ABSTRACT
Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that causes debilitating musculoskeletal pain and inflammation and can persist for months to years after acute infection. Although studies of humans and experimentally infected animals suggest that CHIKV infection persists in musculoskeletal tissues, the mechanisms for this remain poorly understood. To evaluate this further, we isolated CHIKV from the serum of persistently infected Rag1−/− mice at day 28. When inoculated into naive wild-type (WT) mice, this persistently circulating CHIKV strain displayed a capacity for earlier dissemination and greater pathogenicity than the parental virus. Sequence analysis revealed a nonsynonymous mutation in the E2 glycoprotein (E2 K200R) and a deletion within the 3′ untranslated region (3′-UTR). The introduction of these changes into the parental virus conferred enhanced virulence in mice, although primary tropism for musculoskeletal tissues was maintained. The E2 K200R mutation was largely responsible for enhanced viral dissemination and pathogenicity, although these effects were augmented by the 3′-UTR deletion. Finally, studies with Irf3/Irf7−/− and Ifnar1−/− mice suggest that the E2 K200R mutation enhances viral dissemination from the site of inoculation independently of interferon regulatory factor 3 (IRF3)-, IRF7-, and IFNAR1-mediated responses. As our findings reveal viral determinants of CHIKV dissemination and pathogenicity, their further study should help to elucidate host-virus interactions that determine acute and chronic CHIKV infection.
IMPORTANCE CHIKV is a globally spreading, mosquito-transmitted virus that causes debilitating acute and chronic musculoskeletal disease in humans. The viral genetic determinants that dictate the severity of acute and chronic diseases are not understood. To improve our understanding of CHIKV pathogenesis, we evaluated a CHIKV strain isolated from the serum of chronically infected immunocompromised mice. Sequence analysis of this persistent CHIKV strain identified two mutations, an amino acid change in the E2 viral attachment protein and a deletion within the 3′-UTR of the viral genome. We identified roles for these mutations in the enhancement of viral dissemination from the inoculation site and in disease severity. These data improve our understanding of the viral determinants of CHIKV pathogenesis and adaptive changes that occur during viral persistence.
KEYWORDS: alphavirus, chikungunya virus, viral pathogenesis, virulence determinants
INTRODUCTION
Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that causes epidemics of incapacitating musculoskeletal inflammatory disease in humans. Since 2004, CHIKV has infected millions of people and expanded into Europe, the Middle East, and the Pacific region (1). In 2013, local transmission of CHIKV occurred in the Western Hemisphere on islands in the Caribbean (2). Since then, CHIKV has caused more than 1.8 million cases in the Americas in more than 40 countries (3). Acute CHIKV disease is characterized by high fever with severe joint pain, joint swelling, and muscle pain (4). Severe outcomes, including death, occur in neonates, the elderly, and those with underlying medical comorbidities (5, 6). Rheumatologic disease signs and symptoms can last for months to years in up to 60% of infected individuals (7–14), which causes a substantial economic burden and loss of quality of life (15, 16). Vaccines and antiviral agents are not approved for CHIKV infection, and treatment is currently limited to managing symptoms with analgesics and anti-inflammatory drugs (17).
Although the mechanisms by which CHIKV infection leads to chronic disease remain poorly characterized, CHIKV antigen and RNA have been detected in synovial and muscle tissue biopsy specimens collected from patients during the chronic phase of disease (18, 19). Consistent with these data, long-term persistence of CHIKV infection and chronic joint disease occur in experimentally infected nonhuman primates (NHPs) and mice (20–24), suggesting that residual virus or viral products in tissues may promote chronic inflammation.
Previous studies demonstrated that adaptive immune responses control CHIKV infectivity during the persistent phase but are unable to clear CHIKV RNA from joint-associated tissue (24, 25). Moreover, CHIKV evades neutralizing antibody responses as a necessary step to establish persistence (23). CHIKV likely uses multiple evasion mechanisms to establish and maintain persistence, as persistent CHIKV infection develops in the setting of sustained innate and adaptive antiviral immune responses. For example, in addition to being essential for the control of CHIKV infection during the acute phase (26), the type I interferon (IFN) response is maintained for many weeks in humans with chronic CHIKV disease and in mice persistently infected with CHIKV (18, 25, 27). Furthermore, CHIKV-specific antibody and T cell responses endure for weeks to months postinfection (p.i.) (21, 28–30). Thus, we hypothesized that the persistence of CHIKV infection requires the acquisition of adaptive mutations to escape host selective pressures.
To explore this hypothesis, we isolated virus from ankle tissue and serum of B cell- and T cell-deficient Rag1−/− mice on day 28 following infection with the Asian CHIKV strain AF15561. Unexpectedly, plaque-purified CHIKV from the serum of chronically infected Rag1−/− mice displayed enhanced acute pathogenicity when inoculated into naive wild-type (WT) mice. Sequence analysis identified polymorphisms in the viral E2 glycoprotein and the 3′ untranslated region (3′-UTR) that, when introduced into the parental CHIKV strain AF15561, conferred enhanced virulence in WT mice. Reverse genetic analysis showed that while mutations in both E2 and the 3′-UTR contributed to enhanced pathogenicity, an enhanced capacity to disseminate more rapidly from the site of inoculation was linked to a single K200R mutation in E2. Studies in immunocompromised mice revealed that the E2 K200R mutation enhanced the ability of CHIKV to disseminate in a manner that was independent of IFN regulatory factor 3 (IRF3)-, IRF7-, and IFNAR1-dependent antiviral responses. Overall, our studies provide new insight into the viral determinants of CHIKV dissemination and virulence.
RESULTS
Isolation of a CHIKV strain with enhanced pathogenicity in WT mice.
We hypothesized that during persistent infection, CHIKV acquires adaptive mutations to facilitate persistence in specific tissues. To investigate this idea, we took advantage of the observations that Rag1−/− and Rag2−/− mice, in contrast to WT mice, support persistent viremia for weeks to months after CHIKV infection but do not succumb to disease (24, 25, 31). To assess whether infectious CHIKV circulating in the serum of Rag1−/− mice had adapted to host pressures during persistence, WT mice were inoculated in the left rear footpad with 20 μl of serum collected from five individual viremic Rag1−/− mice at 28 days postinfection (dpi). In parallel, WT mice were inoculated in the left rear footpad with 103 PFU of AF15561 stock virus or 20 μl of a clarified right-ankle tissue homogenate collected from five individual CHIKV-infected Rag1−/− mice at 28 dpi. The inoculated sera or ankle tissue homogenates each contained between 10 and 20 PFU of infectious virus, as measured by a direct plaque assay (23). Mice were monitored daily for virus-induced changes in weight as a measure of disease. Four of five mice inoculated with serum from persistently infected Rag1−/− mice failed to gain weight through day 10 p.i. compared with those inoculated with AF15561 (P < 0.001) (Fig. 1A). In contrast, and consistent with data from previous studies, WT mice infected with CHIKV AF15561 gained weight throughout the course of infection (Fig. 1A). Mice inoculated with ankle tissue homogenates from CHIKV-infected Rag1−/− mice showed weight gain similar to that of AF15561-infected mice (P > 0.05). Furthermore, mice inoculated with serum from persistently infected Rag1−/− mice exhibited more severe musculoskeletal disease signs, including hind limb grip loss, altered gait, and difficulty righting (data not shown). These data suggested that CHIKV AF15561 in the circulation, but not ankle tissue, of Rag1−/− mice at 4 weeks postinfection had acquired adaptive mutations that enhanced acute pathogenesis in naive WT mice.
FIG 1.
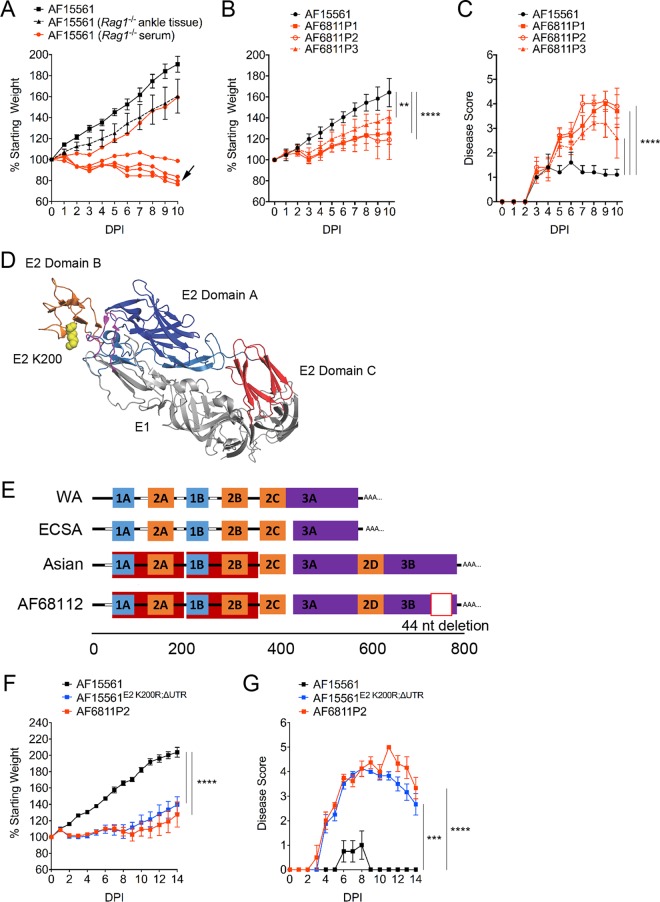
Mutations in E2 and the 3′-UTR enhance CHIKV pathogenicity in mice. (A) WT C57BL/6J mice were inoculated with 103 PFU of AF15561 (n = 5), 20 μl of clarified right ankle tissue homogenate (10 to 20 PFU) collected at day 28 p.i. from 5 individual chronically infected Rag1−/− mice (n = 5), or 20 μl of serum (10 to 20 PFU) collected from 5 individual chronically infected Rag1−/− mice (n = 5; data from individual mice are graphed separately) in the left rear footpad. The percent starting body weight was determined daily. The black arrow indicates the sample used for plaque purification of the virus. (B and C) WT C57BL/6J mice (n = 6 to 7/group) were inoculated with 10 PFU of AF15561 or plaque-purified AF15561 viral isolates derived from the serum of a chronically infected Rag1−/− mouse (AF6811P1 to AF6811P3) in the left rear footpad. The percent starting body weight (B) and disease score (C) were determined daily. Data are derived from two independent experiments. P values were determined by two-way ANOVA with Bonferroni's multiple-comparison test. **, P < 0.01; ****, P < 0.0001. (D) CHIKV E2-E1 heterodimer showing the position of E2 residue 200 (PDB accession no. 3N42) (32). E1 is shown in light gray, the E1 fusion loop is shown in magenta, E2 domain A is shown in dark blue, E2 domain B is shown in orange, and E2 domain C is shown in red. The lysine residue at E2 position 200 is shown in yellow. (E) Schematic of the 3′-UTR structures of West African (WA); East, Central, and South African (ECSA); and Asian genotype CHIKV strains, including the AF6811P2 Asian strain plaque isolated from the serum of chronically infected Rag1−/− mice. UTR sequence features were annotated according to methods described previously by Chen et al. (65). Direct repeats are labeled and illustrated by different-colored blocks. Sequence gaps in the alignment are indicated by white blocks. (F and G) WT C57BL/6J mice were inoculated with 103 PFU of AF15561 (n = 4), AF6811P2 (n = 4), or AF15561E2 K200R;ΔUTR (n = 4) in the left rear footpad. The percent starting body weight (F) and disease score (G) were determined daily. P values were determined by two-way ANOVA with Bonferroni's multiple-comparison test. ***, P < 0.001; ****, P < 0.0001.
One alternative explanation for the enhanced disease observed in WT mice inoculated with serum from persistently infected Rag1−/− mice was the coinoculation of factors present in the serum of chronically infected Rag1−/− mice that enhanced virus-induced disease. To eliminate this possibility, we plaque purified CHIKV from one of the Rag1−/− serum samples that caused severe disease in WT mice (Fig. 1A, arrow). Stocks of individual plaque-purified isolates AF6811P1 to AF6811P6 were generated by a single amplification on Vero cells. WT mice were inoculated with 10 PFU of AF15561 or plaque-purified isolates AF6811P1 to -P3 and monitored for virus-induced effects on weight gain. In contrast to mice inoculated with AF15561, mice inoculated with the plaque-purified viruses exhibited reduced weight gain (Fig. 1B) and more severe musculoskeletal disease signs (Fig. 1C). As the plaque-purified viral isolates retained the enhanced virulence of Rag1−/− mouse serum-derived AF15561, other serum factors were not responsible for the more severe disease signs.
Sequencing analysis identifies mutations in E2, E1, and the 3′-UTR.
We sequenced the complete viral genome of plaque-purified isolate AF6811P2 both at the consensus level and by Illumina-based deep sequencing. Alignment of sequence reads obtained by both methods to the reference CHIKV AF15561 genome identified three mutations in the genome of AF6811P2: a lysine (K)-to-arginine (R) substitution at E2 position 200 (E2 K200R), a synonymous A-to-G mutation in the E1 gene (nucleotide [nt] 10506), and a 44-nt deletion in the 3′-UTR (Table 1). The deep-sequencing results indicated that these mutations were present in 98.5% or more of the sequence reads, suggesting that the AF6811P2 plaque-purified virus was clonal. We also sequenced E2 position 200 and the 3′-UTR in the other five plaque-purified isolates and found that each isolate contained an E2 K200R mutation, with the exception of AF6811P3, which had a mutation to glutamine at this position (E2 K200Q) (Table 2). A 44-nt deletion identical to the deletion present in the 3′-UTR of AF6811P2 was present in three of the other plaque isolates but notably was absent in two others (Table 2), suggesting that this deletion was selected after the mutation at E2 position 200. E2 residue 200 is located in domain B and, based on the crystal structure of the pE2-E1 heterodimer (32), is predicted to make contacts with E1 residues 63 and 95 located in the fusion loop of E1 (Fig. 1D). Analysis of sequence variation among the 335 available CHIKV genomes in the NIAID ViPR database (33) revealed that 333/335 genomes (>99%) code for a K residue at position 200 in E2. The remaining two genomes, isolated from a patient in Thailand in 1995 (SV0444-95) and from a mosquito from the Ivory Coast in 1993 (ArA 30548), encode an R residue at this site, suggesting that the K200R mutation occurs in nature. The 44-nt deletion occurs in a duplicated region of the 3′-UTR that is unique to Asian genotype viruses (Fig. 1E). The 44-nt sequence is also highly conserved, with CHIKV strains across all genotypes encoding one or two copies of this sequence in the 3′-UTR (see Table S1 in the supplemental material).
TABLE 1.
Mutations identified in AF6811P2a
| Nucleotide position(s) | Genome region | Mutation (WT→AF6811P2) | Amino acid change (WT→AF6811P2) |
|---|---|---|---|
| 9140 | E2 | A→G | K200R |
| 10506 | E1 | A→T | Synonymous |
| 11921–11964 | 3′-UTR | Deletion | NA |
Mutations were identified both at the consensus level and by Illumina deep sequencing. Sequences were aligned against the sequence of the AF15561 reference genome (GenBank accession no. EF452493). NA, not applicable.
TABLE 2.
Analysis of mutations at E2 position 200 and the 3′-UTR deletion in plaque-purified isolatesa
| Plaque isolate | Mutation at E2 position 200 | Presence of 3′-UTR deletion |
|---|---|---|
| AF6811P1 | K200R | Absent |
| AF6811P2 | K200R | Present |
| AF6811P3 | K200Q | Absent |
| AF6811P4 | K200R | Present |
| AF6811P5 | K200R | Present |
| AF6811P6 | K200R | Present |
Shown are data from sequence analyses of 6 plaque isolates from the serum of chronically infected Rag1−/− mice at E2 position 200 and the 44-nucleotide deletion identified in the 3′-UTR.
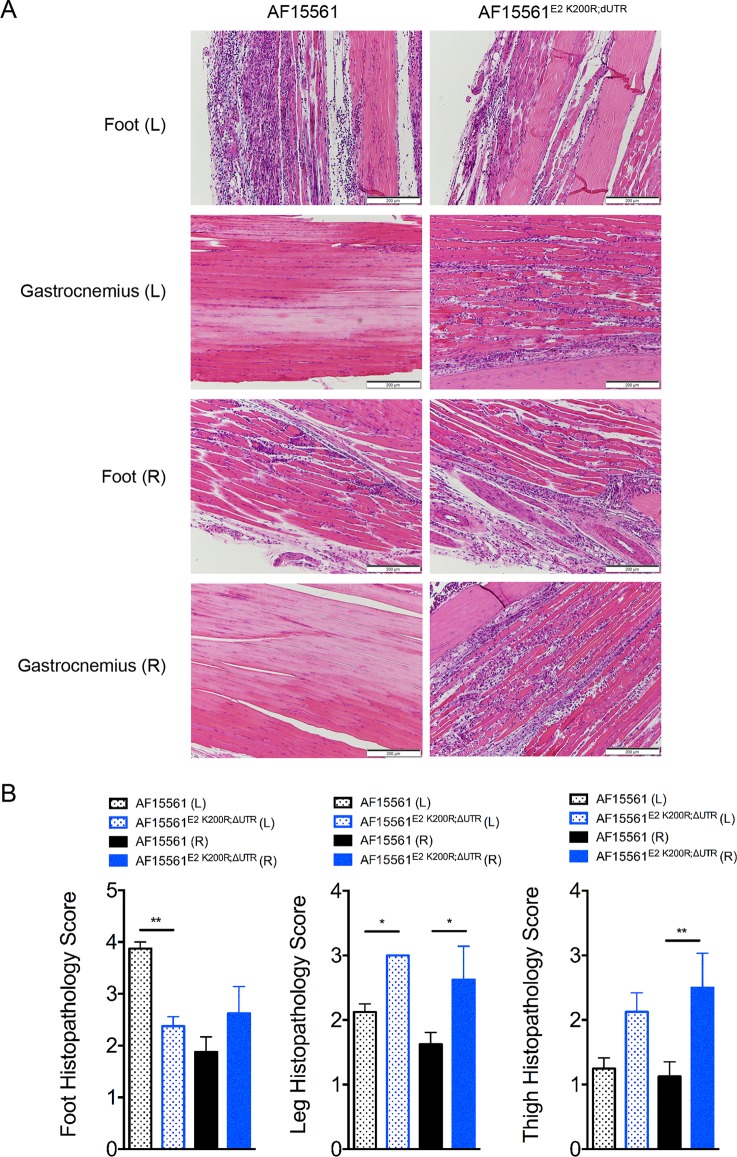
To confirm that these mutations were responsible for the enhanced virulence of the adapted AF6811P2 strain, the E2 K200R mutation and the 3′-UTR deletion were introduced into the plasmid carrying the parental AF15561 cDNA genome to generate AF15561E2 K200R;ΔUTR. WT mice were inoculated in the footpad with 103 PFU of WT strain AF15561, AF6811P2, or AF15561E2 K200R;ΔUTR, and mice were monitored for weight gain and musculoskeletal disease (Fig. 1F and G). Similar to AF6811P2-infected mice, AF15561E2 K200R;ΔUTR-infected mice exhibited diminished weight gain compared with mice inoculated with AF15561 (Fig. 1F), and remarkably, 1 of 4 mice in both groups succumbed to infection by day 10 p.i. Additionally, both AF6811P2- and AF15561E2 K200R;ΔUTR-infected mice exhibited more severe signs of musculoskeletal disease than did mice inoculated with AF15561 (Fig. 1G). At 7 dpi, WT mice inoculated with AF15561E2 K200R;ΔUTR also displayed more severe inflammation and injury in tissues distal to the site of inoculation, including the ipsilateral and contralateral gastrocnemius muscles (leg) and thighs (Fig. 2A and B). Notably, mice infected with AF6811P2 and AF15561E2 K200R;ΔUTR did not develop more severe swelling of the inoculated foot than did AF15561-infected mice (data not shown). These data indicate that an E2 K200R mutation and a deletion of 44 nt in the 3′-UTR enhance systemic disease and injury in musculoskeletal tissues distal to the site of inoculation.
FIG 2.
Mutations in E2 and the 3′-UTR enhance musculoskeletal tissue injury and inflammation. WT C57BL/6J mice were inoculated with 103 PFU of AF15561 or AF15561E2 K200R;ΔUTR in the left rear footpad. (A) At 7 dpi, 5-μm paraffin-embedded sections were generated from the ipsilateral and contralateral hind limbs and stained with hematoxylin and eosin. Images are representative of results for 8 mice per group. (B) Tissues in the foot, leg, and thigh from the left (L) and right (R) limbs (n = 8/group) were scored in a blind manner based on the following scale: 0 for no inflammation, 1 for >5 areas of small clusters of leukocytes, 2 for leukocytes forming larger clusters to thin tracts through tissue with multiple affected sites, 3 for clusters and tracts of leukocytes coalescing into at least one large area that displaces/replaces tissue, with or without necrosis and with or without mineralization, and 4 for leukocytes that are in aggregates that are large enough to replace >40% of normal tissue. Data are derived from results from two independent experiments. P values were determined by a Kruskal-Wallis test with Dunn's multiple-comparison test. *, P < 0.05; **, P < 0.01.
AF15561E2 K200R;ΔUTR displays enhanced dissemination and infection at early times.
Based on the failure of mice infected with AF15561E2 K200R;ΔUTR to gain weight early after infection, we hypothesized that these mutations enhanced viral replication in tissues at early time points p.i. To evaluate viral dissemination and tissue burdens, WT mice were inoculated in the left rear footpad with either AF15561 or AF15561E2 K200R;ΔUTR, and the amount of infectious virus proximal or distal to the site of inoculation was quantified at 1 and 3 dpi. Both AF15561 and AF15561E2 K200R;ΔUTR achieved similar titers at 1 and 3 dpi in the left ankle tissue, which is proximal to the inoculation site (Fig. 3A and B). In contrast to the left ankle, AF15561E2 K200R;ΔUTR rapidly disseminated to distant tissues and achieved higher titers than those in AF15561-infected mice by 1 dpi (Fig. 3A) (right ankle, 3,400-fold [P < 0.0001]; right quadriceps, 460-fold [P < 0.0001]; spleen, 38-fold [P < 0.0001]; liver, 28-fold [P < 0.001]; brain, 11-fold [P < 0.0001]). Similarly, AF15561E2 K200R;ΔUTR-infected mice had a higher level of viremia than did AF15561-infected mice at both 1 dpi (1,000-fold; P < 0.001) and 3 dpi (1,050-fold; P < 0.01) (Fig. 3A and B). Together, these data suggest that the E2 K200R mutation and the 44-nt deletion in the 3′-UTR enhance the ability of CHIKV to disseminate from the site of inoculation.
FIG 3.
Mutations in E2 and the 3′-UTR enhance viral dissemination and tissue burdens in WT mice. WT C57BL/6 mice were inoculated with 103 PFU of AF15561 or AF15561E2 K200R;ΔUTR in the left rear footpad. At 1 dpi (n = 9 mice/group) (A) or 3 dpi (n = 5 mice/group) (B), mice were euthanized, and the amount of infectious virus in the indicated tissues was quantified by a focus-forming assay. Dashed lines indicate the limit of detection. Data are derived from results from two independent experiments. P values were determined by Mann-Whitney tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
To investigate if the E2 K200R and the 3′-UTR mutations altered viral replication in cells in vitro, we performed multistep viral growth analysis in murine fibroblasts and differentiated murine C2C12 muscle cells, key cell type targets in vertebrate hosts (26, 34), and in mosquito-derived C6/36 cells. Virus yields from murine fibroblasts and murine C2C12 muscle cells inoculated with AF15561 or AF15561E2 K200R;ΔUTR at a low multiplicity of infection (MOI) (0.1 and 0.01 focus-forming units [FFU]/cell, respectively) were similar at 12, 24, and 48 h postinoculation (hpi) (Fig. 4A and B). The yields of infectious virus were reduced in C6/36 mosquito cells infected with AF15561E2 K200R;ΔUTR (Fig. 4C); however, the magnitude of these differences was small, only 2- to 3-fold. These data, together with the data demonstrating no differences in viral loads at the site of inoculation of mice (Fig. 3A and B), suggest that the E2 K200R and 3′-UTR mutations predominantly impact viral dissemination in vivo.
FIG 4.
AF15561 and AF15561E2 K200R;ΔUTR replicate similarly in vitro. Mouse embryo fibroblasts (A), differentiated C2C12 murine muscle cells (B), or C6/36 Aedes albopictus mosquito cells (C) were inoculated with AF15561 or AF15561E2 K200R;ΔUTR at an MOI of 0.1 FFU/cell (mouse embryo fibroblasts) or 0.01 FFU/cell (C2C12 and C6/36 cells). At 0 hpi (input) and 1, 6, 12, 24, 48, and 72 hpi, the amount of infectious virus present in culture supernatants was quantified by a focus-forming assay. Data are representative of results from two independent experiments. P values were determined by two-way ANOVA with Bonferroni's multiple-comparison test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
The adaptive mutations enhance early viral dissemination independent of IRF3, IRF7, and IFNAR1.
The type I IFN pathway has an important role in restricting CHIKV replication in mice (26, 35–38). Mice deficient in the transcription factors that induce type I IFN (IRF3 and IRF7) or in a required subunit of the type I IFN receptor (IFNAR1) are more susceptible to CHIKV infection, with elevated viral burdens in many tissues, analogous to our results with WT mice infected with the AF15561E2 K200R;ΔUTR mutant virus. We hypothesized that the enhanced viral burdens observed at early times after infection of WT mice with AF15561E2 K200R;ΔUTR might be due to greater antagonism of the type I IFN response. To explore this possibility, we compared AF15561 and AF15561E2 K200R;ΔUTR infections in Irf3−/− Irf7−/− double-knockout (DKO) mice, which are compromised for the production of type I IFN following CHIKV infection (36, 37). Regardless of the mouse strain, AF15561 and AF15561E2 K200R;ΔUTR achieved similar titers in the left ankle tissue after ipsilateral footpad inoculation (Fig. 5A). In both WT and Irf3−/− Irf7−/− DKO mice, AF15561E2 K200R;ΔUTR rapidly disseminated and achieved higher viral loads than those in AF15561-infected mice in the right ankle (234-fold [WT] and 239-fold [Irf3−/− Irf7−/− DKO]; P < 0.05 and P < 0.01, respectively), the right quadriceps (230-fold [WT] and 2,800-fold [Irf3−/− Irf7−/− DKO]; P < 0.05 and P < 0.001, respectively), and serum (1,500-fold [WT] and 370-fold [Irf3−/− Irf7−/− DKO]; P < 0.05 and P < 0.01, respectively) (Fig. 5A). Quantification of IFN-α levels in the sera of naive and CHIKV-infected mice at 1 dpi confirmed that Irf3−/− Irf7−/− DKO mice failed to produce type I IFN in response to CHIKV infection (Fig. 5B). In addition, we found that IFN-α levels in the sera of WT mice infected with AF15561E2 K200R;ΔUTR at 1 dpi were higher than those in the sera of AF15561-infected WT mice (Fig. 5B), demonstrating that AF15561E2 K200R;ΔUTR infection induces a robust type I IFN response. Consistent with the findings for Irf3−/− Irf7−/− DKO mice, AF15561E2 K200R;ΔUTR rapidly disseminated and achieved higher viral loads than did parental strain AF15561 in Ifnar1−/− mice (Fig. 5C). These data suggest that the E2 K200R mutation and the 44-nt deletion in the 3′-UTR enhance the capacity of the virus to rapidly disseminate from the inoculation site by a mechanism that is independent of the evasion or suppression of the type I IFN response, although a role for IRF5-dependent antiviral responses (39) cannot be ruled out.
FIG 5.
Mutations in E2 and the 3′-UTR enhance viral dissemination in Irf3−/− Irf7−/− DKO and Ifnar1−/− mice. (A and B) WT and Irf3−/− Irf7−/− DKO C57BL/6 mice were inoculated with 103 PFU of AF15561 or AF15561E2 K200R;ΔUTR in the left rear footpad. (A) At 1 dpi, the amount of infectious virus in the indicated tissues was quantified by a focus-forming assay. (B) The amount of IFN-α in serum of mock-infected (n = 3/group), AF15561-infected (n = 8-9/group), or AF15561E2 K200R;ΔUTR-infected (n = 8 to 9/group) mice was quantified by an ELISA. (C) Ifnar1−/− C57BL/6 mice were inoculated with 103 PFU of AF15561 or AF15561E2 K200R;ΔUTR in the left rear footpad. At 1 dpi, the amount of infectious virus in the indicated tissues was quantified by a focus-forming assay. Dashed lines indicate the limit of detection. Data are derived from results from two independent experiments. P values were determined by Mann-Whitney tests (A and C) or one-way ANOVA with Tukey's multiple-comparison test (B). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Both the E2 K200R mutation and the 3′-UTR deletion contribute to enhanced disease severity.
To evaluate the contribution of each mutation individually to the disease phenotype observed in WT mice infected with AF15561E2 K200R;ΔUTR, we constructed AF15561 strains encoding either the E2 K200R mutation (AF15561E2 K200R) or the 3′-UTR deletion (AF15561ΔUTR). WT mice were inoculated with AF15561, AF15561E2 K200R;ΔUTR, AF15561E2 K200R, or AF15561ΔUTR and monitored daily for morbidity and musculoskeletal disease (Fig. 6A and B). Mice infected with AF15561E2 K200R failed to gain weight (P < 0.0001) and developed more severe musculoskeletal disease (P < 0.0001) than did AF15561-infected mice, suggesting that the E2 K200R mutation contributes to enhanced pathogenesis in mice. In contrast, mice infected with AF15561ΔUTR gained weight similarly to AF15561-infected mice (P > 0.05) and exhibited similar mild musculoskeletal disease signs, suggesting that this deletion alone is not sufficient to enhance virulence. However, at later times postinfection (days 9 to 14 p.i.), mice infected with AF15561E2 K200R more readily gained weight (Fig. 6A) and displayed less musculoskeletal disease than did AF15561E2 K200R;ΔUTR-infected mice (days 6 to 14 p.i.) (Fig. 6B), suggesting that when combined with the E2 K200R mutation, the 3′-UTR deletion enhances the pathogenicity of CHIKV in mice.
FIG 6.
Both the E2 K200R mutation and the deletion in the 3′-UTR contribute to enhanced viral pathogenicity in mice. (A and B) WT C57BL/6J mice were inoculated with 103 PFU of AF15561 (n = 8), AF15561E2 K200R;ΔUTR (n = 8), AF15561E2 K200R (n = 8), or AF15561ΔUTR (n = 8) in the left rear footpad. The percent starting body weight (A) and disease score (B) were determined daily. Data are derived from results from two independent experiments. P values were determined by two-way ANOVA with Bonferroni's multiple-comparison test. **, P < 0.01; ***, P < 0.001. The P values displayed are for AF15561E2 K200R;ΔUTR versus AF15561E2 K200R. (C) WT C57BL/6J mice were inoculated with 103 PFU of AF15561 (n = 6), AF15561E2 K200R;ΔUTR (n = 6), AF15561E2 K200R (n = 6), or AF15561ΔUTR (n = 6) in the left rear footpad. At 1 dpi, mice were euthanized, and the amount of infectious virus in the indicated tissues was quantified by a focus-forming assay. Data are derived from results from two independent experiments. P values were determined by a Kruskal-Wallis test with Dunn's multiple-comparison test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The E2 K200R mutation enhances viral dissemination in mice.
We next evaluated the contribution of each mutation to the pattern of early viral dissemination. WT mice were inoculated in the left rear footpad with 103 PFU of AF15561, AF15561E2 K200R;ΔUTR, AF15561E2 K200R, or AF15561ΔUTR. At 1 dpi, as expected, similar viral titers were detected in the left ankle regardless of the viral strain (Fig. 6C). In contrast, both AF15561E2 K200R;ΔUTR and AF15561E2 K200R rapidly disseminated to distant tissues and achieved higher viral loads in the right ankle and quadriceps than those in mice infected with AF15561 (Fig. 6C). Similarly, AF15561E2 K200R;ΔUTR- and AF15561E2 K200R-infected mice had a higher level of viremia than did AF15561-infected mice (Fig. 6C). No differences in viral loads in tissues were detected in mice infected with AF15561ΔUTR compared with AF15561-infected mice. Collectively, these data suggest that the E2 K200R mutation enhances the capacity of the virus to disseminate from the site of inoculation, whereas the deletion in the 3′-UTR is dispensable for these effects.
DISCUSSION
Several studies have established that mice deficient in adaptive immune responses develop persistent CHIKV infection in multiple tissues for weeks (23–25, 28, 31, 40). Despite sustaining chronic viral replication for as long as 500 days, Rag1−/− mice exhibit only mild to moderate signs of joint pathology along with modestly elevated serum levels of tumor necrosis factor (TNF), interleukin-6 (IL-6), and IFN-γ (24, 25). Thus, the isolation of an adapted, more pathogenic, strain of CHIKV from the serum of a Rag1−/− mouse was unexpected. However, the isolation of a neurovirulent strain of Sindbis virus from the brains of persistently infected severe combined immune deficiency (scid) mice was previously reported (41), suggesting that persistent replication in immunodeficient mice can select for more pathogenic alphaviruses. Our findings are in contrast to those of a previous study in which CHIKV isolated from the serum of Rag1−/− mice at 100 dpi did not show enhanced fitness when inoculated into WT mice (25). Similarly, CHIKV isolated from the kidney or brain of Rag1−/− mice at 4 to 6 weeks postinfection showed a limited number of coding changes, all within the nonstructural proteins (40), although the effects of these mutations on CHIKV replication and pathogenesis were not evaluated. It is possible that differences in the mouse models (young versus adult mice), founder strains of CHIKV (East, Central, and South African [ECSA] versus Asian genotype), and sources of infectious virus for the inoculum (serum in our study or cell culture passage in the previous study [25]) may account for these differences. Indeed, in our own studies, one mouse receiving serum-derived AF15561 exhibited less severe musculoskeletal disease and weight loss, suggesting that AF15561 circulating in the serum of Rag1−/− mice may be heterogeneous. This is supported by our sequencing results for six plaque-purified isolates from the serum of one chronically infected Rag1−/− mouse, which displayed heterogeneity at E2 residue 200 and in the 3′-UTR.
Although AF6811P2 was isolated from serum, which tissues or cell types in Rag1−/− mice shed CHIKV into the circulation are unknown. In contrast to serum, inoculation of ankle tissue homogenates, which contained infectious CHIKV, from persistently infected Rag1−/− mice did not result in more severe disease signs, suggesting that discrete populations of CHIKV with various fitnesses exist within the same host. Further studies of these populations may provide insight into tissue-specific selective pressures exerted on CHIKV during acute and chronic infections.
The host pressures that selected for the adaptive mutations are unknown. Rag1−/− mice can limit CHIKV replication in the absence of adaptive immune responses (24, 25), likely through type I IFN and other innate immune responses. Mice deficient in the type I IFN response rapidly succumb to CHIKV infection within days of infection (26, 35–38), demonstrating an essential requirement for this immune arm to control early CHIKV replication and dissemination. Passage of the related Semliki Forest virus (SFV) in mice resulted in a strain of SFV (L10) that was resistant to control by the type I IFN response (42). Accordingly, an attractive hypothesis was that the increased pathogenicity and replicative capacity of the adapted CHIKV strain in mice was due to greater resistance to the host type I IFN response. However, our studies with Irf3−/− Irf7−/− DKO and Ifnar1−/− mice suggest that the enhanced viral dissemination conferred by the E2 K200R mutation is independent of the antagonism of type I IFN antiviral responses mediated via IFNAR1. The role that increased resistance to type I IFN responses may play at later stages of the pathogenic sequence remains to be investigated, as Irf3−/− Irf7−/− DKO and Ifnar1−/− mice infected with CHIKV rapidly succumb to infection.
Our analyses of tissue viral burden, tissue histopathology, and disease signs in mice indicate that the AF6811P2 and AF15561E2 K200R;ΔUTR CHIKV strains retain tropism for musculoskeletal tissues and cause musculoskeletal disease. In addition, mice infected with AF15561E2 K200; ΔUTR developed viremia with a higher magnitude and a longer duration than did mice infected with most WT CHIKV strains, a feature that is consistent with CHIKV-infected humans and nonhuman primates, which also develop a remarkably high level of viremia (20, 43–48). The higher viral loads in the quadriceps muscle of AF15561E2 K200R;ΔUTR-infected mice likely contribute to more severe musculoskeletal disease, as disease severity correlates with increased viral burdens in muscle tissue in CHIKV- and Ross River virus-infected mice (34, 38, 49).
Our data indicate that the E2 K200R mutation is sufficient to enhance CHIKV dissemination and pathogenicity in mice. However, the mechanisms by which this mutation influences viral dissemination and pathogenesis remain to be determined. E2 residue K200 is highly conserved among strains of all three CHIKV genotypes, with 333/335 CHIKV genomes in the ViPR database (33) encoding a K residue at this position. The remaining two strains encoded an R residue at this position. The CHIKV E2 glycoprotein forms a dimer with the E1 glycoprotein, and three dimers form one of the 80 spikes on the surface on the virion (32). E2 is highly exposed on the virion and is responsible for receptor binding while also being the primary target of neutralizing antibody responses (28, 30, 50). Based on the X-ray crystal structure of the CHIKV E2-E1 heterodimer, E2 residue K200 makes van der Waals contacts with highly conserved E1 residues (C63 and F95) located near or in the conserved E1 fusion loop (32), suggesting that the K200R mutation might alter E2-E1 interactions at this site (Fig. 1D). E2-E1 interactions in related alphaviruses have been shown to regulate pH-mediated structural changes (51, 52) that are a requisite for the entry of the virus into the cytoplasm (53, 54). Mutations in E2 can also influence virion assembly and release in a cell- and host-specific manner (55–57), dictate viral tropism and pathogenesis in vivo (58–62), and modulate vector competence (63, 64). Further studies are needed to examine whether the E2 K200R mutation influences CHIKV cellular tropism, the pH of fusion, virus assembly and release, or other unknown mechanisms.
Our comparison of AF15561 containing the 3′-UTR deletion alone, AF15561ΔUTR, and the double mutant strain AF15561E2 K200R;ΔUTR suggests that the 44-nt deletion in the 3′-UTR also contributes to enhanced virulence in mice. The CHIKV 3′-UTR contains direct-repeat elements formed by duplications within the 3′-UTR (65). Asian genotype viruses (e.g., AF15561) encode a divergent 3′-UTR compared to those of strains of the West African and ECSA genotypes. This divergent 3′-UTR likely was formed by a large deletion in the 3′-UTR and the subsequent accumulation of adaptive mutations and duplications due to founder effects, as the Asian genotype diverged from an ECSA genotype virus (65). The recent identification of a novel duplication in Asian-American CHIKV isolates suggests that the Asian genotype 3′-UTR is still under selective pressures (66). The 44-nt deletion identified in the virus persistently circulating in Rag1−/− mice occurs in a region of the 3′-UTR, element 3B, that is unique to Asian genotype viruses (Fig. 1E). Thus, the effects of the 44-nt deletion on pathogenicity may be constrained to Asian genotype viruses. However, the 3A element, present in all genotypes of CHIKV, contains a similar sequence that differs by only 1 to 4 nt from the 44-nt deletion in 3B. Further studies are needed to determine if the 3B deletion uniquely affects CHIKV pathogenicity in mice or whether manipulation of the 3A element, present in all CHIKV genotypes, can result in similar effects. The function of the 3′-UTR in alphaviral replication and pathogenesis is largely unknown, as alphaviruses exhibit significant heterogeneity in this region (67–69). Modulation of the 3′-UTR often results in a fitness cost in mosquito cells or whole mosquitoes, whereas large deletions are tolerated in mammalian cells (70–72). However, the 3′-UTR is important for the stability of the viral genome and likely interacts with host factors to facilitate viral replication (73, 74). CHIKV and other alphavirus 3′-UTRs interact with the host HuR protein, which protects the viral RNA from degradation and increases virus yields (75–77). The viral 3′-UTR also can impact viral tropism and host innate immune responses to RNA virus infection. A microRNA (miRNA) binding site in the 3′-UTR of eastern equine encephalitis virus for a myeloid cell-specific miRNA restricts viral replication in myeloid cells, which delays the induction of the type I IFN response (72). West Nile virus, an unrelated flavivirus, also encodes a miRNA in its 3′-UTR that enhances replication in mosquito cells (78), and an RNA secondary structure in flavivirus 3′-UTRs can influence viral replication and innate responses to infection (79). Given these diverse functions of the 3′-UTR, it remains to be determined if the deletion in the 3′-UTR alleviates restriction by a host factor, alters interactions with host proteins that facilitate viral replication, or modulates other pathways to enhance viral pathogenicity in mice.
Our isolation of a CHIKV strain with enhanced virulence in mice and the identification of the E2 K200R mutation and the 3′-UTR deletion defined new determinants of CHIKV virulence. Understanding the mechanisms by which these mutations in E2 and the 3′-UTR contribute to enhanced dissemination and acute and chronic pathogenesis in mice of different ages may provide insight for controlling CHIKV infection and preventing persistence. Finally, infection of mice by this strain recapitulates the high level viremia and more severe disseminated acute musculoskeletal disease reported for humans, which could lead to improvements in the mouse model of CHIKV infection and disease. These advancements may have utility for defining mechanisms of acute and chronic disease pathogenesis and for evaluating vaccine candidates and host- and virus-targeted therapeutics.
MATERIALS AND METHODS
Ethics statement.
This study was conducted in accordance with recommendations in the Guide for the Care and Use of Laboratory Animals (80) and AVMA guidelines for the euthanasia of animals (88). All animal experiments conducted at the University of Colorado Anschutz Medical Campus were performed with the approval of the Institutional Animal Care and Use Committee (IACUC) at the University School of Medicine (assurance no. A3269-01) under protocol B-86514(10)1E. For studies with Ifnar1−/− mice, the protocols were approved by the IACUC at the Washington University School of Medicine (assurance no. A3381-01).
Viruses.
CHIKV strain AF15561 (GenBank accession no. EF452493) was isolated from a human patient during an outbreak of CHIKV in Thailand in 1962 and passaged twice in Vero cells (81, 82). A cDNA clone of AF15561 was generated from 181/25 cDNA by using site-directed mutagenesis as previously described (58). Virus stocks were generated by the electroporation of BHK-21 cells with in vitro-transcribed RNA as previously described (24). Cell culture supernatants from electroporated cells were collected, clarified by centrifugation, and aliquoted. Stock virus titers were quantified by a plaque assay on BHK-21 or Vero cells.
Cells.
BHK-21 cells (ATCC CCL10) were grown in α-minimal essential medium (MEM-α) (Gibco) supplemented with 10% bovine calf serum (HyClone), 10% tryptose phosphate broth, penicillin and streptomycin, and 0.29 mg/ml l-glutamine. Vero cells (ATCC CCL81) were grown in Dulbecco's modified Eagle medium (DMEM)–F-12 medium (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Lonza), nonessential amino acids (Life Technologies), sodium bicarbonate, penicillin and streptomycin, and 0.29 mg/ml l-glutamine. Mouse embryo fibroblasts and C2C12 murine myoblasts were cultured in DMEM supplemented with 10% FBS. C2C12 cells were differentiated into myotubes by culturing in fusion medium (DMEM–2% horse serum) for 5 to 6 days. Aedes albopictus C6/36 cells were cultivated in Leibovitz-15 medium (Invitrogen) supplemented with 10% FBS, 10 mM HEPES, and antibiotics (penicillin and streptomycin) at 29°C.
Mouse experiments.
WT C57BL/6J mice and congenic Rag1−/− or Irf3−/− Irf7−/− DKO mice were bred in specific-pathogen-free facilities at the University of Colorado Anschutz Medical Campus. Ifnar1−/− mice were bred in specific-pathogen-free facilities at Washington University. All mouse infection studies were performed in animal biosafety level 3 laboratories. Three- to four-week-old mice were used for all studies, with the exception of experiments with Ifnar1−/− mice, which used 5-week-old animals. Unless otherwise indicated, mice were inoculated in the left rear footpad with 103 PFU of the virus in diluent (phosphate-buffered saline [PBS] supplemented with 1% FBS) in a volume of 10 μl. Mock-infected animals received diluent alone. Mice were monitored for disease signs and weighed at 24-h intervals. In some experiments, mice were scored for musculoskeletal disease as previously described (83), where a score of 1 indicates a mild deficit in the hind paw gripping ability of the injected foot only; 2 indicates a mild deficit in the bilateral hind paw gripping ability; 3 indicates a bilateral loss of gripping ability, with mild bilateral hind limb paresis and an altered gait that is not readily observable; 4 indicates a bilateral loss of gripping ability, moderate bilateral hind limb paresis, observable altered gait, and difficulty righting self; 5 indicates a bilateral loss of gripping ability, severe bilateral hind limb paresis, altered gait, and an inability to right self; and 6 indicates a moribund state. On the termination day of each experiment, mice were euthanized by sedation with isoflurane followed by thoracotomy, blood was collected, and mice were perfused by intracardiac injection of 1× PBS or 4% paraformaldehyde, depending on the experiment. PBS-perfused tissues were removed by dissection and homogenized in TRIzol reagent (Life Technologies) for RNA isolation or PBS–1% FBS for determining tissue titers by using a MagNA Lyser instrument (Roche).
Selection of viral plaque isolates.
Biological clones were isolated from the serum of a viremic Rag1−/− mouse infected 28 days previously with WT strain AF15561 by plaque purification. Serum was mixed with PBS plus 1% FBS and adsorbed to Vero cells for 1 h. Following adsorption, cells were overlaid with immunodiffusion agarose (MP Biomedicals) and incubated for 40 h to allow plaque formation. An agarose plug was removed from individual plaques by using a pipette tip, and the virus was amplified by a single passage on Vero cells.
Viral genome sequencing.
RNA was isolated from stocks of plaque-purified isolates by using TRIzol reagent (Life Technologies) according to the manufacturer's protocol. To generate cDNA, a 1:1 mix of oligo(dT) and random hexamer primers was used with SuperScript IV reverse transcriptase (Life Technologies). PCR was performed with primers spanning the viral genome and Q5 high-fidelity polymerase (New England BioLabs [NEB]). PCR amplicons were purified by using a PCR cleanup kit (Qiagen) and subjected to Sanger sequencing. Sequences were mapped to the AF15561 genome by using Geneious software (Biomatters). The AF6811P2 genome was also deep sequenced by using Illumina-based sequencing. Isolated viral RNA (500 ng) was reverse transcribed by using 500 ng of random primer 9 (NEB) and SuperScript II reverse transcriptase (Thermo Fisher Scientific). Briefly, the reaction mixture was equilibrated at 25°C for 2 min before the addition of 200 U of SuperScript II reverse transcriptase. The reverse transcription reaction mixture was incubated at 25°C for 10 min and at 42°C for 180 min and then inactivated at 70°C for 15 min. Double-stranded cDNA was prepared by using the NEBNext mRNA Second Strand Synthesis module (NEB). Sequencing libraries were prepared from the double-stranded cDNA by using the Nextera XT DNA library preparation kit (Illumina). Residual nucleotides were removed by using Agencourt AMPure XP beads (Beckman Coulter) at a DNA-to-bead ratio of 0.6:1. Library size and quality were measured by using a 2100 Bioanalyzer (Agilent Technologies) and quantified with a Qubit fluorometer by using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific). Sequencing reactions were performed on a MiSeq desktop sequencer (Illumina). Reads were sorted and aligned, and mutations were characterized by using Bowtie2 (84) and SAMtools (85). Results were visualized by using Integrative Genomics Viewer (86).
Construction of AF15561E2 K200R, AF15561ΔUTR, and AF15561E2 K200R;ΔUTR.
To insert the E2 K200R substitution, the AF15561 cDNA clone was mutated by site-directed mutagenesis (Agilent). Restriction digestion was performed with XmaI and XhoI to release a 1,035-bp fragment containing the desired mutation, and this fragment was ligated into an XmaI- and XhoI-digested unmutagenized plasmid. The ligated fragment was sequenced to confirm that only the desired mutation was present. To insert the ΔUTR mutation into the AF15561 or AF15561E2 K200R backbone, a multistep cloning approach was used. cDNA from the AF6811P2 stock or WT AF15561 was generated by the isolation of viral RNA using TRIzol reagent (Life Technologies), and cDNA was generated by using SuperScript IV reverse transcriptase with a 1:1 mix of oligo(dT) and random hexamer primers. This cDNA was utilized as a template for PCR using primers CHIKV_10699F (5′-GCACCATCTGGCTTCAAGTA-3′) and CHIKV_12036R (5′-GAAATATTAAAAACAAAATAACATCTCCTA-3′) and Q5 high-fidelity polymerase (New England BioLabs). This PCR amplified a region from the 3′ end of E1 to the 3′-UTR immediately preceding the poly(A) tail and contains restriction sites for SgrDI (CHIKV nucleotide position 10864) and SspI (CHIKV nucleotide position 12032). The PCR product was cloned into the TOPO-II Zero Blunt vector (Life Technologies). To insert the AF6811P2 3′-UTR containing the 44-nt deletion into AF15561 or AF15561E2 K200R plasmids, the viral cDNA-carrying plasmids and the TOPO vector containing the AF6811P2 3′-UTR were subjected to sequential digestions with SgrDI (Thermo Fisher) and SspI (New England BioLabs). Fragments were ligated by using T4 DNA ligase (New England BioLabs). Due to a second SspI site in the pSinRep5 plasmid 200 bp downstream of the CHIKV poly(A) tail, this strategy resulted in the loss of the poly(A) tail and 200 bp of the plasmid backbone. To restore the poly(A) tail and plasmid backbone sequence, an unmutagenized AF15561 plasmid was digested with SspI to produce an ∼200-bp fragment containing the authentic 3′-UTR and plasmid backbone. The cDNA plasmids lacking the poly(A) tail and 200 bp of the plasmid backbone were then linearized with SspI. The 200-bp fragment was then ligated into the linearized plasmids. Restoration of the poly(A) tail and plasmid backbone was confirmed by sequencing.
In vitro virus replication.
Triplicate wells were inoculated with CHIKV at the indicated MOIs. Viruses were adsorbed onto cells for 1 h at 37°C. Wells were then washed three times with 1 ml of room-temperature 1× PBS. Growth medium was then added to each well, and cells were incubated at 37°C. For cumulative growth analysis, 100-μl samples of culture supernatants were removed at various times p.i., and an equal volume of fresh growth medium was added to maintain a constant volume within each well. Samples were stored at −80°C for analysis by a focus-forming assay.
Plaque and focus-forming assays.
To quantify the amount of infectious virus in samples, either a plaque assay or a focus-forming assay was used. For plaque assays, BHK-21 cells were seeded into 6-well dishes. Samples containing the virus were serially diluted in a solution containing PBS, 1% FBS, 1× Ca2+, and 1× Mg2+ and adsorbed onto the cells for 1 h at 37°C. Cells were overlaid with 1% immunodiffusion agarose (MP Biomedicals), and plaques were allowed to form for 36 to 40 h at 37°C. Plaques were visualized with a neutral red stain, and titers were enumerated as plaques per milliliter of serum or gram of tissue. For focus-forming assays, Vero cells were seeded into 96-well plates. Samples containing virus were serially diluted in DMEM–F-12 medium plus 2% FBS and allowed to adsorb onto the cells for 2 h at 37°C. At the end of 2 h, the sample was removed, and cells were overlaid with 0.5% methylcellulose in MEM-α plus 10% FBS and incubated at 37°C for 16 to 18 h. Cells were then fixed with 1% paraformaldehyde and probed with CHIKV-specific monoclonal antibody CHK-11 (87) at 500 ng/ml in Perm Wash (1× PBS, 0.1% saponin, 0.1% bovine serum albumin [BSA]). The antibody was allowed to bind for 2 h at room temperature. Next, cells were washed with Perm Wash, secondary goat anti-mouse IgG conjugated to horseradish peroxidase at a 1:2,000 dilution in Perm Wash was applied, and the mixture was incubated for 2 h at room temperature. Cells were washed with Perm Wash, and antigen-positive cells were visualized with the TrueBlue substrate (Fisher). Foci were counted with a CTL Biospot analyzer and Biospot software (Cellular Technology). Titers were calculated as the number of foci per milliliter of serum or gram of tissue.
Type I IFN ELISA.
The IFN-α level in sera was measured by using a mouse IFN-α enzyme-linked immunosorbent assay (ELISA) (Pestka Biomedical Laboratories) (49).
Histopathological analysis.
At 7 dpi, mice were sacrificed and perfused by intracardiac injection of 4% paraformaldehyde (pH 7.3), and the indicated tissues were dissected and fixed in 4% paraformaldehyde (pH 7.3). Tissues were embedded in paraffin, and 5-μm sections were prepared. Tissue sections were stained with hematoxylin and eosin (H&E) and evaluated by light microscopy. A board-certified veterinary anatomic pathologist scored the presence, distribution, and severity of histological lesions in muscle, joint space, synovium, tendon, and periarticular tissue in a blind manner. The following scoring system was used: 0 for no inflammation, 1 for >5 areas of small clusters of leukocytes, 2 for leukocytes forming larger clusters to thin tracts through tissue with multiple affected sites, 3 for clusters and tracts of leukocytes coalescing into at least one large area that displaces/replaces tissue, with or without necrosis and with or without mineralization, and 4 for leukocytes that are in aggregates large enough to replace >40% of normal tissue, with or without necrosis and with or without mineralization.
Statistical analysis.
All data were analyzed by using GraphPad Prism 6 software. Data were evaluated for statistically significant differences by using a two-tailed, unpaired t test; Mann-Whitney tests; one-way analysis of variance (ANOVA) followed by Tukey's multiple-comparison test; two-way ANOVA followed by a Bonferroni multiple-comparison test, or a Kruskal-Wallis test with Dunn's multiple-comparison test. A P value of <0.05 was considered statistically significant.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by Public Health Service grants R01 AI08725 and U19 AI09680 (T.E.M.), R01 AI104972 and R01 AI114816 (M.S.D.), and R01 AI03311 and R21 AI123811 (N.J.M.) from the National Institute of Allergy and Infectious Diseases. D.W.H. was supported by Public Health Service grant T32 AI052066 from the National Institute of Allergy and Infectious Diseases. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.00816-17.
REFERENCES
- 1.Weaver SC, Forrester NL. 2015. Chikungunya: evolutionary history and recent epidemic spread. Antiviral Res 120:32–39. doi: 10.1016/j.antiviral.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. 2014. Chikungunya in the Americas. Lancet 383:514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- 3.Pan American Health Organization. 2015. Number of reported cases of chikungunya fever in the Americas, by country or territory. Pan American Health Organization, Washington, DC: http://www.paho.org/hq/index.php?option=com_topics&view=article&id=343& Accessed 15 May 2017. [Google Scholar]
- 4.Suhrbier A, Jaffar-Bandjee MC, Gasque P. 2012. Arthritogenic alphaviruses—an overview. Nat Rev Rheumatol 8:420–429. doi: 10.1038/nrrheum.2012.64. [DOI] [PubMed] [Google Scholar]
- 5.Economopoulou A, Dominguez M, Helynck B, Sissoko D, Wichmann O, Quenel P, Germonneau P, Quatresous I. 2009. Atypical Chikungunya virus infections: clinical manifestations, mortality and risk factors for severe disease during the 2005-2006 outbreak on Reunion. Epidemiol Infect 137:534–541. doi: 10.1017/S0950268808001167. [DOI] [PubMed] [Google Scholar]
- 6.Tandale BV, Sathe PS, Arankalle VA, Wadia RS, Kulkarni R, Shah SV, Shah SK, Sheth JK, Sudeep AB, Tripathy AS, Mishra AC. 2009. Systemic involvements and fatalities during Chikungunya epidemic in India, 2006. J Clin Virol 46:145–149. doi: 10.1016/j.jcv.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 7.Simon F, Parola P, Grandadam M, Fourcade S, Oliver M, Brouqui P, Hance P, Kraemer P, Ali Mohamed A, de Lamballerie X, Charrel R, Tolou H. 2007. Chikungunya infection: an emerging rheumatism among travelers returned from Indian Ocean islands. Report of 47 cases. Medicine (Baltimore) 86:123–137. doi: 10.1097/MD/0b013e31806010a5. [DOI] [PubMed] [Google Scholar]
- 8.Brighton SW, Prozesky OW, de la Harpe AL. 1983. Chikungunya virus infection. A retrospective study of 107 cases. S Afr Med J 63:313–315. [PubMed] [Google Scholar]
- 9.Borgherini G, Poubeau P, Jossaume A, Gouix A, Cotte L, Michault A, Arvin-Berod C, Paganin F. 2008. Persistent arthralgia associated with chikungunya virus: a study of 88 adult patients on Reunion Island. Clin Infect Dis 47:469–475. doi: 10.1086/590003. [DOI] [PubMed] [Google Scholar]
- 10.Larrieu S, Pouderoux N, Pistone T, Filleul L, Receveur MC, Sissoko D, Ezzedine K, Malvy D. 2010. Factors associated with persistence of arthralgia among Chikungunya virus-infected travellers: report of 42 French cases. J Clin Virol 47:85–88. doi: 10.1016/j.jcv.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, Pierre V. 2009. Post-epidemic Chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl Trop Dis 3:e389. doi: 10.1371/journal.pntd.0000389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schilte C, Staikovsky F, Couderc T, Madec Y, Carpentier F, Kassab S, Albert ML, Lecuit M, Michault A. 2013. Chikungunya virus-associated long-term arthralgia: a 36-month prospective longitudinal study. PLoS Negl Trop Dis 7:e2137. doi: 10.1371/journal.pntd.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rodriguez-Morales AJ, Calvache-Benavides CE, Giraldo-Gomez J, Hurtado-Hurtado N, Yepes-Echeverri MC, Garcia-Loaiza CJ, Patino-Barbosa AM, Sabogal-Roman JA, Patino-Valencia S, Hidalgo-Zambrano DM, Vasquez-Serna H, Jimenez-Canizales CE. 2016. Post-chikungunya chronic arthralgia: results from a retrospective follow-up study of 131 cases in Tolima, Colombia. Travel Med Infect Dis 14:58–59. doi: 10.1016/j.tmaid.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Gerardin P, Fianu A, Malvy D, Mussard C, Boussaid K, Rollot O, Michault A, Gauzere BA, Breart G, Favier F. 2011. Perceived morbidity and community burden after a Chikungunya outbreak: the TELECHIK survey, a population-based cohort study. BMC Med 9:5. doi: 10.1186/1741-7015-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soumahoro MK, Boelle PY, Gauzere BA, Atsou K, Pelat C, Lambert B, La Ruche G, Gastellu-Etchegorry M, Renault P, Sarazin M, Yazdanpanah Y, Flahault A, Malvy D, Hanslik T. 2011. The Chikungunya epidemic on La Reunion Island in 2005-2006: a cost-of-illness study. PLoS Negl Trop Dis 5:e1197. doi: 10.1371/journal.pntd.0001197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gopalan SS, Das A. 2009. Household economic impact of an emerging disease in terms of catastrophic out-of-pocket health care expenditure and loss of productivity: investigation of an outbreak of chikungunya in Orissa, India. J Vector Borne Dis 46:57–64. [PubMed] [Google Scholar]
- 17.Simon F, Javelle E, Cabie A, Bouquillard E, Troisgros O, Gentile G, Leparc-Goffart I, Hoen B, Gandjbakhch F, Rene-Corail P, Franco JM, Caumes E, Combe B, Poiraudeau S, Gane-Troplent F, Djossou F, Schaerverbeke T, Criquet-Hayot A, Carrere P, Malvy D, Gaillard P, Wendling D, Societe de Pathologie Infectieuse de Langue Francaise. 2015. French guidelines for the management of chikungunya (acute and persistent presentations). November 2014. Med Mal Infect 45:243–263. doi: 10.1016/j.medmal.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 18.Hoarau JJ, Jaffar Bandjee MC, Krejbich Trotot P, Das T, Li-Pat-Yuen G, Dassa B, Denizot M, Guichard E, Ribera A, Henni T, Tallet F, Moiton MP, Gauzere BA, Bruniquet S, Jaffar Bandjee Z, Morbidelli P, Martigny G, Jolivet M, Gay F, Grandadam M, Tolou H, Vieillard V, Debre P, Autran B, Gasque P. 2010. Persistent chronic inflammation and infection by chikungunya arthritogenic alphavirus in spite of a robust host immune response. J Immunol 184:5914–5927. doi: 10.4049/jimmunol.0900255. [DOI] [PubMed] [Google Scholar]
- 19.Ozden S, Huerre M, Riviere J-P, Coffey LL, Afonso PV, Mouly V, de Monredon J, Roger J-C, El Amrani M, Yvin J-L, Jaffar M-C, Frenkiel M-P, Sourisseau M, Schwartz O, Butler-Browne G, Desprès P, Gessain A, Ceccaldi P-E. 2007. Human muscle satellite cells as targets of chikungunya virus infection. PLoS One 2:e527. doi: 10.1371/journal.pone.0000527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Labadie K, Larcher T, Joubert C, Mannioui A, Delache B, Brochard P, Guigand L, Dubreil L, Lebon P, Verrier B, de Lamballerie X, Suhrbier A, Cherel Y, Le Grand R, Roques P. 2010. Chikungunya disease in nonhuman primates involves long-term viral persistence in macrophages. J Clin Invest 120:894–906. doi: 10.1172/JCI40104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Messaoudi I, Vomaske J, Totonchy T, Kreklywich CN, Haberthur K, Springgay L, Brien JD, Diamond MS, DeFilippis VR, Streblow DN. 2013. Chikungunya virus infection results in higher and persistent viral replication in aged rhesus macaques due to defects in anti-viral immunity. PLoS Negl Trop Dis 7:e2343. doi: 10.1371/journal.pntd.0002343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhrlaub JL, Pulko V, DeFilippis VR, Broeckel R, Streblow DN, Coleman GD, Park BS, Lindo JF, Vickers I, Anzinger JJ, Nikolich-Žugich J. 2016. Dysregulated TGF-β production underlies the age-related vulnerability to chikungunya virus. PLoS Pathog 12:e1005891. doi: 10.1371/journal.ppat.1005891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hawman DW, Fox JM, Ashbrook AW, May NA, Schroeder KMS, Torres RM, Crowe JE, Dermody TS, Diamond MS, Morrison TE. 21 July 2016. Pathogenic chikungunya virus evades B cell responses to establish persistence. Cell Rep doi: 10.1016/j.celrep.2016.06.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hawman DW, Stoermer KA, Montgomery SA, Pal P, Oko L, Diamond MS, Morrison TE. 2013. Chronic joint disease caused by persistent Chikungunya virus infection is controlled by the adaptive immune response. J Virol 87:13878–13888. doi: 10.1128/JVI.02666-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poo YS, Rudd PA, Gardner J, Wilson JA, Larcher T, Colle MA, Le TT, Nakaya HI, Warrilow D, Allcock R, Bielefeldt-Ohmann H, Schroder WA, Khromykh AA, Lopez JA, Suhrbier A. 2014. Multiple immune factors are involved in controlling acute and chronic chikungunya virus infection. PLoS Negl Trop Dis 8:e3354. doi: 10.1371/journal.pntd.0003354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Couderc T, Chretien F, Schilte C, Disson O, Brigitte M, Guivel-Benhassine F, Touret Y, Barau G, Cayet N, Schuffenecker I, Despres P, Arenzana-Seisdedos F, Michault A, Albert ML, Lecuit M. 2008. A mouse model for Chikungunya: young age and inefficient type-I interferon signaling are risk factors for severe disease. PLoS Pathog 4:e29. doi: 10.1371/journal.ppat.0040029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson JA, Prow NA, Schroder WA, Ellis JJ, Cumming HE, Gearing LJ, Poo YS, Taylor A, Hertzog PJ, Di Giallonardo F, Hueston L, Le Grand R, Tang B, Le TT, Gardner J, Mahalingam S, Roques P, Bird PI, Suhrbier A. 2017. RNA-Seq analysis of chikungunya virus infection and identification of granzyme A as a major promoter of arthritic inflammation. PLoS Pathog 13:e1006155. doi: 10.1371/journal.ppat.1006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lum FM, Teo TH, Lee WW, Kam YW, Renia L, Ng LF. 2013. An essential role of antibodies in the control of Chikungunya virus infection. J Immunol 190:6295–6302. doi: 10.4049/jimmunol.1300304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoarau JJ, Gay F, Pelle O, Samri A, Jaffar-Bandjee MC, Gasque P, Autran B. 2013. Identical strength of the T cell responses against E2, nsP1 and capsid CHIKV proteins in recovered and chronic patients after the epidemics of 2005-2006 in La Reunion Island. PLoS One 8:e84695. doi: 10.1371/journal.pone.0084695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kam YW, Lee WW, Simarmata D, Harjanto S, Teng TS, Tolou H, Chow A, Lin RT, Leo YS, Renia L, Ng LF. 2012. Longitudinal analysis of the human antibody response to Chikungunya virus infection: implications for serodiagnosis and vaccine development. J Virol 86:13005–13015. doi: 10.1128/JVI.01780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teo TH, Lum FM, Claser C, Lulla V, Lulla A, Merits A, Renia L, Ng LF. 2013. A pathogenic role for CD4+ T cells during Chikungunya virus infection in mice. J Immunol 190:259–269. doi: 10.4049/jimmunol.1202177. [DOI] [PubMed] [Google Scholar]
- 32.Voss JE, Vaney MC, Duquerroy S, Vonrhein C, Girard-Blanc C, Crublet E, Thompson A, Bricogne G, Rey FA. 2010. Glycoprotein organization of Chikungunya virus particles revealed by X-ray crystallography. Nature 468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 33.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, Hunt V, Liu M, Kumar S, Zaremba S, Gu Z, Zhou L, Larson CN, Dietrich J, Klem EB, Scheuermann RH. 2012. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res 40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rohatgi A, Corbo JC, Monte K, Higgs S, Vanlandingham DL, Kardon G, Lenschow DJ. 2014. Infection of myofibers contributes to increased pathogenicity during infection with an epidemic strain of chikungunya virus. J Virol 88:2414–2425. doi: 10.1128/JVI.02716-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schilte C, Couderc T, Chretien F, Sourisseau M, Gangneux N, Guivel-Benhassine F, Kraxner A, Tschopp J, Higgs S, Michault A, Arenzana-Seisdedos F, Colonna M, Peduto L, Schwartz O, Lecuit M, Albert ML. 2010. Type I IFN controls chikungunya virus via its action on nonhematopoietic cells. J Exp Med 207:429–442. doi: 10.1084/jem.20090851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rudd PA, Wilson J, Gardner J, Larcher T, Babarit C, Le TT, Anraku I, Kumagai Y, Loo YM, Gale M Jr, Akira S, Khromykh AA, Suhrbier A. 2012. Interferon response factors 3 and 7 protect against Chikungunya virus hemorrhagic fever and shock. J Virol 86:9888–9898. doi: 10.1128/JVI.00956-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schilte C, Buckwalter MR, Laird ME, Diamond MS, Schwartz O, Albert ML. 2012. Cutting edge: independent roles for IRF-3 and IRF-7 in hematopoietic and nonhematopoietic cells during host response to Chikungunya infection. J Immunol 188:2967–2971. doi: 10.4049/jimmunol.1103185. [DOI] [PubMed] [Google Scholar]
- 38.Gardner CL, Burke CW, Higgs ST, Klimstra WB, Ryman KD. 2012. Interferon-alpha/beta deficiency greatly exacerbates arthritogenic disease in mice infected with wild-type chikungunya virus but not with the cell culture-adapted live-attenuated 181/25 vaccine candidate. Virology 425:103–112. doi: 10.1016/j.virol.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnes BJ, Richards J, Mancl M, Hanash S, Beretta L, Pitha PM. 2004. Global and distinct targets of IRF-5 and IRF-7 during innate response to viral infection. J Biol Chem 279:45194–45207. doi: 10.1074/jbc.M400726200. [DOI] [PubMed] [Google Scholar]
- 40.Seymour RL, Adams AP, Leal G, Alcorn MD, Weaver SC. 2015. A rodent model of chikungunya virus infection in RAG1−/− mice, with features of persistence, for vaccine safety evaluation. PLoS Negl Trop Dis 9:e0003800. doi: 10.1371/journal.pntd.0003800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Levine B, Griffin DE. 1993. Molecular analysis of neurovirulent strains of Sindbis virus that evolve during persistent infection of scid mice. J Virol 67:6872–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deuber SA, Pavlovic J. 2007. Virulence of a mouse-adapted Semliki Forest virus strain is associated with reduced susceptibility to interferon. J Gen Virol 88:1952–1959. doi: 10.1099/vir.0.82264-0. [DOI] [PubMed] [Google Scholar]
- 43.Simmons G, Bres V, Lu K, Liss NM, Brambilla DJ, Ryff KR, Bruhn R, Velez E, Ocampo D, Linnen JM, Latoni G, Petersen LR, Williamson PC, Busch MP. 2016. High incidence of chikungunya virus and frequency of viremic blood donations during epidemic, Puerto Rico, USA, 2014. Emerg Infect Dis 22:1221–1228. doi: 10.3201/eid2207.160116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanciotti RS, Kosoy OL, Laven JJ, Panella AJ, Velez JO, Lambert AJ, Campbell GL. 2007. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parola P, de Lamballerie X, Jourdan J, Rovery C, Vaillant V, Minodier P, Brouqui P, Flahault A, Raoult D, Charrel RN. 2006. Novel chikungunya virus variant in travelers returning from Indian Ocean islands. Emerg Infect Dis 12:1493–1499. doi: 10.3201/eid1210.060610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Panning M, Grywna K, van Esbroeck M, Emmerich P, Drosten C. 2008. Chikungunya fever in travelers returning to Europe from the Indian Ocean region, 2006. Emerg Infect Dis 14:416–422. doi: 10.3201/eid1403.070906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Appassakij H, Khuntikij P, Kemapunmanus M, Wutthanarungsan R, Silpapojakul K. 2013. Viremic profiles in asymptomatic and symptomatic chikungunya fever: a blood transfusion threat? Transfusion 53:2567–2574. doi: 10.1111/j.1537-2995.2012.03960.x. [DOI] [PubMed] [Google Scholar]
- 48.Chow A, Her Z, Ong EK, Chen JM, Dimatatac F, Kwek DJ, Barkham T, Yang H, Renia L, Leo YS, Ng LF. 2011. Persistent arthralgia induced by Chikungunya virus infection is associated with interleukin-6 and granulocyte macrophage colony-stimulating factor. J Infect Dis 203:149–157. doi: 10.1093/infdis/jiq042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stoermer Burrack KA, Hawman DW, Jupille HJ, Oko L, Minor M, Shives KD, Gunn BM, Long KM, Morrison TE. 2014. Attenuating mutations in nsP1 reveal tissue-specific mechanisms for control of Ross River virus infection. J Virol 88:3719–3732. doi: 10.1128/JVI.02609-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weger-Lucarelli J, Aliota MT, Kamlangdee A, Osorio JE. 2015. Identifying the role of E2 domains on alphavirus neutralization and protective immune responses. PLoS Negl Trop Dis 9:e0004163. doi: 10.1371/journal.pntd.0004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roussel A, Lescar J, Vaney MC, Wengler G, Wengler G, Rey FA. 2006. Structure and interactions at the viral surface of the envelope protein E1 of Semliki Forest virus. Structure 14:75–86. doi: 10.1016/j.str.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 52.Glomb-Reinmund S, Kielian M. 1998. fus-1, a pH shift mutant of Semliki Forest virus, acts by altering spike subunit interactions via a mutation in the E2 subunit. J Virol 72:4281–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. 2010. Structural changes of envelope proteins during alphavirus fusion. Nature 468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibbons DL, Vaney MC, Roussel A, Vigouroux A, Reilly B, Lepault J, Kielian M, Rey FA. 2004. Conformational change and protein-protein interactions of the fusion protein of Semliki Forest virus. Nature 427:320–325. doi: 10.1038/nature02239. [DOI] [PubMed] [Google Scholar]
- 55.Jupille HJ, Medina-Rivera M, Hawman DW, Oko L, Morrison TE. 2013. A tyrosine-to-histidine switch at position 18 of the Ross River virus E2 glycoprotein is a determinant of virus fitness in disparate hosts. J Virol 87:5970–5984. doi: 10.1128/JVI.03326-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li ML, Liao HJ, Simon LD, Stollar V. 1999. An amino acid change in the exodomain of the E2 protein of Sindbis virus, which impairs the release of virus from chicken cells but not from mosquito cells. Virology 264:187–194. doi: 10.1006/viro.1999.9971. [DOI] [PubMed] [Google Scholar]
- 57.Snyder AJ, Sokoloski KJ, Mukhopadhyay S. 2012. Mutating conserved cysteines in the alphavirus E2 glycoprotein causes virus-specific assembly defects. J Virol 86:3100–3111. doi: 10.1128/JVI.06615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ashbrook AW, Burrack KS, Silva LA, Montgomery SA, Heise MT, Morrison TE, Dermody TS. 2014. Residue 82 of the Chikungunya virus E2 attachment protein modulates viral dissemination and arthritis in mice. J Virol 88:12180–12192. doi: 10.1128/JVI.01672-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ryman KD, Gardner CL, Burke CW, Meier KC, Thompson JM, Klimstra WB. 2007. Heparan sulfate binding can contribute to the neurovirulence of neuroadapted and nonneuroadapted Sindbis viruses. J Virol 81:3563–3573. doi: 10.1128/JVI.02494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gardner CL, Choi-Nurvitadhi J, Sun C, Bayer A, Hritz J, Ryman KD, Klimstra WB. 2013. Natural variation in the heparan sulfate binding domain of the eastern equine encephalitis virus E2 glycoprotein alters interactions with cell surfaces and virulence in mice. J Virol 87:8582–8590. doi: 10.1128/JVI.00937-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gardner CL, Hritz J, Sun C, Vanlandingham DL, Song TY, Ghedin E, Higgs S, Klimstra WB, Ryman KD. 2014. Deliberate attenuation of chikungunya virus by adaptation to heparan sulfate-dependent infectivity: a model for rational arboviral vaccine design. PLoS Negl Trop Dis 8:e2719. doi: 10.1371/journal.pntd.0002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kinney RM, Chang GJ, Tsuchiya KR, Sneider JM, Roehrig JT, Woodward TM, Trent DW. 1993. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J Virol 67:1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsetsarkin KA, Weaver SC. 2011. Sequential adaptive mutations enhance efficient vector switching by chikungunya virus and its epidemic emergence. PLoS Pathog 7:e1002412. doi: 10.1371/journal.ppat.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S. 2009. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One 4:e6835. doi: 10.1371/journal.pone.0006835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen R, Wang E, Tsetsarkin KA, Weaver SC. 2013. Chikungunya virus 3′ untranslated region: adaptation to mosquitoes and a population bottleneck as major evolutionary forces. PLoS Pathog 9:e1003591. doi: 10.1371/journal.ppat.1003591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stapleford KA, Moratorio G, Henningsson R, Chen R, Matheus S, Enfissi A, Weissglas-Volkov D, Isakov O, Blanc H, Mounce BC, Dupont-Rouzeyrol M, Shomron N, Weaver S, Fontes M, Rousset D, Vignuzzi M. 2016. Whole-genome sequencing analysis from the chikungunya virus Caribbean outbreak reveals novel evolutionary genomic elements. PLoS Negl Trop Dis 10:e0004402. doi: 10.1371/journal.pntd.0004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pfeffer M, Kinney R, Kaaden O. 1998. The alphavirus 3′-nontranslated region: size heterogeneity and arrangement of repeated sequence elements. Virology 240:100–108. doi: 10.1006/viro.1997.8907. [DOI] [PubMed] [Google Scholar]
- 68.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev 58:491–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hyde JL, Chen R, Trobaugh DW, Diamond MS, Weaver SC, Klimstra WB, Wilusz J. 2015. The 5′ and 3′ ends of alphavirus RNAs—non-coding is not non-functional. Virus Res 206:99–107. doi: 10.1016/j.virusres.2015.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kuhn R, Hong Z, Strauss JH. 1994. Mutagenesis of the 3′ nontranslated region of Sindbis virus RNA. J Virol 64:1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Moreno M, Sanz MA, Carrasco L. 2016. A viral mRNA motif at the 3′-untranslated region that confers translatability in a cell-specific manner. Implications for virus evolution. Sci Rep 6:19217. doi: 10.1038/srep19217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trobaugh DW, Gardner CL, Sun C, Haddow AD, Wang E, Chapnik E, Mildner A, Weaver SC, Ryman KD, Klimstra WB. 2014. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature 506:245–248. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuhn RJ, Griffin DE, Zhang H, Niesters HG, Strauss JH. 1992. Attenuation of Sindbis virus neurovirulence by using defined mutations in nontranslated regions of the genome RNA. J Virol 66:7121–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Garneau NL, Sokoloski KJ, Opyrchal M, Neff CP, Wilusz CJ, Wilusz J. 2008. The 3′ untranslated region of Sindbis virus represses deadenylation of viral transcripts in mosquito and mammalian cells. J Virol 82:880–892. doi: 10.1128/JVI.01205-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dickson AM, Anderson JR, Barnhart MD, Sokoloski KJ, Oko L, Opyrchal M, Galanis E, Wilusz CJ, Morrison TE, Wilusz J. 2012. Dephosphorylation of HuR protein during alphavirus infection is associated with HuR relocalization to the cytoplasm. J Biol Chem 287:36229–36238. doi: 10.1074/jbc.M112.371203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barnhart MD, Moon SL, Emch AW, Wilusz CJ, Wilusz J. 2013. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep 5:909–917. doi: 10.1016/j.celrep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sokoloski KJ, Dickson AM, Chaskey EL, Garneau NL, Wilusz CJ, Wilusz J. 2010. Sindbis virus usurps the cellular HuR protein to stabilize its transcripts and promote productive infections in mammalian and mosquito cells. Cell Host Microbe 8:196–207. doi: 10.1016/j.chom.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hussain M, Torres S, Schnettler E, Funk A, Grundhoff A, Pijlman GP, Khromykh AA, Asgari S. 2012. West Nile virus encodes a microRNA-like small RNA in the 3′ untranslated region which up-regulates GATA4 mRNA and facilitates virus replication in mosquito cells. Nucleic Acids Res 40:2210–2223. doi: 10.1093/nar/gkr848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chapman EG, Costantino DA, Rabe JL, Moon SL, Wilusz J, Nix JC, Kieft JS. 2014. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344:307–310. doi: 10.1126/science.1250897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 81.Harrison VR, Eckels KH, Bartelloni PJ, Hampton C. 1971. Production and evaluation of a formalin-killed Chikungunya vaccine. J Immunol 107:643–647. [PubMed] [Google Scholar]
- 82.Levitt NH, Ramsburg HH, Hasty SE, Repik PM, Cole FE, Lupton HW. 1986. Development of an attenuated strain of chikungunya virus for use in vaccine production. Vaccine 4:157–162. doi: 10.1016/0264-410X(86)90003-4. [DOI] [PubMed] [Google Scholar]
- 83.Jupille HJ, Oko L, Stoermer KA, Heise MT, Mahalingam S, Gunn BM, Morrison TE. 2011. Mutations in nsP1 and PE2 are critical determinants of Ross River virus-induced musculoskeletal inflammatory disease in a mouse model. Virology 410:216–227. doi: 10.1016/j.virol.2010.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pal P, Dowd KA, Brien JD, Edeling MA, Gorlatov S, Johnson S, Lee I, Akahata W, Nabel GJ, Richter MK, Smit JM, Fremont DH, Pierson TC, Heise MT, Diamond MS. 2013. Development of a highly protective combination monoclonal antibody therapy against Chikungunya virus. PLoS Pathog 9:e1003312. doi: 10.1371/journal.ppat.1003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.American Veterinary Medical Association. 2013. AVMA guidelines for the euthanasia of animals: 2013 edition. AVMA, Schaumburg, IL. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.