ABSTRACT
Malaria is one of the leading causes of infectious disease in travelers returning from the tropics. The diagnosis of malaria is typically performed by examining Giemsa-stained thick and thin peripheral blood smears, which is time consuming, labor intensive, and requires high levels of proficiency. Alternatively, loop-mediated isothermal amplification (LAMP) is a new molecular method, which is rapid, sensitive, and requires less capital equipment and technological training. We conducted a retrospective study comparing two formats of a commercial LAMP assay (Meridian illumigene malaria [M] and malaria Plus [MP]) versus reference microscopy on archived blood specimens (n = 140) obtained from unique returning travelers suspected of having malaria. Discrepant results were resolved by either repeat testing or a laboratory developed ultrasensitive real-time PCR method. On initial testing, the Meridian illumigene M and MP kits had sensitivities of 97.3% (95% confidence interval [CI], 90.7 to 99.7%) and 100.0% (95.1 to 100.0%) and specificities of 93.8% (84.8 to 98.3%) and 91.5% (81.3 to 97.2%), respectively, versus reference microscopy. We project a significant cost reduction in low prevalence settings where malaria is not endemic with LAMP-based malaria screening given the excellent negative predictive value achieved with LAMP.
KEYWORDS: diagnostics, LAMP, malaria
INTRODUCTION
Malaria is a common disease globally. The World Health Organization estimates the Plasmodium parasite infected 212 million people in 2015 (1). Malaria is also one of the most common infectious diseases found in travelers returning from the tropics. Between 2004 and 2014, CanTravNet surveillance, a network of travel clinics in large urban centers, reported that malaria was the most common specific cause of fever in returning travelers (2). Malaria is often preventable during travel using readily available chemoprophylaxis or personal protective measures such as insecticide-treated bed nets and clothing (3). Plasmodium falciparum is the most virulent species for malaria and must be promptly diagnosed to prevent infectious complications or death due to a delay in diagnosis, particularly in returning travelers (4). Malaria-related hospitalizations in the United States are more frequent than once thought according to a recent study which identified 22,029 hospitalizations in a 15-year period, with an average length of stay of 4.36 days at a cost of US$25,789 (5).
Malaria diagnosis in most clinical microbiology laboratories continues to rely on microscopic examination of Giemsa-stained thick and thin peripheral blood smears, often in combination with rapid diagnostic tests (RDTs), which detect the circulating parasite antigen. However, this method is time consuming, labor intensive, and challenging in areas where malaria is not endemic, as it requires continuous proficiency training where positivity rates are low. Microscopy may also miss malaria infections with low peripheral blood parasitemia levels (such as asymptomatic cases and those involving pregnancy or cerebral malaria), as it cannot reliably detect parasite densities <100 parasites/μl depending on the technologist's experience, which is often limited in areas where malaria is not endemic (6, 7). In fact, a four-year retrospective study in our region demonstrated that microscopy is only able to detect 90.4% (150/166) of high-risk P. falciparum cases compared with that of quantitative PCR (qPCR) (8). Numerous studies have demonstrated that molecular testing using PCR has increased the analytical sensitivity compared with that of microscopy (9–12). The limit of detection (LOD) of PCR is lower than that of microscopy, and can detect at or below 1 parasite/μl depending on the specific molecular approach used (e.g., nested PCR or real-time PCR) (11). Both microscopy and molecular testing are time consuming and challenging to perform in smaller diagnostic laboratories or as near-patient tests. RDTs rely on antigen detection from a drop of blood and are easy to perform. However, RDTs lack analytical sensitivity with limits of detection higher than 100 parasites/μl, resulting in low sensitivities reported especially for non-falciparum Plasmodium species (12). Molecular-based isothermal tests for field diagnosis of malaria have recently been well studied in resource-limited settings with acceptable clinical results (12–24).
Loop-mediated isothermal amplification (LAMP) is a one-step amplification technique requiring specialized primers, which utilize self-recurring strand displacement to amplify DNA at a single temperature parameter (25). Sensitivity and specificity are dependent on primer development, species, sample type, target gene choice (e.g., mitochondrial DNA or ribosomal DNA), extraction methods (RNA, DNA, or both), and blood source (filter paper blood dots or whole blood). Reported analytical sensitivities vary from study to study but typically are at 1 parasite per μl or less (22, 26–33). Limits of detection of 0.3 to 2 parasites/μl for P. falciparum and 0.1 for P. vivax μl have been reported, although the manufacturer's product insert reports even lower limits of detection (see Materials and Methods) (34). The commercially available illumigene malaria LAMP assay (Meridian Bioscience) was tested in Senegal, coupled with molecular confirmation in a reference laboratory in the United States, and yielded promising results in terms of assay performance, ease of use, reagent stability, and simplified extraction technology (7). However, no data have yet been published on the performance of the illumigene malaria test in a large clinical laboratory in a setting where malaria is not endemic. Moreover, no information exists on how LAMP technology would be most efficiently used within current diagnostic malaria algorithms in settings where malaria is not endemic or on the potential impact on turnaround time, cost, and feasibility in a 24-h 7-day laboratory service. The illumigene malaria LAMP assay has the potential to allow for more sensitive detection of Plasmodium parasites, with a rapid turnaround time, with decreased capital investment, and without requiring molecular specialist expertise.
This study retrospectively assessed collected Plasmodium samples (n = 140) from returning travelers between 2003 and 2014 using the illumigene malaria LAMP method and compared the results with those from reference microscopy. Results that were initially discrepant were resolved by an in-house validated real-time PCR (9). A revised, cost-effective alternative diagnostic algorithm is proposed based on our finding that incorporates the routine use of LAMP to provide more-sensitive screening of possible malaria cases than current rapid methods provide.
RESULTS
Initial detection of malaria using routine microscopy versus LAMP.
A total of 140 patient blood samples were studied using both LAMP methods (Table 1). The performance of routine microscopy versus the M/MP LAMP assays is shown in Table 2. Thick and thin microscopy was performed on all 140 blood samples, and 76 (54%) of the samples were positive for Plasmodium parasites in asexual stages. P. falciparum was the most common species and occurred as a monoinfection in 38 (27%) of the samples. Monoinfections with P. vivax, P. ovale, and P. malariae also occurred in 25 (18%), 8 (6%), and in 1 (1%), respectively. Four samples had dual mixed infections: 2 with P. falciparum and P. malariae and 2 other samples with P. vivax and P. ovale. Compared with that of microscopy, the M kit had a sensitivity of 97.3% (95% confidence interval [CI], 90.7 to 99.7%) and a specificity of 93.8% (84.8 to 98.3%). The MP kit compared with reference microscopy had a sensitivity of 100.0% (95.1 to 100.0%) and a specificity of 91.5% (81.3 to 97.2%). There was no significant difference between the initial performance of the M and MP assays, suggesting that DNA sample extraction was equivalent for both kit methods.
TABLE 1.
Thick and thin film microscopy results for patient samples enrolled in the study
| Result | No. of samples |
|||||||
|---|---|---|---|---|---|---|---|---|
| P. falciparum | P. falciparum/P. malariae mixed | P. vivax | P. ovale | P. vivax/P. ovale mixed | P. malariae | Negative | Total | |
| Positive | 38 | 2 | 25 | 8 | 2 | 1 | 76 | |
| Negative | 64 | 64 | ||||||
| Total | 38 | 2 | 25 | 8 | 2 | 1 | 64 | 140 |
TABLE 2.
Performance of the illumigene malaria M and MP kits
| Resulta |
illumigene malaria kit |
|
|---|---|---|
| M | MP | |
| True positive (n) | 73 | 73 |
| False positive (n) | 4 | 5 |
| False negative (n) | 2 | 0 |
| True negative (n) | 60 | 54 |
| Invalid resultb (n) | 1 | 8 |
| Totalc (n) | 139 | 132 |
| Sensitivity (% [95% CI]) | 97.3 (90.7–99.7) | 100.0 (95.1–100.0) |
| Specificity (% [95% CI]) | 93.8 (84.8–98.3) | 91.5 (81.3–97.2) |
| Negative predictive valued (%) | 99.8 | 100 |
| Positive predictive valued (%) | 45.2 | 38.2 |
| Invalid rate (%) | <0.01 | 5.7 |
Results were compared with those from reference microscopy.
illumigene result was recorded as “invalid” due to a failed run. “Empty well” and instrument errors were rerun from a fresh extract as per the product insert and included in the performance calculations.
Invalid results were not included in the sensitivity and specificity calculations.
Both negative and positive predictive values are calculated based on a prevalence of malaria of 0.05 (5%) in the returning traveler population.
Resolution of discrepant results with illumigene M kit.
There were 6 discrepancies between the M kit and microscopy (Table 3). Of these, 2 occurrences were detected with microscopy alone and 4 cases were detected with the M kit only. Both of the false-negative M kit results were positive on repeat testing. Of the 4 samples that were positive by the M kit and negative by microscopy, 3 were positive by qPCR, suggesting they were true-positive results. The remaining specimen was negative with repeat testing.
TABLE 3.
Discrepant resolution by repeating illumigene M test or in-house qPCR to detect Plasmodium spp.
| Study IDa | Resultb |
||||
|---|---|---|---|---|---|
| Microscopy |
illumigene malaria test |
In-house qPCR | Final | ||
| Original | Repeat | ||||
| N02 | P. vivax/P. ovalec | NEG | POS | POS (P. ovale) | TP |
| N17 | P. vivax | NEG | POS | POS (P. vivax) | TP |
| N93 | NEG | POS | Not doned | POS (P. falciparum) | TP |
| N55 | NEG | POS | NEG | NEG | TN |
| S32 | NEG | POS | Not doned | POS (P. falciparum) | TP |
| S62 | NEG | POS | Not doned | POS (P. falciparum) | TP |
| N119 | P. ovale | Invalid | Invalid | POS (P. ovale) | Invalid |
ID, identifier.
NEG, negative; POS, positive; TP, true positive; TN, true negative.
P. vivax/P. ovale implies the microscopist was unable to distinguish the two species and gave a slash call result.
illumigene test was not repeated if the real-time qPCR agreed with original M kit result.
Resolution of discrepant results with illumigene MP kits.
There were 4 discrepancies between the results of the MP kit and microscopy, all due to false positives (Table 4). Of the 5 that were falsely positive, qPCR confirmed two were actually positive. Two of the falsely positive tests were negative on repeat testing, in line with the microscopy result, and one remained falsely positive due to insufficient sample volume to repeat the test.
TABLE 4.
Discrepant resolution by repeating illumigene MP test and in-house qPCR to detect Plasmodium spp.
| Study IDa | Resultb |
||||
|---|---|---|---|---|---|
| Microscopy |
illumigene malaria Plus test |
In-house qPCR | Final | ||
| Original | Repeat | ||||
| N118 | NEG | POS | Not donee | NEG | FP |
| N55 | NEG | POS | NEG | NEG | TN |
| N34 | NEG | POS | NEG | NEG | TN |
| S32 | NEG | POS | Not donec | POS (P. falciparum) | TP |
| S62 | NEG | POS | Not donec | POS (P. falciparum) | TP |
| N17 | P. vivax | Invalid | POS | Not doned | TP |
| N23 | P. vivax | Invalid | POS | Not doned | TP |
| N57 | NEG | Invalid | NEG | Not doned | TN |
| N59 | NEG | Invalid | NEG | Not doned | TN |
| N75 | NEG | Invalid | NEG | Not doned | TN |
| N80 | NEG | Invalid | Invalid | NEG | Invalid |
| N93 | NEG | Invalid | POS | POS (P. falciparum) | TP |
| S82 | P. ovale | Invalid | POS | Not doned | TP |
ID, identifier.
NEG, negative; POS, positive; TP, true positive; FP, false positive; TN, true negative.
illumigene test not repeated if real-time PCR agreed with original MP kit result.
qPCR was not done if illumigene MP repeat result was concordant with microscopy.
Insufficient sample volume to repeat test.
Resolution of invalid results.
Of note, there were 8/140 (5.7%) occurrences where the MP kit results were considered “invalid” by the instrument and 1/140 (<0.01%) invalid results on the M kit. When repeated from freshly thawed new aliquots as per the product insert, 7 were resolved with successful runs, while 1 sample remained invalid in each kit. An invalid result is where the reaction is completed and the final result is “invalid.” Both types of invalid results were resolved after repeat testing from a freshly thawed specimen with particular attention to sample mixing. The invalid rate was reduced to 1 in 140 (<0.01%) samples with both kits. Invalid results were not included in the initial sensitivity and specificity calculations.
Cost analysis.
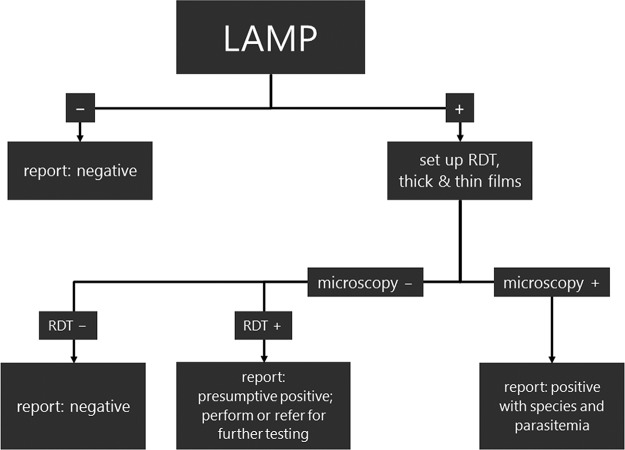
A detailed cost analysis (Table 5) was performed to evaluate the impact of a revised malaria testing algorithm (Fig. 1). The current algorithm relies on both thin and thick film microscopy coupled with RDT (BinaxNOW malaria test) to evaluate EDTA blood submitted for malaria testing with repeat microscopy on negatives. The RDT serves as an adjunct to confirm microscopic results. This algorithm is typical in the setting where malaria is not endemic. Costs were attributed to medical laboratory assistant and technologist times for preparing and reading smears and performing RDTs and to consumables and kits. The costs were based on current labor charges and benefits in our setting for 2017. The kit and consumable costs were based on approximate prices. The major finding is that by reducing the need for repeat microscopy on negative initial results (3 times every 6 to 8 h), labor costs are reduced dramatically. Therefore, overall, in spite of an increase in material costs with the LAMP algorithm, a per-patient cost savings of US$13.19 is obtained.
TABLE 5.
Cost analysis performed on current malaria testing algorithm at our institution compared to a proposed workflow where LAMP is conducted as the initial screena
| Malaria testing algorithm | No. of specimens per year | MLA FTE | MLT FTE | Cost (US$) |
|||
|---|---|---|---|---|---|---|---|
| Yearly |
Per specimen | ||||||
| Materials | Labor | Total | |||||
| Current | |||||||
| Malaria thick and thick film microscopy and RDT to screen, repeat thick and thin film microscopy on negatives | 1,776 | 0.03 | 0.98 | 12,497.06 | 82,531.76 | 95,029.06 | 53.51 |
| Proposedb | |||||||
| LAMP to screen and report negatives as final, work-up for positives only with RDT and thick and thin film microscopy | 1,440 | 0.02 | 0.24 | 35,947.06 | 22,118.82 | 58,065.88 | 40.32 |
| Cost/FTE differential | 336 | 0.01 | 0.74 | 23,450.00 | 60,412.94 | 36,963.18 | 13.19 |
| ↓ | ↓ | ⇊ | ⇈ | ⇊ | ⇊ | ↓ | |
All values are in US dollars as of April 2017. Yearly costs are based on 2017 estimates for labor and materials. MLA, medical laboratory assistant; MLT, medical laboratory technologist; FTE, full-time equivalent; RDT, rapid diagnostic test (BinaxNOW malaria test); ↓, decreased costs; ↑, increased costs. Assumptions made include 4 thick and thick films made per patient and 3 microscopy repeat tests performed per patient when negative. Testing from patients with an initial positive microscopy was also repeated until parasitemia cleared. A positivity rate of less than 5% is assumed.
See Fig. 1 for proposed workflow.
FIG 1.
A proposed algorithm for malaria testing using loop-mediated amplification (LAMP) technology as a screen is presented. In the new algorithm, no repeat testing is required to rule out malaria due to the excellent sensitivity. A microbiologist consultation is recommended when LAMP results are positive and results for other tests are negative. Positives require confirmation for species and parasitemia using traditional methods. In a setting where malaria is not endemic, positives are rare and thus a cost-savings is projected due to the reduced labor associated with making thick and thin peripheral blood films. Validation of this proposed algorithm would require a prospective trial design.
DISCUSSION
We have conducted the first diagnostic study in North America with illumigene Malaria—a commercially available LAMP-based malaria test—on returning travelers. LAMP-based methods for the detection of malaria have been available on a research-use-only basis for over a decade (28). Results from studies performed both in settings where malaria is endemic and where it is not endemic have suggested that LAMP is more sensitive than rapid diagnostic tests (RDTs) such as the BinaxNOW malaria test, especially for the non-falciparum species (13, 17, 30, 35). LAMP appears to perform on an equal footing with other molecular techniques such as nested PCR (35). LAMP may be especially useful for the screening of low-level parasitemia due to its analytical sensitivity (17). LAMP has the distinct advantage of relying on a strand-displacing enzyme that performs at a single temperature (usually 62°C to 65°C), tolerates less-stringent nucleic acid extraction methods, and can be detected with simple turbidity measurements (25). At least two LAMP kits are now commercially available for malaria diagnosis, the Loopamp malaria “Pan/Pf” detection kit (Eiken Chemical Co. Japan) and the illumigene malaria assay tested here, both targeting mitochondrial DNA of Plasmodium spp. for genus-level identification. Of note, the Loopamp malaria Pan/Pf detection kit can distinguish P. falciparum (using 18S rRNA) from other species, whereas the illumigene malaria test cannot, and has demonstrated excellent sensitivity in published studies (13, 30, 34). Identifying P. falciparum is clinically useful because of this organism's high pathogenic potential. Both kits rely on lyophilized reagents that enable these assays to be performed without the refrigeration of reagents, a major advantage in areas where malaria is endemic. The illumigene assay comes in two versions: the M kit, which relies on simple lysis filtration for extraction, and the MP kit, which has an extra gravity-flow column. Neither format requires centrifugation or thermal treatment, which again is a major advantage in situations where sophisticated equipment is a barrier. The only published study to date of the illumigene malaria assay is by Lucchi and colleagues (34), who demonstrated a sensitivity of 97.2% for both extraction methods and a specificity of 87.7% with the simplified extraction (M kit) and a specificity of 93.8% for the MP kit in comparison to a PCR gold standard. The work was performed in a clinical laboratory in Senegal (a resource-limited setting in Africa where malaria is endemic). Their study also showed no difference in performance between banked retrospective specimens and fresh specimens when using the illumigene malaria assay.
Our study was performed in a clinical laboratory in North America using fresh-frozen specimens from returning travelers. All species except P. knowlesi are represented in our sample set. Our results demonstrate that excellent sensitivity was achieved by both simplified (M, 97.3%) and gravity-flow (MP, 100%) extraction kits in comparison to that of reference microscopy. A high invalid rate of 5.7% was observed with the MP kit. The reasons for this are unclear, but we speculate this may have to do with the resin beds and filters that may be disturbed during transportation, affecting performance and/or the amount of heme present in the thawed sample used here. We noted that many of our frozen specimens had considerable erythrocyte lysis. A prospective study using fresh specimens may shed light on this issue. Repeat testing with the same assay lot and instrument resolved 7 of 8 MP kit invalid results and gave the correct identification in relation to that from qPCR, which suggests the technologist's level of training may also play a factor.
Both M (99.8%) and MP (100%) kits achieved excellent negative predictive values, assuming a 5% prevalence in returning febrile travelers. False positives were noted with both M and MP kits in comparison to that with microscopy, even after discrepant analysis with qPCR. While contamination has been suggested to be a significant risk with LAMP technology, this was not observed in our study with strict adherence to decontamination and a one-way flow format, where closed-tube reaction products are discarded in a location different from where the extraction takes place (15). The assay is simple to perform and can be completed in 60 min. By comparison, the malaria RDTs used in our laboratory (BinaxNOW malaria test; Alere, Waltham, MA) can be completed in 25 min. The excellent sensitivity and negative predictive value in a setting of low prevalence and the ease of performance suggest that this test may be best used as a screen for malaria. Positives would require further confirmation with microscopy given the lower positive predictive value and inability to determine the identity to the species level or to quantify infections. Our previous study demonstrated that LAMP methodology is superior to that of RDTs, especially in terms of sensitivity—RDT sensitivity was on the order of 70% for non-falciparum species and 90% for P. falciparum in a similar archived sample set (35). RDTs rely on antigens which vary between species and thus affect test performance. Deletions in key genes encoding diagnostic antigens are a growing concern (12).
Limitations of this study include the retrospective nature of the design. Discrepant resolution was performed only on a subset of specimens using qPCR, which may introduce bias. Frozen samples may not perform in the same way as fresh samples, although our in-house experiments suggest no loss in signal after four freeze-thaw cycles with both kits, even at a parasitemia of <0.1%. Other limitations include not having evaluated the impact of “empty well” errors, instrument errors, and invalid results in real time. Delays due to repeat testing could be significant, as physicians expect a rapid turnaround (<2 h) for this critical test. Furthermore, we did not have P. knowlesi specimens to test in our bank. Determinations of whether an infection results from pathogens at a specific sexual stage (gametocytes) cannot be made with the assay evaluated here. Therefore, microscopy is still required. Infections with gametocytes only do not require treatment and often reflect previous antimalarial therapy.
For LAMP, the major outstanding question is whether a negative result with LAMP implies no further testing is required. Lucchi and colleagues obtained a limit of detection of 2 parasites/μl with the illumigene malaria assay, which is far superior to those of microscopy and RDT and is in keeping with other reports on LAMP technology (22, 27–32, 34, 41). In our setting, a highly sensitive screen, excellent LOD, and ease of use would mean no repeat testing is required and no peripheral blood film examination is required on over 95% of our specimens, based on current positivity rates. Our current algorithm for malaria testing relies on 4 thick and thin slides being made together with a rapid diagnostic (BinaxNOW malaria) test for testing. The RDT result is released as a preliminary result until thin and thick films are read for final confirmation of species and parasitemia. Initial negatives are repeated every 6 to 8 h on average 3 times to ensure no parasites are seen, as per CDC recommendations (36). While this is prudent given the limitations of microscopy, this adds a significant labor cost.
Given the significant gain in analytical sensitivity over RDTs and microscopy reported in the literature, we proposed an alternative algorithm for malaria screening in a setting where malaria is not endemic (Fig. 1). In a four-year review conducted in our jurisdiction, a sensitivity of 90.4% (150/166) for the detection of P. falciparum was achieved using our current algorithm (microscopy and RDT) compared with that from qPCR (8). This poses a significant mortality risk given the risk of fulminant disease with this species. The LAMP approach we propose, with its ease of sample preparation, rapid result (60 min), and improved negative predictive value compared with that from traditional RDTs, would be used as a screen and reported as a final result if it is negative and a preliminary genus-level result if it is positive. Presumptive positives (<5% of samples in most settings where malaria is not endemic) would then be confirmed with thin and thick film microscopy for determination of species and parasitemia. A rapid diagnostic test can also be used on positives if microscopy expertise is not present to distinguish P. falciparum from other species. If the revised algorithm were to be implemented, this would amount to a significant cost savings of US$13.19 per malaria test in the setting where malaria is not endemic. A prospective trial in this regard would be of great value in cementing the utility of this approach, resolving the issue of repeat testing and obtaining regulatory approval for use of this assay in North America.
MATERIALS AND METHODS
Patient population and sample collection.
Calgary Laboratory Services (CLS) collected samples and performed all of the described malaria testing. CLS is a large centralized microbiology laboratory service covering 1.3 million people. All of the previously archived stored study samples (n = 140 blood samples) were collected between 2003 and 2014 from individual returning travelers presenting in our health care region with acute febrile symptoms soon after being in a region where malaria is endemic. Specimens were stored at −80°C. The demographic details and countries visited by our catchment population have been described previously (37). In brief, the majority of patients were in the 20 to 44 years age group and were visiting friends and relatives, with the majority of P. falciparum cases imported from sub-Saharan Africa and P. vivax cases from the Indian subcontinent. Table 1 summarizes the malaria species present in this sample set. P. falciparum and P. vivax are the predominant species, followed by P. ovale and P. malariae. The relative contribution of each species is typical of most North American laboratories. No P. knowlesi was present in the sample set, although the assay is designed to detect this fifth species. Blood aliquot samples were stored in cryovials at −80°C until being enrolled into this study.
Microscopy and illumigene malaria assay.
Microscopy was performed using standard Giemsa-stained thick and thin peripheral films at the time of initial diagnosis (38). Aliquots of freshly thawed venous EDTA whole blood were tested with the illumigene malaria (M) and illumigene malaria PLUS (MP) (Meridian Bioscience, Cincinnati, OH) assays, which are CE marked and Health Canada-approved for diagnostic use. Both LAMP assays use different proprietary centrifugation-free techniques for DNA extraction that are included as part of their kit: either a simple filtration method (M assay) or a gravity-driven gel filtration column (MP assay). Amplification is carried out using the illumipro-10 incubator/reader that detects the production of magnesium pyrophosphate, which in turn increases the turbidity of the samples. The M/MP LAMP assays detect the presence or absence of Plasmodium spp., but cannot differentiate to the species level. The manufacturer claims increased analytical sensitivity with the MP assay (0.063 P. vivax parasites/μl and 0.25 P. falciparum parasites/μl) versus the M assay (0.125 P. vivax parasites/μl and 2 P. falciparum parasites/μl). The product insert reports a sensitivity of 100% (M and MP kits) and specificities of 89.3% (M kit) and 82.7% (MP kit) versus microscopy. The assay has only been verified on purified genomic DNA of P. knowlesi. M and MP LAMP assays were run in parallel on single blood samples strictly according to the manufacturer's instructions. According to the manufacturer's product insert, if the illumipro-10 has an “empty well” error on the instrument panel prior to amplification, an “invalid” result reported after the amplification, or “instrument error” result, then the specimen should be reextracted from the beginning and rerun.
Resolution of discrepancies.
Discrepant tests occurred when the initial results of microscopy and either the M/MP LAMP assay results were different. If an invalid, empty well, or instrument error occurred, repeat M/MP LAMP assays were performed using another frozen aliquot of the patient's blood sample. A fully validated in-house real-time PCR (qPCR) method was subsequently used to verify discrepant results that were not resolved by repeat LAMP testing (9, 39). The qPCR assay has a sensitivity and specificity of 100% in monoinfections (27). Primers and probes target the 18S rRNA gene and comprise a genus-specific target coupled with a species-specific target which enables identification to the species level (27). The assay has been further optimized for the detection of P. ovale and P. malariae as described previously (40).
Cost analysis.
A cost comparison was performed for our current testing algorithm, which includes microscopy and rapid diagnostic tests (BinaxNOW malaria test), with a proposed algorithm with LAMP as the screening test (Fig. 1). The assumption made is that LAMP with its negative predictive value approaching 100% would permit a final negative result to be reported. Only positives would require further work-up by microscopy for identification to the species level and quantitative parasitemia. In our current algorithm, negative initial microscopy results are repeated every 6 to 8 h up to a maximum of 3 times as per CDC recommendations (36).
Statistical analysis.
Data were entered into Microsoft Excel (Microsoft Professional Office 2010; Microsoft, Seattle, WA), and diagnostic test performance was determined by standard statistical methods executed using SISA online statistical software (http://www.quantitativeskills.com/sisa/).
ACKNOWLEDGMENTS
Kits for this study were kindly provided by Somagen Canada. We also thank Gisele Peirano and Abu Naser Mohon who assisted with sample collection.
The study was partly funded by Calgary Laboratory Services, Calgary, Canada.
REFERENCES
- 1.World Health Organization. Malaria. World Health Organization, Geneva, Switzerland: http://www.who.int/malaria/en/ Accessed 6 August 2017. [Google Scholar]
- 2.Boggild AK, Geduld J, Libman M, Yansouni CP, McCarthy AE, Hajek J, Ghesquiere W, Vincelette J, Kuhn S, Freedman DO, Kain KC. 2016. Malaria in travellers returning or migrating to Canada: surveillance report from CanTravNet surveillance data, 2004–2014. CMAJ Open 4:E352–E358. doi: 10.9778/cmajo.20150115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Public Health Agency of Canada. About CATMAT. Public Health Agency of Canada, Ottawa, Canada: http://www.phac-aspc.gc.ca/tmp-pmv/catmat-ccmtmv/index-eng.php Accessed 6 August 2017. [Google Scholar]
- 4.McCarthy AE, Morgan C, Prematunge C, Geduld J. 2015. Severe malaria in Canada, 2001–2013. Malar J 14:151. doi: 10.1186/s12936-015-0638-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khuu D, Eberhard ML, Bristow BN, Javanbakht M, Ash LR, Shafir SC, Sorvillo FJ. 2017. Malaria-related hospitalizations in the United States, 2000–2014. Am J Trop Med Hyg 97:213–221. doi: 10.4269/ajtmh.17-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulletin of the World Health Organization. 1988. Malaria diagnosis: memorandum from a WHO meeting. Bull World Health Organ 66:575–594. [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization. 2000. New perspectives in malaria diagnosis: report of a joint WHO/USAID informal consultation. 25 to 27 October 1999, document no WHO/CDS/RBM/2000.14;WHO/MAL/2000.1091. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 8.Shokoples S, Mukhi SN, Scott AN, Yanow SK. 2013. Impact of routine real-time PCR testing of imported malaria over 4 years of implementation in a clinical laboratory. J Clin Microbiol 51:1850–1854. doi: 10.1128/JCM.00195-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khairnar K, Martin D, Lau R, Ralevski F, Pillai DR. 2009. Multiplex real-time quantitative PCR, microscopy and rapid diagnostic immuno-chromatographic tests for the detection of Plasmodium spp: performance, limit of detection analysis and quality assurance. Malar J 8:284. doi: 10.1186/1475-2875-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Snounou G, Viriyakosol S, Zhu XP, Jarra W, Pinheiro L, do Rosario VE, Thaithong S, Brown KN. 1993. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol 61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- 11.Hofmann N, Mwingira F, Shekalaghe S, Robinson LJ, Mueller I, Felger I. 2015. Ultrasensitive detection of Plasmodium falciparum by amplification of multicopy subtelomeric targets. PLoS Med 12:e1001788. doi: 10.1371/journal.pmed.1001788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maltha J, Gillet P, Jacobs J. 2013. Malaria rapid diagnostic tests in endemic settings. Clin Microbiol Infect 19:399–407. doi: 10.1111/1469-0691.12151. [DOI] [PubMed] [Google Scholar]
- 13.Polley SD, Gonzalez IJ, Mohamed D, Daly R, Bowers K, Watson J, Mewse E, Armstrong M, Gray C, Perkins MD, Bell D, Kanda H, Tomita N, Kubota Y, Mori Y, Chiodini PL, Sutherland CJ. 2013. Clinical evaluation of a loop-mediated amplification kit for diagnosis of imported malaria. J Infect Dis 208:637–644. doi: 10.1093/infdis/jit183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel JC, Lucchi NW, Srivastava P, Lin JT, Sug-Aram R, Aruncharus S, Bharti PK, Shukla MM, Congpuong K, Satimai W, Singh N, Udhayakumar V, Meshnick SR. 2014. Field evaluation of a real-time fluorescence loop-mediated isothermal amplification assay, RealAmp, for the diagnosis of malaria in Thailand and India. J Infect Dis 210:1180–1187. doi: 10.1093/infdis/jiu252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sema M, Alemu A, Bayih AG, Getie S, Getnet G, Guelig D, Burton R, LaBarre P, Pillai DR. 2015. Evaluation of non-instrumented Nucleic acid amplification by loop-mediated isothermal amplification (NINA-LAMP) for the diagnosis of malaria in Northwest Ethiopia. Malar J 14:44. doi: 10.1186/s12936-015-0559-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tegegne B, Getie S, Lemma W, Mohon AN, Pillai DR. 2017. Performance of loop-mediated isothermal amplification (LAMP) for the diagnosis of malaria among malaria suspected pregnant women in Northwest Ethiopia. Malar J 16:34. doi: 10.1186/s12936-017-1692-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aydin-Schmidt B, Xu W, Gonzalez IJ, Polley SD, Bell D, Shakely D, Msellem MI, Bjorkman A, Martensson A. 2014. Loop-mediated isothermal amplification (LAMP) accurately detects malaria DNA from filter paper blood samples of low density parasitaemias. PLoS One 9:e103905. doi: 10.1371/journal.pone.0103905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aydin-Schmidt B, Morris U, Ding XC, Jovel I, Msellem MI, Bergman D, Islam A, Ali AS, Polley S, Gonzalez IJ, Martensson A, Bjorkman A. 2017. Field evaluation of a high-throughput loop mediated isothermal amplification test for the detection of asymptomatic plasmodium infections in Zanzibar. PLoS One 12:e0169037. doi: 10.1371/journal.pone.0169037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris U, Khamis M, Aydin-Schmidt B, Abass AK, Msellem MI, Nassor MH, Gonzalez IJ, Martensson A, Ali AS, Bjorkman A, Cook J. 2015. Field deployment of loop-mediated isothermal amplification for centralized mass-screening of asymptomatic malaria in Zanzibar: a pre-elimination setting. Malar J 14:205. doi: 10.1186/s12936-015-0731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook J, Aydin-Schmidt B, Gonzalez IJ, Bell D, Edlund E, Nassor MH, Msellem M, Ali A, Abass AK, Martensson A, Bjorkman A. 2015. Loop-mediated isothermal amplification (LAMP) for point-of-care detection of asymptomatic low-density malaria parasite carriers in Zanzibar. Malar J 14:43. doi: 10.1186/s12936-015-0573-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdul-Ghani R, Al-Mekhlafi AM, Karanis P. 2012. Loop-mediated isothermal amplification (LAMP) for malarial parasites of humans: would it come to clinical reality as a point-of-care test? Acta Trop 122:233–240. doi: 10.1016/j.actatropica.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 22.Mohon AN, Elahi R, Khan WA, Haque R, Sullivan DJJ, Alam MS. 2014. A new visually improved and sensitive loop mediated isothermal amplification (LAMP) for diagnosis of symptomatic falciparum malaria. Acta Trop 134:52–57. doi: 10.1016/j.actatropica.2014.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Britton S, Cheng Q, Grigg MJ, Poole CB, Pasay C, William T, Fornace K, Anstey NM, Sutherland CJ, Drakeley C, McCarthy JS. 2016. Sensitive detection of Plasmodium vivax using a high-throughput, colourimetric loop mediated isothermal amplification (HtLAMP) platform: a potential novel tool for malaria elimination. PLoS Negl Trop Dis 10:e0004443. doi: 10.1371/journal.pntd.0004443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perera RS, Ding XC, Tully F, Oliver J, Bright N, Bell D, Chiodini PL, Gonzalez IJ, Polley SD. 2017. Development and clinical performance of high-throughput loop-mediated isothermal amplification for detection of malaria. PLoS One 12:e0171126. doi: 10.1371/journal.pone.0171126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. 2000. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res 28:E63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, Kachur SP, Barnwell JW, Udhayakumar V. 2010. Real-time fluorescence loop-mediated isothermal amplification for the diagnosis of malaria. PLoS One 5:e13733. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han E-T, Watanabe R, Sattabongkot J, Khuntirat B, Sirichaisinthop J, Iriko H, Jin L, Takeo S, Tsuboi T. 2007. Detection of four Plasmodium species by genus- and species-specific loop-mediated isothermal amplification for clinical diagnosis. J Clin Microbiol 45:2521–2528. doi: 10.1128/JCM.02117-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poon LLM, Wong BWY, Ma EHT, Chan KH, Chow LMC, Abeyewickreme W, Tangpukdee N, Yuen KY, Guan Y, Looareesuwan S, Peiris JSM. 2006. Sensitive and inexpensive molecular test for falciparum malaria: detecting Plasmodium falciparum DNA directly from heat-treated blood by loop-mediated isothermal amplification. Clin Chem 52:303–306. doi: 10.1373/clinchem.2005.057901. [DOI] [PubMed] [Google Scholar]
- 29.Polley SD, Mori Y, Watson J, Perkins MD, Gonzalez IJ, Notomi T, Chiodini PL, Sutherland CJ. 2010. Mitochondrial DNA targets increase sensitivity of malaria detection using loop-mediated isothermal amplification. J Clin Microbiol 48:2866–2871. doi: 10.1128/JCM.00355-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hopkins H, Gonzalez IJ, Polley SD, Angutoko P, Ategeka J, Asiimwe C, Agaba B, Kyabayinze DJ, Sutherland CJ, Perkins MD, Bell D. 2013. Highly sensitive detection of malaria parasitemia in a malaria-endemic setting: performance of a new loop-mediated isothermal amplification kit in a remote clinic in Uganda. J Infect Dis 208:645–652. doi: 10.1093/infdis/jit184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buates S, Bantuchai S, Sattabongkot J, Han E-T, Tsuboi T, Udomsangpetch R, Sirichaisinthop J, Tan-ariya P. 2010. Development of a reverse transcription-loop-mediated isothermal amplification (RT-LAMP) for clinical detection of Plasmodium falciparum gametocytes. Parasitol Int 59:414–420. doi: 10.1016/j.parint.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Lau Y-L, Fong M-Y, Mahmud R, Chang P-Y, Palaeya V, Cheong F-W, Chin L-C, Anthony CN, Al-Mekhlafi AM, Chen Y. 2011. Specific, sensitive and rapid detection of human Plasmodium knowlesi infection by loop-mediated isothermal amplification (LAMP) in blood samples. Malar J 10:197. doi: 10.1186/1475-2875-10-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemleu S, Guelig D, Eboumbou Moukoko C, Essangui E, Diesburg S, Mouliom A, Melingui B, Manga J, Donkeu C, Epote A, Texier G, LaBarre P, Burton R, Ayong L. 2016. A field-tailored reverse transcription loop-mediated isothermal assay for high sensitivity detection of Plasmodium falciparum infections. PLoS One 11:e0165506. doi: 10.1371/journal.pone.0165506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucchi NW, Gaye M, Diallo MA, Goldman IF, Ljolje D, Deme AB, Badiane A, Ndiaye YD, Barnwell JW, Udhayakumar V, Ndiaye D. 2016. Evaluation of the Illumigene Malaria LAMP: a robust molecular diagnostic tool for malaria parasites. Sci Rep 6:36808. doi: 10.1038/srep36808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mohon AN, Lee LD-Y, Bayih AG, Folefoc A, Guelig D, Burton RA, LaBarre P, Chan W, Meatherall B, Pillai DR. 2016. NINA-LAMP compared to microscopy, RDT, and nested PCR for the detection of imported malaria. Diagn Microbiol Infect Dis 85:149–153. doi: 10.1016/j.diagmicrobio.2015.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centers for Disease Control and Prevention. Treatment of malaria: guidelines for clinicians(United States). Centers for Disease Control and Prevention, Atlanta, GA: https://www.cdc.gov/malaria/diagnosis_treatment/clinicians1.html. Accessed 6 August 2017. [Google Scholar]
- 37.Lee CS, Gregson DB, Church D, Laupland KB, Eckhardt R, Ross T, Chan W, Pillai DR. 2013. Population-based laboratory surveillance of imported malaria in metropolitan Calgary, 2000–2011. PLoS One 8:e60751. doi: 10.1371/journal.pone.0060751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia L. 2007. Diagnostic medical parasitology, 5th ed ASM Press, Washingtion, DC. [Google Scholar]
- 39.Shokoples SE, Ndao M, Kowalewska-Grochowska K, Yanow SK. 2009. Multiplexed real-time PCR assay for discrimination of Plasmodium species with improved sensitivity for mixed infections. J Clin Microbiol 47:975–980. doi: 10.1128/JCM.01858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phuong M, Lau R, Ralevski F, Boggild AK. 2014. Sequence-based optimization of a quantitative real-time PCR assay for detection of Plasmodium ovale and Plasmodium malariae. J Clin Microbiol 52:1068–1073. doi: 10.1128/JCM.03477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Britton S, Cheng Q, McCarthy JS. 2016. Novel molecular diagnostic tools for malaria elimination: a review of options from the point of view of high-throughput and applicability in resource limited settings. Malar J 15:88. doi: 10.1186/s12936-016-1158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]