ABSTRACT
Our understanding of archaeal virus diversity and structure is just beginning to emerge. Here we describe a new archaeal virus, tentatively named Metallosphaera turreted icosahedral virus (MTIV), that was isolated from an acidic hot spring in Yellowstone National Park, USA. Two strains of the virus were identified and were found to replicate in an archaeal host species closely related to Metallosphaera yellowstonensis. Each strain encodes a 9.8- to 9.9-kb linear double-stranded DNA (dsDNA) genome with large inverted terminal repeats. Each genome encodes 21 open reading frames (ORFs). The ORFs display high homology between the strains, but they are quite distinct from other known viral genes. The 70-nm-diameter virion is built on a T=28 icosahedral lattice. Both single particle cryo-electron microscopy and cryotomography reconstructions reveal an unusual structure that has 42 turret-like projections: 12 pentameric turrets positioned on the icosahedral 5-fold axes and 30 turrets with apparent hexameric symmetry positioned on the icosahedral 2-fold axes. Both the virion structural properties and the genome content support MTIV as the founding member of a new family of archaeal viruses.
IMPORTANCE Many archaeal viruses are quite different from viruses infecting bacteria and eukaryotes. Initial characterization of MTIV reveals a virus distinct from other known bacterial, eukaryotic, and archaeal viruses; this finding suggests that viruses infecting Archaea are still an understudied group. As the first known virus infecting a Metallosphaera sp., MTIV provides a new system for exploring archaeal virology by examining host-virus interactions and the unique features of MTIV structure-function relationships. These studies will likely expand our understanding of virus ecology and evolution.
KEYWORDS: Archaea, archaeal virus, crenarchaeal, cryo-electron microscopy, environmental virology, extremophiles, viruses in extreme environments
INTRODUCTION
The recognition of Archaea as a separate domain of cellular life is relatively recent. Consequently, our knowledge of archaeal cell biology, biochemistry, genetics, and viruses lags far behind of that of bacteria and eukaryotes. Viruses infecting members of the Archaea are relatively understudied. Of the >6,000 viral isolates infecting prokaryotes, only 120 infect archaeal hosts (1–5). Most of these archaeal viruses infect just a few genera. Of the 41 viral isolates infecting members of the phylum Crenarchaeaota, 16 infect the species of just one genus, Sulfolobus, and 11 infect Acidianus species (2, 3). More viruses have been isolated from a single species of eukaryotes, Homo sapiens, than from an entire domain of life, Archaea (6). For full understanding of the virosphere, a more comprehensive understanding of archaeal virus diversity and function is necessary.
Despite the limited number of known archaeal viruses, they encompass a wide range of genetic content and morphological diversity (1, 2). Most predicted open reading frames (ORFs) in archaeal viral genomes have little similarity to known proteins, and virion morphology differs more among the viruses of Archaea than among those of Bacteria and Eukarya (2, 7, 8). Archaeal virions range from the icosahedral and head-and-tail phage-like particles seen in bacterial and eukaryotic viruses to unusual bottle, lemon, and spindle shapes unique to archaeal viruses (2, 7, 8). The evolution of these unusual viruses remains a major unresolved question.
Archaeal virions with icosahedral morphology are found in both the Crenarchaeaota and Euryarchaeota phyla of Archaea (2). Sulfolobus turreted icosahedral viruses 1 and 2 (STIV 1 and STIV 2) are among the better-characterized icosahedral archaeal viruses. The STIV major structural protein consists of a double β-barrel jelly roll fold common among icosahedral viruses, forming a pseudo-T=31, 74-nm-diameter virus particle with turret-like projections extending 13 nm at each 5-fold axis (9). STIV also contains an internal lipid bilayer derived from a subset of host cell membranes (9, 10). Other known icosahedral archaeal viruses, which are found replicating in halophilic archaeal hosts, are structurally similar to STIV, sharing high triangulation numbers, an internal lipid bilayer, and the double β-barrel fold of their major capsid proteins (11–14). One surprise of the high-resolution structure of STIVs was that the double β-barrel fold of their major capsid proteins is highly structurally similar to those of adenovirus and phage PRD1, demonstrating a likely evolutionary link between archaeal viruses and viruses from the two other domains of life (9).
We describe here a new archaeal virus isolated from a hot spring located in Yellowstone National Park (YNP), USA. The virion is built on an unusual icosahedral structure with turret-like projections; it packages a small, linear double-stranded DNA (dsDNA) genome quite distinct from that of any previously known virus. This virus likely represents the founding member of a new family of archaeal viruses.
RESULTS
Virus and host isolation and characterization.
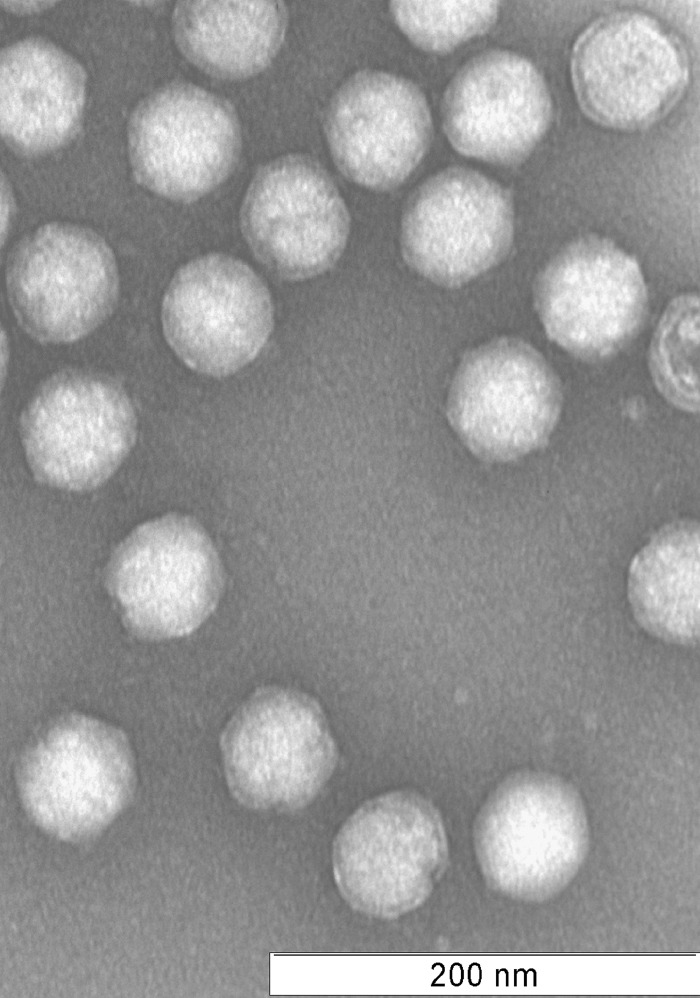
Spherical 65- to 70-nm-diameter virus particles were initially visualized in primary enrichment cultures established from hot spring samples. Similar virus particles continued to be observed after three successive rounds of serial dilution. The virus was purified on Cs2SO4 buoyant density gradients at a density of 1.225 g/cm3, within range of other archaeal viruses (9, 15–17). Negative staining of purified particles revealed virions consisting of two spherical layers and uniformly distributed projections protruding from the virion surface (Fig. 1).
FIG 1.
Negative-stain electron micrograph of purified virions. The 70-nm particles exhibit icosahedral morphology with turret-like protrusions across the capsid. The particles display at least two layers, with ring-like structures surrounding each capsid.
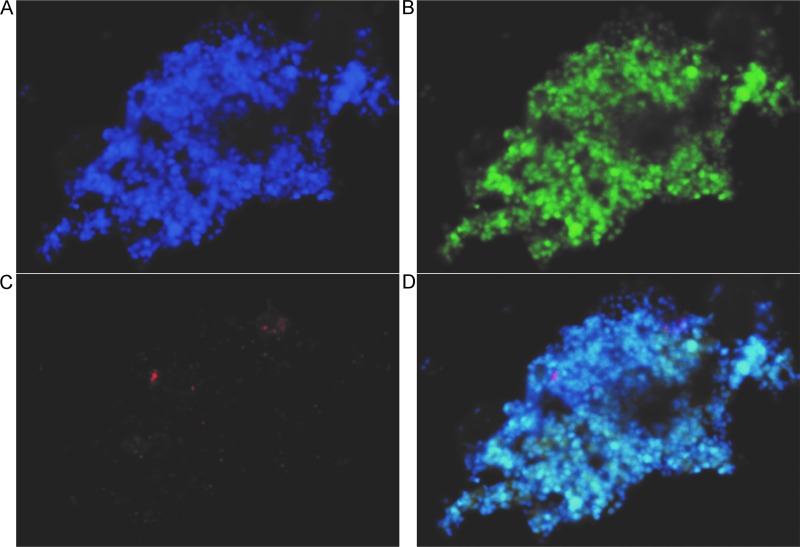
To identify the virus host, cells from cultures producing virus after three successive rounds of serial dilution were collected and subjected to PCR-based amplification, cloning, and sequencing of the near-full-length 16S rRNA gene. Of the 18 resulting sequences, 17 matched with 99% identity to the 16S rRNA gene of Metallosphaera yellowstonensis MK1, a strain previously isolated from Yellowstone National Park (18). The one remaining sequence aligned with 99% identity to an unclassified Sulfolobus species (19–21). After an additional five rounds of passaging the culture, recloning and sequencing of the 16S rRNA gene resulted in 16 clones matching Metallosphaera yellowstonensis and 1 sequence aligning to an Acidianus brierleyi strain, DSM 1651 (NCBI RefSeq no. NR_028246.1). To further test Metallosphaera yellowstonensis as a viral host, cells from virus-producing cultures were dually labeled with fluorescent probes specific to the virus genome and to Metallosphaera 16S rRNA by use of a direct viral fluorescence in situ hybridization (FISH) assay (Fig. 2) (22). We observed colocalization of both the virus DNA (red) and Metallosphaera cell rRNA (green) probes. Dual labeling for the Acidianus cell type failed to show colocalization with the viral genome. Controls of viral and Metallosphaera probes in STIV and Sulfolobus solfataricus culture failed to fluoresce, demonstrating probe specificity. Both 16S rRNA gene sequencing data and catalyzed reporter deposition (CARD)-FISH analysis indicated that a Metallosphaera-type organism was the virus host. Furthermore, the cultures required pyrite for growth, a common feature of Metallosphaera organisms, which are capable of autotrophic growth using pyrite as an electron donor (18, 23). Unfortunately, a virus-free culture of Metallosphaera yellowstonensis strain MK1 (kindly provided by W. Inskeep, Montana State University) was not susceptible to virus infection.
FIG 2.
Direct viral FISH images of virus-host cultures. (A) Total nucleic acids are stained blue with DAPI. (B) Metallosphaera rRNA is labeled in green. (C) Viral nucleic acids are labeled in red. (D) Overlay shows the colocalization of total nucleic acids, Metallosphaera rRNA, and viral DNA and indicates that a Metallosphaera organism is the viral host. Bar, 5 μm.
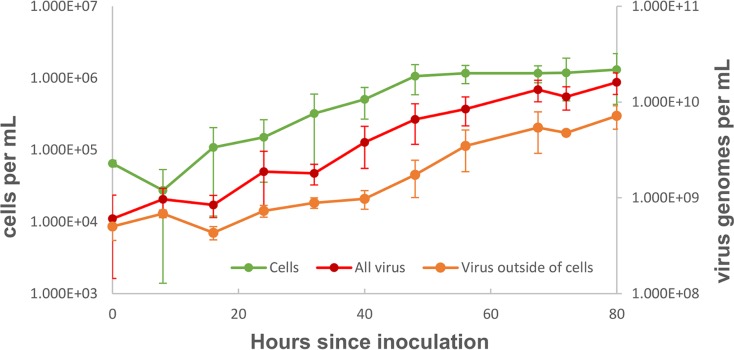
Despite numerous attempts, we were unable to obtain a virus-free isolate in order to complete Koch's postulates, indicating that the cellular host is likely chronically infected by the virus. Virus production closely mirrors cellular growth (Fig. 3). The ratio of extracellular virus particles to cells remains relatively constant over time (∼1,000:1). Notably, cell numbers do not decline during virus production, as is typical for many lytic viruses, suggesting that the virus chronically infects its host. Taken together, these results indicate that the virus is chronically infecting a host closely related to Metallosphaera yellowstonensis MK1. Based on its host identification and virion morphology (described in further detail below), we have named this virus Metallosphaera icosahedral turreted virus (MITV).
FIG 3.
Growth curve of virus and cells in enrichment culture. During the 80 h postpassage, virus genomes and cell counts increase by more than an order of magnitude. No cell decline is observed with virus genome increases, suggesting a chronic infection.
Virus genome.
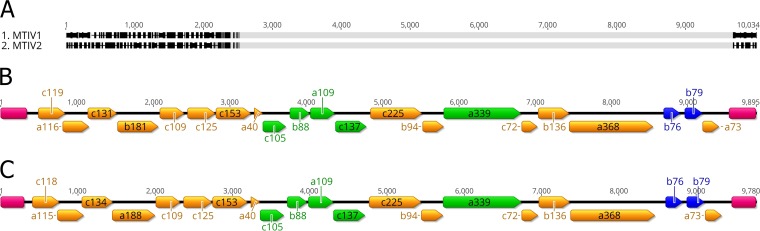
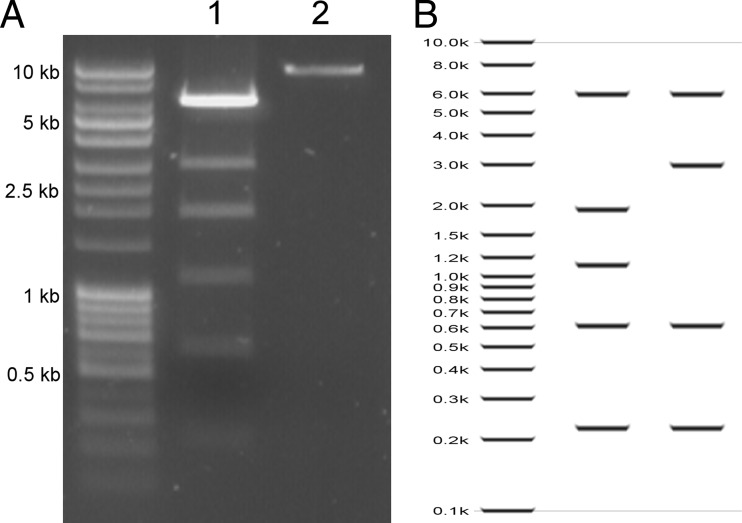
The sequences of nucleic acids extracted from purified virions were assembled into two major contigs of 9,780 and 9,895 bp (Fig. 4). Each contig likely represents the (nearly) full-length linear genome of one of two strains of MITV, referred to here as MITV1 and MITV2. The overall nucleotide identity between the two strains is 89%; the major differences between the strains are the sequences that compose inverted terminal repeats (ITRs) and five ORFs encoded on the 5′ end of each genome. The two genomes have similar GC contents (54.0% and 53.4%, respectively), which are similar to that of the closest known cellular host, Metallosphaera yellowstonensis (GC content, 49%) (18). Restriction endonuclease mapping and primer extension assays of isolated viral genomes confirmed that each MITV strain packages a dsDNA linear genome of ∼9.8 kb (Fig. 5).
FIG 4.
MTIV genomes. The two assembled viral genomes, MTIV1 and MTIV2, are 89% identical at the nucleotide level. (A) The nucleotide differences between MTIV1 and MTIV2 are located at the ends of the genomes (black bars). (B and C) Each strain encodes 21 predicted ORFs, which are represented by filled colored arrows. (B) MTIV1; (C) MTIV2. Only two ORFs with significant similarity to known proteins were identified in each genome (b76 and b79 [blue]) (see the text). Virion structural proteins were identified by mass spectrometry and are color-coded green. Both MTIV1 and MTIV2 genomes are capped with inverted terminal repeats (pink).
FIG 5.
EcoRI digest of MTIV DNA. (A) EcoRI digestion of DNA extracted from MTIV virions resulted in six bands on a 0.1% agarose gel (lane 1). In the no-enzyme negative control (lane 2), one band at approximately 10 kb was observed. (B) In silico EcoRI digests of MTIV2 (left) and MTIV1 (right) illustrate how the combination of the two strains would result in the banding pattern observed in panel A.
Both MTIV strains possess large ITRs (Fig. 4). MTIV1 contains a 318-bp ITR, whereas MTIV2 contains a 355-bp ITR. There is no significant similarity in sequence or predicted structures between the ITRs of the two viral strains. Both ITRs lack obvious palindromic or hairpin sequences, and the ends of both ITRs consist of ∼10 bp of homopolymeric tracks of guanine or cytosine. Like those of other linear dsDNA viruses, these ITRs are likely involved in genome replication (24).
The genome of each MTIV strain encodes 21 highly similar ORFs (Table 1). Most of the predicted proteins are small (10 to 15 kDa). In both genomes, all ORFs are coded on one strand (Fig. 4). MITV1 is predicted to be 91% coding, and MITV2 is predicted to be 90% coding, proportions typical of the coding efficiency of viruses. All predicted ORFs use standard start codons: (16 AUG, 4 GUG, and 1 UUG codon). Most start codons (13 of 21) are preceded 12 to 25 bp upstream by at least 5 bp of TATA-like archaeal promoter sequence. Polycistronic mRNAs could potentially be produced to translate ORFs b88 and a109, which were identified as major structural proteins (described below). Sixteen ORFs are identical in the two genomes. Predicted protein pairwise identity for the remaining ORFs ranges from 37.9% to 84.4%. These variable ORFs are in the initial coding section of each genome and are nearly identical in length, although several of the predicted proteins have a few extra amino acids on their C termini. Overall, genome synteny and identity suggest that these are two strains of the same virus with similar gene products and functions.
TABLE 1.
MTIV genome annotations
| ORF name in MTIV1 | Mol mass (kDa) of predicted protein in MTIV1 | ORF name in MTIV2 | Mol mass (kDa) of predicted protein in MTIV2 | % identity |
Start codon | Annotation | |
|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | ||||||
| c118 | 13.20 | c119 | 13.41 | 66.8 | 53.8 | AUG | |
| a115 | 13.23 | a116 | 13.82 | 57.3 | 37.9 | AUG | |
| c134 | 16.02 | c131 | 15.28 | 63.4 | 43.5 | AUG | |
| a188 | 21.52 | b181 | 20.94 | 60.6 | 51.1 | AUG | |
| c109 | 12.87 | c109 | 13.08 | 83.9 | 84.4 | AUG | |
| c125 | 14.94 | c125 | 14.94 | 100.0 | 100.0 | AUG | |
| c153 | 16.81 | c153 | 16.81 | 100.0 | 100.0 | AUG | |
| a40 | 4.82 | a40 | 4.82 | 100.0 | 100.0 | AUG | |
| c105 | 12.10 | c105 | 12.10 | 100.0 | 100.0 | GUG | Structural protein |
| b88 | 9.61 | b88 | 9.61 | 100.0 | 100.0 | AUG | Structural protein |
| a109 | 11.20 | a109 | 11.20 | 100.0 | 100.0 | GUG | Structural protein |
| c137 | 14.70 | c137 | 14.70 | 100.0 | 100.0 | GUG | Structural protein |
| c225 | 23.54 | c225 | 23.54 | 100.0 | 100.0 | AUG | |
| b94 | 10.45 | b94 | 10.45 | 100.0 | 100.0 | AUG | |
| a339 | 32.93 | a339 | 32.93 | 100.0 | 100.0 | UUG | Structural protein |
| c72 | 7.98 | c72 | 7.98 | 100.0 | 100.0 | AUG | |
| b136 | 14.55 | b136 | 14.55 | 100.0 | 100.0 | AUG | |
| a368 | 38.48 | a368 | 38.48 | 100.0 | 100.0 | AUG | |
| b76 | 7.67 | b76 | 7.67 | 100.0 | 100.0 | AUG | Acidianus rod-shaped virus hypothetical protein |
| b79 | 9.40 | b79 | 9.40 | 100.0 | 100.0 | AUG | Coiled-coil protein |
| a73 | 8.39 | a73 | 8.39 | 100.0 | 100.0 | GUG | |
The putative viral proteins encoded by the two genomes display little similarity to known viral or cellular proteins. BLAST (August 2016 release), HHpred (25, 26), and Pfam (27) queries failed to find homologous proteins in the public databases for all but two predicted proteins. BLASTp matched b76 with 54% homology across 89% of the ORF to a hypothetical Acidianus rod-shaped virus protein (E value, 6e−16). HHpred analysis, which looks for similarity based on a hidden Markov model and includes secondary-structure predictions, identified an ORF common to both genomes: ORF b79 hit with >90% probability to a variety of coiled-coil proteins, including bacteriophage P22 tail needle proteins, chromosomal segregation ATPases, dynein chains, and flagellar export proteins. This result indicates that b79 is likely a coiled-coil protein but fails to pinpoint its biological function. Overall, MITV genome annotation failed to predict homologous proteins, putative functions, or relationships to known viruses for most predicted ORFs.
Virion proteins.
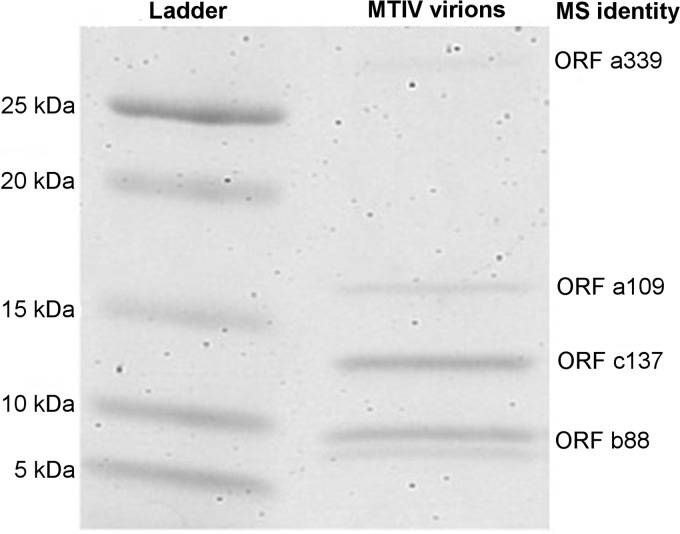
SDS-PAGE analysis of purified virus revealed five bands of approximately 9 kDa, 10 kDa, 14 kDa, 16 kDa, and 32 kDa (Fig. 6). Assuming that the 9-kDa protein is a modified product of the 10-kDa protein, densitometric analysis of the four remaining bands identified a ratio of 20:20:5:2 with respect to ascending size. In-gel trypsin digestion of these bands followed by liquid chromatography-mass spectrometry allowed identification of the viral ORFs encoding these proteins (data not shown). Each band matched spectra to peptides from the predicted ORFs b88, a109, and c137. These ORFs are all adjacent to each other within the conserved region of the viral genomes (Fig. 4). However, in each band, most spectra could be assigned to a single ORF. For the 9- or 10-kDa band, the majority of spectra hit to the predicted 9.60-kDa protein encoded by ORF b88. For the 14-kDa band, the majority of spectra hit to the predicted 14.69-kDa protein encoded by ORF c137. For the 16-kDa band, the majority of spectra hit to the predicted 11.19-kDa protein encoded by ORF a109. For the 32-kDa band, the majority of spectra hit to the predicted 32.91-kDa protein encoded by ORF a339, which is downstream from the cluster of other structural proteins (Fig. 4). The 14-kDa band also contained 17 spectra matching to four peptides from the predicted 12.09-kDa protein encoded by c105, which is adjacent on the 5′ end to the cluster of structural proteins (Fig. 4). These data suggest that the b88, a109, c137, and a339 gene products are major structural proteins of MITV virions, while ORF c105 plays a minor role in the virion. Unlike most major icosahedral capsid proteins (28), secondary-structure predictions for two of the putative structural proteins are predominately α-helical (b88, 83% α-helical; a109, 47% α-helical). In contrast, the c137 protein is predicted to be 60% β-strands, while the a339 product is predicted to be 50% disordered.
FIG 6.
Protein gel of purified virions. The 4-to-20% SDS-PAGE gel of purified virus revealed four major bands at approximately 10, 14, 16, and 32 kDa, with a doublet at the 10-kDa band. By densitometric analysis, these bands are in a 20:20:5:2 ratio. Liquid chromatography followed by mass spectrometry identified the ORF in each band to which most spectra matched (data not shown).
Virion structure.
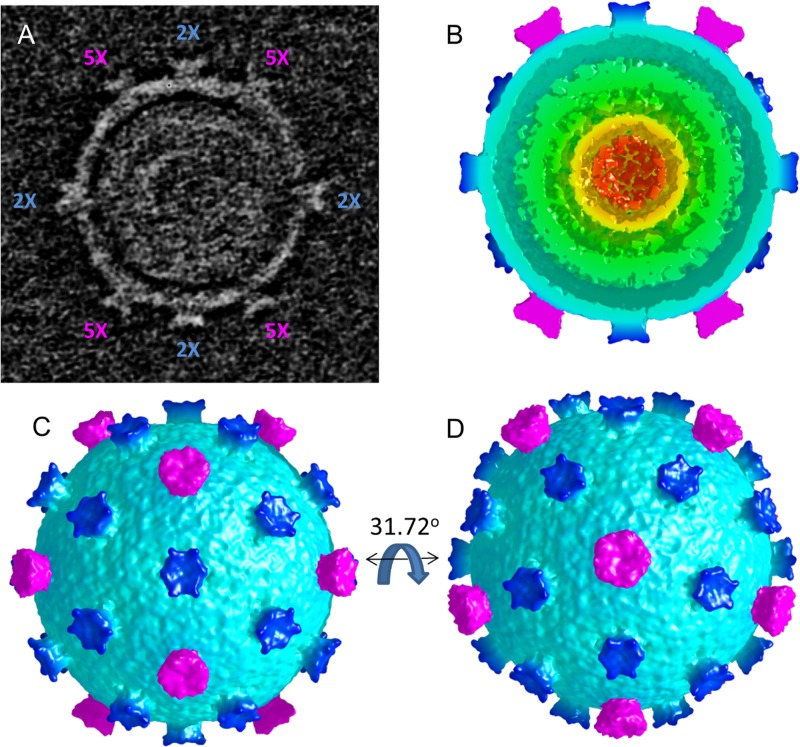
To further investigate virion morphology, both cryo-electron tomography and single-particle analysis were performed. The tomographic work was performed first, since undamaged virus was difficult to purify with high yields. The reconstructed tomogram of a single virion revealed a two-layered spherical shell with turret-like projections extending from the outer surface. Initial segmentation revealed both 5-fold and 6-fold symmetric, turret-like projections. The positions of the 12 5-fold symmetric turrets indicated icosahedral symmetry, allowing the virion to be icosahedrally averaged (Fig. 7). Using two independent half-sets of icosahedral symmetry operators suggests a nominal resolution of ∼3.7 nm, as judged by Fourier shell correlation (FSC; cutoff, 0.143), between the averaged tomographic structure and the 2.2-nm structure from single-particle analysis (see below). The averaged particle shows an outer shell diameter of approximately 70 nm through which extend 12 5-fold symmetric and 30 apparently 6-fold symmetric projections that are coincident with the icosahedral 2-fold axes. The outer shell is 6 to 7 nm thick, and the inner shell is ∼10 nm thick.
FIG 7.
Cryo-electron tomogram of a single, icosahedrally averaged MTIV particle. (A) A single slice through the center of the unaveraged particle rotated into a standard icosahedral orientation (2-fold axes along x, y, and z). The locations of the 2-fold (2×) (magenta) and 5-fold (5×) (blue) axes lying in the central plane are indicated. (B) A single hemisphere of the icosahedrally averaged particle (in a standard orientation; comparable to panel A) reveals the outer surface layer (cyan) surrounding an inner shell (green) that, in turn, encases three additional layers of concentric density (green, yellow, orange) indicative of packaged DNA. (C) A surface view down the icosahedral 2-fold axis (standard orientation). The turrets at the icosahedral 5-fold axes (magenta) and the approximately 6-fold symmetric turrets on the 2-fold axes (dark blue) are shown. (D) The particle is rotated 31.72° about the horizontal axis to show the view down an icosahedral 5-fold axis.
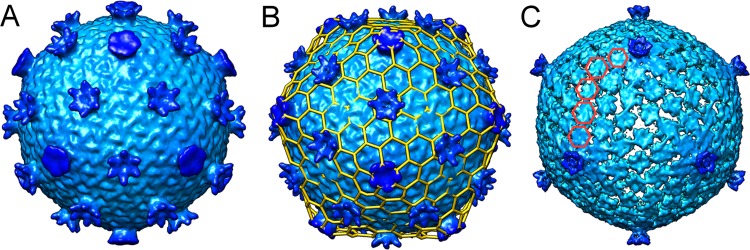
As the purification protocol improved, we generated a small amount of material suitable for preliminary single-particle electron microscopy (EM) analysis. A map calculated from images of 65 virion particles revealed a more-detailed view of this unusual virion morphology (Fig. 8). At 22 Å resolution, the 69.2-nm-diameter outer shell is built on a T=28 icosahedral lattice (Fig. 8B). Importantly, the T=28 lattice places a hexon on each of the icosahedral 2-fold axes. Thus, the T=28 icosahedral symmetry is consistent not only with the observation of 12-penton, 9-nm turrets that mark the 5-fold axes but also with 30-hexon, 10-nm turrets located on the icosahedral 2-fold axes. Overall, the morphologies of the 5-fold and 6-fold turrets appear distinct from each other, but this speculation will need to be confirmed by higher-resolution MTIV structures. Interestingly, at high contour levels, the hexon turrets disappear while the penton turrets remain, suggesting that the hexon turrets lack full occupancy on individual particles (Fig. 8C). In agreement with this, lower occupancy at the hexon turrets is also seen in the averaged tomographic reconstruction of the single virion. These types of transient surface features are reminiscent of many large virus particles found among bacterial and eukaryotic viruses that incorporate cementing or glue proteins that give the capsid increased physical strength or stability (29–31). Overall, structural analysis reveals an unusual virion morphology and gene content not seen previously among archaeal viruses.
FIG 8.
Single-particle cryo-electron microscopy reconstruction of MTIV. (A) Surface view of MTIV at 22 Å resolution showing the two types of turret-like projections extending above the main capsid surface. (B) T=28 icosahedral lattice superimposed on the surface of MTIV. (C) Reduced-contour view of MTIV showing only the 12 turret-like projections at the 5-fold axes.
DISCUSSION
We have isolated an archaeal virus from an acidic, high-temperature hot spring in YNP that is novel in both virion morphology and genome content. The double-shelled 70-nm virion exhibits an icosahedral morphology built on a T=28 icosahedral lattice, with 42 turret-like projections, 12 pentameric turrets positioned on the icosahedral 5-fold axes and 30 turrets with apparent hexameric symmetry positioned on the icosahedral 2-fold axes. The linear dsDNA 9.8-kb genome encodes 21 potential proteins, the vast majority of which show no significant similarity to known proteins. This virus chronically infects a crenarchaeal Metallosphaera species; it is the first virus isolated to infect this genus of Archaea. We propose naming the new virus Metallosphaera turreted icosahedral virus (MTIV) and submit that it represents the founding member of a new archaeal viral family.
At only 9.8 kbp, the MTIV genome is one of the smaller archaeal virus genomes. It is roughly comparable in size to the 5.2-kbp circular genome of Aeropyrum pernix bacilliform virus 1, the 9.7-kbp circular genome of Halogeometricum pleomorphic virus 1, the 8.7-kbp circular genome of Halorubrum virus 3, and the 8-kbp circular genome of Haloarcula hispanica virus 1 (2). Among known icosahedral archaeal viruses, STIV packages a 17.7-kb genome within a 74-nm virion, while SH1 packages a 30.9-kb genome inside a 79-nm virion (9, 32). The presence of long, >300-bp ITRs unique to each MTIV strain suggests that these structures are involved in genome replication, possibly similarly to ITRs in the linear genomes of adenovirus and some herpesviruses (23, 33). It is not known whether MTIV, like adenovirus, uses a protein as a primer for DNA replication.
We identified two different genomes for MTIV present in the enrichment cultures, which likely represent two different strains of MTIV. Approximately 89% of these two genomes are identical at the DNA sequence level. The remaining 11% in sequence divergence is found predominately in each genome's unique ITR sequences. At present, we do not know if these two different genomes are replicating within different cells in the Metallosphaera species population, are separately packed into separate capsids, or are copackaged within the same virion. Our inability to cure the cultures of either or both of these viruses and the nature of host-virus growth curves indicate that the cultures are chronically infected with both strains of MTIV. There is no indication that MTIV integrates into its host chromosome.
Both MTIV genomes encode 21 proteins. Most predicted proteins are identical in the two strains, but the first 5 ORFs encoded on the 5′ end of the genome range in amino acid identity from 43.5 to 84.4%. At present, it is not known why this is the only cluster of divergent viral proteins, but we speculate that they could play a role in avoiding host defense mechanisms. Since both genomes persist in a near-1:1 ratio in culture supernatants, even after multiple passages, they are likely equally fit under our culture conditions.
The MTIV virion is structurally similar to known archaeal viruses in several respects. The T=28, 70-nm virion uses a high triangulation number, like the pseudo-T=31 and T=28 virions of STIV and SH1, respectively (9, 32). In visualizations of the high-contour reconstruction levels, the virus has 12 turret-like projections protruding from each of the 5-fold axes. Although only 9 nm, they are reminiscent of STIV's 13-nm turrets. In STIV, these are suggested to serve as a tunnel for DNA in the virion-packaging or -unpackaging event (34, 35). Additionally, initial cryo-electron tomography analysis suggests a bilayer at the virion's inner surface reminiscent of STIV's internal lipid bilayer. The putative major capsid proteins are small—9, 13, and 16 kDa—less than half the size of STIV's 37-kDa major capsid protein (36). From the ratios of structural proteins visualized on SDS-PAGE gels, it is tempting to speculate that the 10-kDa and 14-kDa bands make up the majority of the underlying T=28 protein shell, that the 32-kDa protein contributes to the 12 5-fold turrets, and that the 16-kDa protein makes up the 20 6-fold related turret structures. In contrast to other archaeal icosahedral viruses, secondary-structure predictions do not suggest a double β-jelly roll fold for any of the major capsid proteins. Using Phyre2, b88 and a109 are predicted to be α-helical, while the c137 and a339 proteins are predicted to be composed of 60% β-strands and to be 50% disordered, respectively. Phyre2 found no significant similarity to known protein structures for any of these MTIV proteins.
Visualizing the single-particle analysis virion reconstruction at low contour levels reveals hexon, turret-like projections on the icosahedral 2-fold axes. It remains unclear whether these extra turrets are present only in a subset of virions and are potentially a result of different viral strains or whether they are cement proteins without full occupancy. The tomographic reconstruction suggests the latter, since the hexon turrets are present in the averaged tomogram only at reduced contour levels. Cement proteins have been reported on other icosahedral viruses, including adenovirus, T4 phage, and bacteriophage L, but not yet in a virus that infects Archaea (29–31). These cement proteins are hypothesized to confer further structural stability. In bacteriophage L, these cement proteins exhibit highly discriminatory binding: the cement protein Dec binds only to the HK-97-like major capsid protein at the quasi-3-fold axis nearest the icosahedral 2-fold axes (30).
Classical virology techniques and culturing limitations of Archaea implicitly limited the types of archaeal viruses discovered. Cell types can host only a finite number of viruses, so archaeal viral diversity can only extend so far when limited to cell cultures of a few genera. MTIV is the first known virus to infect a Metallosphaera cell. Simultaneously, it exhibits a significant lack of genetic homology, even among archaeal viruses, and structural data thus far point toward a novel morphology. Therefore, MTIV indicates that significant diversity remains to be found among archaeal viruses, and studying viruses infecting other genera may lead to the discovery of that diversity.
MATERIALS AND METHODS
Virus isolation and growth.
Hot spring water samples were collected from CHANN041, an acidic (pH 2.1), high temperature (75 to 82°C) thermal feature in Yellowstone National Park (44°65.329′N, 110°48.47′W). Within 24 h of collection, a primary enrichment culture was established by collecting cells from 1 liter of a hot spring sample on an in-line 0.8-μm filter (Millipore, Billerica, MA), gently washing cells from the filter, and resuspending cells in 5 ml of a minimal salt medium [0.4 g/liter (NH4)2SO4, 0.4 g/liter K2HPO4, 0.4 g/liter MgSO4·7H2O, 0.2 g/liter yeast extract (pH 2.5)]. The resulting material was diluted 10-fold into 25 ml of this medium supplemented with 1% (wt/vol) pyrite and was incubated aerobically in a shaking oil bath at 70°C and 130 rpm. After 96 h of incubation, samples were stained with 2% uranyl acetate and were screened for the presence of virus-like particles (VLPs) using a Leo912AB transmission electron microscope (TEM). Using the same growth conditions, primary enrichment cultures displaying VLPs were passaged three times by 10-fold dilution, followed by three serial dilutions to extinction. At each passage, the serially diluted material was screened for VLPs by TEM. Following serial dilutions, the culture-producing virus was maintained by passage of a 10-fold dilution using the conditions described above.
Virus purification.
Virus was collected from cultures during maximal VLP accumulation as estimated by TEM visualization (∼80 h postpassage). Cells were removed by filtration through 0.22-μm in-line filters (Millipore, Darmstadt, Germany), and the resulting virus filtrate was concentrated 10-fold to 100-fold by use of a Corning (Corning, NY) Spin-X UF concentrator (molecular weight cutoff [MWCO], 100,000) to a volume of ∼10 ml. Virus was subsequently purified by banding on a Cs2SO4 continuous density gradient (30% [wt/vol] in culture medium lacking pyrite [pH 2.5]), and fractions were dialyzed into culture medium without yeast extract and were screened for VLPs by TEM or by a quantitative PCR (qPCR) assay (described below). Fractions containing VLPs were pooled and further concentrated 10- to 100-fold to approximately 100 μl in dialysis tubing by dehydration with 40% polyethylene glycol (MWCO, 20,000) in culture medium lacking yeast extract and pyrite.
Virus genome extraction, sequencing, assembly, and analysis.
Nucleic acids were extracted from purified virions by treatment with 0.1% SDS and proteinase K, followed by extraction with Tri reagent and precipitation with ethanol (37). The purified viral DNA was sequenced using the Illumina MiSeq v3 system with 250-nt paired-end reads at the University of Illinois Champaign-Urbana Sequencing Center. The paired reads were trimmed with Trimmomatic, v0.35.0, and reads were subsampled and assembled using the Geneious 9.1.5 de novo assembler (38, 39). Genome assemblies were confirmed by using Geneious 9.1.5 to map all reads onto assembled genomes and by selective PCR amplification, using primers based on the assembled genome, followed by DNA sequencing of the resultant PCR products (38). To confirm the sequences of the viral genome ends, adapters described by Hamilton et al. (40) were ligated onto the viral DNA. Primers unique to each end (for MTIV1, 5′-TGACTCCCCCATCCTCATCA-3′; for MTIV2, 5′-AGTCTCTTGGGGATCTCGCT-3′) and the forward adapter primer were used to PCR amplify the unique genome ends for each viral genome. The resulting PCR products were cloned into the PCR 2.1 plasmid (TOPO-TA cloning kit; Invitrogen, Carlsbad, CA), purified with a PureYield plasmid miniprep kit (Promega, Madison, WI), and subjected to Sanger sequencing.
Open reading frames (ORFs) were identified on the assembled genomes using a combination of Glimmer3 (41), Geneious 9.1.5 (38), and hand curation. The genome and translated ORFs were used to search the NCBI RefSeq database for similarity using BLASTn, BLASTx, and BLASTp (August 2016 release). HHpred (25, 26), Phyre2 (42), and Pfam (27) were also used to search for similarity to translated ORFs.
qPCR assay development.
Primer3 was used to develop qPCR assays for each of the two assembled viral genomes (43, 44). Primers resulting in 150- to 200-bp products were designed for the conserved region of both assembled genomes (F, 5′-ACTACGTCGCCTCTTCTCCT-3′; R, 5′-ATACTGGGCGTACTCCTGGT-3′) and for the unique regions of MTIV1 (F, 5′-TTGATGTGGTGGGCAGGATC-3′; R, 5′-AGTCTCTTGGGGATCTCGCT-3′) and MTIV2 (F, 5′-GGTCAGCCTAACCCTGCATT-3′; R, 5′-ACACCTCACACGGAACATCC-3′). PCR products were cloned, confirmed by DNA sequencing, purified, and used as standards in subsequent qPCR assays. The qPCR assay was performed using SsoFast EvaGreen supermix (Bio-Rad, Hercules, CA) in a Rotor-Gene Q cycler (Qiagen, Hilden, Germany) and was used to track virus during culturing and purification.
Culture growth curve.
Samples of virus-host cultures passaged with a 10-fold dilution were taken every 8 h and were rapidly cooled to 4°C. Within 24 h of collection, cells were stained with SYBR gold stain (Invitrogen, Carlsbad, CA) and were counted using a BD Accuri C6 cytometer (BD Biosciences, San Jose, CA). Fluorescence events were filtered first against a blank medium control and then for cell clumps by plotting side-scattering height over side-scattering area and eliminating nonlinear results. The virus copy number was measured by consensus qPCR both in the unadulterated sample and in the supernatant after cells were pelleted in a microcentrifuge.
Major coat protein identification.
Major proteins associated with purified virions were displayed on a 4-to-20% SDS-PAGE gel. Following staining with Coomassie brilliant blue R-250 (Fisher Scientific, Pittsburgh, PA), major protein bands were excised, subjected to in-gel trypsin digestion (45), and analyzed on a Bruker maXis Impact mass spectrometer. Peaks were used to search a database containing all predicted viral genome ORFs and the Metallosphaera yellowstonensis proteome (NCBI RefSeq no. NZ_AHKJ00000000.1) using X! Tandem (46) and PeptideShaker 1.7.1 (47). Spectrum matches were filtered for m/z errors of <2.
Sequencing of the 16S rRNA gene.
The near-full-length 16S rRNA gene region was amplified from cultures by PCR amplification using a universal archaeal forward primer (5′-TTCCGGTTGATCCCGCCGGA-3′) and reverse primer (5′-GACGGGCGGTGAGTACA-3′). The resulting 1,390-bp product was cloned into the PCR 2.1 vector, purified, and subjected to Sanger sequencing. Sequences were analyzed with the NCBI RefSeq database using BLASTn (August 2016 release).
Host-virus CARD-FISH.
Cultured cells producing virus were grown to the late-log-growth phase, concentrated 100-fold by low-speed centrifugation, and fixed with 1% (wt/vol) paraformaldehyde. Two probes, one specific to the 16S rRNA region of Metallosphaera spp. (5′-TGGGCGCCCCCCGACGGGATC-3′) and one specific to the 16S rRNA region of Acidianus brierleyi (5′-ACCTCTAGACAGTATTAGCCT-3′), were designed. Primers designed for the MTIV genomes using Primer3 (43, 44; also data not shown) were used to amplify 10 DNA probes of approximately 200 to 500 bp from the viral DNA. PCR products were labeled using the Ulysis Alexa Fluor 594 nucleic acid labeling kit (Thermo Fisher Scientific, Waltham, MA) following the protocol described in reference 22. These probes were hybridized to fixed cells so as to differentially fluorescently label the virus and cells via a modified version of the direct-gene FISH method (22). In the modified procedure, cells were dried onto slides and were dehydrated in 50, 80, and 96% ethanol, respectively, for 3 min each, followed by air drying. Cells were then permeabilized in 50 μl of 50 mM glucose–20 mM Tris (pH 7.5)–10 mM EDTA–0.2% Tween 20 and were incubated on ice for 1 h. After permeabilization, cells were washed with 1× PBS for 5 min and with water for 1 min. Native peroxidases were then deactivated with 0.2 N HCl for 10 min, followed by washing with 1× PBS for 5 min, water for 1 min, and 96% EtOH for 1 min, followed by air drying. The hybridization buffer was optimized for 16S rRNA specificity as described previously (3, 48), and the viral probes were designed to hybridize at the same formamide concentration using the protocol described in reference 22. Viral primers were added to the hybridization buffer to a final concentration of 62 pg/μl. Probes were denatured at 85°C for 45 min and were then transferred to 46°C for 2 h. Five minutes after the transfer to 46°C, the 16S rRNA probes were added to a final concentration of 0.16 ng/μl. After hybridization, cells were washed at 48°C for 15 min in washing buffer. Cells were then incubated in 1× PBS for 15 min, followed by a 10-min incubation at 37°C in amplification buffer with 0.0015% H2O2 and 0.33 μg/ml Alexa Fluor 488 Tyramide (Thermo Fisher Scientific, Waltham, MA). Cells were washed at 46°C for 10 min, followed by 1 min in H2O and 1 min in EtOH, followed by air drying. Cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and were mounted with VectaShield antifade mounting medium (Vector Laboratories, Burlingame, CA).
Tomographic data collection.
Purified virions were dialyzed into 5 mM citrate buffer (pH 3.0) and were frozen on Quantifoil R 2/1 holey carbon grids (copper, 200 mesh) (Quantifoil, Großlöbichau, Germany) in liquid propane-ethane using a Vitrobot Mark III system (FEI, Hillsboro, OR). Tilt series were taken using a Titan Krios transmission electron microscope (FEI, Hillsboro, OR) operated at 300 kV and equipped with a Volta phase plate, a Quantum postcolumn imaging energy filter (Gatan, Pleasanton, CA), and a K2 Summit camera (Gatan, Pleasanton, CA) operated in dose-counting mode (49). Tilt series images were collected using SerialEM software (50). Individual frames of images acquired with the K2 camera were aligned in DigitalMicrograph software (Gatan, Pleasanton, CA). Acquisition parameters were as follows: magnification, ×35,700; tilt range, ±60°; tilt increment, 2°; total dose, ∼60 e−/Å2; pixel size, 0.14 nm; defocus value with phase plate, −0.25 μm. The Volta phase plate was operated as described previously (51, 52).
Tomographic reconstruction.
Tomograms were reconstructed using IMOD 4.7 (53). Contrast transfer function (CTF) corrections were not performed. Gold nanoparticles were used as fiducial markers for the alignment of tilt series projection images. The aligned image stack was binned by a factor of 2. The radial filter options were left at their default values (cutoff, 0.35; falloff, 0.05). The resulting tomogram was surface modeled in IMOD, and the icosahedral symmetry revealed was used to average the particle 40-fold using Particle Estimation for Electron Tomography (PEET) (54, 55). Alternatively, the particle underwent 60-fold averaging with strict icosahedral symmetry using AVE (56, 57).
Single-particle cryo-EM analysis.
Cryo-electron microscopy (cryo-EM) specimens were prepared by taking 2.5-μl aliquots of an MTIV preparation (0.1 μg protein/ml in 5 mM citrate buffer [pH 3.0]) and applying them to C-flat perforated carbon grids (Protochips, Inc., Morrisville, NC) that were plasma-cleaned. Cryo-specimens were prepared by manual face-on blotting and vitrification in liquid ethane and were imaged using an Arctica transmission electron microscope (FEI, Hillsboro, OR) operating at 200 kV, at a magnification of ×36,000 (resulting in image sampling at 1.2 Å/pixel), and with defocus values between −2.3 and −4.1 μm. Images were recorded using Leginon, on a K2 Summit direct electron detector (Gatan, Pleasanton, CA) operated in counting mode. Dose-fractioned frames were aligned (58), and contrast transfer function parameters for each image were determined using the CTFFIND4 program (59). Single virion EM images were analyzed using the RELION program (60).
Accession number(s).
The complete sequences of MTIV1 and MTIV2 have been submitted to GenBank under accession numbers MF443783 and MF443784, respectively.
ACKNOWLEDGMENTS
This work was supported by NSF grant DEB-4W4596 to M.J. Y., and it was performed under Yellowstone National Park research permit YELL-2016-SCI-5090. Funding was also provided by National Science Foundation, grant MCB-1413534, to C.M.L. The M. J. Murdock Charitable Trust also funded instrumentation on which cryo-electron microscopy was performed.
Special thanks to Susan Brumfield for assistance with electron microscopy.
REFERENCES
- 1.Ackermann H-W, Prangishvili D. 2012. Prokaryote viruses studied by electron microscopy. Arch Virol 157:1843–1849. doi: 10.1007/s00705-012-1383-y. [DOI] [PubMed] [Google Scholar]
- 2.Dellas N, Snyder JC, Bolduc B, Young MJ. 2014. Archaeal viruses: diversity, replication, and structure. Annu Rev Virol 1:399–426. doi: 10.1146/annurev-virology-031413-085357. [DOI] [PubMed] [Google Scholar]
- 3.Hochstein R, Amenabar MJ, Munson-McGee JH, Boyd ES, Young MJ. 2016. Acidianus tailed spindle virus: a new archaeal large tailed spindle virus discovered by culture-independent methods. J Virol 90:3458–3468. doi: 10.1128/JVI.03098-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rensen EI, Mochizuki T, Quemin E, Schouten S, Krupovic M, Prangishvili D. 2016. A virus of hyperthermophilic archaea with a unique architecture among DNA viruses. Proc Natl Acad Sci U S A 113:2478–2483. doi: 10.1073/pnas.1518929113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, Wang J, Liu Y, Wang Y, Zhang Z, Oksanen HM, Bamford DH, Chen X. 2015. Identification and characterization of SNJ2, the first temperate pleolipovirus integrating into the genome of the SNJ1-lysogenic archaeal strain. Mol Microbiol 98:1002–1020. doi: 10.1111/mmi.13204. [DOI] [PubMed] [Google Scholar]
- 6.International Committee on Taxonomy of Viruses (ICTV). 2017. Virus taxonomy: the classification and nomenclature of viruses. The online (10th) report of the ICTV. https://talk.ictvonline.org/ictv-reports/ictv_online_report/.
- 7.Prangishvili D, Forterre P, Garrett RA. 2006. Viruses of the Archaea: a unifying view. Nat Rev Microbiol 4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 8.Snyder JC, Bolduc B, Young MJ. 2015. 40 years of archaeal virology: expanding viral diversity. Virology 479–480:369–378. doi: 10.1016/j.virol.2015.03.031. [DOI] [PubMed] [Google Scholar]
- 9.Rice G, Tang L, Stedman K, Roberto F, Spuhler J, Gillitzer E, Johnson JE, Douglas T, Young M. 2004. The structure of a thermophilic archaeal virus shows a double-stranded DNA viral capsid type that spans all domains of life. Proc Natl Acad Sci U S A 101:7716–7720. doi: 10.1073/pnas.0401773101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu C-Y, Wang K, Gan L, Lanman J, Khayat R, Young MJ, Jensen GJ, Doerschuk PC, Johnson JE. 2010. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure 18:1579–1586. doi: 10.1016/j.str.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Porter K, Kukkaro P, Bamford JK, Bath C, Kivelä HM, Dyall-Smith ML, Bamford DH. 2005. SH1: a novel, spherical halovirus isolated from an Australian hypersaline lake. Virology 335:22–33. doi: 10.1016/j.virol.2005.01.043. [DOI] [PubMed] [Google Scholar]
- 12.Jaakkola ST, Penttinen RK, Vilén ST, Jalasvuori M, Rönnholm G, Bamford JK, Bamford DH, Oksanen HM. 2012. Closely related archaeal Haloarcula hispanica icosahedral viruses HHIV-2 and SH1 have nonhomologous genes encoding host recognition functions. J Virol 86:4734–4742. doi: 10.1128/JVI.06666-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Porter K, Tang S-L, Chen C-P, Chiang P-W, Hong M-J, Dyall-Smith M. 2013. PH1: an archaeovirus of Haloarcula hispanica related to SH1 and HHIV-2. Archaea 2013:456318. doi: 10.1155/2013/456318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mei Y, Chen J, Sun D, Chen D, Yang Y, Shen P, Chen X. 2007. Induction and preliminary characterization of a novel halophage SNJ1 from lysogenic Natrinema sp. F5. Can J Microbiol 53:1106–1110. doi: 10.1139/W07-072. [DOI] [PubMed] [Google Scholar]
- 15.Bath C, Dyall-Smith ML. 1998. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J Virol 72:9392–9395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Häring M, Peng X, Brügger K, Rachel R, Stetter KO, Garrett RA, Prangishvili D. 2004. Morphology and genome organization of the virus PSV of the hyperthermophilic archaeal genera Pyrobaculum and Thermoproteus: a novel virus family, the Globuloviridae. Virology 323:233–242. doi: 10.1016/j.virol.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 17.Häring M, Rachel R, Peng X, Garrett RA, Prangishvili D. 2005. Viral diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J Virol 79:9904–9911. doi: 10.1128/JVI.79.15.9904-9911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozubal MA, Dlakic M, Macur RE, Inskeep WP. 2011. Terminal oxidase diversity and function in “Metallosphaera yellowstonensis”: gene expression and protein modeling suggest mechanisms of Fe(II) oxidation in the Sulfolobales. Appl Environ Microbiol 77:1844–1853. doi: 10.1128/AEM.01646-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy S, Gradnigo J, Johnson T, Payne S, Lipzen A, Martin J, Schackwitz W, Moriyama E, Blum P. 2015. Complete genome sequence of Sulfolobus solfataricus strain 98/2 and evolved derivatives. Genome Announc 3(3):e00549-15. doi: 10.1128/genomeA.00549-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaubert C, Danioux C, Oberto J, Cortez D, Bize A, Krupovic M, She Q, Forterre P, Prangishvili D, Sezonov G. 2013. Genomics and genetics of Sulfolobus islandicus LAL14/1, a model hyperthermophilic archaeon. Open Biol 3:130010. doi: 10.1098/rsob.130010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grogan D, Palm P, Zillig W. 1990. Isolate B12, which harbours a virus-like element, represents a new species of the archaebacterial genus Sulfolobus, Sulfolobus shibatae, sp. nov. Arch Microbiol 154:594–599. doi: 10.1007/BF00248842. [DOI] [PubMed] [Google Scholar]
- 22.Barrero-Canosa J, Moraru C, Zeugner L, Fuchs BM, Amann R. 2017. Direct-geneFISH: a simplified protocol for the simultaneous detection and quantification of genes and rRNA in microorganisms. Environ Microbiol 19:70–82. doi: 10.1111/1462-2920.13432. [DOI] [PubMed] [Google Scholar]
- 23.Peeples TL, Kelly RM. 1993. Bioenergetics of the metal/sulfur-oxidizing extreme thermoacidophile, Metallosphaera sedula. Fuel 72:1619–1624. doi: 10.1016/0016-2361(93)90345-3. [DOI] [Google Scholar]
- 24.Challberg MD, Kelly TJ. 1979. Adenovirus DNA replication in vitro: origin and direction of daughter strand synthesis. J Mol Biol 135:999–1012. doi: 10.1016/0022-2836(79)90524-2. [DOI] [PubMed] [Google Scholar]
- 25.Alva V, Nam S-Z, Söding J, Lupas AN. 2016. The MPI bioinformatics toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44:W410–W415. doi: 10.1093/nar/gkw348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Söding J, Biegert A, Lupas AN. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bamford DH, Grimes JM, Stuart DI. 2005. What does structure tell us about virus evolution? Curr Opin Struct Biol 15:655–663. doi: 10.1016/j.sbi.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Qin L, Fokine A, O'Donnell E, Rao VB, Rossmann MG. 2010. Structure of the small outer capsid protein, Soc: a clamp for stabilizing capsids of T4-like phages. J Mol Biol 395:728–741. doi: 10.1016/j.jmb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang L, Gilcrease EB, Casjens SR, Johnson JE. 2006. Highly discriminatory binding of capsid-cementing proteins in bacteriophage L. Structure 14:837–845. doi: 10.1016/j.str.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Reddy VS, Nemerow GR. 2014. Structures and organization of adenovirus cement proteins provide insights into the role of capsid maturation in virus entry and infection. Proc Natl Acad Sci U S A 111:11715–11720. doi: 10.1073/pnas.1408462111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jäälinoja HT, Roine E, Laurinmäki P, Kivelä HM, Bamford DH, Butcher SJ. 2008. Structure and host-cell interaction of SH1, a membrane-containing, halophilic euryarchaeal virus. Proc Natl Acad Sci U S A 105:8008–8013. doi: 10.1073/pnas.0801758105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davison AJ. 2007. Chapter 2. Comparative analysis of the genomes. In Arvin A, Campadelli-Fiume G, Mocarski E, et al. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, MA. [PubMed] [Google Scholar]
- 34.Happonen LJ, Oksanen E, Liljeroos L, Goldman A, Kajander T, Butcher SJ. 2013. The structure of the NTPase that powers DNA packaging into Sulfolobus turreted icosahedral virus 2. J Virol 87:8388–8398. doi: 10.1128/JVI.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dellas N, Snyder JC, Dills M, Nicolay SJ, Kerchner KM, Brumfield SK, Lawrence CM, Young MJ. 2015. Structure-based mutagenesis of Sulfolobus turreted icosahedral virus B204 reveals essential residues in the virion-associated DNA-packaging ATPase. J Virol 90:2729–2739. doi: 10.1128/JVI.02435-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khayat R, Tang L, Larson ET, Lawrence CM, Young M, Johnson JE. 2005. Structure of an archaeal virus capsid protein reveals a common ancestry to eukaryotic and bacterial viruses. Proc Natl Acad Sci U S A 102:18944–18949. doi: 10.1073/pnas.0506383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malke H. 1990. Book review. J. Sambrock [sic], E. F. Fritsch and T. Maniatis, Molecular cloning, a laboratory manual (second edition), volumes 1, 2 and 3. J Basic Microbiol 30:623. doi: 10.1002/jobm.3620300824. [DOI] [Google Scholar]
- 38.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamilton MB, Pincus EL, Di Fiore A, Fleischer RC. 1999. Universal linker and ligation procedures for construction of genomic DNA libraries enriched for microsatellites. Biotechniques 27:500–507. [DOI] [PubMed] [Google Scholar]
- 41.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley LA, Stefans M, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3—new capabilities and interfaces. Nucleic Acids Res 40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koressaar T, Remm M. 2007. Enhancements and modifications of primer design program Primer3. Bioinformatics 23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- 45.Shevchenko A, Tomas H, Havli J, Olsen JV, Mann M. 2006. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 46.Craig R, Beavis RC. 2004. TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 47.Vaudel M, Burkhart JM, Zahedi RP, Oveland E, Berven FS, Sickmann A, Martens L, Barsnes H. 2015. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat Biotechnol 33:22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- 48.Munson-McGee JH, Field EK, Bateson M, Rooney C, Stepanauskas R, Young MJ. 2015. Nanoarchaeota, their Sulfolobales host, and Nanoarchaeota virus distribution across Yellowstone National Park hot springs. Appl Environ Microbiol 81:7860–7868. doi: 10.1128/AEM.01539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Danev R, Nagayama K. 2010. Phase plates for transmission electron microscopy. Methods Enzymol 481:343–369. doi: 10.1016/S0076-6879(10)81014-6. [DOI] [PubMed] [Google Scholar]
- 50.Mastronarde DN. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Fukuda Y, Laugks U, Lučić V, Baumeister W, Danev R. 2015. Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol 190:143–154. doi: 10.1016/j.jsb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 52.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. 2014. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci U S A 111:15635–15640. doi: 10.1073/pnas.1418377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kremer JR, Mastronarde DN, McIntosh JR. 1996. Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 54.Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR. 2006. The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 55.Heumann JM, Hoenger A, Mastronarde DN. 2011. Clustering and variance maps for cryo-electron tomography using wedge-masked differences. J Struct Biol 175:288–299. doi: 10.1016/j.jsb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kleywegt GJ, Zou J-Y, Kjeldgaard M, Jones TA. 2006. Around O, p 353–356. In Rossmann MG, Arnold E (ed), International tables for crystallography, vol F. Crystallography of biological macromolecules, 1st online ed. International Union of Crystallography, Chester, England. [Google Scholar]
- 57.Ten Eyck LF, Watenpaugh KD. 2012. Introduction to refinement, p 459–465. In Arnold E, Himmel DM, Rossmann MG (ed), International tables for crystallography, vol F Crystallography of biological macromolecules, 2nd online ed. International Union of Crystallography, Chester, England. [Google Scholar]
- 58.Li X, Zheng S, Agard DA, Cheng Y. 2015. Asynchronous data acquisition and on-the-fly analysis of dose fractionated cryoEM images by UCSFImage. J Struct Biol 192:174–178. doi: 10.1016/j.jsb.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rohou A, Grigorieff N. 2015. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J Struct Biol 192:216–221. doi: 10.1016/j.jsb.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Scheres SHW. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol 180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]