ABSTRACT
Baboons naturally infected with simian T lymphotropic virus (STLV) are a potentially useful model system for the study of vaccination against human T lymphotropic virus (HTLV). Here we expanded the number of available full-length baboon STLV-1 sequences from one to three and related the T cell responses that recognize the immunodominant Tax protein to the tax sequences present in two individual baboons. Continuously growing T cell lines were established from two baboons, animals 12141 and 12752. Next-generation sequencing (NGS) of complete STLV genome sequences from these T cell lines revealed them to be closely related but distinct from each other and from the baboon STLV-1 sequence in the NCBI sequence database. Overlapping peptides corresponding to each unique Tax sequence and to the reference baboon Tax sequence were used to analyze recognition by T cells from each baboon using intracellular cytokine staining (ICS). Individual baboons expressed more gamma interferon and tumor necrosis factor alpha in response to Tax peptides corresponding to their own STLV-1 sequence than in response to Tax peptides corresponding to the reference baboon STLV-1 sequence. Thus, our analyses revealed distinct but closely related STLV-1 genome sequences in two baboons, extremely low heterogeneity of STLV sequences within each baboon, no evidence for superinfection within each baboon, and a ready ability of T cells in each baboon to recognize circulating Tax sequences. While amino acid substitutions that result in escape from CD8+ T cell recognition were not observed, premature stop codons were observed in 7% and 56% of tax sequences from peripheral blood mononuclear cells from animals 12141 and 12752, respectively.
IMPORTANCE It has been estimated that approximately 100,000 people suffer serious morbidity and 10,000 people die each year from the consequences associated with human T lymphotropic virus (HTLV) infection. There are no antiviral drugs and no preventive vaccine. A preventive vaccine would significantly impact the global burden associated with HTLV infections. Here we provide fundamental information on the simian T lymphotropic virus (STLV) naturally transmitted in a colony of captive baboons. The limited viral sequence heterogeneity in individual baboons, the identity of the viral gene product that is the major target of cellular immune responses, the persistence of viral amino acid sequences that are the major targets of cellular immune responses, and the emergence in vivo of truncated variants in the major target of cellular immune responses all parallel what are seen with HTLV infection of humans. These results justify the use of STLV-infected baboons as a model system for vaccine development efforts.
KEYWORDS: intracellular cytokine staining, next generation sequencing, Papio anubis, Tax, simian T-cell leukemia virus
INTRODUCTION
Human T lymphotropic virus (HTLV) and simian T lymphotropic virus (STLV) are members of the Deltaretrovirus genus of the Retrovirus family (1). HTLV-1 was the first retrovirus to be directly associated with a human malignancy, adult T cell leukemia (ATL) (2–4). HTLV-1 is also the causative agent of HTLV-1-associated myelopathy (HAM), tropical spastic paraparesis (TSP), uveitis, arthropathy, and infective dermatitis (4–6). It is estimated that 10 million to 20 million people are infected with HTLV-1 worldwide (3). However, the lifetime risk of developing ATL in an infected individual is less than 5%, with the risk being slightly higher for men (5, 7).
HTLV-1 is believed to have originated from the cross-species transmission of simian T lymphotropic virus 1 (STLV-1) from infected nonhuman primates to humans (8, 9). Consequently, HTLV-1 is closely related to the STLV found in some species of nonhuman primates (8). STLV is thought to have made the jump to humans on multiple occasions in different geographical regions, leading to the multiple subtypes of HTLV-1 seen around the globe (2, 10). Both HTLV-1 and STLV-1 establish lifelong persistent infections in their hosts (2, 3, 6, 11). Some nonhuman primates that are naturally infected with STLV-1 eventually develop diseases like those caused by HTLV-1, including ATL (12–14). As with HTLV-1, most infections of nonhuman primates with STLV-1 are asymptomatic, with only about 1% to 2% of animals developing STLV-associated leukemia, which can share clinical and pathological features with ATL in humans (12–14). These similarities make nonhuman primates infected with STLV-1 an attractive model for the study of HTLV-1 biology and approaches to vaccination.
STLV-1 can be found in over 20 species of Asian and African primates (8, 15, 16). Due to the high frequencies of STLV-1 infection in both wild and captive baboons, we propose that baboons are a potentially useful model system for such studies. A recent study analyzed the cellular immune response to STLV-1 in infected olive baboons (Papio anubis) (17). This study utilized peptides based on a baboon SLTV-1 reference sequence found in the NCBI database. Similar to the findings of other studies with HLTV and STLV, Tax was found to be the immunodominant target of CD8+ T lymphocytes in baboons (18–20). The goal of the present study was to document sequence polymorphisms that may be present among baboon STLV-1 strains and to investigate whether there may be selection for virus variants that escape recognition by CD8+ T cell responses.
Currently, there is only one full-length STLV-1 sequence derived from a baboon in the NCBI database (GenBank accession number JX987040.1). Here we expand the number of full-length, baboon-derived STLV-1 sequences from one to three. Although no amino acid changes that would alter recognition by Tax-specific CD8+ T lymphocytes were found, truncating premature stop codons were selectively observed in tax genes present in circulating peripheral blood mononuclear cells (PBMC) but not in continuously growing cell lines derived from the same baboons. Our findings should help to inform and guide the development of vaccine concepts that can be tested experimentally in baboons.
RESULTS
Generation of STLV-1-positive baboon cell lines.
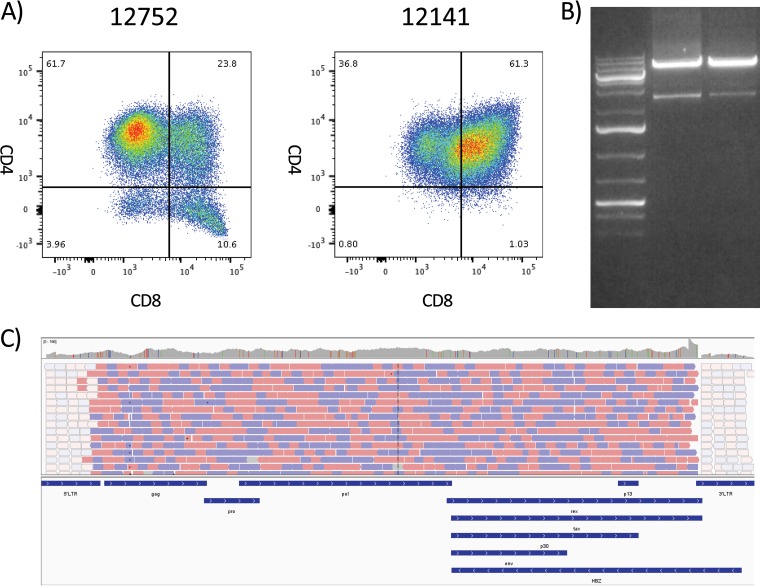
Sixteen STLV-1-positive olive baboons were bled and peripheral blood mononuclear cells (PBMC) were isolated. PBMC were used to generate continuously growing T cell lines as outlined in Materials and Methods. T cell lines from two olive baboons (animals 12752 and 12141) were selected for continued study due to their growth characteristics in culture. Flow cytometry analyses of the cell lines revealed that the cell line generated from animal 12752 was primarily made up of CD3+ CD4+ CD8− T cells (61.7%) (Fig. 1A). Of the remaining cell types, CD4− CD8+ T cells made up 10.6%, CD4+ CD8+ cells made up 23.8%, and CD4− CD8− cells made up 3.9% of the cell line. In contrast, the cell line generated from animal 12141 was primarily composed of double-positive T cells (61.3% CD3+ CD4+ CD8+). CD4+ CD8− T cells made up 36.8%, CD4− CD8+ T cells made up 1.0%, and CD4− CD8− T cells made up 0.8% of the cell line. Neither of these cell lines became interleukin-2 (IL-2) independent.
FIG 1.
PCR amplification and sequencing of the whole STLV-1 genome from STLV-positive T cell lines. Genomic DNA was purified from T cell lines grown out of STLV-positive baboon PBMC samples. (A) The cell line phenotype was determined by flow cytometry, gating on the lymphocyte population, and standard CD3, CD4, and CD8 T cell staining. The cell line of animal 12752 was predominantly made up of CD4+ CD8− T cells, whereas the cell line of animal 12141 displayed a large degree of double-positive CD4+ CD8+ T cells. The numbers in each quadrant represent the percentage of live CD3+ lymphocytes in that quadrant. (B) PCR was performed with an ultra-high-fidelity DNA polymerase with 25 rounds of amplification to minimize PCR error. PCR products were run on an agarose gel, and the 8.5-kb band was excised and purified for NGS. (C) Next-generation sequencing was performed on a HiSeq 2500 desktop sequencer with the assistance of the University of Miami’s John P. Hussman Institute for Human Genomics sequencing core. Sequencing provided almost complete genome coverage, with only a small portion of the LTR being excluded. Animal 12752 provided 21.2 million quality reads, whereas animal 12141 provided 23.5 million quality reads.
PCR amplification of STLV-1 proviral genomes.
In order to better understand the STLV strains circulating among baboons at the Southwest National Primate Research Center (SNPRC) and to document STLV sequence variation within and between baboons, we set out to analyze the STLV genome sequences. Genomic DNA was isolated from the T cell lines of animals 12141 and 12752 and used to amplify the STLV genome sequences. Viral genomes were amplified using primers specific for the 5′ long terminal repeat (LTR) and the 3′ LTR following the terminal end of tax. The forward PCR primer recognized positions 135 to 152 of the U3 region of the 5′ LTR, and the reverse PCR primer recognized positions 64 to 84 of the U3 region of the 5′ LTR. The primer sequences recognizing the 3′ LTR were located downstream of the termination codon for tax, whose sequences extend into the 3′ LTR. This amplification strategy allowed assembly of an almost complete U3-R-U5 LTR sequence. The sequence lacked 51 bp from positions 84 to 134. To avoid the introduction of mutations by PCR amplification, Phusion Hot Start II DNA polymerase, an ultra-high-fidelity DNA polymerase, was used. Phusion Hot Start II DNA polymerase is known to have an error rate 50-fold lower than that of traditional Taq polymerase. To further reduce the introduction of errors by PCR, the numbers of PCR cycles were also kept to a minimum (20 to 25 cycles).
For both cell lines, we were readily able to isolate a band of approximately 9 kb (Fig. 1B). This band was excised and purified for next-generation sequencing (NGS).
Next-generation sequencing of STLV-1 genomes.
NGS was performed by the University of Miami’s John P. Hussman Institute for Human Genomics sequencing core on a HiSeq 2500 desktop sequencer (Illumina). The PCR strategy used to amplify the full-length STLV-1 genomes provided nearly complete genome coverage, with only a 51-bp portion of the LTR from the U3 region being excluded. Following NGS, the terminal portions (∼300 to 500 bp) of the 5′ and 3′ LTR were excluded from analysis due to an inability of the software to determine the precise location of the reads. LTR sequences were PCR amplified from the genomic DNA of the T cell lines and sequenced by Sanger sequencing in order to confirm the sequences recovered by NGS as well as sequence the 51-bp fragment excluded in the full-length PCR. This allowed us to construct full viral genomes for the viruses from both animal 12752 and animal 12141.
Animals 12752 and 12171 provided 21.2 million and 23.5 million pass-filter reads per sample, respectively. When they were aligned to the reference STLV-1 genome (GenBank accession number JX987040.1), the viral genomes recovered from animals 12141 and 12752 were quite distinct. The viral genome recovered from animal 12752 contained 415 nucleotide polymorphisms compared to the sequence of the reference genome. Animal 12141 yielded a viral genome with 110 polymorphisms compared to the sequence of the reference genome. Heterogeneity of greater than 10% was noted at only 6 nucleotide positions of the viral genome from animal 12752 and 13 nucleotide positions of the viral genome from animal 12141. These findings suggest only very limited heterogeneity in both T cell lines and the origin of each cell line from a single STLV strain.
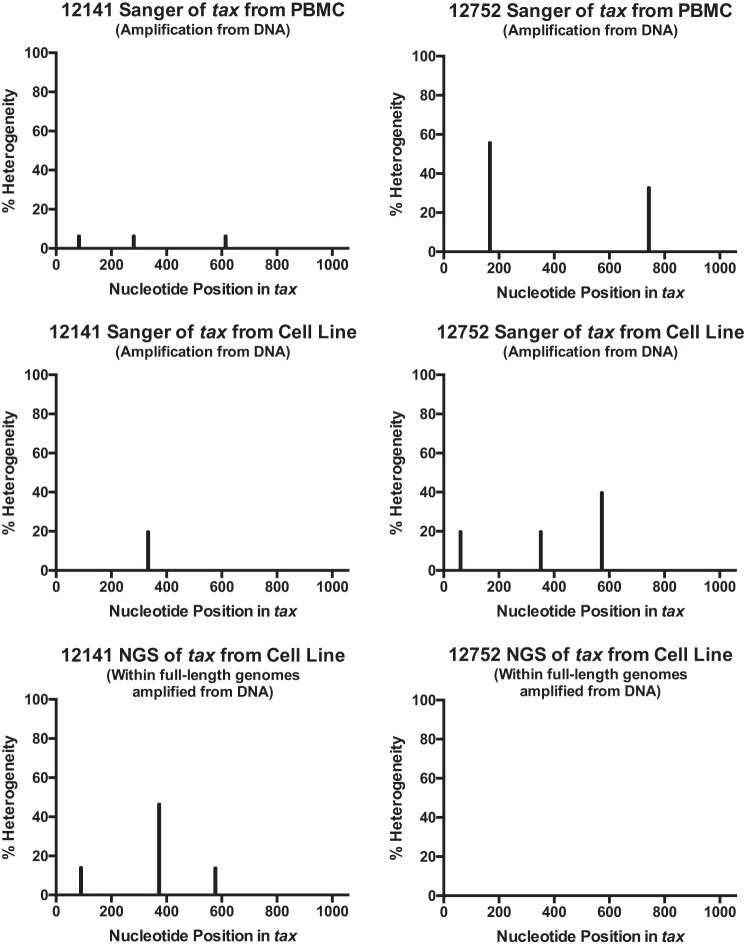
To confirm the accuracy of the NGS results, tax was PCR amplified from the genomic DNA of the T cell lines using primers specific for regions outside the tax coding region and sequenced by traditional Sanger sequencing. The results of NGS and Sanger sequencing of tax were compared. The comparisons showed essentially identical nucleotide sequences and limited sequence heterogeneity at only several locations (Fig. 2 and Table 1). Consensus amino acid sequences were identical for animal 12141. For animal 12752, consensus amino acid sequences were identical at all amino acid positions except amino acid position 191 (Table 1). Since the sequences from the cell lines appeared to correspond to a single viral genome with extremely minimal sequence heterogeneity, we further investigated the extent of heterogeneity and the possibility of superinfection in animals 12752 and 12141 by Sanger sequencing of tax amplified from PBMC isolated 10 months after the initial blood draw. tax was amplified from the genomic DNA isolated from fresh PBMC by PCR using primers specific for regions outside the tax coding sequence and cloned into a sequencing vector, as outlined in Materials and Methods. Nine clones from animal 12752 and 15 clones from animal 12141 were sequenced by Sanger sequencing. Premature stop codons that were absent by the sequencing of tax from the genomes in the cell lines were found in tax sequenced from PBMC (Table 1). Stop codons at position 28 were observed in 7% of the clones from animal 12141. For animal 12752, 56% of the clones displayed premature stop codons (positions 56 and 248). Other than stop codons, the tax sequences from PBMC and the cell lines were nearly identical.
FIG 2.
Analysis of sequence variants. The tax sequences obtained from NGS, Sanger sequencing of tax amplified from T cell lines, and Sanger sequencing of tax amplified from PBMC were analyzed for the presence of sequence variants. The results for variants whose sequences differed from the consensus sequence by each sequencing method were plotted. Five sequences of tax amplified from cell lines were examined. For PBMC from animals 12141 and 12752, 15 and 9 sequences were analyzed, respectively. For NGS, sequence variants with a cutoff of 10% variance were noted.
TABLE 1.
Locations of sequence heterogeneity in taxa
| Animal and amino acid position | Sequence determined by: |
||
|---|---|---|---|
| NGS (cell line) | Sanger sequencing (cell line) | Sanger sequencing (PBMCs) | |
| Animal 12141 | |||
| 28 | 100% W | 100% W | 93.34% W, 6.66% stop codon |
| 94 | 100% T | 100% T | 93.34% T, 6.66% I |
| 125 | 53.3% G, 46.7% R | 20% R, 80% G | 100% G |
| 193 | 86% E, 14% K | 100% E | 100% E |
| 205 | 100% L | 100% L | 93.34% L, 6.66% P |
| Animal 12752 | |||
| 21 | 100% G | 80% G, 20% R | 100% G |
| 56 | 100% W | 100% W | 44% W, 56% stop codon |
| 118 | 100% G | 80% G, 20% R | 100% G |
| 191 | 100% I | 60% M, 40% I | 100% M |
| 248 | 100% W | 100% W | 66% W, 33% stop codonb |
The amino acid sequences obtained by NGS of tax, Sanger sequencing of tax amplified from T cell lines, and Sanger sequencing of tax amplified from PBMC were compared for animals 12141 and 12752. Variants whose sequences differed from the consensus tax sequence obtained by each sequencing method are noted. Five sequences of tax amplified from cell lines were examined. Fifteen and 9 sequences were analyzed from PBMC from animals 12141 and 12752, respectively. For NGS, sequence variants with a 10% variance cutoff were noted.
Sequences with stop codons present at position 248 also contained stop codons at position 56. In total, 56% of clones contained stop codons.
APOBEC3G is a restriction factor that can induce a G-to-A hypermutation during replication of retroviruses via its cytidine deaminase activity. We analyzed our sequence polymorphisms for signatures of APOBEC3G activity. For animal 12752, there were 42 polymorphisms in tax, and 5 involved the G → A change. For animal 12141, there were 18 polymorphisms in tax, and 5 involved the G → A change. For both Sanger sequencing and NGS, the number of G → A polymorphisms did not appear outside the range of numerical expectations. However, all 3 of the premature stop codons in tax sequenced from PBMC resulted from G → A changes. Similar to the finding of Fan et al. with HTLV, these data may be an example of STLV-infected cells exploiting APOBEC deaminases to escape detection by the host immune system through the loss of Tax expression (21).
Comparison of sequences of viruses from animals 12141 and 12752, isolate F88395, HTLV-1, HTLV-2, and STLV-1 TE4.
The sequences obtained by NGS were analyzed to identify regions corresponding to known STLV-1 proteins. More than 90% sequence identity was observed in the alignments of all the protein amino acid sequences of the viruses from animals 12141 and 12752, with the exception of the HBZ and p30 sequences (Table 2). The sequence of the virus from animal 12141 had a higher degree of identity to the sequence of the STLV-1 reference genome than to the sequence of the virus from animal 12752. In contrast to the reference sequence, the sequences of the viruses from both animal 12141 and animal 12752 exhibited a polymorphism from a methionine to an isoleucine in the putative start codon of p13. This polymorphism was also observed in the STLV-1 TE4 strain of Asian origin (GenBank accession number Z46900.1).
TABLE 2.
Amino acid sequence identitya
| Virus sequences compared | % identity |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gag | Pro | Pol | Rex | Tax | Env | HBZ | p30 | p13 | |
| 12752 vs reference | 97.5 | 93.6 | 96.8 | 92.6 | 95.5 | 97.7 | 85 | 85.9 | 90.8 |
| 12752 vs 12141 | 97.9 | 93.2 | 97.2 | 93.7 | 96 | 97.5 | 86.9 | 88 | 94.3 |
| 12752 vs STLV-1 of Asian origin | 93.3 | 88.5 | 91.3 | 84.1 | 90.9 | 93.9 | 71 | 75.4 | 88.5 |
| 12752 vs HTLV-1 | 96.1 | 93.2 | 97.5 | 93.1 | 96.9 | 97.3 | 85 | 86.7 | 95.4 |
| 12752 vs HTLV-2 | 73.5 | 33.3 | 60.3 | 55.8 | 71.4 | 69 | NAb | NA | NA |
| 12141 vs reference | 99.3 | 98.7 | 98.3 | 97.4 | 96.3 | 98.6 | 96.1 | 96.3 | 94.3 |
| 12141 vs HTLV-1 | 97 | 91.9 | 96.7 | 95.2 | 97.2 | 97.7 | 84 | 87.1 | 96.6 |
| 12141 vs HTLV-2 | 73.7 | 33.3 | 60.2 | 55.3 | 71.1 | 69 | NA | NA | NA |
| 12141 vs STLV-1 of Asian origin | 94.5 | 89.3 | 91 | 84.1 | 92.1 | 93.6 | 70 | 75 | 88.5 |
| Reference vs STLV-1 of Asian origin | 93.8 | 88 | 90.2 | 84.1 | 90.7 | 93.6 | 70.5 | 73.4 | 85.1 |
| Reference vs HTLV-1 | 96.3 | 92.3 | 96 | 94.2 | 96 | 98 | 85 | 85.9 | 95.4 |
| Reference vs HTLV-2 | 73.7 | 33.3 | 59.9 | 55.3 | 70.5 | 68.6 | NA | NA | NA |
| HTLV-1 vs STLV-1 of Asian origin | 95.3 | 87.2 | 91.9 | 84.7 | 91.2 | 94.1 | 73.8 | 77 | 89.7 |
| HTLV-1 vs HTLV-2 | 73.7 | 33.8 | 60.2 | 55.3 | 72.2 | 69.2 | NA | NA | NA |
| HTLV-2 vs STLV-1 of Asian origin | 73.9 | 33.3 | 59.6 | 52.1 | 72.2 | 70.1 | NA | NA | NA |
The sequences from animals 12752 and 12141 were recovered by next-generation sequencing and were analyzed to identify the coding regions of STLV-1. Translated sequences from both animals were aligned to the STLV-1 reference amino acid sequence, the NCBI reference HTLV-1 amino acid sequence (accession number NC_001436.1) and HTLV-2 amino acid sequence (accession number NC_001488.1) from the NCBI Viral Genome Database, as well as the amino acid sequence of STLV-1 isolate TE4 from Macaca tonkeana (Asian origin, GenBank accession number Z46900.1).
NA, not applicable. In HTLV-2, p13, p30, and HBZ were not directly comparable to the gene products of STLV-1. Although HTLV-2 has homologous proteins, such as p28 and APH2, the differences in amino acid sequence make a direct comparison not applicable (37).
As expected, the viruses from both animal 12141 and animal 12752 showed a high degree of similarity to HTLV-1. Previous work published on the phylogenetic analysis of HTLV-1, HTLV-2, and HTLV-3 indicated that STLV-1 and HTLV-1 arose from a common ancestor, primate T lymphotropic virus 1 (PTLV-1) (9). These analyses also indicated that HTLV-1 arose from the cross-species transmission of STLV-1 from nonhuman primates to humans (22). This accounts for the very high degree of similarity among our isolates, the reference genome, and HTLV-1. Also predicted by the phylogenetic analysis, the viral proteins of the viruses from animals 12141 and 12752 showed very low levels of similarity to the viral proteins of HTLV-2, with the highest similarities being in the Gag and Tax proteins. For comparisons with the HTLV-2 sequence, HBZ, p30, and p13 were not homologous enough to allow a confident alignment; therefore, their percent similarity was omitted from Table 2. HTLV-2 APH2 and p28 differed from HTLV-1 HBZ and p30 in sequence and function, which made a comparison not possible.
When the baboon STLV-1 isolates were compared to an STLV-1 isolate of Asian origin (isolate TE4 from a macaque), the level of amino acid sequence identity was markedly decreased. The level of amino acid sequence identity for all three baboon isolates fell within the 70 to 95% identity range, with the lowest identity being for HBZ and p30. The STLV-1 TE4 isolate shared a slightly higher level of identity with HTLV-1 than it did with the isolates from either animal 12141 or animal 12752 (Table 2).
CD8+ T cell responses to Tax reference and variant peptide sequences.
Previous studies in humans have observed that immunodominant responses are directed to the Tax protein (18–20, 23). In a recently published study, immunogenic regions of Tax were mapped in individual animals using peptides that correspond to those of the reference genome from isolate F88395 (17). Here, we set out to determine the influence of sequence polymorphisms on recognition of virus by CD8+ T cells from each of the two baboons in our study. During our preliminary analysis, it appeared that regions with sequence polymorphisms tended to be regions identified by Castro et al. to be immunogenic regions of Tax (17). To examine whether these sequence polymorphisms could represent immune escape variants, we constructed variant peptides, which are described in Table 3. These 15-mer peptides overlapped by 11 amino acids.
TABLE 3.
Peptide designa
15-mer peptides overlapping by 11 amino acids were constructed for the reference STLV-1 genome, the full-length genome of a baboon isolate of STLV-1 from the NCBI database (GenBank accession number JX987040.1). Using the sequences isolated from T cell lines of animals 12141 and 12752, variant peptides were generated for any region whose sequence varied from the reference Tax sequence. These differences were pooled as indicated by the outlined sequences. Blue, peptide pools tested for animal 12752; green, peptide pools tested for animal 12141. When both animals possessed the same mutations, this was indicated by the text “SAME as 12752.” By comparison of the reference and variant peptide pools, it was possible to determine if the mutations had any impact on the immunogenicity of that epitope. Sub, subset.
Fresh PBMC from animals 12141 and 12752 were isolated, stimulated with Tax peptides, and stained by intracellular cytokine staining (ICS). ICS assays allow the characterization of the frequency of STLV-1-specific CD8+ T cell responses against the reference peptides as well as peptides that correspond to the sequences obtained from animals 12141 and 12752 (variant peptides). As a negative control, PBMC from an STLV-1-negative animal (animal 15659) were included. Because animal 12869 has a well-documented CD8+ T cell response to Tax (17), PBMC from this animal were included as a positive control.
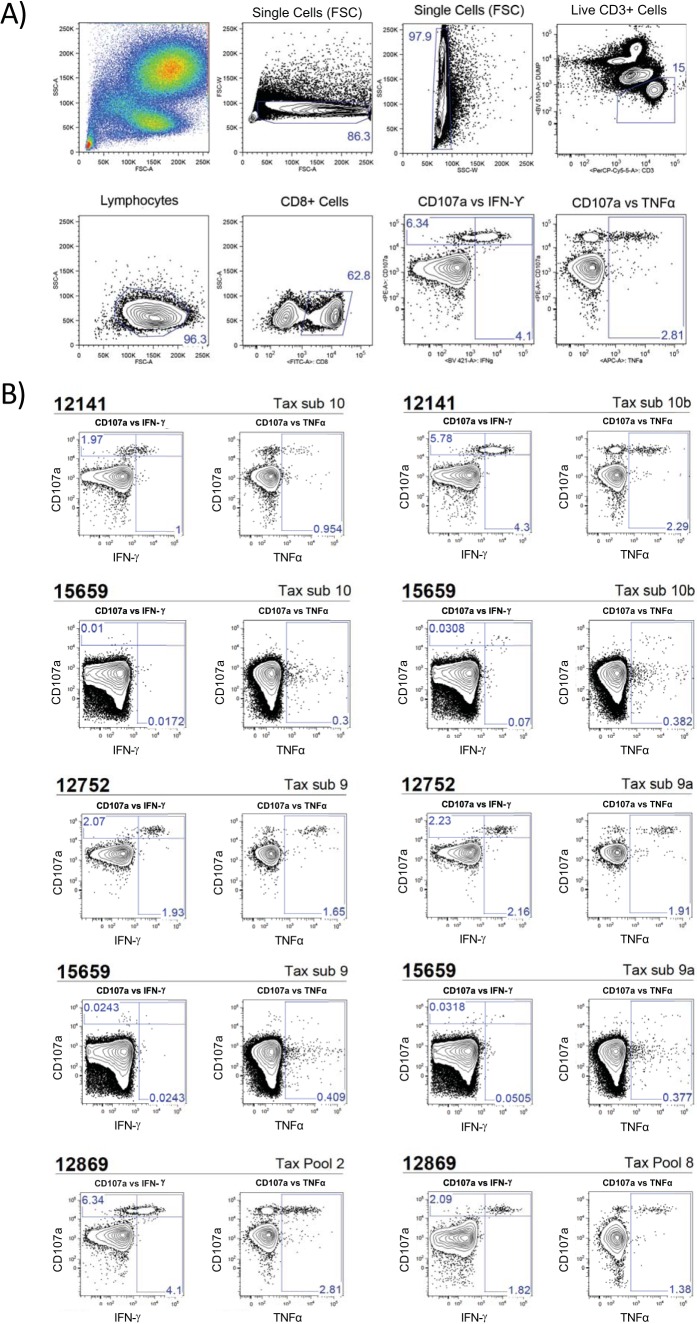
Our ICS assays revealed high-frequency gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) responses directed to Tax in animals 12752 and 12141: Tax subsets 9 and 10, respectively (see Fig. 4). Of interest, animal 12141 had readily detectable ICS responses against both the reference peptide and the variant peptide of Tax subset 10 (Fig. 3 and 4). In animal 12141, 4.3% of CD8+ T cells expressed IFN-γ and 2.29% of CD8+ T cells expressed TNF-α in response to Tax subset 10b. When stimulated with the reference peptides (Tax subset 10), we observed reduced cytokine responses in comparison to the responses observed with simulation with Tax subset 10b: 1% and 0.954% for IFN-γ and TNF-α, respectively. We observed similar results with animal 12752 (Fig. 3 and 4). In animal 12752, 2.16% of CD8+ T cells expressed IFN-γ and 1.91% of CD8+ T cells expressed TNF-α in response to Tax subset 9a. In response to the reference peptide, Tax subset 9 responses were decreased: 1.93% and 1.65% for IFN-γ and TNF-α, respectively. Both animals displayed CD8 T cell responses against their individual Tax sequences higher than those against the matched reference peptide sequences.
FIG 4.
Intracellular cytokine staining of Tax-specific CD8+ T cells. PBMC were freshly isolated from whole blood by Ficoll density centrifugation. Isolated PBMC were incubated with the indicated 15-mer peptide pools for 8 h before performing intracellular cytokine staining for IFN-α and TNF-α. CD107a was also used as a degranulation marker. CD3+ CD8+ cells were analyzed for the expression of both CD107a and IFN-α or TNF-α. Peptide subsets for the reference genome are indicated by the subset number, whereas variant pools for animal 12752 are indicated by the subset number followed by an “a” and variant pools for animal 12141 are followed by a “b.” All variant peptide pools were tested; however, only responding pools were included. As a negative control, PBMC from an STLV-negative animal (animal 15659) were tested with the same peptides used to test PBMC from the STLV-positive animals. (A) Gating strategy used to isolate CD3+ CD8+ T cells for ICS analysis. APC, allophycocyanin. (B) PBMC from animal 12141 were found to be positive for reactivity with Tax subset (sub) 10 as well as variant subset 10b. PBMC from animal 12752 were found to be positive for reactivity with Tax subset 9 and variant subset 9a. PBMC from animal 12869, a known responder, were included as a positive control due to already documented immune responses to Tax pools 2 and 8 (17).
FIG 3.
Tax protein alignments. Tax sequences from NGS were translated into the corresponding amino acid sequence and aligned to the Tax sequence of the NCBI reference genome of STLV-1 using the ClustalW program. Regions of Tax that elicited an immune response by ICS are highlighted in red for animal 12752 and green for animal 12141. Peptides designed to be specific for the reference genome and the variant genome were compared.
DISCUSSION
By combining long-range PCR techniques and next-generation sequencing, we determined the sequences of the STLV-1 isolates from two olive baboons of the Southwest National Primate Research Center colony. Although each STLV-1 isolate was genetically distinct, the sequences shared high degrees of similarity with each other and with the sequence of HTLV-1. Very little heterogeneity within each full-length STLV sequence from each continuously growing cell line was observed. The main difference observed was the presence of stop codons in tax isolated from PBMC that were not present in the sequences from the continuously growing cell lines. With the exception of these stop codons, the tax sequences amplified from PBMC closely matched the tax sequence from each cell line, with very little heterogeneity in the sequence present in each individual baboon being found. Thus, neither baboon showed any evidence of superinfection with a second STLV strain. CD8+ T cell responses against the immunodominant Tax protein recognized peptides corresponding to the circulating Tax sequence at reasonably high frequencies in each baboon.
Our findings are consistent with what is known about HTLV-1. Early studies found that cases of ATL are monoclonal in nature, with a single infecting HTLV-1 species being responsible for infection (24). These clones tend to persist for several years in the same individual, maintaining a stable genetic profile of the virus (25). Taking into account the genetic stability of HTLV-1, maintenance of a high proviral load is likely accomplished by persistent clonal proliferation of infected immortalized cells in vivo and not by viral replication (26–29). Similarly, we observed an analogous genetic stability in our sequencing efforts with STLV-1 isolates from baboons. When Wattel et al. analyzed 100 molecular clones of the envelope from two HTLV-1 carriers, they found almost no genetic variation (27). These findings are consistent with the sequences obtained from both NGS of the continuously growing cell lines and Sanger sequencing of tax from freshly isolated PBMC. Minimal variation in tax was observed, despite the fact that Sanger sequencing of PBMC was performed 10 months after the initial bleed to establish continuously growing cell lines (Table 1).
Escape of HTLV-infected cells from cytotoxic T lymphocyte recognition has been documented in both ATL patients and non-ATL patients (30, 31). During the progression to ATL, cells at some point no longer require Tax expression for continued immortalized cell growth, and reduced Tax expression in such cells may result in reduced HTLV-specific CD8+ T cell killing (23). Premature stop codons in tax can be frequently found in ATL patients, and tax sequences with premature stop codons can also be found as a minority population in many non-ATL patients (30). Our findings with STLV-1 infection of baboons are again consistent with those with HTLV-1 infection of humans. Animal 12752 was found to have premature stop codons in 56% of the tax sequences isolated, suggesting that this animal may be on its way to having an ATL-like disease. Animal 12141 had premature stop codons in 7% of isolated sequences, which suggests that this animal may be further from disease progression.
Although the polymorphisms observed in animals 12141 and 12752 appeared to be located in regions of Tax identified by Castro et al. (17) to be epitopes of CD8+ T cell recognition, we found that T cells from both animals recognized both the reference peptide and the variant peptide at reasonably high frequencies. The correspondence of sequence polymorphisms within the targets of CD8+ T cell recognition suggests the possibility of immune selective pressure. However, any such immune selective pressure may not be evident within the time interval of our measurements. Furthermore, the sequence polymorphisms in the epitope targets that were observed were not sufficient to confer clear escape.
Because of the error-prone nature of reverse transcriptase, viral replication is the main source of sequence heterogeneity in retroviruses. The enormous heterogeneity in HIV sequences present within a single infected individual is a manifestation of the ongoing viral replication. The lack of STLV sequence heterogeneity in the two baboons in our study reinforces the finding that continuous HTLV/STLV replication is not a major factor in the persistence of the virus. As has been suggested previously, the persistence of HTLV/STLV is likely achieved principally through continuous replication of the immortalized cells in which it resides (25–28).
In considering attempts to develop a preventive vaccine against HTLV, many fundamental questions remain. Since Tax and HBZ (SBZ in nonhuman primates) appear to be continuously expressed in order to establish the growth-immortalized state (32–34), it is reasonable to consider the inclusion of Tax and HBZ antigens in any vaccine formulation. But will this be enough? Is it reasonable to seek sterilizing immunity from a vaccine? Will a neutralizing antibody response to the Env protein be required for or facilitate the goal of sterilizing immunity? Will a reduction in the viral load be sufficient for protective effects from a vaccine? Deriving answers to these and other questions would benefit from a suitable animal model to inform and guide the development of vaccine concepts for use in humans.
Due to the similarities of HTLV-1 with STLV-1 and the similarities between the olive baboon and human immune systems, olive baboons appear to be ideally suited to be nonhuman primate models for use in vaccine trials. By utilizing breeding groups with a large female-to-male ratio, one or two infected baboons can be introduced into a colony of vaccinated individuals to examine efficacy in a realistic setting. Our results reported here provide a useful first step toward such a goal.
MATERIALS AND METHODS
Research animals.
The animals used in this study were olive baboons (Papio anubis) housed at the Southwest National Primate Research Center (SNPRC) at the Texas Biomedical Research Institute. They were housed in accordance with the regulations set by the Guide for the Care and Use of Laboratory Animals of the National Research Council (35), as approved by the Texas Biomedical Research Institute Institutional Animal Care and Use Committee.
For this study, we included STLV-1-infected baboons and uninfected animals, which were used as negative controls. STLV-1 serology was performed at SNPRC using the macaque-tracking multiplexed fluorometric immunoassay (MFIA). MFIA is a Luminex bead-based serology test developed by Charles River Laboratories (CRL; Wilmington, MA). The measurement of serology by the MFIA method is comparable to that by the enzyme-linked immunosorbent assay method (36). In brief, the STLV-1 Luminex multiplex assay uses two different bead sets for anti-STLV-1 antibody detection. The first bead set uses HTLV-1 and -2 whole-virus lysate mixes, whereas the second bead set uses a purified, truncated STLV-1 p21 protein (12 kDa) produced in insect cells. Additionally, beads coated with baculovirus were used as nonspecific assay controls. Data analysis was carried out per the manufacturer's protocol, using the Charles River Laboratories MFIA results Excel workbook (37). In summary, an assay score is created on the basis of the reactivity against the virus-specific beads minus the background. A sample was considered positive when that reactivity against both of the virus-specific beads exceeded the cutoff. The assay result was considered inconclusive if there was reactivity against only one set of beads and negative if no reactivity was detected.
Animal 12141 first tested positive for the presence of STLV antibodies in July 2014 at the age of 20 years and 0 months. Animal 12752 was tested for the presence of STLV antibodies in July 2014, but the results were indeterminate. In August 2014, animal 12752 was tested for STLV by PCR and was found to be positive at the age of 19 years and 4 months. Neither animal had been tested prior to the above-mentioned dates. These baboons had never been housed together before they were tested for STLV seropositivity in this study. These baboons were then bled in October 2014 for the isolation of peripheral blood mononuclear cells (PBMC) and the establishment of continuously growing T cell lines, as described below. These animals were bled again in July 2015 for sequencing of virus from freshly isolated PBMC.
Blood collection, processing, and cell line generation.
We isolated PBMC from EDTA-treated blood by density centrifugation. Whole blood was spun at 1,700 rpm for 10 min. Plasma was removed and spun at 3,300 rpm and was then aliquoted into cryotubes for storage.
Dulbecco's phosphate-buffered saline (DPBS) with 2% fetal bovine serum (FBS) was added to double the original blood volume. The reconstituted, diluted whole blood was then layered over lymphocyte separation medium (MP Biomedical) in SepMate-50 lymphocyte separation tubes (Stem Cell Technologies) and centrifuged at 2200 rpm for 20 min. The top layer including the PBMC was poured into a fresh 50-ml conical tube. DPBS with 2% FBS was added to wash the cells, and the cells were centrifuged again at 1,000 rpm for 10 min. The supernatant was decanted, and the cells were resuspended in 2 ml ammonium-chloride-potassium lysis solution (BioWhittaker) and incubated at 37°C for 10 min to lyse any remaining red blood cells. The cells were washed an additional two times with DPBS with 2% FBS as described above. The cells were then counted and resuspended in RPMI 1640 culture medium supplemented with 10% fetal bovine serum (heat inactivated), 1% penicillin-streptomycin, 2.5% HEPES buffer, and 50 units/ml recombinant human IL-2. Cells were stimulated with 5% phytohemagglutinin (PHA) for the first 5 days in culture. After stimulation, cells were washed and resuspended in fresh medium without PHA.
Phenotyping of STLV-1-specific T cell lines.
Established T cell lines from animals 12141 and 12752 were analyzed for their phenotypes by T cell staining and flow cytometric analysis. Cells from each line (1 × 106) were washed once with phosphate-buffered saline (PBS) plus 2% FBS. Cells were then stained at 4°C with antibodies directed against the surface molecules CD3 (peridinin chlorophyll protein [PerCP]-Cy5.5; clone SP34-2; BD Biosciences), CD4 (BV605; clone L200; BD Biosciences), CD8 (fluorescein isothiocyanate [FITC]; clone RPA-T8; BioLegend), CD14 (BV510; clone M5E2; BioLegend), CD16 (BV510; clone 3G8; BioLegend), and CD20 (BV510; clone 2H7; BioLegend). An amine aqua dye (Invitrogen) was used to differentiate between live and dead cells.
We gated on CD3+ T lymphocytes, excluding CD14+, CD16+, CD20+, and dead cells. We then separated T lymphocyte subsets on the basis of their expression of either CD4 or CD8.
Amplification of STLV-1 proviral DNA.
PCR amplification of STLV-1 whole-virus genomes and tax for sequencing was done from genomic DNA. DNA was extracted from either 1.0 × 107 cells from the baboon T cell lines or 1.0 × 107 to 2.0 × 107 PBMC per animal using a QIAamp DNA blood midikit (Qiagen, Valencia, CA, USA). PCR amplification of STLV-1 genomes and the tax gene was done using Phusion Hot Start II high-fidelity polymerase (Life Technologies) according to the manufacturer's protocol. The following primers were used for amplification: STLV-1-tax forward (5′-CCA AGT CCT CCA CCA GCA G-3′) and STLV-1-tax reverse (5′-AGT CTG AGC CGG TGC TGA-3′). The primers used for whole-viral-genome PCRs were STLV-genome forward (5′-TCA GCA GCT CAG ACT −3′) and STLV-genome reverse (5′-TTT TAG ACT TCT GTT TCG CGG-3′). PCRs were performed with 20 to 25 cycles to minimize PCR errors.
To sequence tax, the PCR products were gel purified and cloned into a sequencing vector using a Zero Blunt Topo PCR cloning kit (Life Technologies) according to the manufacturer's specifications. Ligated plasmid was transfected into One Shot TOP10 chemically competent Escherichia coli cells (Life Technologies), 10 clones from each sample were purified using a Zyppy plasmid miniprep kit (Zymo Research), and the plasmid was sent for Sanger sequencing (GenScript USA Inc., Piscataway, NJ, USA) using the T7 and M13 reverse sequencing primers supplied by GenScript. Sequence results were analyzed using MacVector sequence analysis software (v14.0.6).
Next-generation sequencing.
Whole STVL-1 genomes were PCR amplified from the baboon T cell lines as described above. PCR products of between 8.5 and 9.5 kb were purified using a Zymoclean gel DNA recovery kit (Zymo Research). To remove any possible contaminants that could complicate sequencing, the PCR products were further purified before NGS using a ZR DNA sequencing cleanup kit (Zymo Research).
NGS was performed by the University of Miami’s John P. Hussman Institute for Human Genomics sequencing core. Library construction was performed using a NEBNext Ultra DNA library preparation kit for Illumina according to the manufacturer's protocol, starting with 1,000 ng DNA with indexing to allow for multiplexing. Cluster generation took place on an Illumina cBot system according to the manufacturer's recommendations. Sequencing was performed on an Illumina HiSeq 2500 sequencer using an Illumina TruSeq PE cluster kit (v4; catalog number PE-401-4001) and TruSeq SBS kit (v4; 250 Cycle; catalog number FC-401-4003) according to the manufacturer's recommendations. Two samples were multiplexed in one-sixth of a lane of a 2 × 125-bp run, generating 21.2 million and 23.5 million pass-filter reads per sample for animals 12752 and 12141, respectively. These raw reads were aligned to a baboon simian T lymphotropic virus 1 isolate F88395 (NCBI GI:442578041) using the BWA algorithm (38). The resulting BAM files were processed according to the best practice recommendations of the Genome Analysis Toolkit (GATK) (39), including the marking of duplicates, indel realignment, and base quality recalibration. Single nucleotide variants were called using the GATK universal genotyper (40).
Peptide libraries.
STLV-1 peptides specific for Tax were designed on the basis of the reference sequence of an STLV-1 isolate from the olive baboon, isolate F88395 (GenBank accession number JX987040.1). Individual peptides were 15 amino acid residues long and overlapped by 11 amino acids. These peptides were designed to span the complete STLV-1 Tax amino acid sequence.
The amino acid sequences of the Tax proteins from animals 12752 and 12141 were aligned to the STLV-1 reference sequence. Variant peptides were designed in any region of the sequence that differed from the reference sequence. Peptides were synthesized by GenScript USA Inc. (Piscataway, NJ, USA) to a purity of >90%. The peptides were previously titrated to determine the optimal concentration for detection of a low-frequency response (17). Each peptide was used for intracellular cytokine staining (ICS) at a concentration of 10 μM. The complete list of peptides and peptide subsets can be found in Table 3.
ICS assay.
We performed a multiparametric ICS assay by incubating freshly isolated PBMC with the reference peptide and variant peptides corresponding to regions of Tax that differed between the reference genome and the genomes of the isolates from animals 12752 and 12141. For a full list of the peptides tested, refer to Table 3. Cells (1.5 × 106) were incubated in a final volume of 200 μl of RPMI 1640 culture medium supplemented with FBS, penicillin-streptomycin, and l-glutamine for 8 h at 37°C in a 5% CO2 incubator in the presence of anti-CD28 (clone L293; BD Biosciences, San Jose, CA, USA), anti-CD49d (clone 9F10; BD Biosciences), and anti-CD107a phycoerythrin (PE; clone H4A3; BioLegend, San Diego, CA) antibodies, 5.0 μg of brefeldin A (BioLegend), and GolgiStop protein transport inhibitor (BD Biosciences). 15-mer peptides were added at a final concentration of 10 μM each. PBMC costimulated in the absence of 15-mer peptides were used as a negative control.
Cells were then stained at 4°C with antibodies directed against the surface molecules CD3 (PerCP-Cy5.5; clone SP34-2; BD Biosciences), CD4 (BV605; clone L200; BD Biosciences), CD8 (FITC; clone RPA-T8; BioLegend), CD14 (BV510; clone M5E2; BioLegend), CD16 (BV510; clone 3G8; BioLegend), and CD20 (BV510; clone 2H7; BioLegend). An amine aqua dye (Invitrogen) was also included to discriminate between live and dead cells. Cells were then fixed and permeabilized using Cytofix/Cytoperm fixation/permeabilization solution (BD Biosciences) prior to staining with antibodies against IFN-γ (BV421; clone 4S.B3; BioLegend), TNF-α (allophycocyanin; clone MAb11; BD Biosciences), and IL-2 (PE-Cy7; clone MQ1-17H12; BioLegend). For each sample, 250,000 events were acquired using a FACSDiva flow cytometer (v8.0.1) on a special-order research product (SORP) BD LSR II instrument equipped with a 50-mW 405-nm violet laser, a 100-mW 488-nm blue laser, and a 50-mW 640-nm red laser, and data were analyzed with FlowJo (v9.8.5) software.
For data analysis, we first gated on time as a quality control parameter (in which perturbations that may be caused by larger aggregates, sample pressure drops, or air bubbles were excluded). Then we gated on lymphocytes (forward scatter [FSC]-by-side scatter [SSC] plot) and excluded aggregates from the analysis using plots of the forward scatter height (FSC-H) versus forward scatter width (FSC-W) and the side scatter height (SSC-H) versus side scatter width (SSC-W) (data not shown). Subsequently, we gated on CD3+ T lymphocytes, excluding CD14+, CD16+, CD20+, and dead cells. At this stage, we separated T lymphocyte subsets on the basis of their expression of either CD4 or CD8 (excluding those expressing both markers) and conducted our analyses (of IFN-γ, CD107a, TNF-α, and IL-2 expression) within these two compartments. Therefore, we expressed our results as the median frequency of CD8+ T cells expressing the cytolysis marker and/or a cytokine(s). For a graphical representation of the gating strategy, see Fig. 4A.
As controls, we stimulated PBMC from STLV-1-negative animals with the same peptides used to stimulate the PBMC from STLV-1-infected animals. Additionally, we included a sample that was not stimulated for assessment of background response levels. Experiments with Tax were conducted for STLV-infected animals (animals 12752 and 12141). An STLV-1-naive animal (animal 15659) was used as a negative control. One STLV-1-positive animal (animal 12869) with previously mapped Tax peptide responses was included as a positive control.
Accession number(s).
The sequences of the STLV-1 isolates from animals 12752 and 12141 have been submitted to the GenBank database and may be found under accession numbers MF621979 and MF621980, respectively.
ACKNOWLEDGMENTS
We are appreciative of the University of Miami’s John P. Hussman Institute for Human Genomics sequencing core for their assistance in next-generation sequencing and data analysis. We also thank Leydi Guzman for administrative assistance as well as Teresa Giret for advice on and expertise with STLV.
This work was supported by the Sylvester Comprehensive Cancer Center, University of Miami Miller School of Medicine, and HTLV-1 Program Project internal funds. In addition, this investigation used resources that are supported by Southwest National Primate Research Center (SNPRC) grant P51 RR013986 from the National Center for Research Resources, National Institutes of Health, and that are currently supported by the Office of Research Infrastructure Programs through grant P51 OD011133.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
J.M.T., D.M.M., D.I.W., and R.C.D. formulated the experimental plans and interpreted the results. J.M.T., D.M.M., H.S.M., I.C., and W.L. performed the experiments. J.M.T and R.C.D. wrote the manuscript, and D.M.M. and D.I.W. helped to revise it. J.P. organized animal selections, animal use, and blood sampling. G.N.B. provided funding and direction and helped with interpretation.
REFERENCES
- 1.Poiesz BJ, Ruscetti FW, Gazdar AF, Bunn PA, Minna JD, Gallo RC. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci U S A 77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matsuoka M, Jeang KT. 2007. Human T-cell leukaemia virus type 1 (HTLV-1) infectivity and cellular transformation. Nat Rev Cancer 7:270–280. doi: 10.1038/nrc2111. [DOI] [PubMed] [Google Scholar]
- 3.Proietti FA, Carneiro-Proietti AB, Catalan-Soares BC, Murphy EL. 2005. Global epidemiology of HTLV-I infection and associated diseases. Oncogene 24:6058–6068. doi: 10.1038/sj.onc.1208968. [DOI] [PubMed] [Google Scholar]
- 4.Verdonck K, Gonzalez E, Van Dooren S, Vandamme AM, Vanham G, Gotuzzo E. 2007. Human T-lymphotropic virus 1: recent knowledge about an ancient infection. Lancet Infect Dis 7:266–281. doi: 10.1016/S1473-3099(07)70081-6. [DOI] [PubMed] [Google Scholar]
- 5.Goncalves DU, Proietti FA, Ribas JG, Araujo MG, Pinheiro SR, Guedes AC, Carneiro-Proietti AB. 2010. Epidemiology, treatment, and prevention of human T-cell leukemia virus type 1-associated diseases. Clin Microbiol Rev 23:577–589. doi: 10.1128/CMR.00063-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miura M, Yasunaga J, Tanabe J, Sugata K, Zhao T, Ma G, Miyazato P, Ohshima K, Kaneko A, Watanabe A, Saito A, Akari H, Matsuoka M. 2013. Characterization of simian T-cell leukemia virus type 1 in naturally infected Japanese macaques as a model of HTLV-1 infection. Retrovirology 10:118. doi: 10.1186/1742-4690-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Iwanaga M, Watanabe T, Yamaguchi K. 2012. Adult T-cell leukemia: a review of epidemiological evidence. Front Microbiol 3:322. doi: 10.3389/fmicb.2012.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koralnik IJ, Boeri E, Saxinger WC, Monico AL, Fullen J, Gessain A, Guo HG, Gallo RC, Markham P, Kalyanaraman V. 1994. Phylogenetic associations of human and simian T-cell leukemia/lymphotropic virus type I strains: evidence for interspecies transmission. J Virol 68:2693–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Switzer WM, Qari SH, Wolfe ND, Burke DS, Folks TM, Heneine W. 2006. Ancient origin and molecular features of the novel human T-lymphotropic virus type 3 revealed by complete genome analysis. J Virol 80:7427–7438. doi: 10.1128/JVI.00690-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ibrahim F, de The G, Gessain A. 1995. Isolation and characterization of a new simian T-cell leukemia virus type 1 from naturally infected Celebes macaques (Macaca tonkeana): complete nucleotide sequence and phylogenetic relationship with the Australo-Melanesian human T-cell leukemia virus type 1. J Virol 69:6980–6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bomford R, Kazanji M, De The G. 1996. Vaccine against human T cell leukemia-lymphoma virus type I: progress and prospects. AIDS Res Hum Retroviruses 12:403–405. doi: 10.1089/aid.1996.12.403. [DOI] [PubMed] [Google Scholar]
- 12.Allan JS, Leland M, Broussard S, Mone J, Hubbard G. 2001. Simian T-cell lymphotropic viruses (STLVs) and lymphomas in African nonhuman primates. Cancer Invest 19:383–395. doi: 10.1081/CNV-100103133. [DOI] [PubMed] [Google Scholar]
- 13.d'Offay JM, Eberle R, Wolf RF, Kosanke SD, Doocy KR, Ayalew S, Mansfeild KG, White GL. 2013. Simian T-lymphotropic virus-associated lymphoma in 2 naturally infected baboons: T-cell clonal expansion and immune response during tumor development. Comp Med 63:288–294. [PMC free article] [PubMed] [Google Scholar]
- 14.Mahieux R, Pecon-Slattery J, Chen GM, Gessain A. 1998. Evolutionary inferences of novel simian T lymphotropic virus type 1 from wild-caught chacma (Papio ursinus) and olive baboons (Papio anubis). Virology 251:71–84. doi: 10.1006/viro.1998.9377. [DOI] [PubMed] [Google Scholar]
- 15.d'Offay JM, Eberle R, Sucol Y, Schoelkopf L, White MA, Valentine BD, White GL, Lerche NW. 2007. Transmission dynamics of simian T-lymphotropic virus type 1 (STLV1) in a baboon breeding colony: predominance of female-to-female transmission. Comp Med 57:105–114. [PubMed] [Google Scholar]
- 16.Courgnaud V, Van Dooren S, Liegeois F, Pourrut X, Abela B, Loul S, Mpoudi-Ngole E, Vandamme A, Delaporte E, Peeters M. 2004. Simian T-cell leukemia virus (STLV) infection in wild primate populations in Cameroon: evidence for dual STLV type 1 and type 3 infection in agile mangabeys (Cercocebus agilis). J Virol 78:4700–4709. doi: 10.1128/JVI.78.9.4700-4709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castro I, Giret TM, Magnani DM, Maxwell HS, Umland O, Perry JK, Pecotte JK, Brasky KM, Barber GN, Desrosiers RC, Watkins DI. 2016. Cellular immune responses against simian T-lymphotropic virus type 1 target Tax in infected baboons. J Virol 90:5280–5291. doi: 10.1128/JVI.00281-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanon E, Hall S, Taylor GP, Saito M, Davis R, Tanaka Y, Usuku K, Osame M, Weber JN, Bangham CR. 2000. Abundant Tax protein expression in CD4+ T cells infected with human T-cell lymphotropic virus type I (HTLV-I) is prevented by cytotoxic T lymphocytes. Blood 95:1386–1392. [PubMed] [Google Scholar]
- 19.Koenig S, Woods R, Brewah Y, Newell A, Jones G, Boone E, Adelsberger J, Baseler M, Robinson S, Jacobson S. 1993. Characterization of MHC class I restricted cytotoxic T cell responses to Tax in HTLV-1 infected patients with neurologic disease. J Immunol 151:3874–3883. [PubMed] [Google Scholar]
- 20.Bangham CRM, Kermode AG, Hall SE, Daenke S. 1996. The cytotoxic T-lymphocyte response to HTLV-I: the main determinant of disease? Semin Virol 7:41–48. doi: 10.1006/smvy.1996.0006. [DOI] [Google Scholar]
- 21.Fan J, Ma G, Nosaka K, Tanabe J, Satou Y, Koito A, Wain-Hobson S, Vartanian JP, Matsuoka M. 2010. APOBEC3G generates nonsense mutations in human T-cell leukemia virus type 1 proviral genomes in vivo. J Virol 84:7278–7287. doi: 10.1128/JVI.02239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giri A, Slattery JP, Heneine W, Gessain A, Rivadeneira E, Desrosiers RC, Rosen L, Anthony R, Pamungkas J, Iskandriati D, Richards AL, Herve V, McClure H, O'Brien SJ, Franchini G. 1997. The tax gene sequences form two divergent monophyletic lineages corresponding to types I and II of simian and human T-cell leukemia/lymphotropic viruses. Virology 231:96–104. doi: 10.1006/viro.1997.8511. [DOI] [PubMed] [Google Scholar]
- 23.Nomura M, Ohashi T, Nishikawa K, Nishitsuji H, Kurihara K, Hasegawa A, Furuta RA, Fujisawa J, Tanaka Y, Hanabuchi S, Harashima N, Masuda T, Kannagi M. 2004. Repression of Tax expression is associated both with resistance of human T-cell leukemia virus type 1-infected T cells to killing by Tax-specific cytotoxic T lymphocytes and with impaired tumorigenicity in a rat model. J Virol 78:3827–3836. doi: 10.1128/JVI.78.8.3827-3836.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida M, Seiki M, Yamaguchi K, Takatsuki K. 1984. Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proc Natl Acad Sci U S A 81:2534–2537. doi: 10.1073/pnas.81.8.2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etoh K-I, Tamiya S, Yamaguchi K, Okayama A, Tsubouchi H, Ideta T, Mueller N, Takatsuki K, Matsuoka M. 1997. Clonal proliferation of human T-lymphotropic virus type I-infected cells. Cancer Res 57:4862. [PubMed] [Google Scholar]
- 26.Etoh K, Yamaguchi K, Tokudome S, Watanabe T, Okayama A, Stuver S, Mueller N, Takatsuki K, Matsuoka M. 1999. Rapid quantification of HTLV-I provirus load: detection of monoclonal proliferation of HTLV-I-infected cells among blood donors. Int J Cancer 81:859–864. [DOI] [PubMed] [Google Scholar]
- 27.Wattel E, Vartanian JP, Pannetier C, Wain-Hobson S. 1995. Clonal expansion of human T-cell leukemia virus type I-infected cells in asymptomatic and symptomatic carriers without malignancy. J Virol 69:2863–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wodarz D, Bangham RMC. 2000. Evolutionary dynamics of HTLV-I. J Mol Evol 50:448–455. doi: 10.1007/s002390010047. [DOI] [PubMed] [Google Scholar]
- 29.Kubota R, Hanada K, Furukawa Y, Arimura K, Osame M, Gojobori T, Izumo S. 2007. Genetic stability of human T lymphotropic virus type I despite antiviral pressures by CTLs. J Immunol 178:5966–5972. doi: 10.4049/jimmunol.178.9.5966. [DOI] [PubMed] [Google Scholar]
- 30.Furukawa Y, Tara M, Izumo S, Arimura K, Osame M. 2006. HTLV-I viral escape and host genetic changes in the development of adult T cell leukemia. Int J Cancer 118:381–387. doi: 10.1002/ijc.21328. [DOI] [PubMed] [Google Scholar]
- 31.Furukawa Y, Kubota R, Tara M, Izumo S, Osame M. 2001. Existence of escape mutant in HTLV-I tax during the development of adult T-cell leukemia. Blood 97:987–993. [DOI] [PubMed] [Google Scholar]
- 32.Sugata K, Yasunaga J, Mitobe Y, Miura M, Miyazato P, Kohara M, Matsuoka M. 2015. Protective effect of cytotoxic T lymphocytes targeting HTLV-1 bZIP factor. Blood 126:1095–1105. doi: 10.1182/blood-2015-04-641118. [DOI] [PubMed] [Google Scholar]
- 33.Zhao T, Matsuoka M. 2012. HBZ and its roles in HTLV-1 oncogenesis. Front Microbiol 3:247. doi: 10.3389/fmicb.2012.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahieux R. 2015. A vaccine against HTLV-1 HBZ makes sense. Blood 126:1052–1053. doi: 10.1182/blood-2015-06-652040. [DOI] [PubMed] [Google Scholar]
- 35.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 36.Liao Q, Guo H, Tang M, Touzjian N, Lerche NW, Lu Y, Yee JL. 2011. Simultaneous detection of antibodies to five simian viruses in nonhuman primates using recombinant viral protein based multiplex microbead immunoassays. J Virol Methods 178:143–152. doi: 10.1016/j.jviromet.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wunderlich ML, Dodge ME, Dhawan RK, Shek WR. 2011. Multiplexed fluorometric immunoassay testing methodology and troubleshooting. J Vis Exp 2011:3715. doi: 10.3791/3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]