ABSTRACT
The global spread and infective complications of Zika virus (ZKV) and dengue virus (DENV) have made them flaviviruses of public health concern. Serological diagnosis can be challenging due to antibody cross-reactivity, particularly in secondary flavivirus infections or when there is a history of flavivirus vaccination. The virus neutralization assay is considered to be the most specific assay for measurement of anti-flavivirus antibodies. This study describes an assay where the neutralization endpoint is measured by real-time PCR, providing results within 72 h. It demonstrated 100% sensitivity (24/24 ZKV and 15/15 DENV) and 100% specificity (11/11 specimens) when testing well-characterized sera. In addition, the assay was able to determine the correct DENV serotype in 91.7% of cases. The high sensitivity and specificity of the real-time PCR neutralization assay makes it suitable to use as a confirmatory test for sera that are reactive in commercial IgM/IgG enzyme immunoassays. Results are objective and the PCR-based measurement of the neutralization endpoint lends itself to automation so that throughput may be increased in times of high demand.
KEYWORDS: dengue, flavivirus, serology, Zika
INTRODUCTION
Zika virus (ZKV) and dengue virus (DENV) are members of the family Flaviviridae, genus Flavivirus, which also includes yellow fever, Japanese encephalitis, and West Nile viruses. There are at least 4 closely related but serologically distinct DENV, designated DENV serotypes 1 through 4 (1). There is only transient and weak cross-protection among the 4 DENV serotypes, so individuals living in an area where DENV is endemic can be infected up to 4 times (2).
ZKV and DENV have spread across the tropics in Africa, Asia, the Pacific, and the Americas with their vector mosquitoes Aedes aegypti and A. albopictus (3, 4). Cocirculation of ZKV and DENV types 1 through 4 has been documented in both French Polynesia and Brazil (5, 6) and likely occurs in many other tropical countries. Incursions into temperate countries have also occurred, with autochthonous transmission observed in Europe (DENV in southeastern France, Croatia, and Madeira Island) and the United States (DENV and ZKV in Florida) (7–9).
Both ZKV and DENV infection can produce a wide spectrum of illnesses with many asymptomatic or subclinical infections (10, 11). ZKV's potential to cause fetal central nervous system abnormalities, growth restriction, and death (12) has necessitated ZKV testing of potentially exposed pregnant women, among whom nonspecific serological activity may occur, and asymptomatic individuals with low pretest probabilities of positive ZKV serology.
Direct detection of ZKV nucleic acids has limited utility in asymptomatic patients, as the periods of viremia and shedding in urine and saliva are relatively brief, occurring shortly after the onset of symptoms, if present (13–15). Consequently, we must rely on serology to diagnose infection in asymptomatic patients. A number of commercial enzyme immunoassays (EIAs) have been designed to detect IgM and IgG to ZKV and DENV. However, they are known to lack specificity, particularly when it comes to IgG. This tends to be more of a concern during secondary flavivirus infections, when there can be extensive IgG cross-reactivity with other flaviviruses, and in some cases, higher serological activity to the original infecting flavivirus (16, 17). Cross-reactivity can also occur in primary infection, particularly if there is a history of yellow fever, Japanese encephalitis, or tick-borne encephalitis virus vaccination (18, 19).
Virus neutralization assay, most commonly in the form of a plaque reduction neutralization test (PRNT), is considered the most specific assay for measurement of anti-flavivirus antibodies (20). It detects the production of antibodies that functionally inhibit viral infection; significant plaque reduction is generally considered to have occurred when there is a 90% reduction in the number of viral plaques (PRNT90) (20). In recent serological studies of ZKV the serological criteria for ZKV infection included a positive IgM ZKV EIA, ZKV PRNT90 titers ≥20, and a ZKV PRNT90/DENV PRNT90 ratio ≥4 (11, 21). PRNT has a number of limitations, being expensive and labor intensive, requiring skilled scientists and a cell culture facility. It also has a turnaround time of up to 10 days and so does not allow for a real-time diagnosis.
The aims of this study were to develop and validate ZKV and DENV neutralization assays using real-time PCR to measure viral neutralization by antibody, thereby providing a faster turnaround time and developing an assay that is both more objective and scalable.
RESULTS
Optimization of stock virus-Vero cell incubation time.
The duration of incubation of the virus-serum mix and Vero cell monolayer was the main determinant of the turnaround time for the real-time PCR neutralization assay. ZKV and DENV 1 to 4 stock viruses demonstrated adequate viral replication, as measured by real-time PCR, after 48 and 72 h incubation periods, but not 24 h (data not shown). An incubation period of 48 h was chosen as it provided a shorter assay turnaround time.
Example of the ZKV real-time PCR neutralization assay.
An example of the real-time PCR neutralization assay results and analysis for ZKV is shown in Table 1. A convalescent-phase serum specimen collected after PCR-confirmed diagnosis of ZKV infection was tested against ZKV stock virus. Viral replication in the real-time PCR neutralization assay resulted in a real-time PCR cycle threshold that was lower than the background, nonreplicating cycle threshold. Inhibition of viral replication was demonstrated by a real-time PCR cycle threshold that was equal to or greater than the background cycle threshold. The neutralizing antibody titer was the reciprocal of the highest serum dilution, resulting in inhibition of virus replication compared to nonreplicating viral background. Results were considered valid when the virus back titration confirmed that approximately 100 infectious virus particles were present in the neutralization assay (virus nucleic acids were detectable in back titration dilutions of 1/10 and 1/100 but undetectable at a dilution of 1/1000) and when positive and negative real-time PCR controls gave expected results (data not shown).
TABLE 1.
Real-time PCR neutralization assay for ZKV and a convalescent-phase serum specimen collected following a ZKV infection diagnosis by PCRa
| Serum dilution | ZKV-positive serumb | ZKV-negative serum |
|---|---|---|
| 1/20 | 35.4 | 27.1 |
| 1/40 | 35.2 | 28.1 |
| 1/80 | 35.7 | 28.8 |
| 1/160 | 35.2 | 27.0 |
| 1/320 | 35.7 | 26.8 |
| 1/640 | 29.5 | 29.1 |
| 1/1,280 | 29.8 | 29.2 |
| 1/2,560 | 28.6 | 28.7 |
| Background | 33.1 |
There is neutralizing antibody to ZKV in the known positive specimen (titer, 320) but not in the negative specimen (titer, <20).
Real-time PCR cycle thresholds for each neutralization between the known positive serum, a negative serum specimen, and the ZKV stock virus at working dilution. The presence of neutralizing antibody is demonstrated by a cycle threshold greater than or equal to the background cycle threshold (in bold).
Sensitivity of the real-time PCR-based neutralization assay.
Only sera that had been characterized by the gold-standard diagnostic techniques of real-time PCR and/or PRNT were used to assess the sensitivity of the real-time PCR neutralization assay. The results of the real-time PCR ZKV neutralization assay and DENV serotype-specific neutralization assay, using convalescent-phase sera from patients with proven infection, are shown in Tables 2 and 3, respectively. Neutralizing antibody of various titers was detected in all cases, resulting in a sensitivity of 100% for both ZKV and DENV. In addition, when the infecting DENV serotype was known the neutralization assay detected neutralizing antibody to the correct serotype, either alone or at ≥4-fold higher titer compared to other DENV serotypes, in 11 out of 12 tests (91.7%). The infecting DENV serotype was not able to be determined for sample 1 (Table 3).
TABLE 2.
Real-time PCR neutralization assay results for convalescent-phase sera from patients with proven ZKV infection compared to Institut Pasteur neutralization assay results
| Serum sample no.a | ZKV neutralization by |
|
|---|---|---|
| Real-time PCRb | Institut Pasteur assayc | |
| 1 | 640 | 2,560 |
| 2 | 160 | 160 |
| 3 | 640 | 640 |
| 4 | 640 | 1,280 |
| 5 | 1,280 | 1,280 |
| 6 | 320 | 320 |
| 7 | 640 | 1,280 |
| 8 | 160 | 320 |
| 9 | 160 | 320 |
| 10 | 320 | 640 |
| 11 | 640 | ≥2,560 |
| 12 | 320 | 640 |
| 13 | 1,280 | ≥2,560 |
| 14 | 1,280 | 1,280 |
| 15 | 80 | 80 |
| 16 | 320 | 320 |
| 17 | 320 | 320 |
| 18 | 640 | 1,280 |
| 19 | 80 | 80 |
| 20 | ≥2,560 | ≥2,560 |
| 21 | 320 | 320 |
| 22 | 160 | 640 |
| 23 | 320 | 1,280 |
| 24 | 160 | 160 |
Convalescent-phase serum from patient with PCR-proven ZKV infection.
Real-time PCR neutralization titer expressed as the reciprocal of the highest dilution of patient serum that resulted in neutralization of the stock virus.
Institut Pasteur neutralization titer expressed as the reciprocal of the highest dilution of patient serum that resulted in the absence of ZKV-specific cytopathic effect.
TABLE 3.
Real-time PCR neutralization assay results for convalescent-phase sera from patients with proven DENV infection
| Patient sample no. | DENV type as per real-time PCR | Time post PCR detection | Stock virusa |
|||
|---|---|---|---|---|---|---|
| DENV 1 | DENV 2 | DENV 3 | DENV 4 | |||
| 1 | 1 | 20 days | 160 | 80 | 40 | 20 |
| 2 | 1 | 24 days | 20 | <20 | <20 | <20 |
| 3 | 1 | 35 days | 160 | <20 | <20 | <20 |
| 4 | 1 | 18 mo | 40 | <20 | <20 | <20 |
| 5 | 2 | 12 days | <20 | 20 | <20 | <20 |
| 6 | 2 | 44 days | <20 | 320 | <20 | <20 |
| 7 | 2 | 4 mo | <20 | 320 | <20 | <20 |
| 8 | 2 | 6 mo | <20 | 640 | 20 | <20 |
| 9 | 3 | 12 days | <20 | <20 | 20 | <20 |
| 10 | 3 | 18 days | <20 | 20 | 160 | <20 |
| 11 | 3 | 2 years | <20 | <20 | 80 | <20 |
| 12 | 4 | 36 days | 20 | <20 | <20 | 160 |
| 13b | Undetermined | 5 years | 80 | <20 | <20 | <20 |
| 14b | Undetermined | — | 160 | 40 | 20 | <20 |
| 15b | Undetermined | — | 1,280 | 80 | 320 | 20 |
Real-time PCR neutralization titer expressed as the reciprocal of the highest dilution of patient serum that resulted in neutralization of the stock virus. The highest real-time PCR neutralization titer for each assay is indicated (in bold).
DENV serotype had not been determined previously for these specimens. When there was no history of nucleic acid confirmation of DENV infection (—), DENV-neutralizing antibody was detected using the PRNT.
Specificity of the real-time PCR-based neutralization assay.
The specificity of the neutralization assay was assessed using sera positive for a non-ZKV and/or non-DENV flavivirus, either by serological or molecular testing, collected from individuals with a clinical history of infection or vaccination (Table 4). A serum specimen positive for Chikungunya IgG antibodies was also included since this alphavirus is often a differential diagnosis to ZKV and DENV due to its similar geographic distribution and clinical presentation (5, 6, 22). The real-time PCR ZKV and DENV neutralization assays produced a neutralization titer of <20 in 11/11 cases (100%) when the serum was known to be negative for that virus.
TABLE 4.
Specificity testing of ZKV and DENV real-time PCR neutralization assays
| Infection or vaccination type and serum | Verification methodb | Neutralization titer with stock virusa |
||||
|---|---|---|---|---|---|---|
| ZKV | DENV 1 | DENV 2 | DENV 3 | DENV 4 | ||
| Flavivirus infection | ||||||
| ZKV | Real-time PCR | 1,280 | <20 | <20 | <20 | <20 |
| ZKV | Real-time PCR | 160 | <20 | <20 | <20 | <20 |
| DENV 1 | Real-time PCR | <20 | 80 | <20 | <20 | <20 |
| DENV 1 | Real-time PCR | <20 | 160 | 80 | 40 | 20 |
| DENV 2 | Real-time PCR | <20 | <20 | 320 | <20 | <20 |
| DENV 3 | Real-time PCR | <20 | <20 | 20 | 80 | <20 |
| DENV 4 | Real-time PCR | <20 | 20 | <20 | <20 | 160 |
| DENV type unknown | PRNT | <20 | 1,280 | 80 | 320 | 20 |
| Alphavirus infection | ||||||
| Chikungunya | Real-time PCR | <20 | <20 | <20 | <20 | <20 |
| Flavivirus vaccination | ||||||
| YF and JEVc | EIA | <20 | <20 | <20 | <20 | <20 |
| YF | EIA | <20 | <20 | <20 | <20 | <20 |
Neutralization titer expressed as the reciprocal of the highest dilution of patient serum that resulted in neutralization of the stock virus. The highest real-time PCR neutralization titer for each assay is indicated (in bold).
Real-time PCR performed on earlier acute-phase serum specimen. EIA, enzyme immunoassay.
YF, Yellow fever; JEV, Japanese encephalitis virus.
A third specimen from a patient with ZKV infection confirmed by PCR was tested but not included in the specificity data. This specimen was from a patient with likely secondary ZKV infection on a background of previous DENV infection. Cross-reactivity was shown in the gold-standard PRNT, where neutralizing antibody to both DENV and ZKV was detected. The serum specimen demonstrated cross-reactivity in the real-time PCR neutralization assays, with the detection of high-titer neutralizing antibody to both DENV (serotype 1 titer, 160; serotype 2 titer ≥2,560; serotype 3 titer, 160; and serotype 4 titer, 320) and ZKV (titer, 1,280). The highest titer of neutralizing antibody to DENV by ≥4-fold was to DENV serotype 2, making it the most likely previously infecting DENV serotype. These results demonstrate that the real-time PCR neutralization assay is not immune to the difficulties posed by secondary flavivirus infections.
Reproducibility of the real-time PCR-based neutralization assay.
Results were consistent when a selection of known positive serum specimens were tested in the neutralization assay against ZKV and DENV 1 to 4 stock viruses on multiple occasions over a minimum period of 6 months (Table 5).
TABLE 5.
Reproducibility of ZKV and DENV real-time PCR neutralization assays
| Virus type per real-time PCR | Day testedc | Stock virusa |
||||
|---|---|---|---|---|---|---|
| ZKV | DENV type 1 | DENV type 2 | DENV type 3 | DENV type 4 | ||
| ZKV | 1 | 1,280 | —b | — | — | — |
| 29 | ≥2,560 | — | — | — | — | |
| 147 | 1,280 | — | — | — | — | |
| 307 | 1,280 | — | — | — | — | |
| DENV 1 | 1 | — | 40 | <20 | <20 | <20 |
| 68 | — | 80 | <20 | <20 | <20 | |
| 150 | — | 40 | — | — | — | |
| 171 | — | 40 | — | — | — | |
| DENV 2 | 1 | — | <20 | 640 | <20 | <20 |
| 68 | — | <20 | 320 | <20 | <20 | |
| 150 | — | — | 320 | — | — | |
| 197 | — | — | 320 | — | — | |
| DENV 3 | 1 | — | <20 | 20 | 80 | <20 |
| 168 | — | <20 | <20 | 80 | <20 | |
| 194 | — | — | — | 80 | — | |
| DENV 4 | 1 | — | 20 | <20 | <20 | 160 |
| 35 | — | 20 | <20 | 20 | 320 | |
| 148 | — | — | — | — | 160 | |
| 194 | — | — | — | — | 160 | |
Real-time PCR neutralization titer expressed as the reciprocal of the highest dilution of patient serum that resulted in neutralization of the stock virus.
—, assay was not performed.
Relative to the first date tested, in days.
DISCUSSION
This study describes the development and validation of a real-time PCR-based neutralization assay for ZKV and DENV, where the neutralization endpoint is measured by real-time PCR assay instead of by the counting of viral plaques. Few studies have used PCR as an approach to measure virus neutralization assay endpoints (23–25). As far as we are aware, this is the first report of a real-time PCR-based neutralization assay being used in the diagnosis of a flavivirus infection.
The initial steps in the assay, including making virus and serum dilutions and transferring virus-serum mixtures onto cell monolayers, are currently performed manually and so remain relatively labor-intensive. However, the RNA extraction and real-time PCR steps can be automated, allowing throughput to be scaled up in periods of increased demand. In addition, the RNA extraction and real-time PCR protocols described in this study are already used as part of routine clinical diagnostics at the Victorian Infectious Diseases Reference Laboratory (VIDRL) and so the real-time PCR-based neutralization assay can be readily incorporated into the current laboratory work flow. Finally, the high sensitivity of PCR allows the detection of virus replication and the neutralization endpoint to be assessed at 48 h postinfection of the cell monolayer. This results in an overall test turnaround time of less than 72 h.
Real-time PCR amplifications curves were easy to interpret and comparison to background (nonreplicating virus) provided an objective way in which to determine the neutralizing antibody titer. The sensitivity and specificity data generated in this study suggest that the ZKV and DENV real-time PCR neutralization assays will be suitable as confirmatory tests applied to serum specimens that are reactive in an IgM/IgG EIA screen. The sensitivity of the real-time PCR neutralization assay was excellent at 100% for both ZKV and DENV. In addition, the DENV serotype could be correctly discerned in 91.7% of the cases where the infecting DENV serotype was known and in a majority of cases there was minimal or no reactivity with the other DENV serotypes. This provides confidence in reporting the DENV serotype when the neutralizing titer for one serotype is ≥4-fold higher than for the other three DENV serotypes. The specificity of the assay was 100% for ZKV and DENV, respectively, when testing sera from patients with no history of that particular flavivirus infection. A high specificity is important in a neutralization assay as its primary role is to confirm true-positive, and rule out false-positive, EIA results.
The real-time PCR neutralization assay, like PRNT, is expected to have reduced specificity in the setting of secondary flavivirus infections. Significant cross-reactivity was observed in the DENV and ZKV PCR-based neutralization assays on testing a serum specimen collected from a patient with proven current ZKV infection and a background of likely prior DENV infection. The dominant epitopes responsible for highly potent, serotype-specific humoral immunity to DENV appear to be located in the hinge region of the E glycoprotein (26, 27). However, a significant proportion of human anti-DENV antibodies seem to be serotype cross-reactive, with many directed to the prM/M protein and the fusion loop of the E protein (28, 29). During the acute phase of secondary infection, there is a high degree of serotype cross-reactivity among the antibodies produced by memory B cells (30). This means that, when measured by PRNT, high-titer neutralizing antibodies are produced against two, and often all four, DENV as well as against other non-DENV flaviviruses (17). The interaction of virus with serum-neutralizing antibody in the real-time PCR-based neutralization assay occurs in the same manner as in PRNT and so does not eliminate this issue.
Detection of flavivirus-neutralizing antibody activity by PRNT is generally thought to coincide with the detection of seroconversion by commercially available IgM and IgG EIAs (31). For primary ZKV and DENV infection, this is expected to occur from day 10 following the onset of illness (17, 32, 33). The earliest specimen from which DENV-neutralizing antibody was detected in this study was collected 12 days post positive PCR. However, the titer of neutralizing antibody does not appear to be very high at this stage of illness and collection of a convalescent specimen at least 1 month post onset of illness may provide more clear-cut results. A prospective study of children aged between 6 months and 15 years in Thailand showed that while the kinetics of neutralizing antibody produced following primary infection varied somewhat depending on infecting serotype, in general there was increase in neutralizing antibody titer from convalescence to 6 months postinfection (34). Increased antibody titers after this period were attributed to boosting of the immune response due to subsequent infection. This trend is borne out in the data presented in this study, with the highest titers detected between 1 month and 6 months post DENV-positive PCR. Boosting of antibody titers is unlikely to occur in a majority of the Australian population, where acquisition of DENV infection occurs primarily in the setting of travel to tropical countries.
One of the limitations of this study is the small number of samples used. This reflects limited access to well-characterized sera for an infection that is diagnosed relatively uncommonly in the southern Australian population. While our small sample size may have an impact on precision, the use of only well-characterized sera provides the optimal means to compare the real-time PCR neutralization assay to another gold-standard assay and so produce quality sensitivity and specificity data.
The emergence of ZKV has led to the need to diagnose infection in asymptomatic individuals that may be in the convalescent phase of infection. Diagnosis in this setting relies on the use of serology and has prompted the development of a more rapid, gold-standard serological platform for flavivirus diagnosis. It is envisaged that the DENV and ZKV real-time PCR neutralization assays will be performed alongside each other as a confirmatory step for serum specimens that are reactive for ZKV and/or DENV in commercial IgM/IgG EIAs. The demand for this type of testing is expected to continue as these arboviruses continue to spread globally. The real-time PCR-based approach to the virus neutralization assay should be generalizable to other viruses.
MATERIALS AND METHODS
Viruses and cell culture.
Vero cells were grown in 96-well culture plates in minimum essential medium (MEM; Sigma-Aldrich, St. Louis, MO) supplemented with 2% fetal bovine serum (Bovogen, VIC, Australia) (MEM-2).
The African lineage MR766 prototype ZKV strain was kindly provided by PathWest (WA, Australia). DENV serotypes 1 to 3 were isolated from patient specimens at the Victorian Infectious Diseases Reference Laboratory (VIDRL; VIC, Australia). DENV serotype 4 was obtained from the QIMR Berghofer Medical Research Institute (QLD, Australia). All viruses were amplified in Vero cells and MEM-2 to generate virus stocks. Extracellular virus was harvested when the observed cytopathic effect reached 4 or greater and the presence of high-titer virus was confirmed by real-time PCR. Virus stock was stored at −80°C in aliquots until use.
Virus stocks were titrated in 150 μl of MEM-2 and inoculated onto Vero cells. Culture supernatant was harvested at 48 h postinfection and virus assayed by real-time PCR. The endpoint dilution, the highest dilution of stock virus to result in detection of nucleic acid by real-time PCR, was determined for each virus (ZKV and DENV types 1 to 4). The neutralization assay virus working dilution, determined to deliver approximately 100 infective virus particles to each well, was then calculated as 200-fold more concentrated than this endpoint dilution.
Serum samples.
Deidentified human adult diagnostic sera collected between 2011 and 2016 and stored at −20°C were used to assess the performance of the neutralization assay. In order to define the sensitivity and specificity of the neutralization assay, only well-characterized sera were employed. All sera used to test the sensitivity of the neutralization assay had been confirmed as ZKV- or DENV-positive by real-time PCR on a previous specimen from the same patient or by PRNT at a reference laboratory (Pathology West, Westmead, NSW, Australia). Additionally, neutralization titers from the ZKV PCR-based neutralization assay were able to be compared to those produced in a ZKV neutralization assay performed at the Institut Pasteur, Noumea, New Caledonia, and adapted from Gallian et al. (35). Sera used to test the specificity of the real-time neutralization assay had been shown to be negative for the flavivirus of interest and positive for another arbovirus by real-time PCR, EIA, and/or PRNT.
The use of human serum samples was approved by the Melbourne Health Human Research Ethics Committee (QA2017026).
Virus RNA extraction and reverse transcription.
For RNA extraction, 150 μl of virus supernatant was mixed with 120 μl of lysis buffer (ATL buffer, Proteinase K; Qiagen, Hilden, Germany) and incubated at room temperature for 10 min. Total RNA was prepared using a robotic column extraction system (QIAcubeHT; Qiagen) according to the manufacturer's instructions.
Reverse transcription (RT) was performed using the SensiFAST cDNA synthesis kit (Bioline, London, UK) using a reduced reaction volume so that each reaction mixture contained 5 μl of extracted RNA, 2.5 μl of nuclease free water (Promega, Maddison, WI), 2 μl of TransAmp buffer, and 0.5 μl of reverse transcriptase. Reduced reaction volumes had previously been validated (36). RT was performed using the following conditions: 25°C for 10 min, 42°C for 15 min, 85°C for 5 min, and a 4°C hold. The cDNA samples were stored at 4°C until required.
ZKV and DENV serotype-specific real-time PCR.
The ZKV real-time PCR assay amplified a region of the NS5 gene using the forward primer CCTCAAGGAYGGGAGATCCA and reverse primer RGCTCGGCCAATCAGTTCAT, with detection using a TaqMan probe (TGGTCCCYTGCCGCCACCAA). The DENV real-time PCR assay comprised four singleplex TaqMan real-time PCRs using primers and probes for DENV types 1, 2, 3, and 4 and has been described previously (37).
Each real-time PCR mixture contained 3 μl of cDNA, 5.9 μl of nuclease-free water, 10 μl of 2× PerfeCTa qPCR FastMix (Quanta BioSciences, Beverly, MA), 0.36 μl virus-specific primer pool (final concentration, 0.9 μM), and 0.8 μl virus-specific TaqMan probe (final concentration, 0.2 μM). Individual reactions were performed in a 96-well plate using the 7500 Fast real-time PCR system instrument and software (Applied Biosystems, Carlsbad, CA) and the following conditions: 95°C for 20 s, followed by 45 cycles of 95°C for 3 s and 60°C for 30 s. ZKV and serotype-specific DENV positive controls and nucleic-acid-free negative controls were used to verify the performance of each PCR run.
Real-time PCR-based neutralization assay.
Serial 2-fold dilutions of patient sera were incubated with an equal volume of predetermined virus stock working dilution (calculated to deliver 100 infectious particles) for 2 h at 37°C. Subsequently, 150 μl of the virus/serum mixtures were transferred in duplicate to the corresponding wells of Vero cell monolayers and incubated for a further 2 h at 37°C. The supernatant was removed and replaced by 150 μl of MEM-2 and the Vero cell culture plate incubated at 37°C. After 48 h the supernatant was harvested and duplicates combined for subsequent virus RNA extraction, RT, and detection of virus by real-time PCR.
A background sample was included in each assay as a measure of nonreplicating virus background. This was performed using the working dilution of virus incubated on Vero cells in the absence of patient sera. In brief, after the working dilution of virus was incubated on the Vero cell monolayer for 2 h, the supernatant was removed and replaced by 150 μl of MEM-2. This volume of MEM-2 was then collected as the background sample and subjected to virus RNA extraction, RT, and real-time PCR, as described previously. The neutralization titer was determined as the highest serum dilution resulting in inhibition of virus replication compared to background. The inclusion of background samples in each assay also controlled for interassay variability in virus replication.
For each assay, a virus back titration was performed for ZKV and DENV types 1 to 4 in order to show that the amount of infectious virus added to each well was correct. This comprised addition of the virus stock to wells at the working dilution (approximately 100 infectious virus particles per well) and at 1/10, 1/100, and 1/1000 dilutions of the working dilution, in the absence of patient serum.
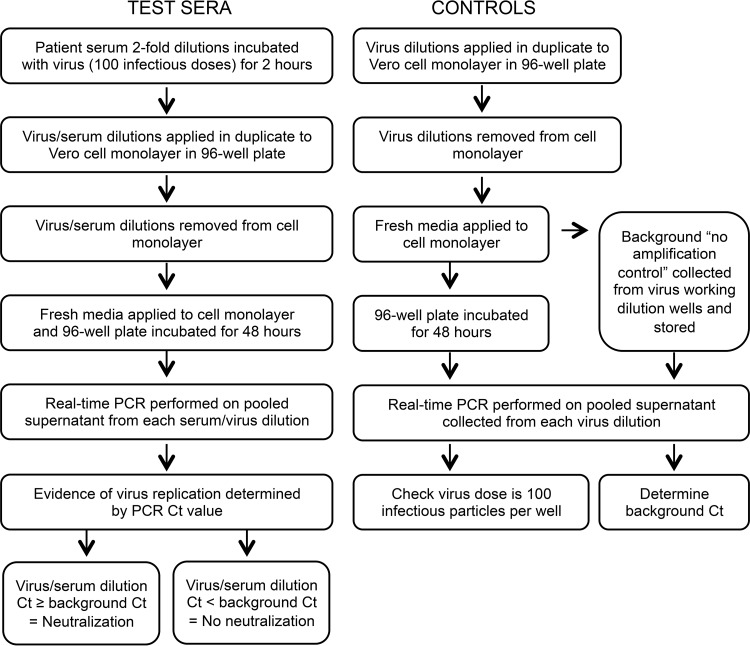
An algorithm demonstrating the steps of the real-time PCR neutralization assay is provided in Fig. 1.
FIG 1.
Real-time-PCR-based neutralization assay algorithm. Virus control dilutions included the working dilution (100 infectious virus particles per well) and 1/10, 1/100, and 1/1000 dilutions. CT, cycle threshold.
ACKNOWLEDGMENTS
Olivia O'Connor for her work performing ZKV neutralization testing and Ann-Claire Gourinat, both at Institut Pasteur, New Caledonia.
There are no potential conflicts of interest relevant to this article.
This research received no specific grant from any funding agency in the public, commercial, or not-for profit sectors.
REFERENCES
- 1.Henchal EA, Putnak JR. 1990. The dengue viruses. Clin Microbiol Rev 3:376–396. doi: 10.1128/CMR.3.4.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gubler DJ. 2002. Epidemic dengue/dengue haemorrhagic fever as a public health, social and economic problem in the 21st century. Trends Microbiol 10:100–103. doi: 10.1016/S0966-842X(01)02288-0. [DOI] [PubMed] [Google Scholar]
- 3.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GRW, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petersen LR, Jamieson DJ, Powers AM, Honein MA. 2016. Zika virus. N Engl J Med 374:1552–1563. doi: 10.1056/NEJMra1602113. [DOI] [PubMed] [Google Scholar]
- 5.Cao-Lormeau VM, Musso D. 2014. Emerging arboviruses in the Pacific. Lancet 384:1571–1572. doi: 10.1016/S0140-6736(14)61977-2. [DOI] [PubMed] [Google Scholar]
- 6.Campos GS, Bandeira AC, Sardi SI. 2015. Zika virus outbreak, Bahia, Brazil. Emerg Infect Dis 21:1885–1886. doi: 10.3201/eid2110.150847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tomasello D, Schlagenhauf P. 2013. Chikungunya and dengue autochthonous cases in Europe, 2007–2012. Travel Med Infect Dis 11:274–284. doi: 10.1016/j.tmaid.2013.07.006. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2010. Locally acquired dengue—Key West, Florida, 2009–2010. MMWR Morb Mortal Wkly Rep 59:577–581. [PubMed] [Google Scholar]
- 9.Likos A, Griffin I, Bingham AM, Stanek D, Fischer M, White S, Hamilton J, Eisenstein L, Atrubin D, Mulay P, Scott B, Jenkins P, Fernandez D, Rico E, Gillis L, Jean R, Cone M, Blackmore C, McAllister J, Vasquez C, Rivera L, Philip C. 2016. Local mosquito-borne transmission of Zika virus—Miami-Dade and Broward Counties, Florida, June–August 2016. MMWR Morb Mortal Wkly Report 65:1032–1038. doi: 10.15585/mmwr.mm6538e1. [DOI] [PubMed] [Google Scholar]
- 10.Tomashek KM, Margolis HS. 2011. Dengue: a potential transfusion-transmitted disease. Transfusion 51:1654–1660. doi: 10.1111/j.1537-2995.2011.03269.x. [DOI] [PubMed] [Google Scholar]
- 11.Duffy MR, Chen TH, Hancock WT, Powers AM, Kool JL, Lanciotti RS, Pretrick M, Marfel M, Holzbauer S, Dubray C, Guillaumot L, Griggs A, Bel M, Lambert AJ, Laven J, Kosoy O, Panella A, Biggerstaff BJ, Fischer M, Hayes EB. 2009. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 360:2536–2543. doi: 10.1056/NEJMoa0805715. [DOI] [PubMed] [Google Scholar]
- 12.Brasil P, Pereira JP, Moreira ME, Ribeiro Nogueira RM, Damasceno L, Wakimoto M, Rabello RS, Valderramos SG, Halai UA, Salles TS, Zin AA, Horovitz D, Daltro P, Boechat M, Raja Gabaglia C, Carvalho de Sequeira P, Pilotto JH, Medialdea-Carrera R, Cotrim da Cunha D, Abreu de Carvalho LM, Pone M, Machado Siqueira A, Calvet GA, Rodrigues Baiao AE, Neves ES, Nassar de Carvalho PR, Hasue RH, Marschik PB, Einspieler C, Janzen C, Cherry JD, Bispo de Filippis AM, Nielsen-Saines K. 2016. Zika virus infection in pregnant women in Rio de Janeiro. N Engl J Med 375:2321–2334. doi: 10.1056/NEJMoa1602412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Musso D, Roche C, Nhan TX, Robin E, Teissier A, Cao-Lormeau VM. 2015. Detection of Zika virus in saliva. J Clin Virol 68:53–55. doi: 10.1016/j.jcv.2015.04.021. [DOI] [PubMed] [Google Scholar]
- 14.Gourinat AC, O'Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. 2015. Detection of Zika virus in urine. Emerg Infect Dis 21:84–86. doi: 10.3201/eid2101.140894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bingham AM, Cone M, Mock V, Heberlein-Larson L, Stanek D, Blackmore C, Likos A. 2016. Comparison of test results for Zika virus in urine, serum, and saliva specimens from persons with travel-associated Zika virus disease—Florida, 2016. MMWR Morb Mortal Wkly Rep 13:475–478. doi: 10.15585/mmwr.mm6518e2. [DOI] [PubMed] [Google Scholar]
- 16.Halstead SB, Rojanasuphot S, Sangkawibha N. 1983. Original antigenic sin in dengue. Am J Trop Med Hyg 32:154–156. doi: 10.4269/ajtmh.1983.32.154. [DOI] [PubMed] [Google Scholar]
- 17.Lanciotti RS, Kosoy OL, Laven JJ, Velez JO, Lambert AJ, Johnson AJ, Stanfield SM, Duffy MR. 2008. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 14:1232–1239. doi: 10.3201/eid1408.080287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allwinn R, Doerr HW, Emmerich P, Schmitz H, Preiser W. 2002. Cross-reactivity in flavivirus serology: new implications of an old finding? Med Microbiol Immunol 190:199–202. doi: 10.1007/s00430-001-0107-9. [DOI] [PubMed] [Google Scholar]
- 19.Yamada K, Takasaki T, Nawa M, Yabe S, Kurane I. 2003. Antibody responses determined for Japanese dengue fever patients by neutralisation and hemagglutination inhibition assays demonstrate cross-reactivity between dengue and Japanese encephalitis viruses. Clin Diagn Lab Immunol 10:725–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roehrig JT, Hombach J, Barrett AD. 2008. Guidelines for plaque-reduction neutralisation testing of human antibodies to dengue viruses. Viral Immunol 21:123–132. doi: 10.1089/vim.2008.0007. [DOI] [PubMed] [Google Scholar]
- 21.Dasgupta S, Reagan-Steiner S, Goodenough D, Russell K, Tanner M, Lewis L, Peterson EE, Powers AM, Kniss K, Meaney-Delman D, Oduyebo T, O'Leary D, Chiu S, Talley P, Hennessey M, Hills S, Cohn A, Gregory C, Zika Virus Response Epidemiology and Laboratory Team. 2016. Patterns in Zika virus testing and infection, by report of symptoms and pregnancy status—United States, January 3–March 5, 2016. MMWR Morb Mortal Wkly Rep 22:395–399. doi: 10.15585/mmwr.mm6515e1. [DOI] [PubMed] [Google Scholar]
- 22.Weaver SC, Lecuit M. 2015. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med 372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 23.Teferedegne B, Lewis AM, Peden K Jr, Murata H. 2013. Development of a neutralization assay for influenza virus using an endpoint assessment based on quantitative reverse-transcription PCR. PLoS One 8:e56023. doi: 10.1371/journal.pone.0056023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varada JC, Teferedegne B, Crim RL, Mdluli T, Audet S, Peden K, Beeler J, Murata H. 2013. A neutralization assay for respiratory syncytial virus using a quantitative PCR-based endpoint assessment. Virol J 10:195–205. doi: 10.1186/1743-422X-10-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramski M, Drozd A, Lichtfuss GF, Dabrowski PW, Ellerbrok H. 2011. Rapid detection of anti-Vaccinia virus neutralizing antibodies. Virol J 8:139–146. doi: 10.1186/1743-422X-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Alwis R, Smith SA, Olivarez NP, Messer WB, Huynh JP, Wahala WM, White LJ, Diamond MS, Baric RS, Crowe JE Jr, de Silva AM. 2012. Identification of human neutralizing antibodies that bind to complex epitopes on dengue virions. Proc Nat Acad Sci U S A 109:7439–7444. doi: 10.1073/pnas.1200566109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teoh EP, Kukkaro P, Teo EW, Lim AP, Tan TT, Yip A, Schul W, Aung M, Kostyuchenko VA, Leo YS, Chan SH, Smith KG, Chan AH, Zou G, Ooi EE, Kemeny DM, Tan GK, Ng JK, Ng ML, Alonso S, Fisher D, Shi PY, Hanson BJ, Lok SM, MacAry PA. 2012. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med 4:139ra83. doi: 10.1126/scitranslmed.3003888. [DOI] [PubMed] [Google Scholar]
- 28.de Alwis R, Beltramello M, Messer WB, Sukupolvi-Petty S, Wahala WM, Kraus A, Olivarez NP, Pham Q, Brien JD, Tsai WY, Wang WK, Halstead S, Kliks S, Diamond MS, Baric R, Lanzavecchia A, Sallusto F, de Silva AM. 2011. In-depth analysis of the antibody response of individuals exposed to primary dengue virus infection. PLoS Negl Trop Dis 5:e1188. doi: 10.1371/journal.pntd.0001188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith SA, Zhou Y, Olivarez NP, Broadwater AH, de Silva AM, Crowe JE Jr. 2012. Persistence of circulating memory B cell clones with potential for dengue virus disease enhancement for decades following infection. J Virol 86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zompi S, Montoya M, Pohl MO, Balmaseda A, Harris E. 2012. Dominant cross-reactive B cell response during secondary acute dengue virus infection in humans. PLoS Negl Trop Dis 6:e1568. doi: 10.1371/journal.pntd.0001568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busch MP, Kleinman SH, Tobler LH, Kamel HT, Norris PJ, Walsh I, Matud JL, Prince HE, Lanciotti RS, Wright DJ, Linnen JM, Caglioti S. 2008. Virus and antibody dynamics in acute West Nile virus infection. J Infect Dis 198:984–993. doi: 10.1086/591467. [DOI] [PubMed] [Google Scholar]
- 32.World Health Organization, Special Programme for Research and Training in Tropical Diseases. 2009. Dengue guidelines for diagnosis, treatment, prevention and control, new ed. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 33.Hu D, Di B, Ding X, Wang Y, Chen Y, Pan Y, Wen K, Wang M, Che X. 2011. Kinetics of non-structural protein 1, IgM and IgG antibodies in dengue type 1 primary infection. Virol J 8:47–50. doi: 10.1186/1743-422X-8-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clapham HE, Rodriguez-Barraquer I, Azman AS, Althouse BM, Salje H, Gibbons RV, Rothman AL, Jarman RG, Nisalak A, Thaisomboonsuk B, Kalayanarooj S, Nimmannitya S, Vaughn DW, Green S, Yoon IK, Cummings DA. 2016. Dengue virus (DENV) neutralizing antibody kinetics in children after symptomatic primary and postprimary DENV infection. J Infect Dis 213:1428–1435. doi: 10.1093/infdis/jiv759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallian P, Cabie A, Richard P, Paturel L, Charrel RN, Pastorino B, Leparc-Goffart I, Tiberghien P, de Lamballerie X. 2017. Zika virus in asymptomatic blood donors in Martinique. Blood 129:263–266. doi: 10.1182/blood-2016-09-737981. [DOI] [PubMed] [Google Scholar]
- 36.Catton M, Druce J, Papadakis G, Tran T, Birch C. 2011. Reality check of laboratory service effectiveness during pandemic (H1N1) 2009, Victoria, Australia. Emerg Infect Dis 17:963–968. doi: 10.3201/eid/1706.101747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santiago GA, Vergne E, Quiles Y, Cosme J, Vazquez J, Medina JF, Medina F, Colon C, Margolis H, Munoz-Jordan JL. 2013. Analytical and clinical performance of the CDC real time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis 7:e2311. doi: 10.1371/journal.pntd.0002311. [DOI] [PMC free article] [PubMed] [Google Scholar]