ABSTRACT
Influenza A virus (IAV) replication relies on an intricate interaction between virus and host cells. How the cellular proteins are usurped for IAV replication remains largely obscure. The aim of this study was to search for novel and potential cellular factors that participate in IAV replication. ZBTB25, a transcription repressor of a variety of cellular genes, was identified by an RNA interference (RNAi) genomic library screen. Depletion of ZBTB25 significantly reduced IAV production. Conversely, overexpression of ZBTB25 enhanced it. ZBTB25 interacted with the viral RNA-dependent RNA polymerase (RdRp) protein and modulated its transcription activity. In addition, ZBTB25 also functioned as a viral RNA (vRNA)-binding protein, binding preferentially to the U-rich sequence within the 5′ untranslated region (UTR) of vRNA. Both protein-protein and protein-RNA interactions involving ZBTB25 facilitated viral RNA transcription and replication. In addition, ZBTB25 suppressed interferon production, further enhancing viral replication. ZBTB25-associated functions required an intact zinc finger domain and posttranslational SUMO-1 modification of ZBTB25. Furthermore, treatment with disulfiram (a zinc ejector) of ZBTB25-overexpressing cells showed significantly reduced IAV production as a result of reduced RNA synthesis. Our findings indicate that IAV usurps ZBTB25 for IAV RNA synthesis and serves as a novel and potential therapeutic antiviral target.
IMPORTANCE IAV-induced seasonal influenza causes severe illness and death in high-risk populations. However, IAV has developed resistance to current antiviral drugs due to its high mutation rate. Therefore, development of drugs targeting cellular factors required for IAV replication is an attractive alternative for IAV therapy. Here, we discovered a cellular protein, ZBTB25, that enhances viral RdRp activity by binding to both viral RdRp and viral RNA to stimulate viral RNA synthesis. A unique feature of ZBTB25 in the regulation of viral replication is its dual transcription functions, namely, promoting viral RNA transcription through binding to the U-rich region of vRNA and suppressing cellular interferon production. ZBTB25 contains a zinc finger domain that is required for RNA-inhibitory activity by chelating zinc ions. Disulfiram treatment disrupts the zinc finger functions, effectively repressing IAV replication. Based on our findings, we demonstrate that ZBTB25 regulates IAV RNA transcription and replication and serves as a promising antiviral target for IAV treatment.
KEYWORDS: IAV replication, ZBTB25, influenza A virus
INTRODUCTION
Influenza A virus (IAV) infection results in seasonal and pandemic influenza in humans and accounts for severe morbidity and mortality every year. IAV, belonging to the Orthomyxoviridae family, is an enveloped and negative-sense RNA virus. Its eight-segmented genome encodes at least 12 viral proteins, including two envelope glycoproteins, hemagglutinin (HA) and neuraminidase (NA), and three polymerase subunits of virus-specific RNA polymerase, namely, polymerase acidic protein (PA) and polymerase basic protein 1 and 2 (PB1 and PB2), as well as nucleoprotein (NP), matrix protein 1 (M1), ion channel protein M2, and two nonstructural proteins 1 and 2 (NS1 and NS2) (1). At the beginning of viral infection, viral HA protein first binds to the sialic acid-containing receptors located on the host cell surface and triggers internalization of the viral particle through endocytosis (2). The viral ribonucleoprotein (vRNP) complexes comprising viral RNA (vRNA), the NP, and viral RNA-dependent RNA polymerase (RdRp) are released after the viral particle fuses with the late endosome. vRNP is subsequently translocated into the nucleus to drive transcription and replication of the influenza virus genome by RdRp (3). IAV RNA transcription involves a cap-snatching process (4, 5). During this process, PB2 first binds to the 5′ cap of the host mRNA. PA subsequently cleaves the host mRNA to generate 10- to 13-nucleotide capped RNA fragments (6), and then PB1 elongates the viral mRNA by RNA polymerization using the viral negative-sense RNA as a template (7). For viral RNA replication, RdRp first uses the viral RNA as a template to generate a replicative intermediate cRNA and then uses cRNA as the template to produce vRNA in a primer-independent process (8). During the late stage of the IAV life cycle, vRNP, M1, and viral envelope proteins assemble the viral particles, which are then released from the cell surface to produce viral progeny. In recent decades, development of drugs targeting viral proteins, including NA, M2, and HA, has been actively pursued. However, IAV developed drug resistance to the current antiviral drugs very quickly because of the high mutability of RNA viruses (9). Therefore, there is an urgent need to identify cellular proteins that are involved in IAV replication and are suitable as antiviral targets.
In this study, we identified the cellular transcription corepressor ZBTB25 as being involved in IAV replication. ZBTB25 was initially identified as a transcription repressor highly enriched in T cells and functions as a negative regulator of the activity of nuclear factor of activated T cells (NFAT) (10). ZBTB25 belongs to the BTB/POZ (broad complex, tramtrack, bric-a-brac/pox virus and zinc finger [ZF]) (DNA-binding zinc finger motifs) transcription factor family, with 60 such genes being encoded in the human genome (11–13). The BTB/POZ domain consisting of approximately 120 amino acids has been reported to mediate protein-protein interactions, and several studies have shown that the BTB domains of some BTB-ZF proteins are responsible for homo- as well as hetero-oligomerization of BTB-ZF proteins (14, 15). BTB-ZF transcription factors generally interact with their cognate DNA sequences via their zinc finger motifs, the majority of which are the Krüppel-like C2H2 type (16), resulting in transcriptional repression via the BTB domain and recruitment of corepressor complexes such as NCoR1 (nuclear receptor coreceptor 1), SMRT (silencing mediator of retinoid and thyroid receptor), and BCoR (BCL6 coreceptor) (17–20). Whether ZBTB family members are involved in regulation of IAV RNA synthesis has not been reported.
Recently, more and more studies have provided evidence that protein posttranslational modification with SUMO (small ubiquitin-like modifier) plays an important role in the regulation of viral replication, either through direct modification of viral proteins or through modulation of host cellular proteins involved in antiviral defense, such as via interferon (IFN) production and the interferon-activated JAK/STAT pathway (21, 22). SUMOylation participates in numerous biological processes, including alteration of protein-protein interactions, subcellular localization, transcription factor transactivation, protein stability, gene transcription, DNA repair, chromatin remodeling, and signal transduction events (23). SUMO proteins belong to the ubiquitin-like protein family and are covalently linked to a lysine residue within the consensus sequence (ΨKXE, where Ψ represents a hydrophobic residue) of the target protein via a multienzymatic cascade of E1 activating enzyme, E2 conjugating enzyme, and E3 ligase (24). Conversely, SUMO-specific proteases (SENPs) catalyze deconjugation of SUMO from substrate proteins using their isopeptidase activity (25, 26).
The first defense of the innate immune system in host cells against virus infection is to trigger interferon (IFN) synthesis and establish an antiviral state. IFN has been demonstrated to inhibit every stage of viral replication, including entry, transcription, RNA stability, initiation of translation, maturation, assembly, and release (27). IAV infection in the host cells provokes an antiviral response involving the expression of type I and II IFNs and IFN-stimulated genes (ISGs). The secreted IFN activates the JAK/STAT/ISGF3 pathways and subsequently induces the production of ISGs, including RIG-1, MDA5, IRF7, STAT1, and many others (28–30).
In this study, we showed that ZBTB25 modulates RdRp activity through direct interaction with viral RdRp proteins and also binds to viral RNA to stimulate viral RNA synthesis. The zinc finger domain and posttranslational SUMOylation of ZBTB25 are both required for viral RNA binding and RdRp complex recruitment. In addition, ZBTB25 suppresses IAV-induced IFN induction to create an environment conducive to IAV replication. Furthermore, treatment with a zinc ejector, disulfiram (DSF), which disrupts zinc finger domain functionality, blocked ZBTB25-mediated IAV replication. We conclude that ZBTB25 may represent a class of positive factors for IAV replication and serve as a potential therapeutic target for IAV treatment.
RESULTS
Identification of ZBTB25 as a positive regulator of IAV replication.
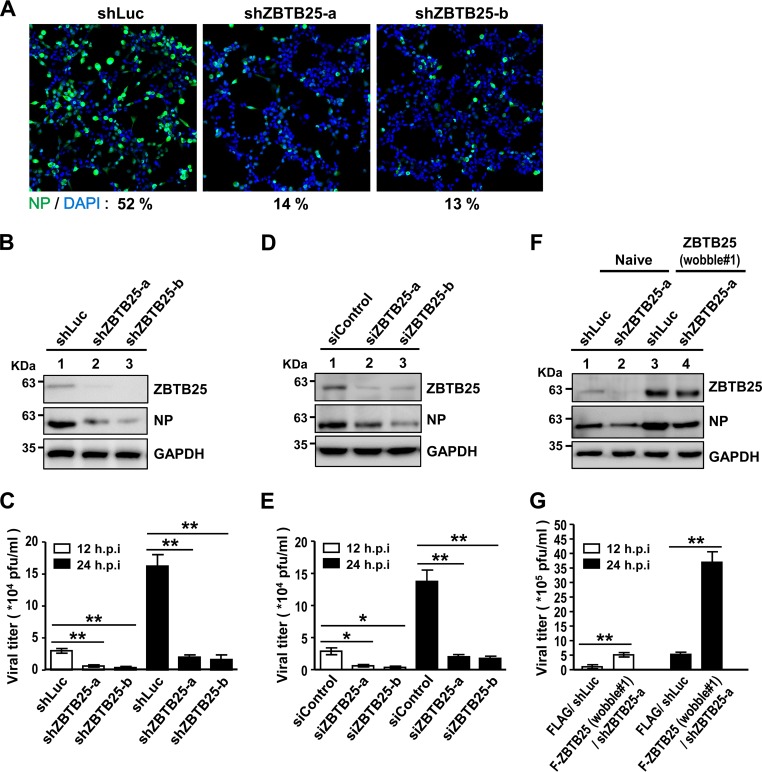
To identify cellular regulators involved in IAV replication, our group previously utilized a high-throughput pooled RNA interference (RNAi) library screen to search for potential candidate genes (31–33). In this study, a secondary RNAi screening was performed to confirm these genes. The candidate genes were initially selected according to the following two criteria: (i) that they show a significant reduction of nuclear NP fluorescence signals in short hairpin RNA (shRNA) knockdown (KD) cells compared to the signals of the control cells at 8 h postinfection (hpi) and (ii) that there are at least two hits of distinct shRNAs targeting the same gene with a similar phenotype. Based on these criteria, two ZBTB25 shRNA clones were identified (Fig. 1A) and selected for further studies. To verify the effect of ZBTB25 on IAV replication, lung adenocarcinoma A549 cells with stable knockdown of the endogenous ZBTB25 or an shRNA targeting luciferase (shLuc) control were generated using a lentivirus-based shRNA approach. Two shZBTB25 clones (shZBTB25-a and -b) produced very low ZBTB25 levels based on immunoblotting (Fig. 1B) and quantitative reverse transcription-PCR (qRT-PCR) analysis (Fig. 2A). Correspondingly, they produced very small amounts of viral NP (Fig. 1B) and viral particles after IAV infection (Fig. 1C). Similar data were obtained using small interfering RNA (siRNA) oligomers (siZBTB25-a and -b) to knock down ZBTB25 expression, with the amounts of viral NP protein and viral titers being significantly reduced in these cells (Fig. 1D and E). We then performed a rescue experiment using a wobble ZBTB25 mutant that expressed wild-type (WT) ZBTB25 but that could not be targeted by shZBTB25-a. As shown in Fig. 1F, the wobble ZBTB25 mutant restored ZBTB25 expression levels and also viral NP protein levels. Reciprocal experiments showed that overexpression of the wobble ZBTB25 mutant in ZBTB25 knockdown cells dramatically enhanced IAV replication (Fig. 1G). Taken together, these results suggest that ZBTB25 plays a positive role during IAV replication. However, Western blotting studies revealed very little change in the expression patterns of ZBTB25 at various postinfection times (data not shown).
FIG 1.
Identification of ZBTB25 as a positive regulator of IAV replication. (A) A549 cells with stable knockdown of ZBTB25 were infected with influenza A/WSN/33 virus at an MOI of 1 for 8 h and processed for immunofluorescence staining with anti-NP antibody (green) and DAPI. (B) Western blot analysis of A549 cells in which the expression of endogenous ZBTB25 was knocked down using a lentivirus-based shRNA approach. (C) The virus titer in ZBTB25 knockdown A549 cells generated by shRNA at 12 and 24 hpi after IAV infection according to a plaque assay. (D) Western blot analysis of A549 cells in which the expression of endogenous ZBTB25 was knocked down using siRNA oligomers. (E) The virus titer in ZBTB25 knockdown A549 cells generated by siRNA oligomers at 12 and 24 hpi after IAV infection using plaque assays. (F) Western blot analysis of overexpression of the ZBTB25 wobble mutant in ZBTB25 knockdown A549 cells. GAPDH served as the loading control. (G) Plaque assay of overexpression of the ZBTB25 wobble mutant in ZBTB25 knockdown A549 cells at 12 and 24 hpi after IAV infection. The data are presented as the means ± SD (n = 3; *, P < 0.05; **, P < 0.01, by Student's t test). F-ZBTB25, Flag-tagged ZBTB25.
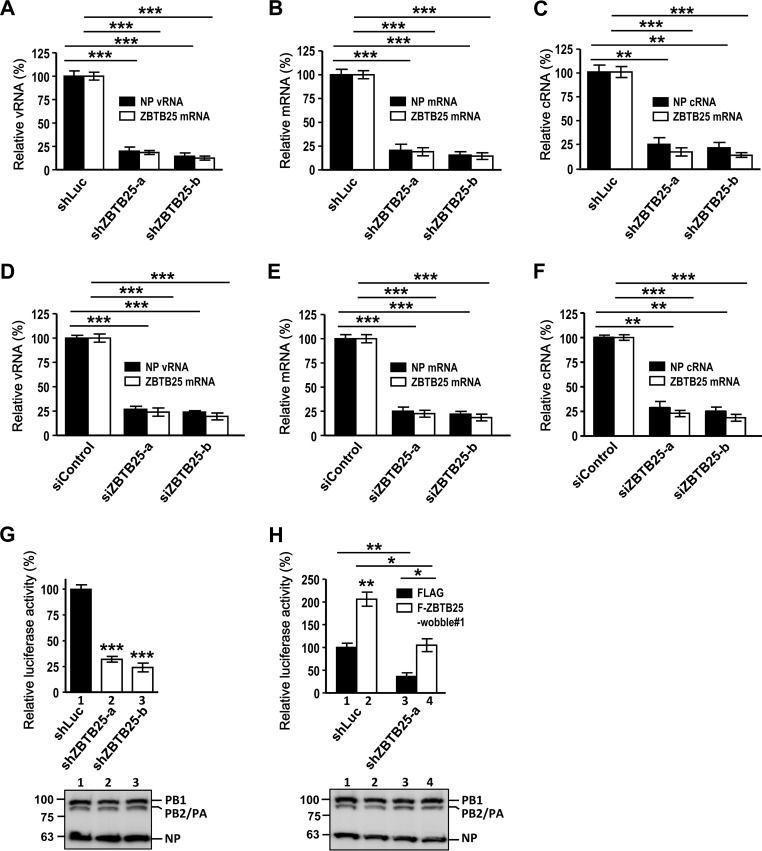
FIG 2.
ZBTB25 regulates IAV replication via modulation of RdRp activity. ZBTB25 knockdown A549 cells generated by shRNAs (A to C) or by siRNA oligomers (D to F) were infected with IAV at an MOI of 1, and cellular RNA was measured by qRT-PCR at 8 hpi. The expression levels of viral RNA (A and D), viral mRNA (B and E), and viral cRNA (C and F) of NP and ZBTB25 mRNA were measured and normalized by GAPDH mRNA. Expression relative to that of the shLuc control is shown. (G to H) ZBTB25 knockdown A549 cells generated by shRNAs were cotransfected with pPolI-Luc and Renilla luciferase-expressing plasmid in the absence (G) or presence (H) of a Flag-tagged ZBTB25 wobble mutant, and luciferase activity was measured at 24 h posttransfection. Renilla luciferase was used as an internal control to normalize transfection efficiency, and the relative expression is shown. The data are presented as the means ± SD (n = 3; *, P < 0.05; **, P < 0.01; and ***, P < 0.001, by Student's t test). The protein levels of RdRp complex and NP were also shown below the graphs.
Next, we investigated how ZBTB25 participates in IAV replication. We first studied whether ZBTB25 is involved in viral entry. ZBTB25 knockdown cells were incubated with IAV for 30 min and then washed with acidic phosphate-buffered saline (PBS) to remove the attached, but not yet internalized, virus particles. Viral M1 protein, which is a sensitive marker for IAV internalization (34), was detected as an indicator of internalized virus particles by Western blotting. Compared to the levels in the control shLuc cells, no significant change in the amount of internalized M1 was observed in ZBTB25 knockdown cells, indicating that ZBTB25 is not involved in the entry step of the IAV life cycle (data not shown).
ZBTB25 regulates IAV replication through modulation of viral RdRp activity.
We then examined replication and transcription of viral RNA by a qRT-PCR method using NP vRNA-, mRNA-, and cRNA-specific probes since these three RNA species have distinct structures and sequences. As demonstrated in Fig. 2A to C, the amounts of all NP vRNA, mRNA, and cRNA were significantly lower in the shZBTB25 clones at 8 hpi than in the controls. Similar data were obtained by using siRNA oligomers to knock down the expression of ZBTB25 (Fig. 2D to F). These results indicate that ZBTB25 participates in both RNA transcription and replication of IAV. We further tested the possibility that ZBTB25 regulates viral RdRp activities directly by performing a minireplicon assay. Knockdown of endogenous ZBTB25 by shRNA resulted in the reduction of RdRp activity by approximately 70% (Fig. 2G), whereas overexpression of the ZBTB25 wobble mutant restored the viral RdRp activity in these cells (Fig. 2H). Together, these results suggest that ZBTB25 regulates viral RdRp activity.
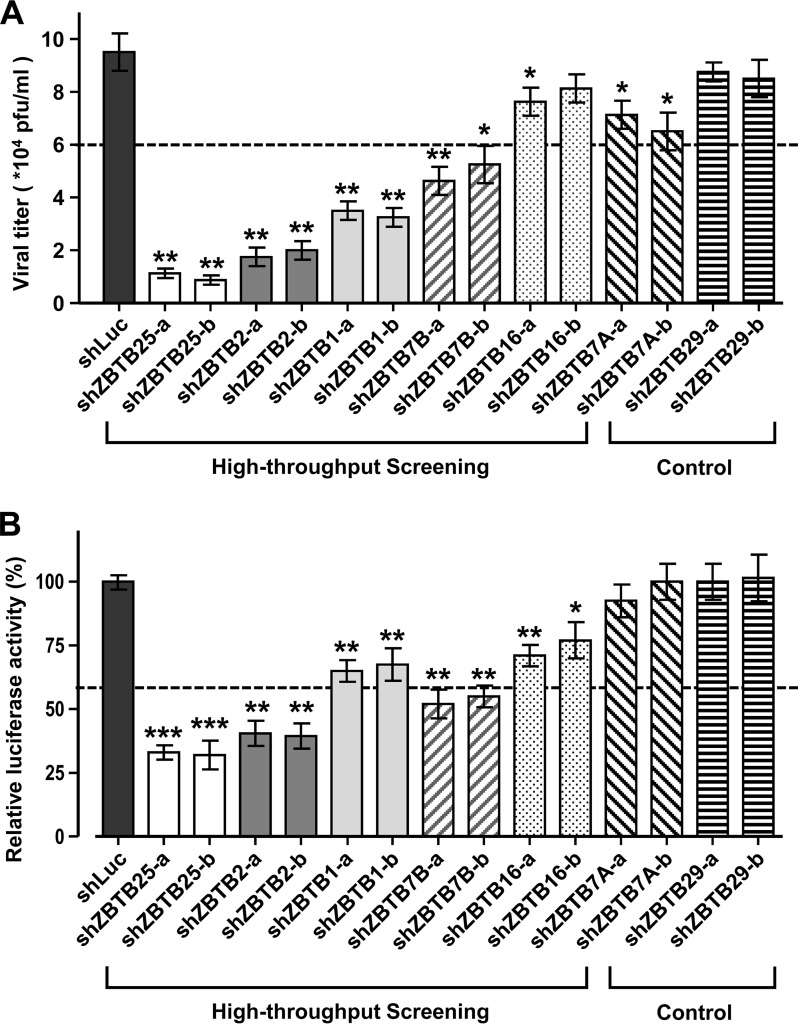
Since the ZBTB family consists of 60 members, we wondered whether any other ZBTB family members participate in IAV replication (Table 1). As indicated in Fig. 3A and B, our RNAi library screening detected several other ZBTB members but only two, namely, ZBTB25 and ZBTB2, exhibited consistent reductions in viral titers and RdRp activities. Knockdown of other ZBTB members, such as ZBTB1 and ZBTB7B, only slightly decreased the viral titer and RdRp activity. Thus, among ZBTB family members, only ZBTB25 and ZBTB2 specifically inhibit IAV replication.
TABLE 1.
Targeted sequences for knockdown of ZBTB family members
| shRNA or siRNA | Sequence |
|---|---|
| shLuc | 5′-GCGGTTGCCAAGAGGTTCCAT-3′ |
| shZBTB25-a | 5′-GCTTCCATTCTGGAAAGTAAT-3′ |
| shZBTB25-b | 5′-CCAACCTGACATATTCAGCTA-3′ |
| shZBTB2-a | 5′-GCAGATCATCAGTTGAGACAA-3′ |
| shZBTB2-b | 5′-CCCAGTGTTAGCTTCCATCAA-3′ |
| shZBTB1-a | 5′-GCAGTCCGAAATAAGAGATAT-3′ |
| shZBTB1-b | 5′-GCAATGAACTACCTACAGCTA-3′ |
| shZBTB7A-a | 5′-CCACTGAGACACAAACCTATT-3′ |
| shZBTB7A-b | 5′-CCGTTTGCATATGCAATGCTA-3′ |
| shZBTB7B-a | 5′-CCGCCTCTCTCTAGCTCGATT-3′ |
| shZBTB7B-b | 5′-CCTGACCTGATGGCCTACCTA-3′ |
| shZBTB16-a | 5′-GAATGCACTTACTGGCTCATT-3′ |
| shZBTB16-b | 5′-GTGGACAGTTTGATGACCATA-3′ |
| shZBTB29-a | 5′-CTGTGCAAGAAACGCCTCAAG-3′ |
| shZBTB29-b | 5′-GTCGTGCGACAAGAGCTACAA-3′ |
| siZBTB25-a | 5′-GGUCAUAAAUUCCCUCGAA-3′ |
| siZBTB25-b | 5′-GUGCAGUCCUCAAAUUUAU-3′ |
FIG 3.
ZBTB25 and ZBTB2 show specificity for IAV replication. (A) The virus titers in A549 cells with shRNA-generated knockdown of ZBTB members at 24 hpi after IAV infection, according to plaque assay. (B) A549 cells with shRNA-generated knockdown of ZBTB members were cotransfected with pPolI-Luc and a Renilla luciferase-expressing plasmid, and their luciferase activities were measured at 24 h posttransfection. Renilla luciferase was used as an internal control to normalize transfection efficiency, and the relative expression is shown. The data are presented as the means ± SD (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student's t test).
ZBTB25 interacts with all three subunits of the viral RdRp complex in vitro.
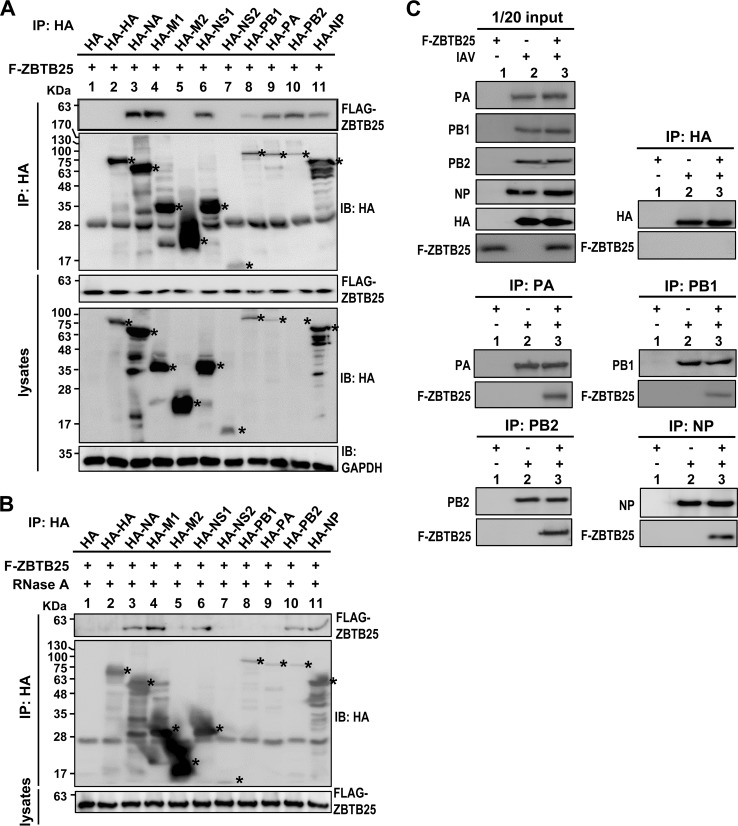
Since ZBTB25 can enhance RdRp activity, we hypothesized that ZBTB25 may interact with components of the viral RdRp complex. We performed an in vitro coimmunoprecipitation assay between ZBTB25 and individual viral proteins. ZBTB25 and HA-tagged viral proteins were first expressed individually in HEK293T cells, cell lysates were immunoprecipitated (IP) with anti-HA antibody, and the precipitated ZBTB25 was detected by anti-Flag antibody. The data showed that ZBTB25 was pulled down by NA, M1, NS1, PB1, PA, PB2, and NP but not by HA, M2, or NS2 (Fig. 4A). This result is consistent with the previous report that NP interacted with ZBTB25 in a yeast two-hybrid screening (35). We also performed the same experiments using RNase A-treated cellular lysates. Essentially the same set of viral proteins, except PA and PB1, were precipitated with ZBTB25 (Fig. 4B). These results suggest that these viral proteins interact directly with ZBTB25. We next determined whether these protein-protein associations occurred in IAV-infected cells. We focused on the components of the RdRp complex. Flag-ZBTB25 was transfected into HEK293T cells prior to IAV infection, and the lysates were immunoprecipitated using anti-PA, anti-PB1, anti-PB2, anti-NP, or anti-HA antibodies. As indicated in Fig. 4C, ZBTB25 coprecipitated with PA, PB1, PB2, and NP but not with HA. These results suggest that ZBTB25 interacts with several components of the viral RdRp complex either directly or through RNA.
FIG 4.
The association of ZBTB25 with distinct viral protein species. (A and B) HEK293T cells were cotransfected with Flag-ZBTB25 and the plasmids expressing individual HA-tagged viral proteins as indicated. The cell lysates were left untreated (A) or treated with 100 U of RNase A (B) at 37°C for 1 h. IP analysis was conducted using anti-HA antibody. The viral proteins and coprecipitated ZBTB25 were detected using anti-HA and anti-Flag antibodies, respectively. Asterisks mark the positions of individual viral proteins. (C) Flag-ZBTB25 was introduced into HEK293T cells prior to IAV infection, and the lysates were pulled down by anti-PA, anti-PB1, anti-PB2, anti-NP, and anti-HA (hemagglutinin) antibodies at 10 h postinfection. These immunocomplexes were analyzed by Western blotting using antibodies against individual viral proteins and Flag-ZBTB25. GAPDH served as the loading control. IB, immunoblotting.
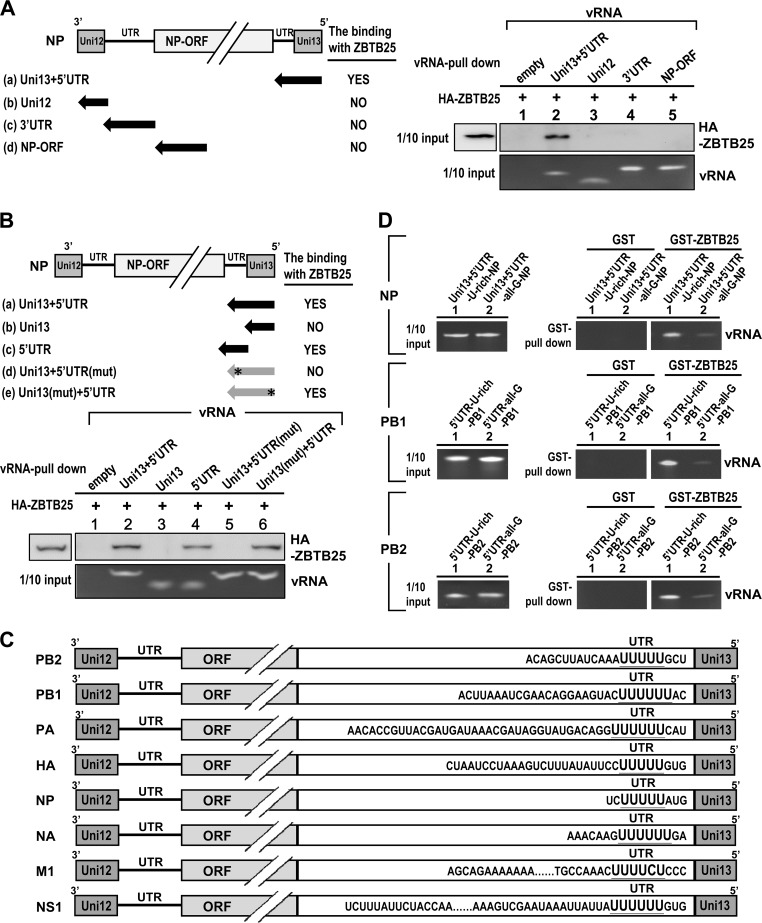
ZBTB25 is a viral RNA-binding protein and recognizes the U-rich sequence within the 5′ UTR of vRNA.
Another possibility is that ZBTB25 may bind viral RNA directly, thereby enabling formation of the RdRp complex. An in vitro RNA-protein binding assay was used to test this possibility. The HA-tagged ZBTB25 protein was synthesized by an in vitro-coupled transcription and translation (TNT) system and incubated with biotin-labeled viral RNAs that contain the various regulatory elements of vRNA for RNA replication, including the 3′- and 5′-terminal 12 and 13 nucleotides (named Uni12 and Uni13, respectively). These sequences are highly conserved among the eight vRNA segments, whereas those of the segments of the adjoining untranslated region (UTR) are variable. The biotin-labeled viral RNA was incubated with HA-ZBTB25 and then pulled down by streptavidin-conjugated beads. The results showed that ZBTB25 proteins bound to a fragment consisting of Uni13 plus the 5′ UTR (Uni13+5′ UTR) and the 5′ UTR alone, but not Uni12, Uni13, the 3′ UTR, or the open reading frame (ORF) encoding NP (NP-ORF) (Fig. 5A and B). Reciprocal experiments using recombinant glutathione S-transferase (GST)-ZBTB25 fusion proteins and various RNA segments also showed that Uni13+5′ UTR vRNA was pulled-down by GST-ZBTB25 fusion proteins (data not shown). These results indicate that ZBTB25 binds with the 5′ UTR but not Uni13, Uni12, or the 3′ UTR. We aligned these 5′ UTRs of eight-segmented genomes and showed that each contains a stretch of U-rich sequence (Fig. 5C). It has been proposed that the viral RdRp usually stutters at the stretch of five to seven uridine residues. A poly(A) tail is subsequently added to the tail of mRNA (36–39). Based on this reasoning, we determined whether ZBTB25 binds to the U-rich sequence. The 5′ UTRs of NP, PB1, and PB2 vRNAs were selected for binding studies. As shown in Fig. 5D, all of the vRNA 5′ UTRs containing the U-rich sequence were precipitated by GST-ZBTB25 fusion proteins but not by GST alone. When the U-rich sequence was replaced with an all-G sequence (Table 2), GST-ZBTB25 proteins pulled down much less of the 5′ UTR–all-G vRNAs. Taken together, these results suggest that ZBTB25 binds to the 5′ UTR of vRNA through recognition of U-rich sequences.
FIG 5.
ZBTB25 is a viral RNA binding protein and recognizes the U-rich sequence within 5′ UTR vRNA. (A and B) In vitro RNA-protein pulldown assay of ZBTB25-vRNA binding activity. Schematic representation of the structure of NP vRNA constructs used in the viral RNA binding assay (black arrows). In constructs d and e of panel B, asterisks mark the positions of vRNAs with U-to-G mutations. HA-ZBTB25 proteins were generated by an in vitro transcription-coupled translation system and reacted with biotin-labeled viral RNAs as shown in the right panel of panel A and the lower panel of B. The biotin-labeled viral RNAs were precipitated by streptavidin-conjugated beads, and coprecipitated HA-ZBTB25 was detected by Western blotting using anti-HA antibody. The input RNA constructs were analyzed by denaturing agarose gel electrophoresis and detected by autoradiography. (C) Schematic representations of the alignment of the IAV eight-segment genome including PB2, PB1, PA, HA, NP, NA, M1, and NS1. (D) GST-ZBTB25 fusion proteins were individually incubated with the chemically synthesized 5′ UTRs of NP, PB1, and PB2 containing U-rich or all-G sequences and pulled down by glutathione (GSH) agarose beads, followed by RNA analysis using denaturing agarose gel electrophoresis and detected by autoradiography. Uni12, unique 12-nucleotide conserved sequence; Uni13, unique 13-nucleotide conserved sequence; UTR, untranslated region; ORF, open reading frame.
TABLE 2.
RNA list for RNA-protein interaction studies
| RNA source and name | Sequencea |
|---|---|
| MDBio, Inc. RNAsb | |
| Uni13+5′ UTR | 5′-AGUAGAAACAAGGGUAUUUUUCU-3′ |
| Uni12 | 5′-UACCCUGCUUUUGCU-3′ |
| 3′ UTR | 5′-GAUUUCGAUGUCACUCUGUGAGUGAUUAUC-3′ |
| NP-3′ ORF | 5′-GUAAGAUCGUUUGGUGCCUUUGGUCGCCAU-3′ |
| Uni13+5′ UTR | 5′-AGUAGAAACAAGGGUAUUUUUCU-3′ |
| Uni13 | 5′-AGUAGAAACAAGGGU-3′ |
| 5′ UTR | 5′-GGGUAUUUUUCUCUA-3′ |
| Uni13+5′ UTR(mut) | 5′-AGUAGAAACAAGGGUAGGGGGCU-3′ |
| Uni13(mut)+5′ UTR | 5′-AGUAGGGGCAAGGGUAUUUUUCU-3′ |
| Dharmacon RNAs | |
| NP-Uni13+5′ UTR (U-rich) | 5′-AGUAGAAACAAGGGUAUUUUUCU-3′ |
| NP-Uni13+5′ UTR (all-G) | 5′-AGUAGAAACAAGGGUAGGGGGCU-3′ |
| PB1-5′ UTR (U-rich) | 5′-CAUUUUUUCAUGAAGGACAAGCUAAAUUCA-3′ |
| PB1-5′ UTR (all-G) | 5′-CAGGGGGGCAUGAAGGACAAGCUAAAUUCA-3′ |
| PB2-5′ UTR (U-rich) | 5′-UCGUUUUUAAACUAUUCGACA-3′ |
| PB2-5′ UTR (all-G) | 5′-UCGGGGGGAAACUAUUCGACA-3′ |
Underlining indicates substituted residues in the MDBio, Inc., RNAs and all-G regions in the Dharmacon RNAs.
All viral RNAs from MDBio, Inc., were biotin labeled at the 5′ end.
ZBTB25 is SUMOylated at residue lysine 204 and deconjugated by the SUMO deconjugation enzyme SENP2.
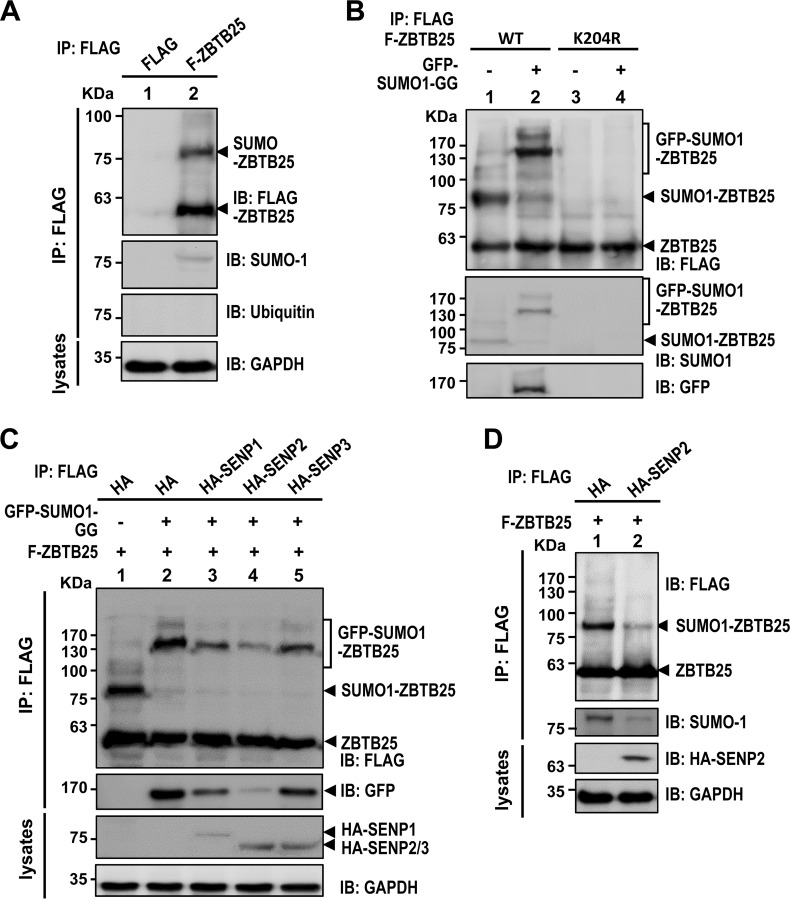
The Cys2-His2 zinc finger domain of ZBTB family members has been reported to bind to DNA, RNA, or proteins (36–39). Several studies have demonstrated that some RNA-binding proteins can be SUMOylated and that this posttranslational modification may influence their RNA-binding abilities and RNA processing (40). We wondered whether ZBTB25 could be SUMOylated. As shown in Fig. 5A, HEK293T cells overexpressing Flag-tagged ZBTB25 revealed a slow-migrating band of about 80 kDa in addition to the expected 50-kDa band, and this slow-migrating band was recognized by anti-SUMO-1 antibody but not by anti-ubiquitin antibody (Fig. 6A). The size of this slow-migrating band (∼80 kDa) is equivalent to 1 molecule of SUMO-1 conjugated to ZBTB25. This result suggests that ZBTB25 can be modified by SUMO-1 but is not ubiquitinylated. We next determined which amino acid residues within ZBTB25 are responsible for SUMO-1 modification. We identified a conserved SUMO conjugation motif, ΨKXE (24), located at lysine 204 and constructed a K204R mutant. As shown in Fig. 6B, the K204R mutant did not yield the SUMO-1-conjugated band (lane 3 versus lane 1). Conjugation of the wild type but not the K204R mutant with enhanced green fluorescent protein (EGFP)–SUMO-1 yielded a protein of 140 kDa, which could be detected by either anti-GFP or anti-SUMO-1 antibodies (Fig. 6B, lane 4 versus lane 2), proving that residue lysine 204 is the SUMO-1 conjugation site. Furthermore, the amount of SUMO-1-conjugated ZBTB25 was reduced by treatment with any of the three SUMO-deconjugating enzymes (SENPs), particularly, SENP2 (Fig. 6C). Finally, endogenous SUMO-1-conjugated ZBTB25 was significantly reduced upon SENP2 overexpression (Fig. 6D). Taken together, these data showed that ZBTB25 can be SUMO-1 modified at lysine 204 and that SENP2 is the major deconjugating enzyme. Interestingly, the amount of SUMOylated ZBTB25 was higher in the virus-infected cells than in uninfected cells. The significance of this finding is not clear (data not shown).
FIG 6.
ZBTB25 is SUMOylated at residue lysine 204 and deconjugated by the SUMO deconjugation enzyme SENP2. (A) HEK293T cells were transfected with Flag-tagged ZBTB25 WT or a control vector and then subjected to IP analysis using anti-Flag antibodies and immunoblotted using different antibodies, as indicated. (B) HEK293T cells were cotransfected with Flag-ZBTB25 WT or the K204R mutant in the presence or absence of GFP–SUMO-1–GG, and IP was performed using anti-Flag antibody. (C) HEK293T cells were transfected with Flag-tagged ZBTB25 WT with or without EGFP–SUMO-1–GG in the presence of HA-SENP1, HA-SENP2, or HA-SENP3 and then subjected to IP analysis using anti-Flag antibodies. (D) IP analysis of HEK293T cells transfected with Flag-tagged ZBTB25 WT in the presence or absence of HA-tagged SENP2 using anti-Flag antibody.
Both the zinc finger domain and SUMOylation are required for ZBTB25-induced IAV replication.
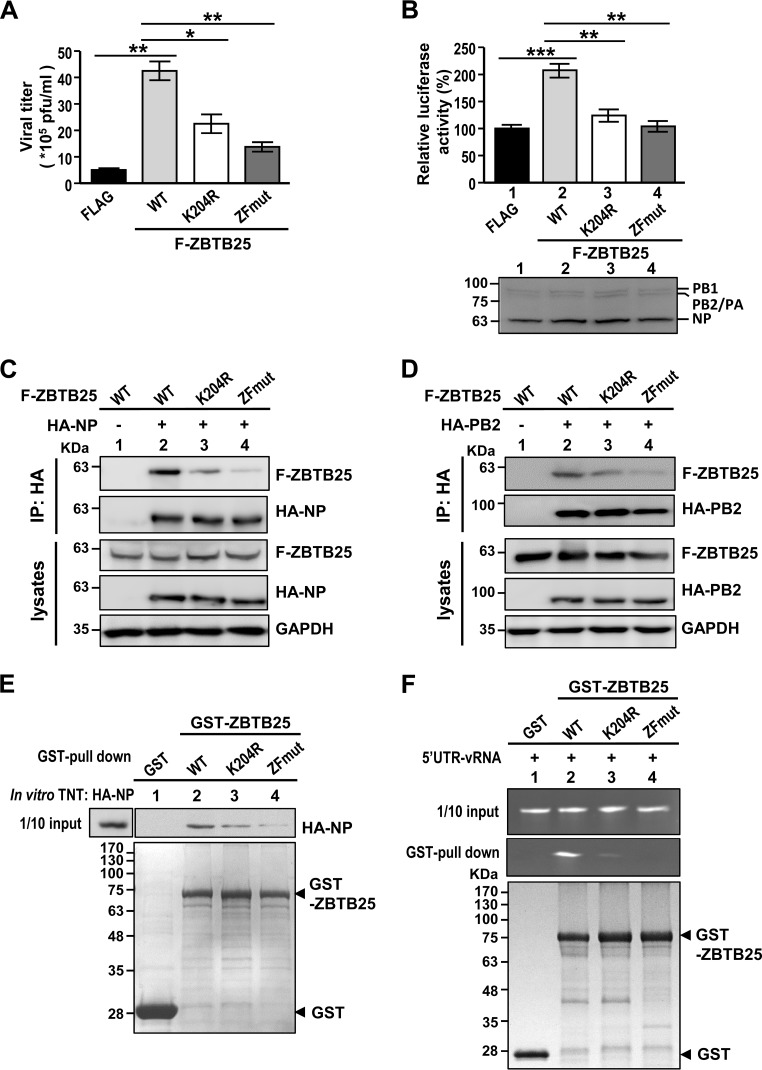
We next investigated the roles of the zinc finger domain and SUMOylation modification in ZBTB25-regulated IAV replication. We found that cells transduced with a SUMOylation mutant (K204R) or a double-cysteine mutant (C240S/C351S-ZFmut) that has an inactivated zinc finger domain (ZFmut) produced a significantly lower virus titer than those transfected with ZBTB25 WT (wild type) (Fig. 7A). However, the two mutants still produced slightly higher virus titers than the titer of the Flag control, suggesting that these two domains work independently. Furthermore, while ZBTB25 WT enhanced the RdRp activity of IAV in the minireplicon assay, the ZBTB25 K204R and ZFmut mutants failed to induce RdRp activities (Fig. 7B). Correspondingly, interaction of ZBTB25 with components of the RdRp complex (NP and PB2) was much weaker for these two mutants, particularly for the ZF mutant (Fig. 7C to E). Both GST-ZBTB25-K204R and -ZFmut mutant proteins also bound much less viral 5′ UTR RNA (Fig. 7F). Together, these results suggest that disruption of the zinc finger domain or removal of SUMO-1 modification interfered with the binding of ZBTB25 to the RdRp and RNA components of the viral RNP complex. Finally, mutation of the BTB/POZ domain also reduced binding of ZBTB25 to NP, suggesting that the various domains of ZBTB25 are involved in viral RNA replication (data not shown).
FIG 7.
Both the zinc finger domain and SUMOylation play important roles in ZBTB25-induced IAV replication. (A) Plaque assay of overexpression of Flag-tagged ZBTB25 WT or the K204R or ZF mutant in A549 cells at 24 h after IAV infection (MOI of 0.2). (B) HEK293T cells were cotransfected with pPolI-Luc and Renilla luciferase in the presence of Flag-tagged ZBTB25 WT or the K204R or ZF mutant, and their luciferase activities were measured at 24 h posttransfection. The protein levels of RdRp complex and NP in each reaction product were detected by Western blotting. The Pol I-driven luciferase activity was normalized by Renilla luciferase expression, and the relative expression is shown. The data are presented as the means ± SD (n = 3; *, P < 0.05; **, P < 0.01; and ***, P < 0.001, by Student's t test). (C and D) HA-NP or HA-PB2 was individually introduced into HEK293T cells in the presence of Flag-tagged ZBTB25 WT or the K204R or ZF mutant and then subjected to IP analysis using anti-HA antibody. GAPDH served as the loading control. (E) HA-NP proteins synthesized by an in vitro transcription and translation system were incubated with purified fusion proteins of GST-ZBTB25-WT or the -K204R or -ZF mutant and then subjected to GST pulldown analysis. These immunocomplexes were analyzed by Western blotting using anti-HA antibody. (F) The recombinant GST-ZBTB25-WT, -K204R, or -ZF mutant fusion protein was purified, followed by incubation with the 5′ UTR of vRNA. GST fusion proteins were then precipitated by glutathione (GSH) agarose beads and subjected to RNA analysis by denaturing agarose gel electrophoresis and detected by autoradiography.
Disulfiram treatment inhibits ZBTB25-mediated IAV replication.
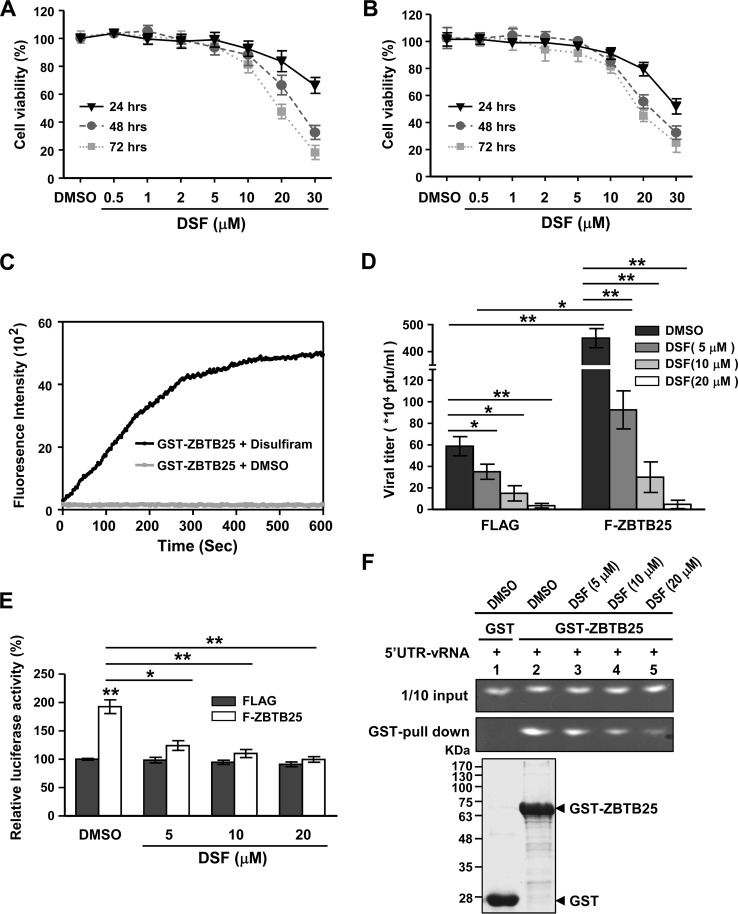
Our finding that a zinc finger protein is involved in the regulation of IAV RNA replication suggests a new approach to anti-influenza virus strategies, namely, targeting a cellular cofactor. Disulfiram (DSF), a zinc ejector used to block zinc finger domain activity, was tested for its potential anti-IAV effects. Cytotoxicity of DSF was first tested on A549 (Fig. 8A) and HEK293T cells (Fig. 8B); the result showed that DSF had only minor cytotoxicity at concentrations of up to 20 μM DSF after 24 h of treatment but significant cytotoxicity at a concentration of 30 μM. We first confirmed that ZBTB25 is indeed a zinc-binding protein. As shown in Fig. 8C, addition of DSF to the recombinant GST-ZBTB25 protein resulted in rapid release of Zn2+ ions, as detected by changes in fluorescence of the zinc-specific fluorophore FluoZin-3. Thus, ZBTB25 is indeed a zinc-containing molecule, and DSF ejected zinc ions from ZBTB25. We then evaluated the effects of DSF on ZBTB25-enhanced IAV viral titers. As shown in Fig. 8D, DSF treatment (5 to 20 μM) significantly lowered virus titers in the presence or absence of overexpression of ZBTB25 in a dose-dependent manner. This result suggests that ZBTB25 and/or other zinc-containing molecules participate in IAV RNA replication. Significantly, DSF treatment inhibited ZBTB25-enhanced RdRp activity in minireplicon assays (Fig. 8E) and, proportionally, also inhibited binding between ZBTB25 and the viral 5′ UTR (Fig. 8F). These results indicate that the zinc-binding activity is a critical component of viral RdRp. Combined, these data suggest that DSF can be used to suppress IAV replication, verifying the concept that cellular factors can be targets of antiviral agents.
FIG 8.
Disulfiram treatment inhibits ZBTB25-mediated IAV replication. (A and B) Cell viability under different doses of DSF treatment in A549 (A) and HEK293T cells (B) by detection of CCK-8 activity. (C) Recombinant GST-ZBTB25 fusion proteins were incubated with disulfiram (DSF), and the release of zinc ions from GST-ZBTB25 was monitored by the fluorescence signal of the Zn-specific fluorophore, FluoZin-3. (D) A549 cells overexpressing Flag-ZBTB25 or a Flag control at 24 h after IAV infection and simultaneously treated with different doses of DSF. The viral titer was detected by plaque assay. (E) HEK293T cells were cotransfected with pPolI-Luc and Renilla luciferase in the presence of Flag-ZBTB25 WT or a Flag control and simultaneously treated with different doses of DSF. Luciferase activity was measured and normalized by Renilla luciferase expression. Relative expression is shown. The data are presented as the means ± SD (n = 3; *, P < 0.05; **, P < 0.01, by Student's t test). (F) Recombinant GST-ZBTB25 fusion proteins were incubated with the 5′ UTR of vRNA in the presence of different doses of DSF, as indicated. GST fusion proteins were then precipitated by glutathione (GSH) agarose beads; the precipitated proteins were subjected to RNA analysis by denaturing agarose gel electrophoresis and detected by autoradiography.
ZBTB25 also suppresses IFN-β1 induction to facilitate IAV replication in a zinc finger domain- and SUMOylation modification-dependent manner.
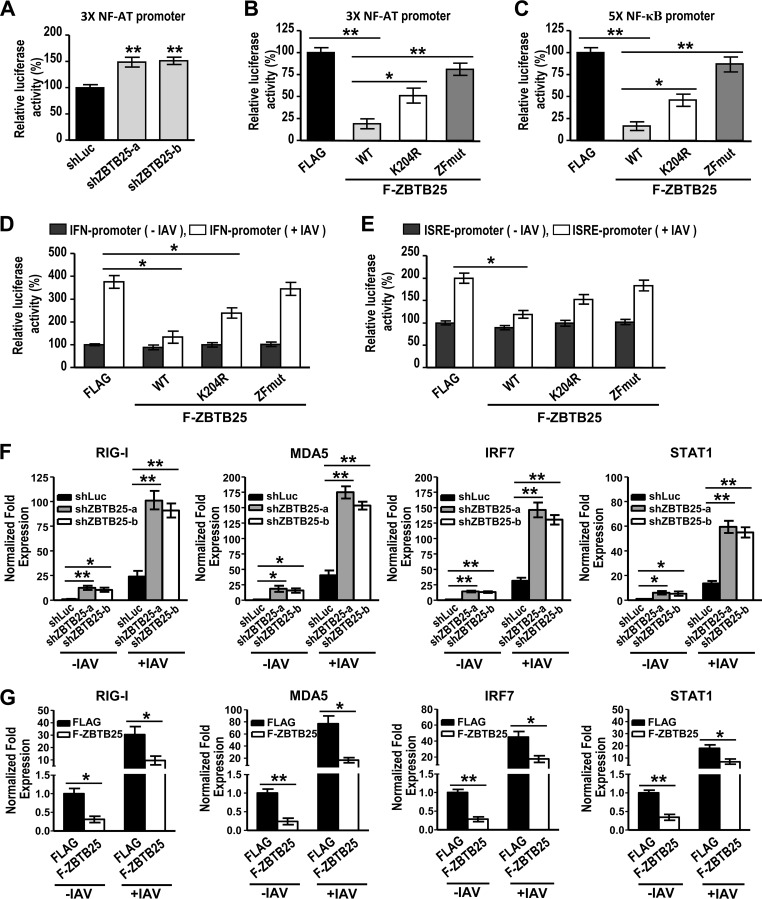
Since ZBTB25 is a cellular transcription repressor, we explored the possibility that ZBTB25 influences IAV replication through regulation of cellular transcription. ZBTB25 has been reported to inhibit NFAT expression (10). We first examined whether ZBTB25 regulates the reporter activity of pGL3-NFAT luciferase driven by a construct containing three binding sites for NFAT (3XNFAT). The result showed that ZBTB25 knockdown by shRNA resulted in increased luciferase activity (Fig. 9A). Conversely, overexpression of ZBTB25 resulted in decreased luciferase activity. The reductions in luciferase activity were proportionally less for the ZBTB25 K204R and ZFmut mutants (Fig. 9B). NFAT has been reported to coordinate with NF-κB to regulate IFN-γ gene transcription (41). The NF-κB RelA subunit is crucial for early IFN-β expression and resistance to RNA virus replication (42). We therefore tested whether ZBTB25 influences NF-κB reporter activity. As shown in Fig. 9C, overexpression of ZBTB25 resulted in decreased reporter activity of a luciferase construct containing five copies of NF-κB binding sites (5XNF-κB-luciferase). Since ZBTB25 can modulate the reporter activities of both NFAT and NF-κB, it is possible that ZBTB25 regulates IAV-induced IFN-β expression. As expected, overexpression of ZBTB25 suppressed IAV-induced IFN-β expression while its mutants induced proportionally less IFN-β (Fig. 9D). A similar observation was made using interferon-stimulating response element (ISRE)-driven luciferase (Fig. 9E). Furthermore, qRT-PCR measurements of gene expression levels of the interferon-stimulated genes RIG-1, MDA5, IRF7, and STAT1 showed that all of them were stimulated by downregulation of ZBTB25 (Fig. 9F). Conversely, overexpression of ZBTB25 significantly inhibited expression of these genes (Fig. 9G). These data suggest that ZBTB25 not only directly stimulates viral RdRp activity to promote IAV replication but also downregulates IFN-β1 induction and ISG expression to indirectly facilitate IAV production.
FIG 9.
ZBTB25 also suppresses IFN-β1 induction to facilitate IAV replication in a zinc finger domain- and SUMOylation modification-dependent manner. (A and B) 3XNFAT-Luc and Renilla luciferase were transfected into A549 cells with knockdown of endogenous ZBTB25 by shRNAs or with overexpression of the Flag-ZBTB25 WT or K204R or ZF mutant, as indicated. (C) 5XNF-κB-Luc and Renilla luciferase were transfected into A549 cells by overexpressing Flag-ZBTB25 WT or the K204R or ZF mutant. (D and E) IFN-β1-Luc or ISRE-Luc, as indicated, combined with Renilla luciferase, was introduced into HEK293T cells in the presence of Flag-ZBTB25 WT or the K204R or ZF mutant and then either left uninfected or infected with IAV. Luciferase activity was measured and normalized by Renilla luciferase expression, and relative expression is shown. (F) Interferon-stimulated gene expression (RIG-1, MDA5, IRF7, and STAT1) measured by quantitative PCR analysis in A549 cells with knockdown of ZBTB25 by shRNA with or without IAV infection. (G) Overexpression of ZBTB25 or a Flag control in A549 cells with or without IAV infection. Gene expression was normalized to GAPDH levels, and the fold change in expression relative to that of the shLuc control is shown. The data are presented as the means ± SD (n = 3; *, P < 0.05; **, P < 0.01, by Student's t test).
DISCUSSION
In this study, we identified a novel cellular protein, ZBTB25, that plays a crucial role in IAV replication. We show that ZBTB25 promotes IAV RNA transcription through direct binding to the U-rich sequence within the 5′ UTR of vRNA (43–45) and also via recruitment of the RdRp complex (PA, PB1, and PB2) and NP through protein-protein interactions. The structure and sequence of the 5′ end of the vRNA are critical for initiation of RNA transcription or replication for many RNA viruses because of the interaction between the 5′ and 3′ ends of viral RNA (46). Indeed, the 5′ and 3′ ends of IAV RNA have been shown to form a panhandle structure. A zinc finger mutant and a SUMOylation-deficient mutant displayed lower vRNA binding ability and less RdRp complex recruitment, exhibiting consequently diminished ZBTB25-induced IAV replication. Furthermore, we discovered that treatment with the zinc ejector disulfiram, which blocked the zinc finger activity of ZBTB25, effectively suppressed IAV production.
The 5′ UTR and 3′ UTR of viral RNA are thought to play an important role in the regulation of viral transcription and, thereby, influence viral replication. Mutations in the 5′ and 3′ ends of vRNA revealed that a stretch of uninterrupted uridines at the 5′ end of the negative-strand RNA is essential for mRNA synthesis (47–50). HuR and La proteins, which bind to U-rich sequences in the 3′ UTR of hepatitis C virus (HCV), promote HCV replication by protecting viral RNA from degradation (51, 52). In addition, SYNCRIP (synaptotagmin-binding cytoplasmic RNA-interacting protein) binds to the 3′ UTR, poly(C)-binding protein 2 binds to the 5′ UTR of HCV RNA, and both promote RNA replication, probably by circularization of the viral RNA (46, 53). HIV-1 Tat protein binds with the transactivation response region (TAR) U-bulge to stimulate transcription (54), and zinc finger protein 134 (hZNF-134) promoted HIV-1 LTR activity and viral propagation (55). Moreover, the 3′ UTR of Sindbis virus RNAs sequesters HuR protein in the cytoplasm, resulting in the dramatic destabilization of cellular mRNAs and significantly altering alternative polyadenylation and splicing processes (56). In contrast, ZAP (zinc finger antiviral protein), another zinc finger domain-containing protein, has been shown to bind to the 3′ UTR of several viral RNAs, including hepatitis C virus (HCV), dengue virus, and HIV (57–59). It recruits exonuclease and endonuclease to degrade mRNA, thereby serving as a negative regulator for RNA viruses. In this study, we have demonstrated that ZBTB25 binds to U-rich sequences within the 5′ UTR of viral RNA to positively promote IAV RNA transcription, emphasizing the crucial role of the 5′ UTR in IAV replication. ZBTB25 may stabilize or unwind the 5′- and 3′-end panhandle structure of IAV RNA. In addition, the U-rich sequence is important for polyadenylation of mRNA in many systems. However, we need more experimental evidence to determine whether ZBTB25 participates in splicing, polyadenylation, mRNA stabilization, mRNA localization, or translation processes of IAV.
It is well-known that cellular transcription repressors are involved in the regulation of DNA virus replication (60–62). However, transcription repressors participating in RNA virus replication have rarely been reported. The functions of ZBTB members are largely unknown; some ZBTB members have been shown to be involved in tumorigenesis, chromatin remodeling, and T-cell and B-cell development and differentiation (11, 63, 64) but not in immune regulation or viral replication. In this study, we have demonstrated that ZBTB25 functions as a transcription repressor to suppress IFN-β1 induction in virus-infected cells, thereby inhibiting the host's innate immunity and creating a favorable environment for IAV replication. In addition, ZBTB25 caused similar levels of enhancement of viral titer in Vero cells (IFN-deficient model system) (65) and A549 cells upon IAV infection, suggesting that ZBTB25-stimulated viral RNA synthesis is not caused solely by ZBTB25-repressed IFN expression (data not shown). We propose that ZBTB25 plays a dual role in IAV replication: first, by directly binding to viral RNA and RdRp to stimulate vRNA transcription and replication and, second, by suppressing host cell innate immunity to provide a beneficial environment for the virus. A ZBTB25-mediated double-acting mechanism may be common among cellular transcription promoters or suppressors of IAV as we previously showed that another transcription repressor (DR1) exhibited these double effects as well (32).
In this study, we demonstrated that ZBTB25 can interact with different viral proteins (Fig. 3A). Interestingly, PB1 and PA lost ZBTB25-binding ability after RNase A treatment (Fig. 4B), suggesting that ZBTB25 interacts with certain viral proteins such as PB1 and PA in an RNA-dependent manner. On the other hand, ZBTB25 directly interacts with NA, M1, NS1, PB2, and NP proteins. Altogether, ZBTB25 is able to interact with different viral proteins, suggesting that ZBTB25 may be involved in other processes of the viral replication cycle. However, further experiments are required to test this hypothesis.
Protein posttranslational modification has been demonstrated to play an important role in regulating the biological functions of proteins. Recently, several studies have indicated that SUMO modification of RNA-binding proteins modulates their RNA-processing functions. Eukaryotic pre-mRNA 3′-end formation is catalyzed by a complex including symplekin (a factor involved in assembly of the complex) and cleavage and polyadenylation specificity factor 73 (CPSF-73; an endonuclease). Both symplekin and CPSF-73 are substrates of SUMO modification, and loss of SUMOylation resulted in marked inhibition of 3′-end processing (40). We found that ZBTB25 can be SUMO-1 modified, and this SUMOylation contributes to the ability of ZBTB25 to bind vRNA and viral proteins, thus further promoting viral transcription and/or replication.
The zinc finger domain is a protein motif that is required for zinc ions to stabilize protein structure. The transcription factor IIIA (TFIIIA) of Xenopus laevis was originally characterized as a zinc finger protein that cooperated with zinc ions to maintain its function (66). DSF is a zinc ejector and initially was used for the treatment of chronic alcoholic patients (67). Recently, DSF has also been applied as an anti-cancer drug (68, 69) and an inhibitor against HIV (70–72). Our lab has also demonstrated that DSF targets to the zinc site of HCV NS5A protein and inhibits HCV replication (73). In all of these cases, DSF targets a viral protein. In contrast, zinc finger-containing ZBTB25 is a cellular protein. Yet DSF is an effective antiviral agent. Thus, DSF is a model for drugs targeting a cellular factor involved in IAV replication. Although ZBTB25 is a C2H2 zinc finger transcription repressor and is involved in multiple cellular functions, the cytotoxic dose of DSF is higher than the dose required for its antiviral activity (Fig. 8A and D). It is curious that DSF can inhibit IAV replication even in the absence of exogenous ZBTB25, suggesting that there are additional zinc-containing cellular molecules involved in IAV replication. This finding is consistent with previous studies showing that increasing the intracellular zinc concentration can suppress the replication of a variety of RNA viruses, including poliovirus and influenza virus (74). Current anti-IAV drugs are designed to target IAV viral proteins and readily develop acquired drug resistance due to the high mutation rate of RNA viruses. Therefore, using cellular factors as antiviral drug targets might be a promising way to treat IAV infection. Overall, our findings indicate that IAV usurps the cellular protein ZBTB25 for IAV RNA replication, and thus ZBTB25 serves as a novel and potential target for antiviral therapy.
MATERIALS AND METHODS
Cell culture.
Human lung adenocarcinoma epithelial cells (A549) were maintained in F-12K medium (Gibco) supplemented with 10% fetal bovine serum (FBS) and antibiotics. Madin-Darby canine kidney (MDCK) cells and human embryonic kidney (HEK293T) cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% FBS and antibiotics.
Short hairpin RNA lentivirus infection and siRNA oligomer transfection.
Endogenous ZBTB25 protein levels were downregulated by short hairpin RNAs (shRNAs) or siRNA oligomers. Two ZBTB25 shRNAs constructed in the pLKO.1 lentiviral vector were obtained from the National RNAi Core Facility (RNAi Core), Academia Sinica, Taiwan, with the targeted sequence of GCTTCCATTCTGGAAAGTAAT and CCAACCTGACATATTCAGCTA (Table 1). For lentivirus infection, A549 cells were infected with lentivirus-containing medium in the presence of Polybrene. Two ZBTB25 siRNA oligomers were purchased from Thermo Scientific Dharmacon, and siRNA oligomers were transfected into cells using RNAiMax reagent (Invitrogen).
Plasmid constructions.
For generation of p3XFLAG-ZBTB25 wild type (WT), the full-length ZBTB25 cDNA was amplified by PCR and inserted into the p3XFLAG vector at the EcoRI and NotI restriction enzyme sites. For ZBTB25 rescue experiments in ZBTB25 knockdown cells, a ZBTB25 wobble mutant was constructed. The cDNA of ZBTB25 WT, 5′-GCTTCCATTCTGGAAAGTAAT-3′, which is targeted by shZBTB25-a, was converted to 5′-GCTTCGATCCTCGAGAGCAAT-3′ without changing the amino acid sequence and inserted into the p3XFLAG-CMV7.1 vector. To construct the SUMOylation-deficient mutant and the zinc finger domain double-cysteine mutant of ZBTB25, a QuikChange II XL site-directed mutagenesis kit (Promega) was used to generate p3XFLAG-ZBTB25-K204R and p3XFLAG-ZBTB25-C240S/C351S using p3XFLAG-ZBTB25 as the template, according to the manufacturer's protocol. For overexpression of viral proteins, the viral cDNAs of HA, NA, M1, M2, NS1, NS2, PB1, PA, PB2, and NP were inserted into pCDNA3.0-HA as described previously (75). For the production of a recombinant GST-ZBTB25 fusion protein, a pGEX-4T1-ZBTB25 construct was generated. The full-length ZBTB25 cDNA was moved from p3XFLAG-ZBTB25 to the pGEX-4T1 vector using EcoRI and NotI restriction sites. The constructs pEGFP–SUMO-1–GG, pCDNA3.0-HA-SENP1, pCDNA3.0-HA-SENP2, and pCDNA3.0-HA-SENP3 were kind gifts from Hsiu-Ming Shih, Academia Sinica, Taiwan.
Antibodies and reagents.
For Western blot analysis, anti-ZBTB25 (ab168116), anti-SUMO-1 (ab11672), and anti-NP (ab20343) antibodies were purchased from Abcam. Anti-GFP (sc-9996) antibodies were obtained from Santa Cruz Biotechnology. Anti-HA (MMS-101R), anti-Flag (M2), anti-ubiquitin (FK2), anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH; GTX100118), and anti-M1 (MCA401) were sourced from Covance, Sigma, Biomol, GeneTex, and AbDSerotec, respectively. For immunoprecipitation assays, anti-PB1 (GTX125923) and anti-PA (GTX118991) antibodies were purchased from GeneTex. Anti-PB2 antibody (sc-17603) and anti-HA (hemagglutinin) antibody (sc-52025) were purchased from Santa Cruz. Alexa Fluor 488-conjugated secondary antibodies used for immunofluorescence were purchased from Molecular Probes (Invitrogen). Radioimmunoprecipitation assay (RIPA) mammalian protein extraction reagent (89901) and anti-HA-conjugated beads (26182) were obtained from Thermo Scientific.
IAV infection.
The A/WSN/33 strain of IAV was used in the study. For IAV infection, cells were washed with phosphate-buffered saline (PBS) and then infected with IAV in minimal essential medium alpha (MEM-alpha) (Gibco) containing 0.5 g/ml tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin (Sigma) at a multiplicity of infection (MOI) of 1 for 1 h. After the virus-containing medium was removed, these infected cells were harvested at 8 h postinfection (hpi) for further analysis.
Immunofluorescence microscopy.
For immunofluorescence staining, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. After incubation with the anti-NP antibody, a secondary antibody-conjugated Alexa Fluor 488 was applied. Cells were counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) for 15 min. Cell images were acquired using a confocal microscope.
Plaque assay.
Virus titer was determined by plaque assay. In brief, MDCK cells were infected with 10-fold serial dilutions of IAV at 37°C for 1 h. After a washing step, cells were overlaid with 0.5% agarose (Lonza) containing MEM-alpha. After 2 days of incubation, the cells were fixed with 3.7% formaldehyde (Sigma) and stained with 1% crystal violet solution (Sigma).
Quantification RT-PCR.
A High Pure RNA isolation kit (Roche) was used to extract total RNA according to the manufacturer's protocol. The cDNAs were synthesized by using a SuperScript III first-strand synthesis system (Invitrogen) with an oligo(dT)20 primer for mRNA, an IAV-specific primer (uni-12, 5′-AGCAAAAGCAGG-3′) for viral RNA, or an IAV-specific primer (uni-13, 5′-AGTAGAAACAAGG-3′) for viral cRNA. Quantitative reverse transcription-PCR (qRT-PCR) assays were performed by using a RealTime ready Universal ProbeLibrary system (Roche). Quantification was further analyzed by LightCycler 480 software (Roche). GAPDH mRNA was used as an internal control. The primers and probes are as follows: IAV_NP segment, 5′-GATGGAGACTGATGGAGAACG-3′ (sense) and 5′-TCATTTTTCCGACAGATGCTC-3′ (antisense) with universal probe 59 (Roche); GAPDH, 5′-AGCCACATCGCTCAGACAC-3′ (sense) and 5′-GCCCAATACGACCAAAT CC-3′ (antisense) with universal probe 60 (Roche); ZBTB25, 5′-CTCCTGCACGGAGGAGTC-3′ (sense) and 5′-AAATTCTCGCTGCATGTTCA-3′ (antisense) with universal probe 67 (Roche). The primers of ISGs for qRT-PCR assays were described in detail in our previous study (32).
Minireplicon assay.
RdRp activity was measured by minireplicon assay. In brief, cells were cotransfected with the pPolI-Luc (where PolI is polymerase I), a Renilla luciferase-expressing plasmid, and four plasmids for expression of the viral proteins PA, PB1, PB2, and NP in the presence or absence of Flag-tagged ZBTB25 WT or mutant, as indicated on the figures and in the figure legends. At 24 h posttransfection, the luciferase activity was measured by using Dual-Glo luciferase (E1960; Promega) according to the manufacturer's protocol. Renilla luciferase activity served as an internal control to normalize transfection efficiency.
Immunoprecipitation assay.
For in vitro immunoprecipitation, cells were transfected with the plasmids as indicated. At 48 h posttransfection, total cell lysates were lysed by using RIPA reagent (Thermo Scientific) and immunoprecipitated with protein G-agarose (Roche) conjugated with the antibodies indicated on the figures and in the figure legends according to the manufacturer's protocol, and subsequently analyzed by Western blotting.
In vitro RNA-protein pulldown assay.
In vitro RNA-protein pulldown assays were used to test the binding ability of ZBTB25 to RNA. An in vitro transcription and translation system (Promega) was used to generate HA-ZBTB25. The commercially synthesized biotin-labeled viral RNAs (MDBio, Inc.) (Table 2), including Uni13+5′ UTR, Uni13, 5′ UTR, Uni12, 3′ UTR, and NP-ORF, were incubated with HA-ZBTB25 in RNA-protein binding (RPB) buffer (Tris-HCl, pH 7.0, 60 mM KCl, 2 mM MgCl2, 1 mM dithiothreitol [DTT], RNase Out [EO0381; Thermo Scientific]) and 5% glycerol. The biotin-labeled viral RNAs were pulled down by streptavidin-conjugated beads (88816; Pierce). After further washing, these precipitated beads were subjected to Western blot analysis using anti-HA antibody.
GST pulldown assay for RNA.
pGEX-4T-1-ZBTB25 or pGEX-4T1 vector alone was introduced into Escherichia coli cells, and the glutathione S-transferase fused with ZBTB25 protein (GST-ZBTB25) and GST alone were purified. Recombinant GST-ZBTB25 fusion proteins and GST alone were incubated with commercially synthesized viral RNAs (Dharmacon) (Table 2) including NP-Uni13+5′ UTR, PB1-5′ UTR, and PB2-5 ′ UTR and reacted with glutathione (GSH) beads in RPB buffer for 2 h at 4°C. After further washing, the precipitated RNAs were separated by denaturing agarose gel electrophoresis and detected by autoradiography.
In vivo SUMOylation assay.
For the in vivo SUMOylation assay, plasmids expressing Flag-tagged ZBTB25 WT or K204R or a Flag-tagged control, together with GFP–SUMO-1–GG, were transiently transduced into HEK293T cells in the presence or absence of HA-SENP1, HA-SENP2, or HA-SENP3, as indicated on the figures and in the figure legends. After 48 h of transfection, the cells were lysed and immunoprecipitated with anti-Flag M2 agarose (Sigma) and subsequently analyzed by Western blotting.
Cell proliferation assay.
The cellular proliferation rates of A549 and HEK293T cells treated with the Zn-ejecting agent, tetraethylthiuram disulfide (disulfiram [DSF]), were measured by Cell Counting Kit-8 (CCK-8) activity (Dojindo Molecular Technologies). Disulfiram was purchased from Sigma and solubilized in 100% dimethyl sulfoxide (DMSO; Sigma). In brief, cells were treated with different doses of disulfiram, as indicated on the figures, and incubated for 24, 48, and 72 h. CCK-8 reagent was added into each well of a plate and incubated for an additional 1 h. After that, the absorbance at 450 nm was measured by a microplate reader.
Zinc ejection assays.
The release of zinc ions from ZBTB25 was determined by the fluorescence emission from the zinc-specific fluorophore FluoZin-3 (Invitrogen/Life Technologies). The recombinant GST-ZBTB25 fusion proteins (10 μM) were reacted with disulfiram (5 μM) and FluoZin-3 (5 μM) at room temperature. Fluorescence emission was monitored by an EnSpire multilabel plate reader (PerkinElmer, USA) at an excitation wavelength of 494 nm and emission wavelength of 516 nm for 10 min.
Luciferase reporter assay.
The 3XNFAT-Luc, 5XNF-κB-Luc, pIFN-B1-Luc (an IFN promoter-driven luciferase), or ISRE-Luc (interferon-sensitive response element [ISRE] promoter-driven luciferase) construct was introduced into HEK293T cells with a Renilla luciferase-expressing plasmid in the presence or absence of p3XFLAG-ZBTB25-WT, -K204R, or -ZFmut. After 6 h of transfection, these cells were either left uninfected or infected with IAV (MOI of 0.2). At 24 h postinfection, their luciferase activities were measured by using a Dual-Glo luciferase assay system. Renilla luciferase activity was used as an internal control.
Statistical analysis.
All bar graphs represent means, and error bars show standard deviations (SD). Student's t tests were used to determine the differences between control and test groups.
ACKNOWLEDGMENTS
We thank the National RNAi Core Facility, Academia Sinica, Taiwan, for providing the lentivector and lentiviral shRNAs and for technical support. We thank Hsiu-Ming Shih at the Institute of Biomedical Sciences, Academia Sinica, Taiwan, for the kind gift of plasmids. We also thank Yi-Ling Lin at the Institute of Biomedical Sciences, Academia Sinica, Taiwan, for the gift of Vero cells and critical suggestions on the manuscript.
REFERENCES
- 1.Bouvier NM, Palese P. 2008. The biology of influenza viruses. Vaccine 26(Suppl 4):D49–D53. doi: 10.1016/j.vaccine.2008.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakadamyali M, Rust MJ, Zhuang X. 2004. Endocytosis of influenza viruses. Microbes Infect 6:929–936. doi: 10.1016/j.micinf.2004.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fodor E. 2013. The RNA polymerase of influenza a virus: mechanisms of viral transcription and replication. Acta Virol 57:113–122. doi: 10.4149/av_2013_02_113. [DOI] [PubMed] [Google Scholar]
- 4.Ulmanen I, Broni BA, Krug RM. 1981. Role of two of the influenza virus core P proteins in recognizing cap 1 structures (m7GpppNm) on RNAs and in initiating viral RNA transcription. Proc Natl Acad Sci U S A 78:7355–7359. doi: 10.1073/pnas.78.12.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eisfeld AJ, Neumann G, Kawaoka Y. 2015. At the centre: influenza A virus ribonucleoproteins. Nat Rev Microbiol 13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dias A, Bouvier D, Crepin T, McCarthy AA, Hart DJ, Baudin F, Cusack S, Ruigrok RW. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914–918. doi: 10.1038/nature07745. [DOI] [PubMed] [Google Scholar]
- 7.Braam J, Ulmanen I, Krug RM. 1983. Molecular model of a eucaryotic transcription complex: functions and movements of influenza P proteins during capped RNA-primed transcription. Cell 34:609–618. doi: 10.1016/0092-8674(83)90393-8. [DOI] [PubMed] [Google Scholar]
- 8.Shapiro GI, Krug RM. 1988. Influenza virus RNA replication in vitro: synthesis of viral template RNAs and virion RNAs in the absence of an added primer. J Virol 62:2285–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shen Z, Lou K, Wang W. 2015. New small-molecule drug design strategies for fighting resistant influenza A. Acta Pharm Sin B 5:419–430. doi: 10.1016/j.apsb.2015.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Benita Y, Cao Z, Giallourakis C, Li C, Gardet A, Xavier RJ. 2010. Gene enrichment profiles reveal T-cell development, differentiation, and lineage-specific transcription factors including ZBTB25 as a novel NF-AT repressor. Blood 115:5376–5384. doi: 10.1182/blood-2010-01-263855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siggs OM, Beutler B. 2012. The BTB-ZF transcription factors. Cell Cycle 11:3358–3369. doi: 10.4161/cc.21277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beaulieu AM, Sant'Angelo DB. 2011. The BTB-ZF family of transcription factors: key regulators of lineage commitment and effector function development in the immune system. J Immunol 187:2841–2847. doi: 10.4049/jimmunol.1004006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellmeier W, Taniuchi I. 2014. The role of BTB-zinc finger transcription factors during T cell development and in the regulation of T cell-mediated immunity. Curr Top Microbiol Immunol 381:21–49. doi: 10.1007/82_2014_374. [DOI] [PubMed] [Google Scholar]
- 14.Hoatlin ME, Zhi Y, Ball H, Silvey K, Melnick A, Stone S, Arai S, Hawe N, Owen G, Zelent A, Licht JD. 1999. A novel BTB/POZ transcriptional repressor protein interacts with the Fanconi anemia group C protein and PLZF. Blood 94:3737–3747. [PubMed] [Google Scholar]
- 15.Kobayashi A, Yamagiwa H, Hoshino H, Muto A, Sato K, Morita M, Hayashi N, Yamamoto M, Igarashi K. 2000. A combinatorial code for gene expression generated by transcription factor Bach2 and MAZR (MAZ-related factor) through the BTB/POZ domain. Mol Cell Biol 20:1733–1746. doi: 10.1128/MCB.20.5.1733-1746.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasai N, Nakao M, Defossez PA. 2010. Sequence-specific recognition of methylated DNA by human zinc-finger proteins. Nucleic Acids Res 38:5015–5022. doi: 10.1093/nar/gkq280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polo JM, Dell'Oso T, Ranuncolo SM, Cerchietti L, Beck D, Da Silva GF, Prive GG, Licht JD, Melnick A. 2004. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat Med 10:1329–1335. doi: 10.1038/nm1134. [DOI] [PubMed] [Google Scholar]
- 18.Ahmad KF, Melnick A, Lax S, Bouchard D, Liu J, Kiang CL, Mayer S, Takahashi S, Licht JD, Prive GG. 2003. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol Cell 12:1551–1564. doi: 10.1016/S1097-2765(03)00454-4. [DOI] [PubMed] [Google Scholar]
- 19.Huynh KD, Fischle W, Verdin E, Bardwell VJ. 2000. BCoR, a novel corepressor involved in BCL-6 repression. Genes Dev 14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 20.Bilic I, Ellmeier W. 2007. The role of BTB domain-containing zinc finger proteins in T cell development and function. Immunol Lett 108:1–9. doi: 10.1016/j.imlet.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Wimmer P, Schreiner S, Dobner T. 2012. Human pathogens and the host cell SUMOylation system. J Virol 86:642–654. doi: 10.1128/JVI.06227-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hannoun Z, Maarifi G, Chelbi-Alix MK. 2016. The implication of SUMO in intrinsic and innate immunity. Cytokine Growth Factor Rev 29:3–16. doi: 10.1016/j.cytogfr.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Gill G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev 18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- 24.Flotho A, Melchior F. 2013. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 25.Kumar A, Zhang KY. 2015. Advances in the development of SUMO specific protease (SENP) inhibitors. Comput Struct Biotechnol J 13:204–211. doi: 10.1016/j.csbj.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hickey CM, Wilson NR, Hochstrasser M. 2012. Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13:755–766. doi: 10.1038/nrm3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blondel D, Maarifi G, Nisole S, Chelbi-Alix MK. 2015. Resistance to Rhabdoviridae infection and subversion of antiviral responses. Viruses 7:3675–3702. doi: 10.3390/v7072794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. 1998. How cells respond to interferons. Annu Rev Biochem 67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Sastre A. 2011. Induction and evasion of type I interferon responses by influenza viruses. Virus Res 162:12–18. doi: 10.1016/j.virusres.2011.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleming SB. 2016. Viral inhibition of the IFN-induced JAK/STAT signalling pathway: development of live attenuated vaccines by mutation of viral-encoded IFN-antagonists. Vaccines (Basel) 4:E23. doi: 10.3390/vaccines4030023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su WC, Chen YC, Tseng CH, Hsu PW, Tung KF, Jeng KS, Lai MM. 2013. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc Natl Acad Sci U S A 110:17516–17521. doi: 10.1073/pnas.1312374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsu SF, Su WC, Jeng KS, Lai MM. 2015. A host susceptibility gene, DR1, facilitates influenza A virus replication by suppressing host innate immunity and enhancing viral RNA replication. J Virol 89:3671–3682. doi: 10.1128/JVI.03610-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Su WC, Hsu SF, Lee YY, Jeng KS, Lai MM. 2015. A nucleolar protein, ribosomal RNA processing 1 homolog B (RRP1B), enhances the recruitment of cellular mRNA in influenza virus transcription. J Virol 89:11245–11255. doi: 10.1128/JVI.01487-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eierhoff T, Ludwig S, Ehrhardt C. 2009. The influenza A virus matrix protein as a marker to monitor initial virus internalisation. Biol Chem 390:509–515. doi: 10.1515/BC.2009.053. [DOI] [PubMed] [Google Scholar]
- 35.Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, Hill DE, Regev A, Hacohen N. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267. doi: 10.1016/j.cell.2009.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brayer KJ, Segal DJ. 2008. Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys 50:111–131. doi: 10.1007/s12013-008-9008-5. [DOI] [PubMed] [Google Scholar]
- 37.Burdach J, O'Connell MR, Mackay JP, Crossley M. 2012. Two-timing zinc finger transcription factors liaising with RNA. Trends Biochem Sci 37:199–205. doi: 10.1016/j.tibs.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Filion GJ, Zhenilo S, Salozhin S, Yamada D, Prokhortchouk E, Defossez PA. 2006. A family of human zinc finger proteins that bind methylated DNA and repress transcription. Mol Cell Biol 26:169–181. doi: 10.1128/MCB.26.1.169-181.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. 2007. Sticky fingers: zinc-fingers as protein-recognition motifs. Trends Biochem Sci 32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Vethantham V, Rao N, Manley JL. 2007. Sumoylation modulates the assembly and activity of the pre-mRNA 3′ processing complex. Mol Cell Biol 27:8848–8858. doi: 10.1128/MCB.01186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sica A, Dorman L, Viggiano V, Cippitelli M, Ghosh P, Rice N, Young HA. 1997. Interaction of NF-κB and NFAT with the interferon-gamma promoter. J Biol Chem 272:30412–30420. doi: 10.1074/jbc.272.48.30412. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Basagoudanavar SH, Wang X, Hopewell E, Albrecht R, Garcia-Sastre A, Balachandran S, Beg AA. 2010. NF-κB RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J Immunol 185:1720–1729. doi: 10.4049/jimmunol.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Te Velthuis AJ, Fodor E. 2016. Influenza virus RNA polymerase: insights into the mechanisms of viral RNA synthesis. Nat Rev Microbiol 14:479–493. doi: 10.1038/nrmicro.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cianci C, Gerritz SW, Deminie C, Krystal M. 2012. Influenza nucleoprotein: promising target for antiviral chemotherapy. Antivir Chem Chemother 23:77–91. doi: 10.3851/IMP2235. [DOI] [PubMed] [Google Scholar]
- 45.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 46.Liu HM, Aizaki H, Choi KS, Machida K, Ou JJ, Lai MM. 2009. SYNCRIP (synaptotagmin-binding, cytoplasmic RNA-interacting protein) is a host factor involved in hepatitis C virus RNA replication. Virology 386:249–256. doi: 10.1016/j.virol.2009.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Robertson JS, Schubert M, Lazzarini RA. 1981. Polyadenylation sites for influenza virus mRNA. J Virol 38:157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo GX, Luytjes W, Enami M, Palese P. 1991. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol 65:2861–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng H, Lee HA, Palese P, Garcia-Sastre A. 1999. Influenza A virus RNA polymerase has the ability to stutter at the polyadenylation site of a viral RNA template during RNA replication. J Virol 73:5240–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li X, Palese P. 1994. Characterization of the polyadenylation signal of influenza virus RNA. J Virol 68:1245–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shwetha S, Kumar A, Mullick R, Vasudevan D, Mukherjee N, Das S. 2015. HuR displaces polypyrimidine tract binding protein to facilitate La binding to the 3′ untranslated region and enhances hepatitis C virus replication. J Virol 89:11356–11371. doi: 10.1128/JVI.01714-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Spangberg K, Wiklund L, Schwartz S. 2001. Binding of the La autoantigen to the hepatitis C virus 3′ untranslated region protects the RNA from rapid degradation in vitro. J Gen Virol 82:113–120. doi: 10.1099/0022-1317-82-1-113. [DOI] [PubMed] [Google Scholar]
- 53.Wang L, Jeng KS, Lai MM. 2011. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J Virol 85:7954–7964. doi: 10.1128/JVI.00339-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. 1990. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J 9:4145–4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Benjamin R, Banerjee A, Balakrishnan K, Sivangala R, Gaddam S, Banerjee S. 2014. Mycobacterial and HIV infections up-regulated human zinc finger protein 134, a novel positive regulator of HIV-1 LTR activity and viral propagation. PLoS One 9:e104908. doi: 10.1371/journal.pone.0104908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barnhart MD, Moon SL, Emch AW, Wilusz CJ, Wilusz J. 2013. Changes in cellular mRNA stability, splicing, and polyadenylation through HuR protein sequestration by a cytoplasmic RNA virus. Cell Rep 5:909–917. doi: 10.1016/j.celrep.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhu Y, Wang X, Goff SP, Gao G. 2012. Translational repression precedes and is required for ZAP-mediated mRNA decay. EMBO J 31:4236–4246. doi: 10.1038/emboj.2012.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller S, Moller P, Bick MJ, Wurr S, Becker S, Gunther S, Kummerer BM. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol 81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 60.Xuan Y, Gong D, Qi J, Han C, Deng H, Gao G. 2013. ZAP inhibits murine gammaherpesvirus 68 ORF64 expression and is antagonized by RTA. J Virol 87:2735–2743. doi: 10.1128/JVI.03015-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo JT, Guo H. 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 9:e1003494. doi: 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.He Q, Li W, Ren J, Huang Y, Huang Y, Hu Q, Chen J, Chen W. 2016. ZEB2 inhibits HBV transcription and replication by targeting its core promoter. Oncotarget 7:16003–16011. doi: 10.18632/oncotarget.7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chevrier S, Corcoran LM. 2014. BTB-ZF transcription factors, a growing family of regulators of early and late B-cell development. Immunol Cell Biol 92:481–488. doi: 10.1038/icb.2014.20. [DOI] [PubMed] [Google Scholar]
- 64.Lee SU, Maeda T. 2012. POK/ZBTB proteins: an emerging family of proteins that regulate lymphoid development and function. Immunol Rev 247:107–119. doi: 10.1111/j.1600-065X.2012.01116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Matskevich AA, Jung JS, Schumann M, Cascallo M, Moelling K. 2009. Vero cells as a model to study the effects of adenoviral gene delivery vectors on the RNAi system in context of viral infection. J Innate Immun 1:389–394. doi: 10.1159/000191408. [DOI] [PubMed] [Google Scholar]
- 66.Hanas JS, Hazuda DJ, Bogenhagen DF, Wu FY, Wu CW. 1983. Xenopus transcription factor A requires zinc for binding to the 5 S RNA gene. J Biol Chem 258:14120–14125. [PubMed] [Google Scholar]
- 67.Wright C, Moore RD. 1990. Disulfiram treatment of alcoholism. Am J Med 88:647–655. doi: 10.1016/0002-9343(90)90534-K. [DOI] [PubMed] [Google Scholar]
- 68.Wang LH, Yang XY, Zhang X, Mihalic K, Fan YX, Xiao W, Howard OM, Appella E, Maynard AT, Farrar WL. 2004. Suppression of breast cancer by chemical modulation of vulnerable zinc fingers in estrogen receptor. Nat Med 10:40–47. doi: 10.1038/nm969. [DOI] [PubMed] [Google Scholar]
- 69.Lun X, Wells JC, Grinshtein N, King JC, Hao X, Dang NH, Wang X, Aman A, Uehling D, Datti A, Wrana JL, Easaw JC, Luchman A, Weiss S, Cairncross JG, Kaplan DR, Robbins SM, Senger DL. 2016. Disulfiram when combined with copper enhances the therapeutic effects of temozolomide for the treatment of glioblastoma. Clin Cancer Res 22:3860–3875. doi: 10.1158/1078-0432.CCR-15-1798. [DOI] [PubMed] [Google Scholar]
- 70.Rasmussen TA, Lewin SR. 2016. Shocking HIV out of hiding: where are we with clinical trials of latency reversing agents? Curr Opin HIV AIDS 11:394–401. doi: 10.1097/COH.0000000000000279. [DOI] [PubMed] [Google Scholar]
- 71.Huang M, Maynard A, Turpin JA, Graham L, Janini GM, Covell DG, Rice WG. 1998. Anti-HIV agents that selectively target retroviral nucleocapsid protein zinc fingers without affecting cellular zinc finger proteins. J Med Chem 41:1371–1381. doi: 10.1021/jm9708543. [DOI] [PubMed] [Google Scholar]
- 72.Elliott JH, McMahon JH, Chang CC, Lee SA, Hartogensis W, Bumpus N, Savic R, Roney J, Hoh R, Solomon A, Piatak M, Gorelick RJ, Lifson J, Bacchetti P, Deeks SG, Lewin SR. 2015. Short-term administration of disulfiram for reversal of latent HIV infection: a phase 2 dose-escalation study. Lancet HIV 2:e520–e529. doi: 10.1016/S2352-3018(15)00226-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lee YM, Duh Y, Wang ST, Lai MM, Yuan HS, Lim C. 2016. Using an old drug to target a new drug site: application of disulfiram to target the Zn-site in HCV NS5A protein. J Am Chem Soc 138:3856–3862. doi: 10.1021/jacs.6b00299. [DOI] [PubMed] [Google Scholar]
- 74.te Velthuis AJ, van den Worm SH, Sims AC, Baric RS, Snijder EJ, van Hemert MJ. 2010. Zn2+ inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog 6:e1001176. doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liao TL, Wu CY, Su WC, Jeng KS, Lai MM. 2010. Ubiquitination and deubiquitination of NP protein regulates influenza A virus RNA replication. EMBO J 29:3879–3890. doi: 10.1038/emboj.2010.250. [DOI] [PMC free article] [PubMed] [Google Scholar]