coli lysate in multiplex microbead immunoassays with E. coli-derived recombinant antigens. J Immunol Methods 435:17–26. doi: 10.1016/j.jim.2016.05.002. [DOI] [PubMed] [Google Scholar]
- 30.Ouchchane L, Rabilloud M, Boire JY. 2009. Sensibilité, spécificité et valeurs prédictives, p 49–78. In Beuscat R, Bénichou J, Roy P (ed), Évaluation des méthodes d'analyse appliquées aux sciences de la vie et de la santé—Biostatistique, vol 2 Omniscience, Paris, France. [Google Scholar]
- 31.Landis JR, Koch GG. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159–174. doi: 10.2307/2529310. [DOI] [PubMed] [Google Scholar]
- 32.Bland JM, Altman DG. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i:307–310. [PubMed] [Google Scholar]
- 33.Bossuyt PM, Reitsma JB, Bruns DE, Gatsonis CA, Glasziou PP, Irwig L, Lijmer JG, Moher D, Rennie D, de Vet HC, Kressel HY, Rifai N, Golub RM, Altman DG, Hooft L, Korevaar DA, Cohen JF, STARD Group. 2015. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 351:h5527. doi: 10.1136/bmj.h5527. [DOI] [PMC free article] [PubMed] [Google Scholar]