ABSTRACT
Inner tegument protein UL37 is conserved among all three subfamilies of herpesviruses. Studies of UL37 homologs from two alphaherpesviruses, herpes simplex virus 1 (HSV-1) and pseudorabies virus (PRV), have suggested that UL37 plays an essential albeit poorly defined role in intracellular capsid trafficking. At the same time, HSV and PRV homologs cannot be swapped, which suggests that in addition to a conserved function, UL37 homologs also have divergent virus-specific functions. Accurate dissection of UL37 functions requires detailed maps in the form of atomic-resolution structures. Previously, we reported the crystal structure of the N-terminal half of UL37 (UL37N) from PRV. Here, we report the crystal structure of HSV-1 UL37N. Comparison of the two structures reveals that UL37 homologs differ in their overall shapes, distributions of surface charges, and locations of projecting loops. In contrast, the previously identified R2 surface region is structurally conserved. We propose that within the N-terminal half of UL37, functional conservation is centered within the R2 surface region, whereas divergent structural elements pinpoint regions mediating virus-specific functions and may engage different binding partners. Together, the two structures can now serve as templates for a structure-guided exploration of both conserved and virus-specific functions of UL37.
IMPORTANCE The ability to move efficiently within host cell cytoplasm is essential for replication in all viruses. It is especially important in the neuroinvasive alphaherpesviruses, such as human herpes simplex virus 1 (HSV-1), HSV-2, and veterinarian pseudorabies virus (PRV), that infect the peripheral nervous system and have to travel long distances along axons. Capsid movement in these viruses is controlled by capsid-associated tegument proteins, yet their specific roles have not yet been defined. Systematic exploration of the roles of tegument proteins in capsid trafficking requires detailed navigational charts in the form of their three-dimensional structures. Here, we determined the crystal structure of the N-terminal half of a conserved tegument protein, UL37, from HSV-1. This structure, along with our previously reported structure of the UL37 homolog from PRV, provides a much needed 3-dimensional template for the dissection of both conserved and virus-specific functions of UL37 in intracellular capsid trafficking.
KEYWORDS: UL37, alphaherpesvirus, crystal structure, herpesvirus, tegument
INTRODUCTION
Successful replication of many viruses hinges on the ability of the capsid to traffic through the host cytoplasm. Since most viruses do not encode their own trafficking machinery, they must hijack host molecular motors to traffic along host microtubules to replicate successfully. This is especially important in the neuroinvasive alphaherpesviruses, which include human herpes simplex virus 1 (HSV-1), HSV-2, and their veterinarian homolog pseudorabies virus (PRV), which infects pigs (1). These viruses initially infect epithelial cells and then spread to the peripheral nervous system to establish a latent infection within the nuclei of neuronal cell bodies. Latent viruses can reactivate and reinfect epithelial cells, typically at the site of initial infection (1). Both during initial infection and reactivation, capsids traffic long distances along neuronal axons, which reinforces the importance of efficient capsid trafficking.
All herpesviruses consist of a double-stranded DNA genome housed within a capsid that is surrounded by an intricate protein layer called the tegument and enclosed by a lipid envelope. The tegument is further subdivided into the inner and outer layers (2). The inner tegument is capsid associated, remains attached to capsid during postentry trafficking from the cellular membrane to the nucleus, and is acquired early during egress (3, 4). In contrast, the outer tegument dissociates from the capsid postentry (5) and is acquired at later stages of egress (6).
Inner tegument protein UL37 is conserved across all herpesviruses and is essential for replication (3, 7–10). It associates with the capsid indirectly by binding another inner tegument protein, UL36 (11, 12). HSV-1 or PRV lacking UL37 shows a postentry delay in capsid transit to the nucleus (13, 14). Additionally, in these viruses, progeny capsids accumulate within the cytoplasm (8, 15), presumably because they cannot reach the site of secondary envelopment at the cytoplasmic membranes derived from trans-Golgi network (3, 15, 16) or early endosomes (17). Indeed, in the absence of UL37, capsids are incapable of traveling long distances in epithelial cells (13, 18). As a result, UL37-null viruses either fail to replicate (HSV-1) or have significantly reduced titers (PRV) (7). These observations implicate UL37 in capsid trafficking within the host cytoplasm during both entry and egress.
While UL37 has a conserved essential function in viral replication in HSV-1 and PRV, the phenotypes of the respective null viruses are distinct, and cell lines expressing PRV UL37 cannot rescue an HSV-1 UL37-null virus and vice versa (7). Thus, in addition to a conserved common function, UL37 homologs have distinct, albeit unknown, virus-specific roles. The ability to dissect functional differences requires detailed structural information. While the crystal structure of the N-terminal half of PRV UL37 is available (19), no structural information is available for HSV-1 UL37. Here, we determined the crystal structure of the N-terminal half of HSV-1 UL37 to provide a three-dimensional template for exploration of the function of UL37 in the viral replication cycle and the functional differences between UL37 homologs. Through structural comparisons, we demonstrate that UL37 homologs differ in their overall shapes, distributions of surface charges, and locations of projecting loops, while the previously identified R2 surface region (19) is structurally conserved. We propose that within the N-terminal half of UL37, functional conservation is limited to the R2 surface region, whereas divergent structural characteristics such as charge distribution and projecting loops pinpoint regions that mediate virus-specific functions and may engage different binding partners.
RESULTS
Overall structure and domain organization.
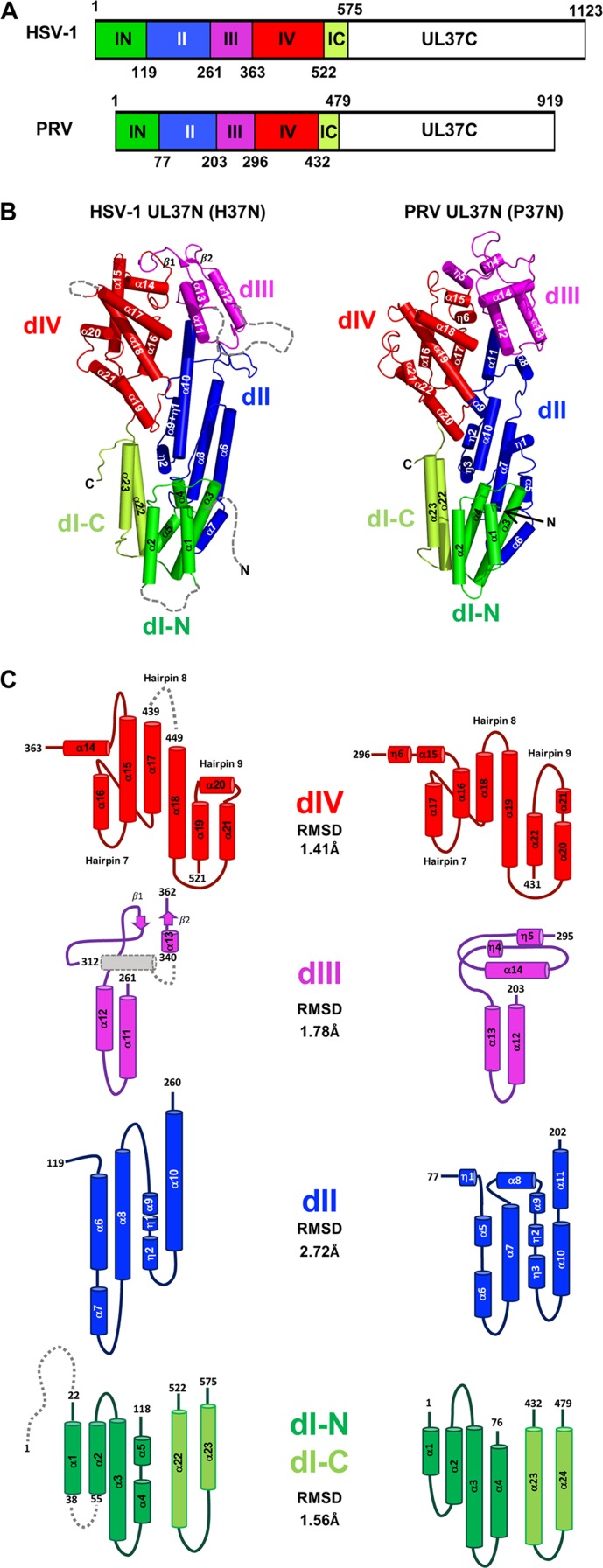
HSV-1 UL37N (H37N) is an elongated molecule with dimensions of 91 Å by 46 Å by 30 Å that resembles a hairpin, with the N and C termini located on one end of the molecule, just like in our previously determined PRV UL37N (P37N) structure (Fig. 1A and B). H37N is composed of 23 α helices and two 310 helices arranged into helical bundles, and two β sheets. The first 21 residues of H37N are not resolved and are predicted to be disordered according to the secondary structure prediction algorithm in PSIPRED (20). Additionally, the linkers between α1 and α2, β1 and α13, and α17 and α18 are unresolved. At 575 residues, H37N is longer in sequence than the 479-residue P37N (Fig. 1A). P37N was previously divided into three domains based on several computer algorithms for domain assignment from protein structure coordinates and visual analysis (19). However, such algorithms yield a very different domain assignment for H37N. Therefore, to maintain consistency in the domain assignment between H37N and P37N, we have divided both proteins into four domains: the discontinuous domain I (dI), dII, dIII, and dIV (Fig. 1 and 2).
FIG 1.
Domain organization in UL37N structures from HSV-1 and PRV. (A) Linear diagram of UL37 from HSV-1 and PRV with domains colored and domain boundaries labeled. (B) Crystal structure of UL37N from HSV-1 (H37N, left) and PRV (P37N, right) colored by domain as in panel A. Secondary structure elements are numbered sequentially. The orientation was chosen to show all secondary structure elements. (C) Topology diagrams of individual domains. Helices are shown as cylinders and beta strands as arrows. The color scheme is the same as in panels A and B.
FIG 2.
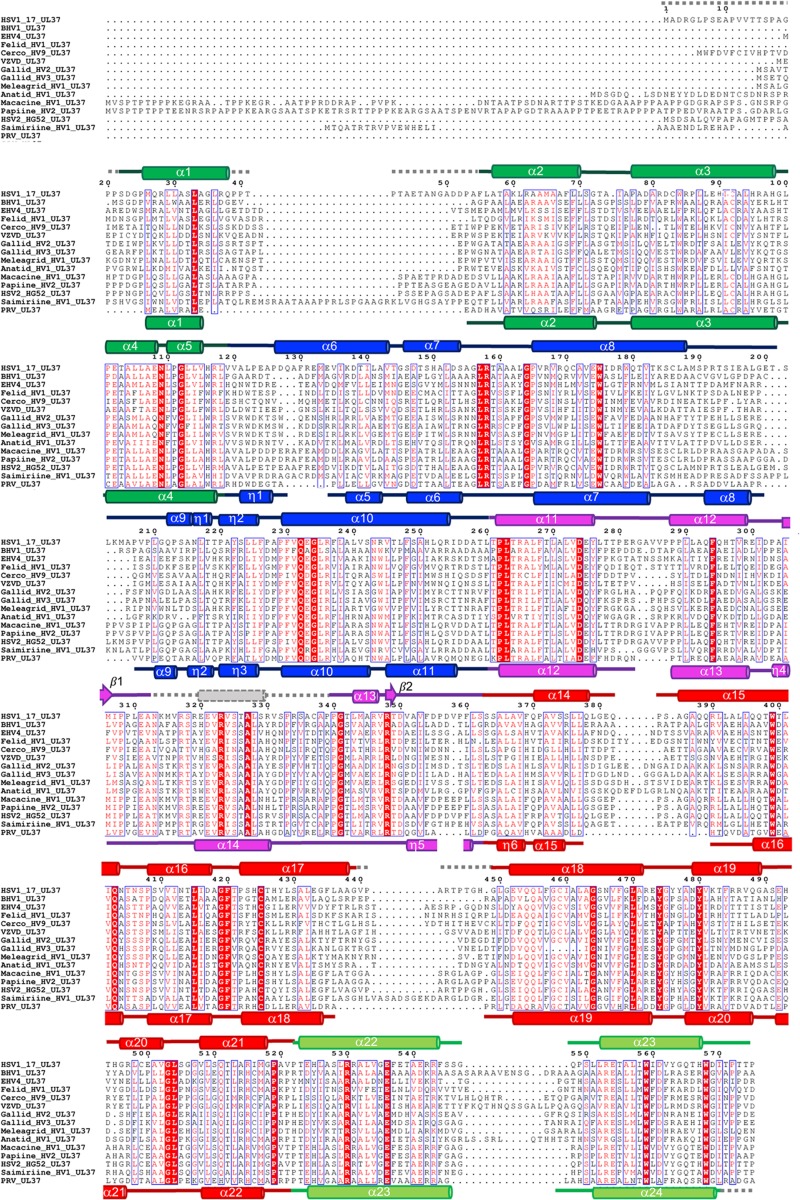
Multiple-sequence alignment of UL37N from 15 alphaherpesviruses. Sequence alignment was generated using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Only the alignment of residues corresponding to residues 1 to 575 of HSV-1 UL37 is shown. Identical residues are shown in white letters on a red background. Similar residues are shown as red letters boxed in blue. Secondary structure derived from the crystal structures of HSV-1 and PRV UL37 homologs is shown above and below the alignment, respectively. Helices are represented by cylinders and beta strands by arrows. Secondary structure elements are labeled and colored by domain using the same color scheme as in Fig. 1. Unresolved residues are represented using gray dashed lines. HSV-1 and PRV UL37N share 29.7% identity and 55.9% similarity. Sequences from the following viruses were used (GenBank Gene ID numbers are given in parentheses): HSV-1_17, herpes simplex virus 1 strain 17 (2703358); BHV1, bovine herpesvirus 1 (1487388); EHV4, equine herpesvirus 4 (1487587); Felid_HV1, felid herpesvirus 1 (8658550); Cerco_HV9, cercopithecine herpesvirus 9 (920525); VZVD, varicella-zoster virus serotype D (1487686); Gallid_HV2, gallid herpesvirus 2 (4811511); Gallid_HV3, gallid herpesvirus 3 (911942); Meleagrid_HV1, meleagrid herpesvirus 1 (918517); Anatid_HV1, anatid herpesvirus 1 (8223349); Macacine_HV1, macacine herpesvirus 1 (1487432); Papiine_HV2, papiine herpesvirus 2 (3850220); HSV2_HG52, herpes simplex virus 2 strain HG52 (1487323); Saimiriine_HV1, saimiriine herpesvirus 1 (9829319); PRV, suid herpesvirus strain Kaplan (2952491).
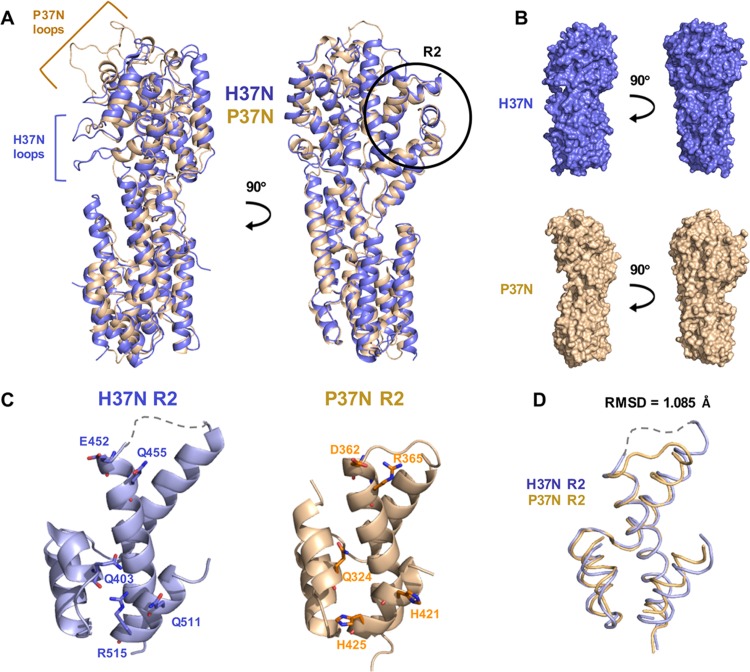
As expected from the 29.7% sequence identity, the H37N and P37N structures are similar and superpose with a root mean square deviation (RMSD) of 1.86 Å over 418 aligned residues. dIV is the most structurally conserved domain, whereas dII is the most structurally divergent (Fig. 1B and C). Most helices superpose well, but there are variations in the lengths of several, with helices α6, α8, α15, and α17 in H37N being longer than their P37N counterparts. Two different sets of loops in H37N and P37N project outward, giving the two molecules distinct appearances (Fig. 3A). H37N has two long loops, between α8 and α9 in dII and between α11 and α12 in dIII, which form finger-like extensions along one margin of the molecule (Fig. 3A). In their place, P37N has a short loop and a short helix, which create a shallow pocket in P37N. In contrast, at the top of the molecule in dIII, H37N has a small two-stranded β sheet whereas P37N has 2 long outward-projecting loops connected by a helix, which gives H37N the appearance of a flat top and P37N that of a pointy top (Fig. 3B).
FIG 3.
Comparison of H37N and P37N structures. H37N is shown in blue and P37N in wheat. (A) Overlay of the two structures in cartoon representation shown in two orientations related by a 90-degree rotation around the vertical axis. (B) Individual structures in surface representation are shown side by side in two orientations related by a 90-degree rotation around the vertical axis. Extended loops are indicated by brackets. The R2 surface region is circled in black and labeled. (C) Close-up views of the R2 surface region in H37N and P37N. The five residues that define the R2 surface region are shown in stick representation and labeled (H37N, Q403/E452/Q455/Q511/R515; P37N, Q324/D362/R365/H421/H425). (D) Close-up of overlaid R2 region backbone with an RMSD of 1.085 Å.
dI.
Domain I (dI) is composed of two discontinuous segments (H37N, residues 22 to 118 and 522 to 575; P37N, residues 1 to 76 and 432 to 479) (Fig. 1). This domain superposes with an RMSD of 1.56 Å over 121 aligned residues. It is composed of three helical hairpins with the up-down topology. The main difference between H37N and P37N lies in hairpin 2, which in H37N consists of a long α3 and two smaller helices, α4 and α5, but in P37N has two longer helices, α3 and α4. Hairpin 3 is made up of two longer antiparallel helices (H37N, α22 and α23; PR37N, α23 and α24) that are discontinuous with the first two hairpins. In both structures, domain I forms a U-shaped groove filled with a 310 helix “plug” from dII (H37N, η2; P37N, η3).
dII.
Domain II (dII) (H37N, residues 119 to 260; P37N, residues 77 to 202) is the most structurally divergent domain, with an RMSD of 2.72 Å over 93 aligned residues. Although in both structures dII is formed by two helical hairpins, 4 and 5, they have different architectures (Fig. 1B and C). In H37N, hairpin 4 consists of a long helix (α6) and a short helix (α7) that run antiparallel to α8. In P37N, hairpin 4 is made up of the two short helices α5 and α6, which run antiparallel to α7. Hairpin 5 is connected to hairpin 4 by a long finger-like loop in H37N but a short helix α8 in P37N. In both structures, one side of hairpin 5 is composed of a short α helix followed by two 310 helices (H37N, α9 and η1 and η2; P37N, α9 and η2 and η3), whereas the other side is made up of one long helix, α10, in H37N but two shorter α helices, α10 and α11, in P37N. At 26 residues, α10 is the longest helix in the H37N structure and resembles a backbone by running straight down the center of the molecule and bridging the top and bottom halves of the structure.
dIII.
Domain III (dIII) (H37N, residues 261 to 362; P37N, residues 203 to 295) is the smallest domain. In H37N, it consists of three α helices, α11 to α13 (plus a putative helix unresolved in the structure), and the only two β strands (β1 and β2). In P37N, dIII consists of three α helices, α12 to α14, and two 310 helices, η4 and η5 (Fig. 1B and C). This domain superposes with an RMSD of 1.78 Å over 58 aligned residues. Helices α11 and α12 in H37N form hairpin 6 (P37N, α12 and α13). The top of dIII is decorated by two β strands in H37N but by two short 310 helices, η4 and η5, in P37N.
dIV.
In both structures, domain IV (dIV) (H37N, residues 363 to 521; P37N, residues 296 to 431) is a helical bundle built around a core helix (H37N, α18; P37N, α19) (Fig. 1B and C) conserved across alphaherpesviruses (Fig. 2). dIV is the most structurally conserved, with an RMSD of 1.41 Å over 115 aligned residues. H37N helix α15 is much longer than its P37N counterpart α16. The last three helices of dIV are arranged differently: in H37N, helices α19 and α21 form a hairpin that displays helix α20 at its tip, whereas in P37N, helices α20 to α22 form a hairpin which creates one short loop at the tip (Fig. 1B and C). The hypothetical nuclear export signal (NES) motif (21) maps to helix α11 and is mostly buried, which suggests either that it is not a functional NES or that UL37N has to undergo a large conformational change to expose the NES.
Electrostatic surface potential.
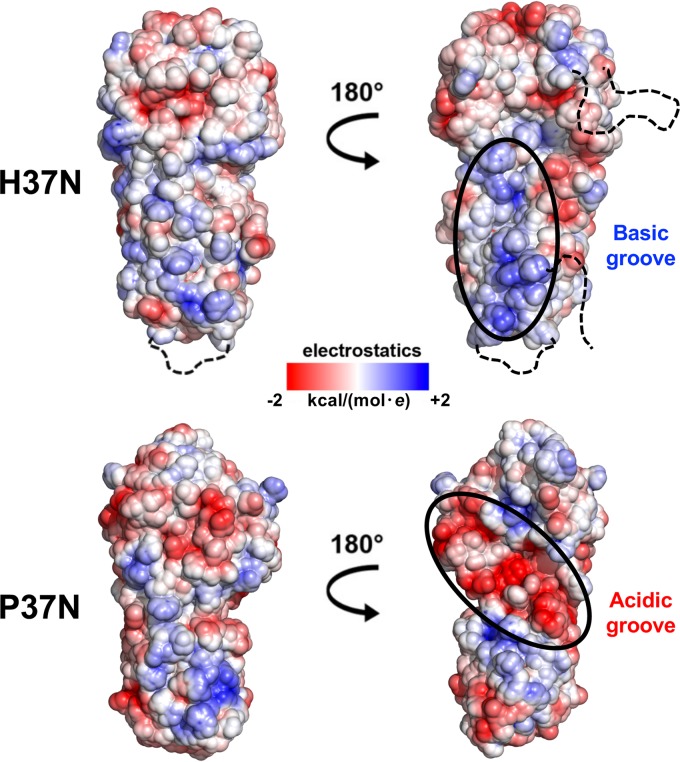
H37N and P37N differ in the distribution of the electrostatic surface potential. P37N has a large negatively charged groove on one side (Fig. 4), which in the crystal structure coordinates two calcium ions (19). These calcium ions participate in crystal contacts, which explains why the presence of calcium was required for P37N crystallization (19). Conversely, H37N has a positively charged groove on the same side (Fig. 4) and did not require calcium for crystallization. These respective grooves of opposite charge could provide binding sites for virus-specific binding partners. The opposite side of UL37N has a more consistent surface charge distribution, with the negatively charged upper half and a positively charged lower half in both homologs.
FIG 4.
Electrostatic surface potential maps of H37N and P37N. Maps were generated using the PBEQ Solver function in the CHARMM program (http://www.charmm-gui.org/?doc=input/pbeqsolver). Charges are on a scale from −2 to +2 kcal/(mol · e). Two orientations are shown related by a 180-degree rotation around the vertical axis. Basic and acidic grooves in H37N and P37N, respectively, are circled in black and labeled. Unresolved loops in H37N are shown as black dashed lines.
Relative functional importance of surface residues.
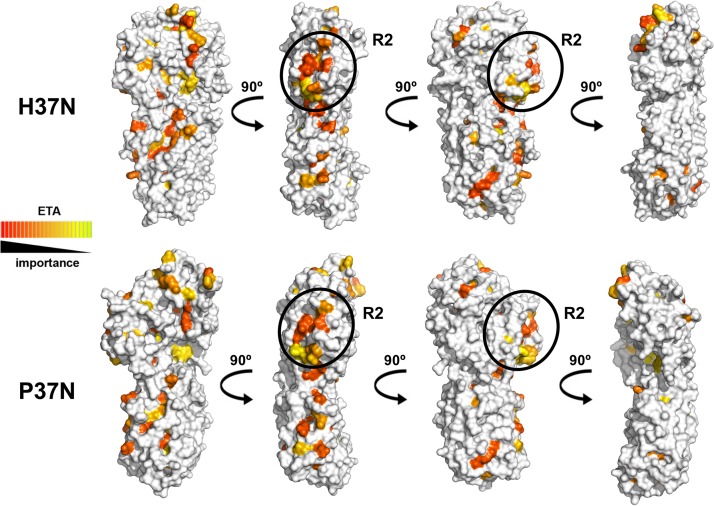
Evolutionary trace analysis (ETA) is a useful tool for identifying the functional importance of a particular residue based on its resistance to mutations over time and clustering on the protein surface with other important residues (22) (http://mammoth.bcm.tmc.edu/uet). Previously, our ETA of P37N led to the successful identification of the functional surface region R2. This region is defined by five point mutations (P37N, Q324A/D362A/R365A/H421A/H425A) that, in combination, lead to reduced cell-cell spread in epithelial cells (19). ETA on H37N confirmed that the largest cluster of functionally important residues is located within the R2 surface region (Fig. 5), whereas the rest of the surface is evolutionarily divergent, with the exception of a small patch on the top of H37N (Fig. 5). The R2 surface region is also structurally conserved (Fig. 3C and D). Therefore, we propose that the R2 surface region mediates a conserved function in alphaherpesviruses, whereas other surface regions of UL37N likely play divergent functional roles.
FIG 5.
Functionally important surface residues in UL37N as identified using universal evolutionary trace analysis (http://mammoth.bcm.tmc.edu/uet/). Individual residues were assigned relative functional importance scores based on resistance to mutations over time and clustering on the protein surface with other important residues. Residues with importance scores in the top 25% were assigned colors from red (more important) to yellow (less important). Four orientations are shown related by a 90-degree rotation around the vertical axis. Areas of the structure corresponding to the R2 region are circled in black and labeled.
DISCUSSION
The inner tegument protein UL37, conserved across all herpesviruses, is essential for replication of HSV-1 and PRV, two of the best-studied alphaherpesviruses. Yet, the lack of UL37 yields distinct phenotypes in HSV-1 versus PRV, and the two homologs are not interchangeable (7). Therefore, in addition to a common function in capsid trafficking, UL37 homologs also have distinct, albeit unknown, virus-specific roles in HSV-1 and PRV.
H37N and P37N have distinct folds and surface properties, including distribution of surface charges and the location of projecting loops. We propose that these divergent structural characteristics pinpoint regions which could allow each homolog to bind distinct binding partners using a similar scaffold. Different sets of binding partners would impart virus-specific functions, helping explain different functional phenotypes of UL37-null viruses. In contrast, the R2 surface region is structurally conserved and likely mediates a common function of UL37.
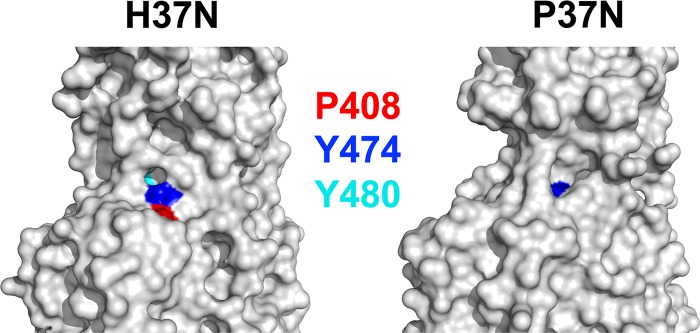
In a recent study, strictly conserved residues in UL37N were mutated with the goal of identifying those necessary for interaction with UL20/gK (23). HSV-1 UL37 variants in which Y474 or Y480 was replaced with alanines failed to complement a UL37-null virus, which suggested that these residues are important for function. On the basis of the homology model of HSV-1 UL37N generated in that study, it was proposed that these two tyrosines, along with another highly conserved residue, P408, form a conserved binding pocket for gK/UL20 (23). In our H37N structure, only P408 and Y474 are surface exposed, while Y480 is completely buried (Fig. 6). Therefore, within this putative binding pocket, Y480 is more likely to play a structural role rather than directly mediating protein-protein interactions. Furthermore, in the P37N structure, only Y384, which is equivalent to H37N Y474, is surface exposed (Fig. 6), and we conclude that this binding pocket likely mediates HSV-1-specific interactions rather than interactions conserved across alphaherpesviruses. The crystal structure of H37N reported here can now accurately inform further mutagenesis efforts. Comparison of the P37N and H37N structures stresses the need for caution in using homology modeling. P37N and H37N share 29.7% identity, which falls within the so-called “twilight zone” where homology modeling is unreliable (24). Moreover, it illustrates how conservation of primary sequence does not necessarily extend into conservation in structure.
FIG 6.
A subset of strictly conserved residues differ in their surface exposure in H37N and P37N structures. Residues Y474, Y480, and P408 in H37N, which are strictly conserved among alphaherpesviruses, were proposed to form a conserved binding pocket for gK/UL20 based on a homology model of H37N (23). A close-up view of the surface in the vicinity of the putative pocket in H37N and P37N illustrates differences in surface exposure of the three strictly conserved residues. Residues are colored as follows: H37N Y474 and P37N Y384, blue; H37N Y480 and P37N Y390, cyan; and H37N P408 and P37N P329, red.
The crystal structure of H37N reported here along with our earlier reported structure of P37N provide detailed three-dimensional templates of the N-terminal halves of UL37 that can now guide the dissection of functional differences between UL37 homologs along with the exploration of many roles of UL37 in the viral replication cycle. Ultimately, a complete description of all activities and interactions of UL37, both conserved and virus specific, awaits the structure of the C-terminal half of UL37, which harbors the deamidase activity of UL37 (25) and has been proposed to engage both viral and cellular binding partners such as UL36 (26), dystonin/BPAG1 (18), and TRAF6 (27).
MATERIALS AND METHODS
Cloning.
The HSV-1 UL37 gene (strain 17), codon optimized for Escherichia coli expression, was synthesized by GeneArt. The N-terminal half of HSV-1 UL37 (referred to as UL37N), which includes residues 1 to 575, was amplified by PCR from the full-length HSV-1 codon-optimized UL37 gene using the primers 5′CGTCAAGGCCGCATGGATCCATGGCAG and 5′ACCCCGACCACTCCGTAGTAGCTCGAGATTTTT (restriction sites are underlined). Residue 570 in HSV-1 UL37 is equivalent to the last resolved residue, 479, in the PRV UL37N structure (19). The PCR product was subcloned into vector pJP4 using the BamHI and XhoI restriction sites to yield plasmid pAK11. The pJP4 plasmid contains a His6-SUMO-PreScission tag in frame with the BamHI restriction site of the multiple-cloning site in a pET24b vector.
Protein expression and purification.
HSV-1 UL37N was expressed as an N-terminal His6-SUMO fusion in low background strain (LOBSTR) E. coli (a gift from Thomas Schwartz). Freshly transformed cells were incubated at 37°C overnight in 5 ml LB starter culture supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol. The starter culture was diluted into 1 liter LB supplemented with 50 μg/ml kanamycin and 34 μg/ml chloramphenicol and then grown at 37°C until the optical density at 600 nm (OD600) reached 0.8 to 1.0. At this point, the temperature was shifted to 16°C and the cells were induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 16 to 20 h. Cells were harvested by centrifugation at 5,000 × g for 40 min, resuspended in 40 ml 50 mM Tris hydrochloride (Tris-HCl) (pH 7.4), 150 mM NaCl, 5% glycerol, 15 mM imidazole, 0.1 mM tris(2-carboxyethyl)phosphine (TCEP), and 1 EDTA-free cOmplete protease inhibitor cocktail tablet (Roche), and lysed using a Microfluidizer. The insoluble fraction was removed by centrifugation of the whole-cell lysate at 13,000 × g for 30 min at 4°C. The soluble lysate was loaded onto a 5-ml Ni-Sepharose 6B FF column (GE Healthcare). The column was subsequently washed with 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 5% glycerol, and 0.1 mM TCEP (buffer A) containing increasing amounts of imidazole from 15 mM to 50 mM. Protein was eluted in buffer A containing 300 mM imidazole. The protein concentration was determined from the absorbance at 280 nm using a calculated extinction coefficient (45,000 M−1 cm−1). Purified recombinant glutathione S-transferase (GST)-tagged HRV3C (PreScission) protease was added to the protein solution at a 1:30 protease-to-protein ratio, and the protein was cleaved overnight at 4°C to remove the His6-SUMO tag. The protease-protein solution was concentrated, the imidazole removed, and the NaCl concentration reduced by buffer exchange into 50 mM Tris-HCl (pH 7.4), 20 mM NaCl, 5% glycerol, and 0.1 mM TCEP (buffer B) in preparation for anion-exchange chromatography using an Ultra-15 50-kDa-cutoff concentrator (Millipore). The protein solution was applied to a HiTrap Q Sepharose FF column (GE Healthcare) for anion-exchange chromatography. The column was washed with buffer B, and then a salt gradient from 20 mM to 500 mM NaCl was applied. Fractions corresponding to 210 mM NaCl containing UL37N separated from His6-SUMO-tagged UL37N and the cleaved His6-SUMO tag were pooled. UL37N was further purified by size exclusion chromatography using a Superdex 200 column in buffer A (GE Healthcare) and concentrated to ∼5.0 mg/ml using an Ultra-15 50-kDa-cutoff concentrator (Millipore). Protein purity was assessed by SDS-polyacrylamide gel electrophoresis (PAGE) and GelCode Blue staining (Thermo Fisher). The final yield was ∼5 mg of pure protein per 1 liter of E. coli culture. All UL37N protein samples used for crystallization and biochemical studies were stored in buffer A with 1× Halt protease inhibitor cocktail (Pierce).
Crystallization and structure determination.
Crystals of HSV-1 UL37N were grown by vapor diffusion at room temperature in hanging drops containing 1 μl protein at ∼5 mg/ml and 1 μl crystallization solution, 8% polyethylene glycol 20000, and 0.1 M bicine, pH 9.0. Needles formed within 24 h and were immediately harvested. During harvesting, crystals were incubated in a solution identical to the well solution plus 20% glycerol for 10 s prior to flash freezing in liquid N2. Diffraction data to 3.5-Å resolution were collected at 100 K on beamline 24-ID-E at the Advanced Photon Source at Argonne National Labs and processed using RAPD software (https://rapd.nec.aps.anl.gov/rapd). Crystals took space group P6122 with 1 molecule per asymmetric unit (Table 1). The structure was determined by molecular replacement using a single copy of the PRV UL37N structure (PDB ID 4K70) (19) as a search template, as implemented in Phaser (28). Prior to refinement, 10% of the data were set aside for cross-validation. A homology model of HSV-1 UL37N, which was generated in Phyre2 and aligned to the molecular replacement solution, was used as the initial model for rebuilding. The model was refined in phenix.refine with iterative rounds of extensive manual rebuilding in Coot (29). Refinement included rigid-body refinement and gradient minimization refinement of XYZ coordinates and individual atomic displacement parameter refinement, all as implemented in phenix.refine (30). Secondary structure and rotamer restraints were used throughout refinement. A composite omit simulated annealing map was used to check the model. The final Rwork is 24.01%, and Rfree is 28.16%. The final model encompasses residues 22 to 575 (unresolved, 1 to 21, 39 to 54, 313 to 339, and 440 to 448). MolProbity as implemented in phenix.refine (30) was used to assess the stereochemical quality of the model throughout the refinement. According to MolProbity, 96.15% of the residues lie in the most favored regions of the Ramachandran plot, and 3.64% lie in the additionally allowed regions. There is one Ramachandran outlier. All relevant crystallographic statistics are listed in Table 1. All software was installed and maintained by SBGrid (31).
TABLE 1.
Data collection and refinement statistics
| Parameter | Valuea |
|---|---|
| Data collection statistics | |
| Space group | P6122 |
| Unit cell a, b, c (Å) | 138.232, 138.232, 164.22 |
| Resolution range (Å) | 164.22–3.51 (3.64–3.51) |
| No. of reflections | |
| Total | 170,104 (16,758) |
| Unique | 12,161 (1,180) |
| Multiplicity | 14.0 (14.1) |
| Completeness (%) | 99.85 (99.49) |
| Mean I/σ〈I〉 | 8.7 (1.20) |
| Wilson B factor (Å2) | 97.01 |
| Rmerge | 0.441 (2.484) |
| Rmeas | 0.457 (2.576) |
| Rpim | 0.119 (0.6695) |
| CC1/2 | 0.993 (0.445) |
| Refinement statistics | |
| Resolution range (Å) | 96.74–3.51 (3.65–3.51) |
| No. of reflections used: | |
| In refinement | 12,107 (1,176) |
| For Rfree | 1,212 (130) |
| Rworkb | 0.2401 (0.3596) |
| Rfreeb | 0.2816 (0.4368) |
| CCwork | 0.861 (0.683) |
| CCfree | 0.828 (0.427) |
| No. of: | |
| Nonhydrogen atoms | 3,806 |
| Macromolecules | 3,804 |
| Ligands | 1 |
| Solvent molecules | 1 |
| Protein residues | 502 |
| RMSD | |
| Bond length (Å) | 0.006 |
| Bond angle (°) | 0.95 |
| Ramachandran plot (%)c | |
| Favored regions | 96.15 |
| Allowed regions | 3.64 |
| Outliers | 0.20 |
| Rotamer outliers (%)c | 0.50 |
| Clashscorec | 13.46 |
| B factor | |
| Avg | 90.99 |
| Macromolecules | 91.00 |
| Ligands | 86.29 |
| Solvent | 66.64 |
Values in parentheses are for the highest-resolution shell.
Rwork and Rfree are defined as ∑||Fobs| − |Fcalc||/∑|Fobs| for the reflections in the working or the test set, respectively.
As determined using MolProbity (molprobity.biochem.duke.edu) (32).
Data availability.
Atomic coordinates and structure factors for the HSV-1 UL37N structure have been deposited to the RCSB Protein Data Bank under accession number 5VYL.
ACKNOWLEDGMENTS
We thank the staff at the NE-CAT (Advanced Photon Source) for help with collecting X-ray diffraction data, Peter Cherepanov (Francis Crick Institute) for the gift of the GST-PreScission protease expression plasmid, and Thomas Schwartz (MIT) for the gift of the E. coli LOBSTR strain. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines. This research used resources of the Advanced Photon Source.
This work was supported by the Investigators in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (E.E.H.), by NIH grant R01 AI056346 (E.E.H.), and by a Faculty Scholar grant from Howard Hughes Medical Institute (E.E.H.). A.L.K. was supported by NIH training grant T32 AI007422. The Northeastern Collaborative Access Team beamlines are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P41 GM103403). The Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility, is operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357.
REFERENCES
- 1.Zaichick SV, Bohannon KP, Smith GA. 2011. Alphaherpesviruses and the cytoskeleton in neuronal infections. Viruses 3:941–981. doi: 10.3390/v3070941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucks MA, O'Regan KJ, Murphy MA, Wills JW, Courtney RJ. 2007. Herpes simplex virus type 1 tegument proteins VP1/2 and UL37 are associated with intranuclear capsids. Virology 361:316–324. doi: 10.1016/j.virol.2006.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandbaumhuter M, Dohner K, Schipke J, Binz A, Pohlmann A, Sodeik B, Bauerfeind R. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell Microbiol 15:248–269. doi: 10.1111/cmi.12075. [DOI] [PubMed] [Google Scholar]
- 4.Henaff D, Remillard-Labrosse G, Loret S, Lippe R. 2013. Analysis of the early steps of herpes simplex virus 1 capsid tegumentation. J Virol 87:4895–4906. doi: 10.1128/JVI.03292-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Granzow H, Klupp BG, Mettenleiter TC. 2005. Entry of pseudorabies virus: an immunogold-labeling study. J Virol 79:3200–3205. doi: 10.1128/JVI.79.5.3200-3205.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antinone SE, Shubeita GT, Coller KE, Lee JI, Haverlock-Moyns S, Gross SP, Smith GA. 2006. The herpesvirus capsid surface protein, VP26, and the majority of the tegument proteins are dispensable for capsid transport toward the nucleus. J Virol 80:5494–5498. doi: 10.1128/JVI.00026-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leege T, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J Gen Virol 90:1560–1568. doi: 10.1099/vir.0.010322-0. [DOI] [PubMed] [Google Scholar]
- 8.Desai P, Sexton GL, McCaffery JM, Person S. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J Virol 75:10259–10271. doi: 10.1128/JVI.75.21.10259-10271.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schipke J, Pohlmann A, Diestel R, Binz A, Rudolph K, Nagel CH, Bauerfeind R, Sodeik B. 2012. The C terminus of the large tegument protein pUL36 contains multiple capsid binding sites that function differently during assembly and cell entry of herpes simplex virus. J Virol 86:3682–3700. doi: 10.1128/JVI.06432-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J Virol 83:105–116. doi: 10.1128/JVI.01032-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klupp BG, Fuchs W, Granzow H, Nixdorf R, Mettenleiter TC. 2002. Pseudorabies virus UL36 tegument protein physically interacts with the UL37 protein. J Virol 76:3065–3071. doi: 10.1128/JVI.76.6.3065-3071.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J Virol 79:9566–9571. doi: 10.1128/JVI.79.15.9566-9571.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luxton GW, Lee JI, Haverlock-Moyns S, Schober JM, Smith GA. 2006. The pseudorabies virus VP1/2 tegument protein is required for intracellular capsid transport. J Virol 80:201–209. doi: 10.1128/JVI.80.1.201-209.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krautwald M, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Translocation of incoming pseudorabies virus capsids to the cell nucleus is delayed in the absence of tegument protein pUL37. J Virol 83:3389–3396. doi: 10.1128/JVI.02090-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klupp BG, Granzow H, Mundt E, Mettenleiter TC. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J Virol 75:8927–8936. doi: 10.1128/JVI.75.19.8927-8936.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pasdeloup D, Beilstein F, Roberts AP, McElwee M, McNab D, Rixon FJ. 2010. Inner tegument protein pUL37 of herpes simplex virus type 1 is involved in directing capsids to the trans-Golgi network for envelopment. J Gen Virol 91:2145–2151. doi: 10.1099/vir.0.022053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollinshead M, Johns HL, Sayers CL, Gonzalez-Lopez C, Smith GL, Elliott G. 2012. Endocytic tubules regulated by Rab GTPases 5 and 11 are used for envelopment of herpes simplex virus. EMBO J 31:4204–4220. doi: 10.1038/emboj.2012.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J Virol 87:2857–2867. doi: 10.1128/JVI.02676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pitts JD, Klabis J, Richards AL, Smith GA, Heldwein EE. 2014. Crystal structure of the herpesvirus inner tegument protein UL37 supports its essential role in control of viral trafficking. J Virol 88:5462–5473. doi: 10.1128/JVI.00163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED Protein Analysis Workbench. Nucleic Acids Res 41:W349–W357. doi: 10.1093/nar/gkt381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe D, Ushijima Y, Goshima F, Takakuwa H, Tomita Y, Nishiyama Y. 2000. Identification of nuclear export signal in UL37 protein of herpes simplex virus type 2. Biochem Biophys Res Commun 276:1248–1254. doi: 10.1006/bbrc.2000.3600. [DOI] [PubMed] [Google Scholar]
- 22.Lichtarge O, Bourne HR, Cohen FE. 1996. An evolutionary trace method defines binding surfaces common to protein families. J Mol Biol 257:342–358. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 23.Chouljenko DV, Jambunathan N, Chouljenko VN, Naderi M, Brylinski M, Caskey JR, Kousoulas KG. 2016. Herpes simplex virus 1 UL37 protein tyrosine residues conserved among all alphaherpesviruses are required for interactions with glycoprotein K, cytoplasmic virion envelopment, and infectious virus production. J Virol 90:10351–10361. doi: 10.1128/JVI.01202-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rost B. 1999. Twilight zone of protein sequence alignments. Protein Eng 12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 25.Zhao J, Zeng Y, Xu S, Chen J, Shen G, Yu C, Knipe D, Yuan W, Peng J, Xu W, Zhang C, Xia Z, Feng P. 2016. A viral deamidase targets the helicase domain of RIG-I to block RNA-induced activation. Cell Host Microbe 20:770–784. doi: 10.1016/j.chom.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bucks MA, Murphy MA, O'Regan KJ, Courtney RJ. 2011. Identification of interaction domains within the UL37 tegument protein of herpes simplex virus type 1. Virology 416:42–53. doi: 10.1016/j.virol.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu X, Fitzgerald K, Kurt-Jones E, Finberg R, Knipe DM. 2008. Herpesvirus tegument protein activates NF-kappaB signaling through the TRAF6 adaptor protein. Proc Natl Acad Sci U S A 105:11335–11339. doi: 10.1073/pnas.0801617105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. 2007. Phaser crystallographic software. J Appl Crystallogr 40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, Sliz P. 2013. Collaboration gets the most out of software. eLife 2:e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC. 2007. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Atomic coordinates and structure factors for the HSV-1 UL37N structure have been deposited to the RCSB Protein Data Bank under accession number 5VYL.