ABSTRACT
The emerging multidrug-resistant pathogenic yeast Candida auris represents a serious threat to global health. Unlike most other Candida species, this organism appears to be commonly transmitted within health care facilities and causes health care-associated outbreaks. To better understand the epidemiology of this emerging pathogen, we investigated the ability of C. auris to persist on plastic surfaces common in health care settings compared with that of Candida parapsilosis, a species known to colonize the skin and plastics. Specifically, we compiled comparative and quantitative data essential to understanding the vehicles of spread and the ability of both species to survive and persist on plastic surfaces under controlled conditions (25°C and 57% relative humidity), such as those found in health care settings. When a test suspension of 104 cells was applied and dried on plastic surfaces, C. auris remained viable for at least 14 days and C. parapsilosis for at least 28 days, as measured by CFU. However, survival measured by esterase activity was higher for C. auris than C. parapsilosis throughout the 28-day study. Given the notable length of time Candida species survive and persist outside their host, we developed methods to more effectively culture C. auris from patients and their environment. Using our enrichment protocol, public health laboratories and researchers can now readily isolate C. auris from complex microbial communities (such as patient skin, nasopharynx, and stool) as well as environmental biofilms, in order to better understand and prevent C. auris colonization and transmission.
KEYWORDS: Candida, Candida auris, Candida parapsilosis, clinical methods, persistence, public health
INTRODUCTION
Invasive candidiasis, severe fungal infection caused by yeast in the genus Candida, is an increasing risk in health care settings that affects the most vulnerable of patients, e.g., those in critical condition or with compromised immune systems such as cancer patients, premature infants, and the elderly. The emerging multidrug-resistant yeast Candida auris was first identified and described in a clinical culture from an external ear canal (1). Subsequently, molecular techniques were used to identify a 1996 bloodstream isolate recovered from Chonnam National University Hospital in South Korea as the earliest C. auris case (2). C. auris infections have been reported in over 15 countries, causing outbreaks in health care facilities (3, 4). Hospital outbreaks of Candida were previously considered uncommon, but outbreaks of C. auris have been reported in Colombia (5), Venezuela (6), Israel (7), and the United Kingdom (8). New phylogenetic analysis of 47 whole-genome sequenced (WGS) patient isolates collected from South Asia (India/Pakistan), Africa, South America, and East Asia (Korea/Japan) show four highly clonal phylogenetic and geographically distinct clades that have emerged independently of one another (9).
There is growing evidence that C. auris has the ability to persistently colonize hospital environments (3, 8), with the propensity for transmission in health care settings (3, 9–11). Although poorly defined for C. auris, environmental survival and persistence properties have been documented for Candida parapsilosis, another colonizer of human skin, which is one of the most common causes of invasive candidiasis (12). Similarly to C. auris, C. parapsilosis has been documented in nosocomial infections from environmental sources (13) and is also known to survive for weeks on plastics, fabrics, and nonporous surfaces (14, 15). Furthermore, infections with C. parapsilosis have increased in prevalence over the past 2 decades in neonatal and intensive care units, where patients are at the highest risk for infection (16, 17). The modes and mechanisms of survival and transmission of C. parapsilosis in hospitals can occur via horizontal transfer through medical devices or other external sources, even without prior colonization (12), and may be similar to those of C. auris (6), making C. parapsilosis an excellent model for comparison with C. auris.
Some strains of C. auris can be resistant to multiple antifungal classes, severely limiting treatment options and making infection prevention guided by rapid detection in health care settings essential. To better understand the epidemiology of this emerging pathogen and to facilitate development of infection control measures, we investigated the ability of C. auris to persist on a textured plastic surface and compared the survival and persistence rates of C. auris on surfaces with those of C. parapsilosis. Patient and environmental specimens often contain a complex community of microorganisms. We set out to develop and validate an enrichment broth procedure that will allow researchers to collect samples from a wide variety of environments and rapidly isolate C. auris for investigation of the modes of transmission in health care facilities and halt further spread. Enrichment for C. auris reduces the chances of false negatives, and aids in isolation of C. auris from the background microbial community. Isolation is necessary to obtain accurate species identification and to perform phylogenetic analysis to assess possible transmission. Here we evaluated two enrichment broths for C. auris using a broad panel of Candida isolates, screening each for their ability to grow at elevated temperature and salinity in order to establish a method which could be used in combination with direct plating to aid in its isolation.
RESULTS
Multidrug-resistant yeast Candida auris can survive for at least 2 weeks on plastic surfaces.
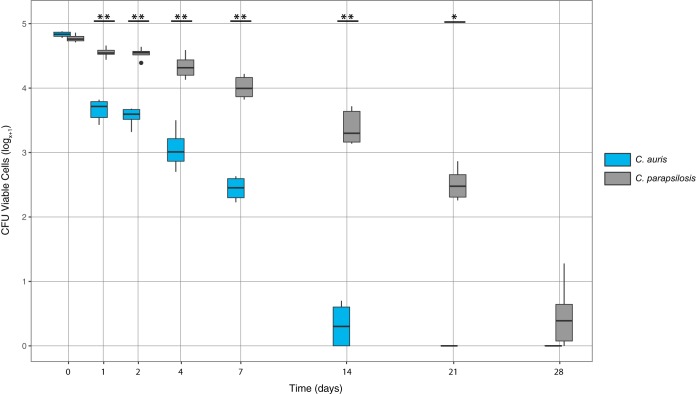
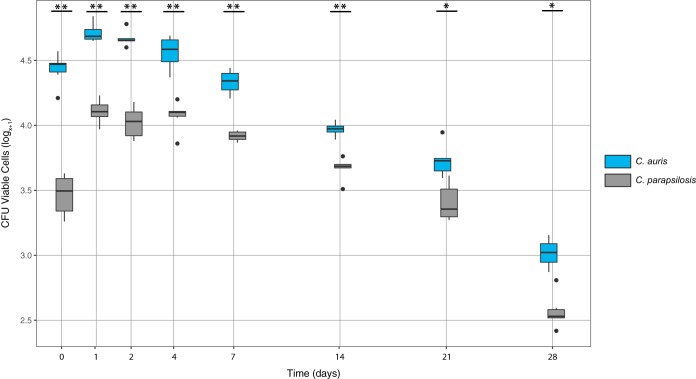
C. auris and C. parapsilosis remained viable for at least 14 and 28 days, respectively, as measured by colony counts (Fig. 1). Interestingly, the esterase activity assay that scans individual cells for viability demonstrated that C. auris cells were viable on surfaces 2 weeks beyond what was detected by culture (Fig. 2), indicating that the cells entered a viable but nonculturable state. Culturable C. parapsilosis cells were recovered at significantly higher numbers for all time points other than time zero (Fig. 1). Conversely, there were significantly more viable C. auris cells detected than C. parapsilosis at all time points when measured by esterase activity (Fig. 2 and Table S1 in the supplemental data).
FIG 1.
The log transformed recovery of viable C. auris (blue) and C. parapsilosis (gray) at each time point as determined by culture. The middle bar within each box represents the median; the top and bottom of the box represent the 75th and 25th quartiles, respectively, and dark circles represent outliers. A single asterisk indicates P < 0.05 and double asterisk indicates P < 0.001 between C. auris and C. parapsilosis for each time point.
FIG 2.
The log transformed recovery of viable C. auris (blue) and C. parapsilosis (gray) at each time point as determined by esterase activity. The middle bar within each box represents the median; the top and bottom of the box represent the 75th and 25th quartiles, respectively, and dark circles represent outliers. A single asterisk indicates P < 0.05 and double asterisk indicates P < 0.001 between C. auris and C. parapsilosis for each time point.
The initial Candida inocula deposited on the plastic coupons dried within 30 min. The transfer and processing of plastic coupons did not reduce the viability of the initial inocula. No significant differences were observed in the number of viable cells recovered from the inocula versus the plastic coupons at T0, and no significant differences were observed between the inocula of C. auris and C. parapsilosis at T0 (P = 0.71; Table S1). Live and chemically fixed formalin-killed cells were analyzed for survival using the esterase activity assay for validation. Less than 0.006% of the chemically killed cells were detected by the ScanRDI, indicating that the assay was fairly specific to live viable cells (Fig. S1). It is also possible that this small subset of cells were mislabeled or not inactivated by the formalin.
Enrichment broth procedure growth results using a broad panel of Candida isolates.
From the panel of Candida isolates, only C. auris had observable growth at an elevated temperature (40°C) and salinity (10%wt/vol) in the Sabouraud (SAB) or yeast nitrogen base (YNB) broths with dulcitol or mannitol as carbon sources. Candida glabrata was the only species other than C. auris to have observable growth in the Salt SAB Broth with dextrose as the carbon source (Table 1). All four clades of C. auris were able to grow under elevated temperature and salinity but the closely related Candida haemulonii, Candida doubushaemulonii, and other more distantly related Candida species were not able to grow (Table 1).
TABLE 1.
Growth results for panel of Candida isolatesa
| Candida species (location of collection) | Growth by strain and type of mediumb |
||||
|---|---|---|---|---|---|
| SAB |
YNB |
||||
| Dextrose | Dulcitol | Mannitol | Dulcitol | Mannitol | |
| C. auris (South Asia) | + | + | + | + | + |
| C. auris (Africa) | + | + | + | + | + |
| C. auris (South America) | + | + | + | + | + |
| C. auris (East Asia) | + | + | + | + | + |
| C. glabrata | + | − | − | − | − |
| C. albicans | − | − | − | − | − |
| C. doubushaemulonii | − | − | − | − | − |
| C. haemulonii | − | − | − | − | − |
| C. parapsilosis | − | − | − | − | − |
| C. tropicalis | − | NAc | NA | NA | NA |
Isolates incubated at 40°C with shaking at 250 rpm in a Sabouraud broth or yeast nitrogen base with either dextrose, dulcitol, or mannitol as the added carbon source.
SAB, Sabouraud broth; YNB, yeast nitrogen base. All media contained 10% NaCl (wt/vol).
NA, not available.
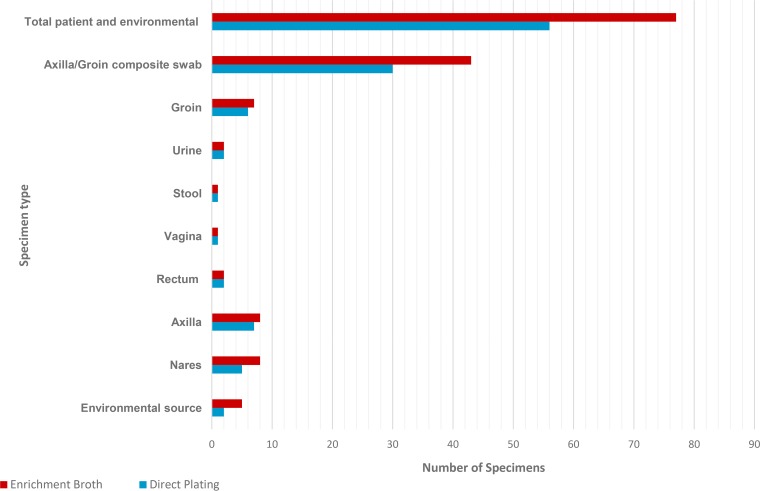
Clinical specimens (n = 72), including specimens from known clinical cases, were simultaneously processed for isolation by directly plating on CHROMagar as well as utilizing the Salt SAB Dex enrichment procedure. With the Salt SAB Dex broth C. auris was successfully isolated from all 72 clinical samples and 5 environmental samples, while with direct plating on CHROMagar Candida only 54 of the clinical samples (73%) and 2 of the environmental samples (40%) were isolated from C. auris (Table 2 and Fig. 3). In addition, multiple Candida species were recovered from direct plating onto CHROMagar plates, while only C. auris, C. glabrata, and some strains of C. parapsilosis were isolated from the enrichment in Salt SAB Dex broth. The enrichment procedure resulted in fewer isolates to process, culture plates used, and consumables needed to obtain a faster specimen identification by MALDI (data not shown).
TABLE 2.
Growth results for clinical samples positive for C. auris that were directly plated on CHROMagar Candida and processed through the enrichment broth procedure for isolation of C. auris
| Specimen type | Positive by CHROMagar Candida (% positive)a | Positive by Salt SAB Dex broth (% positive) |
|---|---|---|
| Vaginal swab | 1 (100) | 1 (100) |
| Stool | 1 (100) | 1 (100) |
| Urine | 2 (100) | 2 (100) |
| Rectal swab | 2 (100) | 2 (100) |
| Environmental swab | 2 (40) | 5 (100) |
| Groin swab | 6 (86) | 7 (100) |
| Nasal swab | 5 (63) | 8 (100) |
| Axilla swab | 7 (87) | 8 (100) |
| Axilla/groin composite swab | 30 (68) | 43 (100) |
| Total specimens tested | 56 (73) | 77 (100) |
Percentage of specimens that were found positive by both methods.
FIG 3.
Isolation of C. auris from clinical and environmental specimens (n = 77) by direct plating on CHROMagar Candida (blue bars) and by processing through the Salt SAB Dex enrichment broth procedure (red bars).
DISCUSSION
C. auris appears to readily spread and cause outbreaks in health care settings. Infection prevention guided by rapid detection is an essential component needed to prevent dissemination. To better understand this organism, we evaluated its ability to survive on plastic surfaces in comparison with that of C. parapsilosis, which is another skin colonizing yeast known to survive and persist on plastics and in hospital environments, causing nosocomial outbreaks (18). Our results demonstrated that both species of Candida are capable of surviving for weeks in the environment in health care settings, raising concerns for the possibility of nosocomial transmission. To further assist with isolation and detection of C. auris in patients and the environment we developed the enrichment broth procedure that facilitates isolation by taking advantage of C. auris' ability to actively grow under high salt and high temperature conditions; growth conditions that are restrictive to most other microbes.
Survival and persistence of Candida on surfaces was evaluated using two complementary independent methods, culture (CFU) and direct observation of viability by measurement of esterase activity with a solid-phase cytometer, a non-growth-based technology. The esterase assay is capable of detecting a single yeast or bacterium and has been recognized by the FDA as an alternative testing method for sterile products (19). As expected, viability detected by measurement of esterase activity was equivalent or slightly higher across all time points than that measured by culture, since the assay detects individual cells rather than clusters of cells that may all grow to form single colonies. Interestingly, C. auris yielded significantly higher metabolically active viable cell counts compared to C. parapsilosis at all time points as measured by esterase activity (Fig. 2), but lower culture-based viable cell counts compared to C. parapsilosis at all time points except time zero as measured by CFU counts (Fig. 1). In relation to previous studies, which only rely on culture-based colony counts, these data highlight the value in using an additional direct viability assay in order to gather a more comprehensive picture of survival and persistence of Candida in the environment. The metabolically active viable but nonculturable (VBNC) cells of C. auris were detected for at least 4 weeks, and colonies were recovered by culture for at least 2 weeks, providing the first estimate of this species' survival time in the environment (Fig. 1 and 2). Similarly, C. parapsilosis colonies were recovered by culture for the full 4 weeks of the studies (Fig. 1), which was twice as long as previous studies observed (14). Traore et al. used only culture based colony counts from direct plating to measure the survival of C. parapsilosis and C. albicans on glass, stainless steel, and fabric surfaces under ambient environmental conditions. Although the Traore et al. study did not test plastic surfaces and deposited a 3-log level higher inoculum on these surfaces (107 versus the 104 cells used for the inoculum here), they demonstrated survival from all surfaces for at least 2 weeks for C. parapsilosis and 2 weeks on fabrics for C. albicans, but only 3 days for C. albicans on glass and stainless steel (14).
In all experiments the measurement of esterase activity detected higher numbers of viable cells of both species of Candida on the second day of incubation than at the time-zero counts. Conversely, the remaining time points showed the total number of viable cells decreasing, as expected, with each successive time point. One possible explanation for this unexpected increase in the number of cells at day 2 is that some fraction of the yeast cells divided after being left overnight on the plastic coupons, which is a known technique of yeast and other single-cell organisms to increase surface to volume ratio. Alternatively, the lower number of recovered cells detected by esterase activity at time zero may be attributed to the higher likelihood of adherence of some of the fresh active cells to the dilution tubes, pipet tips, or the filter column, as opposed to the quiescent viable cells of the later time points. The esterase activity measurements by solid-phase cytometer demonstrated that the assay was highly specific to live viable cells, as only live cells were detected by the instrument (see Fig. S1 in the supplemental material). In spite of the time-zero limitations, the findings clearly demonstrate that a substantial proportion of C. auris cells exist in a viable but nonculturable (VBNC) state over the test period of 28 days (Fig. 2). C. parapsilosis also showed significantly higher numbers at the 14-, 21-, and 28-day time points. Whether these VBNC C. auris cells are capable of reviving and becoming infectious, as has been determined for other pathogens (20–22), is something that requires further examination.
Our results demonstrate that both C. auris and C. parapsilosis can survive and persist outside their host on dry, nonporous surfaces for weeks, emphasizing the importance of infection control for preventing nosocomial transmission. Long environmental persistence of C. auris is particularly troubling considering the ability of C. auris to colonize the skin (8). Since humans shed skin cells at an approximant rate of 106 particles per hour (23, 24), patients shedding C. auris- or C. parapsilosis-colonized skin cells could contribute to prolonged outbreaks with high transmissibility in health care settings. Although, C. parapsilosis currently tends to display lower levels of pathogenicity and antifungal resistance than the emerging multidrug-resistant C. auris, the rise of antifungal resistance among C. parapsilosis is concerning (25, 26).
Because of the technical limitations of testing multiple strains and environmental conditions, only a single clinical isolate of each species was tested under specific temperature and humidity, 25°C and 55 to 57% relative humidity (RH), on plastic surfaces. Therefore, other potential sources of variability have not been evaluated. It is possible that using a higher inoculum or testing different environmental conditions, surfaces, or isolates from a different clade would generate different results, though even survival for 24 h would provide a potential for transmission. The C. auris isolate tested was resistant to fluconazole and falls within the South Asian clade (Table 3). It remains to be seen whether there are some fitness tradeoffs in the level of drug resistance and ability to survive and persist in the environment. Our study was designed to investigate and gather baseline data for the ability of the emerging pathogen C. auris to survive in the environment outside a human host, and quickly disseminate that information to better inform infection control efforts. There are many interesting research avenues that need to be explored to better understand the landscape of survival and persistence of pathogenic yeast outside the human host. Future studies are needed to investigate the landscape of the entire species complex from all four clades of C. auris and how their survival and persistence differs strain to strain and from susceptible to resistant. Researchers with the proper facilities and protocol precautions to handle multidrug-resistant organisms may consider investigating these questions.
TABLE 3.
Isolate metadata for Candida isolates
| Candida species | Strain | Origin | Specimen type | Yr collected |
|---|---|---|---|---|
| C. auris | B11103 | Pakistan | Urine | 2015 |
| C. auris | B11220 | Japan | Auditory Canal | 2008 |
| C. auris | B11221 | South Africa | Blood | 2012 |
| C. auris | B11244 | Venezuela | Blood | 2012 |
| C. parapsilosis | B13522 | USA | Blood | 2008 |
| C. albicans | B10423 | NAa | NA | NA |
| C. glabrata | B12206 | USA | Axilla | 2016 |
| C. doubushaemulonii | B10440 | NA | NA | NA |
| C. haemulonii | B10441 | NA | NA | NA |
NA, not available.
The description of the type strain reported C. auris has an ideal growth temperature of 37-40°C, and is positive for growth in 10% NaCl/5% glucose medium (1). The enrichment broth procedure combines high temperature and high salinity to broadly inhibit other microbes and aid in the isolation of C. auris. However, the enrichment broth procedure cultures C. auris at the upper limits of environmental conditions this organism can tolerate and actively grow, and may require longer incubation periods, which are important to consider when trying to rule out false negatives. Combining the high salt media and the antibiotics gentamicin and chloramphenicol with high temperature (40°C) incubation excludes the vast majority of microbes. The methods outlined here should facilitate the isolation of C. auris from human and environmental sites with complex microbial communities (e.g., stool samples), and allow researchers to gather data essential to determining the prevalence of this pathogen colonizing the skin, gastrointestinal, or other body sites.
We demonstrated that C. auris suspended in a soil load and dried on nonporous plastic surfaces (similar to environments found in health care settings) can persist for at least 2 weeks as determined by culture, remaining metabolically active for at least 4 weeks. C. parapsilosis suspended in a soil load and dried on nonporous plastic surfaces can remain culturable and metabolically active for at least 4 weeks. These data indicate that the survival of these species outside a human host is considerable and allows ample time for transmission within a health care facility. Our results are comparable to the life-history traits of problematic nosocomial Gram-positive bacterial pathogens Staphylococcus aureus or enterococci (27). These findings may help explain why C. auris has caused numerous outbreaks in health care facilities and reinforces the critical need for effective infection control, cleaning, and disinfection practices in affected facilities.
Infection prevention guided by rapid detection in health care settings is essential, because C. auris is difficult to treat as it is commonly resistant to multiple antifungal drug classes. To assist with isolation and identification of C. auris in these settings, we developed and evaluated novel enrichment broths, with which we isolated C. auris from complex microbial communities, such as skin, stool, and environmental surfaces. The simple, low cost, and effective enrichment broth procedure has since become a key tool in controlling the spread of C. auris in the United States and other countries.
MATERIALS AND METHODS
Organisms tested and growth conditions.
The clinical strains used in the survival and persistence experiments were C. auris B11103 and parapsilosis B13522 (accession number SAMN05379581). Cultures were grown overnight on Sabouraud dextrose agar (SAB) supplemented with chloramphenicol and gentamicin at a final concentration of 50 mg/liter (SAB Dex). A single colony was resuspended in 3 ml SAB Dex broth in 15-ml test tubes and grown up to late log growth phase at 30°C and 250 rpm for 48 h in triplicate, meaning each strain was grown in 3 replicate test tubes. The clinical Candida strains used in the enrichment broth experimental evaluation consisted of four C. auris isolates (one from each polygenetic and geographical clade), C. parapsilosis, Candida tropicalis, C. albicans, C. doubushaemulonii, C. haemulonii, and C. glabrata (Tables 1 and 3).
Patient and environmental specimens.
Specimens from patients infected or colonized by C. auris and their surrounding environments were submitted to the CDC as part of the ongoing C. auris outbreak response to evaluate infection control measures at the affected facilities. These specimens included swabs from the axilla/groin (n = 43), axilla (n = 8), nares (n = 8), groin (n = 7), rectum (n = 2), vagina (n = 1), and environmental sources (n = 5). In addition, two urine and one stool sample were submitted. This investigation was considered an urgent public health response and therefore was not subject to review by CDC's Institutional Review Board.
Inoculation of plastic carriers, artificial test soil load, air temperature, and relative humidity.
Plastic square coupons of 1 cm2 were prepared at the Centers for Disease Control and Prevention (CDC) from PVC acrylic alloy (Kydex-T, O-80 thickness, P1 haircell texture). The plastic coupons represented common nonporous environmental surfaces in hospitals. Preparation and sterilization of coupons prior to inoculation consisted of thorough cleaning with a nonbactericidal detergent (Versa-Clean; Fisher Scientific, Pittsburgh, PA), ultrapure, endotoxin-free reverse osmosis water (RO), and 70% ethanol. The coupons were then sterilized by UV light, 20 min each side (Phillips Sterilamp G36T6L, approximately 107_W-s/cm2). Inocula were contained in an artificial test soil (ATS) (Healthmark Industries, Fraser, MI), which was a mixture of 0.180 mg/ml mucin, 0.023 mg/ml hemoglobin, 0.260 mg/ml RPMI, 0.055 mg/ml cellulose, 0.110 mg/ml egg yolk powder, and 0.201 mg/ml albumin. The dried ATS product was suspended in reverse osmosis water, then diluted 1:5 in Butterfield's buffer. Late exponential cultures were pelleted at 2,000 × g for 20 mins at 4°C and resuspended in ATS to a final concentration of 5 × 106 cells per ml. A 10-μl aliquot of the cell suspension was placed on 24 coupons (three for each time period to be tested), and 10 μl of ATS only (no organisms) were placed on 8 coupons as negative controls (one for each time period tested). Three test coupons and one control coupon were processed immediately, and the remaining coupons were placed in a humidity chamber maintained at 25°C and 55 to 57% RH.
Tests for survival on inanimate materials.
Three test coupons and one negative-control coupon were retrieved from the chamber at days 1, 2, 4, 7, 14, 21, and 28 and placed in 15-ml polyethylene tubes with 3 ml of phosphate-buffered saline containing 0.02% Tween 80 (PBST). Coupons were then vortexed for 30 s and immediately sonicated (FS 20, a 40-KHz ultrasonic bath cleaner; Fisher Scientific) for 30 s, with the cycle repeating for a total of three cycles. Preliminary tests were conducted to confirm that vortex and sonication did not reduce viable cell counts. Direct aliquots and serial dilutions of the PBST/cell supernatant were plated in triplicate, the plates the incubated at 36°C for 48 h, and CFUs were counted to determine the number of culturable cells surviving at each time point. A separate aliquot was used for an independent assay of survival, measured by the presence of esterase activity. Esterase activity was determined using a ScanRDI (bioMérieux, Inc., Durham NC) solid-phase cytometer and the instrument's standard viability assay sample preparation protocol (28). The ScanRDI separately counts individual cells that fluoresce if esterase is produced, and the results from the instrument were manually validated cell by cell using epifluorescence microscopy. Preliminary tests were conducted using live and formalin-fixed cells (2.0% final concentration for 30 mins at 4°C) to confirm that the esterase activity assay identified and counted only viable cells (Fig. S1).
Number of tests and analysis of data.
Direct cell count and colony count data were adjusted for dilution, and to account for zeros in the colony count data were modified and then transformed [log10(x + 1)]. Means and standard deviations were calculated, normality of the data was tested using the Shapiro-Wilk statistic, and the differences between time and isolates were determined as significant at P values of < 0.05 by use of the Student t test.
Enrichment broth growth assay and panel of Candida isolates for evaluating enrichment for C. auris.
Two enrichment broths with amino acids, nitrogenous compounds, and 10%, wt/vol sodium chloride were evaluated with separate carbon compounds. The first was the Salt Sabouraud Enrichment Broth (Salt SAB Broth) that consists of 5 g pancreatic digest of casein (Remel, Lenexa, KS, USA), 5 g peptic digest of animal tissue (Neogen, Lansing, MI, USA), and 100 g sodium chloride (NaCl) dissolved in a liter of deionized (DI) water with 20 g of either dextrose, dulcitol/galactitol, or mannitol (Difco, Franklin Lakes, NJ, USA) as the added carbon source. The medium was mixed well, the pH was adjusted to 5.6 ± 2, and the filter was sterilized through a 0.2-μm filter. The second enrichment broth was salt yeast nitrogen base broth (Salt YNB), which consists of 6.8 g yeast nitrogen base (Difco, Franklin Lakes, NJ, USA), and 100 g sodium chloride (NaCl) dissolved in a liter of DI water with either dulcitol/galactitol, or mannitol (Difco, Franklin Lakes, NJ, USA) as the added carbon source. In all media, gentamicin and chloramphenicol were added to a 50-mg/liter final concentration. The medium was mixed well and filter sterilized through a 0.2-μm filter. All Candida isolates were grown to late log phase, pelleted at 2,000 × g for 20 mins at 4°C, and resuspended in PBS to a final concentration of 5 × 106 cells per ml as previously described above. A 10-μl aliquot of the cell suspension (104 cells) was used to inoculate 3 ml of each enrichment broth, and each strain was grown in 3 replicate test tubes incubated at 40°C with shaking with aeration at 250 rpm. Cell density was measured at optical density (OD) 600 using an ELx808 microplate reader (BioTek, USA) at time = 48 and 72 h.
Isolation of C. auris from patient and environmental specimens.
C. auris cultures from patients were obtained using either rayon tip swabs (Fisherfinest Amies Charcoal bacteriology culture collection and transport system; Fisher health care, Ontario, Canada) or nylon-flocked swab (BD Eswab collection and transport system; Becton Dickinson and Company, Sparks, MD). Patient swab specimens were vortexed for 5 s to release the sample from the swab tip and evenly disperse and suspend the patient specimen in the liquid transport medium. Aliquots of 75 μl of transport medium were directly plated on CHROMagar Candida (CHROMagar, Becton Dickinson, San Jose, CA) and incubated at 37°C for 7 days. CHROMagar Candida is a medium proven to support the growth of C. auris and was chosen as a baseline to independently quantify the number of positive patient and environmental samples. Pale and pink colonies were selected, colony purified (see Fig. S2 in the supplemental data), and subjected for identification with matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF). Simultaneously, 100-μl aliquots of transport medium were inoculated into enrichment broth and incubated at 40°C with constant agitation at 250 rpm for 7 days. Aliquots of 100 μl of urine specimens were directly plated on CHROMagar and 100-μl aliquots were inoculated in enrichment broth cultures. Approximately 10 μl of stool was obtained with a sterile inoculation loop and streaked for isolation on CHROMagar, while another 10-μl loop was used to inoculate into enrichment broth, and incubated at 40°C with constant agitation at 250 rpm for 7 days. Enrichment broths were checked daily for signs of growth and a 10-μl sterile loop was used to streak for isolation on CHROMagar Candida. After 7 days, each culture tube with no visible sign of growth was transferred to CHROMagar Candida, and all isolates obtained were subjected to identification via MALDI-TOF.
Environmental samples taken prior to terminal cleaning of the rooms were collected using a standard sponge-wipe sampling protocol (3M sponge stick with neutralized buffer; 3M Health Care, St. Paul, MN) (29). Briefly, 90 ml phosphate-buffered saline containing 0.02% Tween 80 (PBST) was used to express the sample using a Stomacher 400 Circulator (Seward Lab. Systems, Inc., Bohemia NY). The sample was then concentrated via centrifugation, and cultured by simultaneously plating on CHROMagar and enrichment broth (29). The inoculated broths were examined daily and those that appeared turbid were streaked for isolation on CHROMagar Candida and incubated at 37°C. Individual colonies were selected and subjected for MALDI identification using the rapid yeast extraction protocol (30) on the MALDI Bruker Biotyper (Bruker Daltonics). The MicrobeNet MALDI database (Centers for Disease Control and Prevention, Atlanta, GA) (31) was used to assign accurate species identification, which includes all four clades of C. auris and the closely related strains C. haemulonii and C. doubushaemulonii (32).
Supplementary Material
ACKNOWLEDGMENTS
We thank Brendan Jackson, Tom Chiller, Judith Noble-Wang, and Clifford MacDonald for critical reading of the manuscript and helpful discussions. The use of product names in this article does not imply their endorsement by the U.S. Department of Health and Human Services. The findings and conclusions in this article are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00921-17.
REFERENCES
- 1.Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. 2009. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol 53:41–44. doi: 10.1111/j.1348-0421.2008.00083.x. [DOI] [PubMed] [Google Scholar]
- 2.Lee WG, Shin JH, Uh Y, Kang MG, Kim SH, Park KH, Jang HC. 2011. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol 49:3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vallabhaneni S, Kallen A, Tsay S, Chow N, Welsh R, Kerins J, Kemble SK, Pacilli M, Black SR, Landon E, Ridgway J, Palmore TN, Zelzany A, Adams EH, Quinn M, Chaturvedi S, Greenko J, Fernandez R, Southwick K, Furuya EY, Calfee DP, Hamula C, Patel G, Barrett P, Lafaro P, Berkow EL, Moulton-Meissner H, Noble-Wang J, Fagan RP, Jackson BR, Lockhart SR, Litvintseva AP, Chiller TM. 2016. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus—United States, May 2013-August 2016. MMWR Morb Mortal Wkly Rep 65:1234–1237. doi: 10.15585/mmwr.mm6544e1. [DOI] [PubMed] [Google Scholar]
- 4.Clancy CJ, Nguyen MH. 2016. Emergence of Candida auris: an international call to arms. Clin Infect Dis doi: 10.1093/cid/ciw696. [DOI] [PubMed] [Google Scholar]
- 5.Morales-López SE, Parra-Giraldo CM, Ceballos-Garzón A, Martínez HP, Rodríguez GJ, Álvarez-Moreno CA, Rodríguez JY. 2017. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg Infect Dis 23:162. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calvo B, Melo AS, Perozo-Mena A, Hernandez M, Francisco EC, Hagen F, Meis JF, Colombo AL. 2016. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect 73:369–374. doi: 10.1016/j.jinf.2016.07.008. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Ami R, Berman J, Novikov A, Bash E, Shachor-Meyouhas Y, Zakin S, Maor Y, Tarabia J, Schechner V, Adler A. 2017. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis 23:195. doi: 10.3201/eid2302.161486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schelenz S, Hagen F, Rhodes JL, Abdolrasouli A, Chowdhary A, Hall A, Ryan L, Shackleton J, Trimlett R, Meis JF. 2016. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control 5:35. doi: 10.1186/s13756-016-0132-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lockhart SR, Etienne KA, Vallabhaneni S, Farooqi J, Chowdhary A, Govender NP, Colombo AL, Calvo B, Cuomo CA, Desjardins CA, Berkow EL, Castanheira M, Magobo RE, Jabeen K, Asghar RJ, Meis JF, Jackson B, Chiller T, Litvintseva AP. 2016. Simultaneous emergence of multidrug-resistant Candida auris on three continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhary A, Sharma C, Duggal S, Agarwal K, Prakash A, Singh PK, Jain S, Kathuria S, Randhawa HS, Hagen F, Meis JF. 2013. New clonal strain of Candida auris, Delhi, India. Emerg Infect Dis 19:1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakrabarti A, Sood P, Rudramurthy SM, Chen S, Kaur H, Capoor M, Chhina D, Rao R, Eshwara VK, Xess I, Kindo AJ, Umabala P, Savio J, Patel A, Ray U, Mohan S, Iyer R, Chander J, Arora A, Sardana R, Roy I, Appalaraju B, Sharma A, Shetty A, Khanna N, Marak R, Biswas S, Das S, Harish BN, Joshi S, Mendiratta D. 2015. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med 41:285–295. doi: 10.1007/s00134-014-3603-2. [DOI] [PubMed] [Google Scholar]
- 12.Trofa D, Gacser A, Nosanchuk JD. 2008. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 21:606–625. doi: 10.1128/CMR.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez V, Vazquez JA, Barth-Jones D, Dembry L, Sobel JD, Zervos MJ. 1993. Nosocomial acquisition of Candida parapsilosis: an epidemiologic study. Am J Med 94:577–582. doi: 10.1016/0002-9343(93)90207-6. [DOI] [PubMed] [Google Scholar]
- 14.Traore O, Springthorpe VS, Sattar SA. 2002. A quantitative study of the survival of two species of Candida on porous and nonporous environmental surfaces and hands. J Appl Microbiol 92:549–555. doi: 10.1046/j.1365-2672.2002.01560.x. [DOI] [PubMed] [Google Scholar]
- 15.De Bernardis F, Mondello F, San Millan R, Ponton J, Cassone A. 1999. Biotyping and virulence properties of skin isolates of Candida parapsilosis. J Clin Microbiol 37:3481–3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guinea J. 2014. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 20(Suppl 6):S5–S10. doi: 10.1111/1469-0691.12539. [DOI] [PubMed] [Google Scholar]
- 17.Pammi M, Holland L, Butler G, Gacser A, Bliss JM. 2013. Candida parapsilosis is a significant neonatal pathogen: a systematic review and meta-analysis. Pediatr Infect Dis J 32:e206–216. doi: 10.1097/INF.0b013e3182863a1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Asbeck EC, Huang YC, Markham AN, Clemons KV, Stevens DA. 2007. Candida parapsilosis fungemia in neonates: genotyping results suggest healthcare workers hands as source, and review of published studies. Mycopathologia 164:287–293. doi: 10.1007/s11046-007-9054-3. [DOI] [PubMed] [Google Scholar]
- 19.Smith R, Von Tress M, Tubb C, Vanhaecke E. 2010. Evaluation of the ScanRDI(R) as a rapid alternative to the pharmacopoeial sterility test method: comparison of the limits of detection. PDA J Pharm Sci Technol 64:356–363. [PubMed] [Google Scholar]
- 20.Epalle T, Girardot F, Allegra S, Maurice-Blanc C, Garraud O, Riffard S. 2015. Viable but not culturable forms of Legionella pneumophila generated after heat shock treatment are infectious for macrophage-like and alveolar epithelial cells after resuscitation on Candida auris for the clinical microbiology laboratory: not your grandfather's Candida species. Microb Ecol 69:215–224. doi: 10.1007/s00248-014-0470-x. [DOI] [PubMed] [Google Scholar]
- 21.Pinto D, Almeida V, Almeida Santos M, Chambel L. 2011. Resuscitation of Escherichia coli VBNC cells depends on a variety of environmental or chemical stimuli. J Appl Microbiol 110:1601–1611. doi: 10.1111/j.1365-2672.2011.05016.x. [DOI] [PubMed] [Google Scholar]
- 22.Colwell R, Brayton P, Grimes D, Roszak D, Huq S, Palmer L. 1985. Viable but non-culturable Vibrio cholerae and related pathogens in the environment: implications for release of genetically engineered microorganisms. Nat Biotechnol 3:817–820. doi: 10.1038/nbt0985-817. [DOI] [Google Scholar]
- 23.You R, Cui W, Chen C, Zhao B. 2013. Measuring the short-term emission rates of particles in the “personal cloud” with different clothes and activity intensities in a sealed chamber. Aerosol Air Qual Res 13:911–921. [Google Scholar]
- 24.Bhangar S, Adams RI, Pasut W, Huffman J, Arens EA, Taylor JW, Bruns TD, Nazaroff WW. 2016. Chamber bioaerosol study: human emissions of size-resolved fluorescent biological aerosol particles. Indoor Air 26:193–206. doi: 10.1111/ina.12195. [DOI] [PubMed] [Google Scholar]
- 25.Sarvikivi E, Lyytikäinen O, Soll DR, Pujol C, Pfaller MA, Richardson M, Koukila-Kähkölä P, Luukkainen P, Saxén H. 2005. Emergence of fluconazole resistance in a Candida parapsilosis strain that caused infections in a neonatal intensive care unit. J Clin Microbiol 43:2729–2735. doi: 10.1128/JCM.43.6.2729-2735.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moudgal V, Little T, Boikov D, Vazquez JA. 2005. Multiechinocandin-and multiazole-resistant Candida parapsilosis isolates serially obtained during therapy for prosthetic valve endocarditis. Antimicrob Agents Chemother 49:767–769. doi: 10.1128/AAC.49.2.767-769.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neely AN, Maley MP. 2000. Survival of enterococci and staphylococci on hospital fabrics and plastic. J Clin Microbiol 38:724–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costanzo SP, Borazjani RN, McCormick PJ. 2002. Validation of the Scan RDI for routine microbiological analysis of process water. PDA J Pharm Sci Technol 56:206–219. [PubMed] [Google Scholar]
- 29.Shams AM, Rose LJ, Edwards JR, Cali S, Harris AD, Jacob JT, LaFae A, Pineles LL, Thom KA, McDonald LC. 2016. Assessment of the overall and multidrug-resistant organism bioburden on environmental surfaces in healthcare facilities. Infect Cont Hosp Epidemiol 37:1426–1432. doi: 10.1017/ice.2016.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser M, Brown Z, Houldsworth M, Borman AM, Johnson EM. 2016. Rapid identification of 6328 isolates of pathogenic yeasts using MALDI-TOF MS and a simplified, rapid extraction procedure that is compatible with the Bruker Biotyper platform and database. Med Mycol 54:80–88. [DOI] [PubMed] [Google Scholar]
- 31.Janda JM, Lopez DL. 2017. Mini review: new pathogen profiles: Elizabethkingia anophelis. Diagn Microbiol Infect Dis 88:201–205. doi: 10.1016/j.diagmicrobio.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 32.Lockhart SR, Berkow EL, Chow N, Welsh RM. 2017. Candida auris for the clinical microbiology laboratory: not your grandfather's Candida species. Clin Microbiol Newsletter 39:99–103. doi: 10.1016/j.clinmicnews.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.