ABSTRACT
Ebola virus (EBOV) is a member of the Filoviridae family and the cause of hemorrhagic fever outbreaks. The EBOV VP40 (eVP40) matrix protein is the main driving force for virion assembly and budding. Indeed, expression of eVP40 alone in mammalian cells results in the formation and budding of virus-like particles (VLPs) which mimic the budding process and morphology of authentic, infectious EBOV. To complete the budding process, eVP40 utilizes its PPXY L-domain motif to recruit a specific subset of host proteins containing one or more modular WW domains that then function to facilitate efficient production and release of eVP40 VLPs. In this report, we identified additional host WW-domain interactors by screening for potential interactions between mammalian proteins possessing one or more WW domains and WT or PPXY mutant peptides of eVP40. We identified the HECT family E3 ubiquitin ligase WWP1 and all four of its WW domains as strong interactors with the PPXY motif of eVP40. The eVP40-WWP1 interaction was confirmed by both peptide pulldown and coimmunoprecipitation assays, which also demonstrated that modular WW domain 1 of WWP1 was most critical for binding to eVP40. Importantly, the eVP40-WWP1 interaction was found to be biologically relevant for VLP budding since (i) small interfering RNA (siRNA) knockdown of endogenous WWP1 resulted in inhibition of eVP40 VLP egress, (ii) coexpression of WWP1 and eVP40 resulted in ubiquitination of eVP40 and a subsequent increase in eVP40 VLP egress, and (iii) an enzymatically inactive mutant of WWP1 (C890A) did not ubiquitinate eVP40 or enhance eVP40 VLP egress. Last, our data show that ubiquitination of eVP40 by WWP1 enhances egress of VLPs and concomitantly decreases cellular levels of higher-molecular-weight oligomers of eVP40. In sum, these findings contribute to our fundamental understanding of the functional interplay between host E3 ligases, ubiquitination, and regulation of EBOV VP40-mediated egress.
IMPORTANCE Ebola virus (EBOV) is a high-priority, emerging human pathogen that can cause severe outbreaks of hemorrhagic fever with high mortality rates. As there are currently no approved vaccines or treatments for EBOV, a better understanding of the biology and functions of EBOV-host interactions that promote or inhibit viral budding is warranted. Here, we describe a physical and functional interaction between EBOV VP40 (eVP40) and WWP1, a host E3 ubiquitin ligase that ubiquitinates VP40 and regulates VLP egress. This viral PPXY-host WW domain-mediated interaction represents a potential new target for host-oriented inhibitors of EBOV egress.
KEYWORDS: E3 ubiquitin ligase, L-domain, PPXY, VLPs, VP40, WW domain, WWP1, budding, Ebola virus
INTRODUCTION
Filoviruses (Ebola and Marburg) cause sporadic outbreaks of hemorrhagic fever in primates and humans, the severity of which was highlighted in the most recent and largest outbreak of Ebola virus (EBOV) in West Africa in 2014 to 2015. The intersection between filoviral and host proteins that promote viral budding remains of widespread interest since a better understanding of these interactions and the mechanisms that govern them will provide new insights into viral biology and pathogenesis.
The filoviral VP40 matrix protein is the most abundant structural protein, and expression of VP40 alone drives assembly and budding of virus-like particles (VLPs) in mammalian cells (1–10). To facilitate the efficient release (pinching off) of VLPs, EBOV VP40 (eVP40) recruits multiple host proteins via its late (L) budding domains (PPXY and/or PTAP) during the final stages of virion assembly and egress (1, 7, 9–15). For example, the eVP40 PPXY L-domain motif interacts with HECT family E3 ubiquitin (Ub) ligases Nedd4 (1, 6, 13, 16–20) and Itch (21), as well as the cytoskeletal remodeling protein IQGAP1 (12). In general, these virus-host interactions are advantageous for efficient virus production (1, 14, 15, 18, 19, 22–38). The eVP40 PPXY core motif recruits host proteins by interacting specifically with one or more of their modular WW domains (39–47). Indeed, there is an integral degree of specificity in these virus-host interactions as the eVP40 PPXY motif interacts physically and functionally with a highly select subset of WW-domain-bearing host proteins (21, 48, 49). Thus, it is of interest to identify and validate the repertoire of WW-domain-containing host protein interactors that may influence or regulate eVP40-mediated egress.
In this report, we used both a wild-type (WT) and PPXY mutant peptide of eVP40 to screen a glutathione S-transferase (GST) array of 115 mammalian proteins containing one or more Src homology 3 (SH3) or WW domain modules (50). Using this unbiased approach, we identified WWP1 (WW-domain-containing E3 ubiquitin protein ligase 1) as a specific interactor with the eVP40 PPXY motif. WWP1, like Nedd4 and Itch, is a HECT family E3 ubiquitin ligase expressed in a wide array of tissues (e.g., testis, placenta, spinal cord, and retina) and is involved in regulating cellular and viral processes (24, 51–57). We found that all four tandem WW domains WW1 to WW4 [WW1–4] of WWP1 were able to interact with the eVP40 PPXY motif and that WWP1 WW domain 1 appears to be the most important domain for binding to eVP40. In addition, we showed that expression of wild-type WWP1 (WWP1-WT) enhanced egress of eVP40 VLPs and, importantly, that the E3 ligase activity was required for the enhancement of VLP egress. Indeed, expression of the enzymatically inactive WWP1 mutant, WWP1-C890A, did not enhance eVP40 VLP egress although the mutant remained capable of physically interacting with eVP40. Finally, we showed that WWP1-WT ubiquitinated eVP40 and that the presence of higher-molecular-weight (MW) oligomers of eVP40 was reduced significantly in the presence of WWP1-WT but not in the presence of WWP1-C890A. These findings indicate that a functional interaction between eVP40 and WWP1 leads to eVP40 ubiquitination, likely promoting more efficient self-assembly, maturation, and scission of eVP40 VLPs from the plasma membrane (58–61).
RESULTS
Screening of proline-rich reader arrays using eVP40 PPXY peptides.
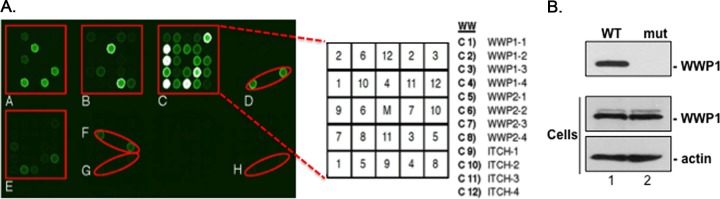
As described previously (21, 49) we fluorescently labeled biotinylated peptides containing either the WT (MRRVILPTAPPEYMEAI) or mutated PPXY motif (MRRVILPTAAAEAMEAI; motif in boldface) and used them to screen a proline-rich reading array composed of 115 WW-domain- and 40 SH3-domain-containing proteins to detect novel host interactors (Fig. 1). We identified a limited subset of WW-domain interactors with the WT eVP40 peptide (Fig. 1A, shown in red boxes or ovals) and no WW-domain interactors with the PPXY mutant peptide. Neither peptide interacted with any of the displayed SH3 domains. We found that the WT peptide interacted with all four of the tandem WW domains within host proteins WWP1 and WWP2, as well as with Itch, as we described previously (Fig. 1A, block C) (21). Here, we will characterize further the physical and functional interactions between eVP40 and WWP1.
FIG 1.
Proline-rich reading array screen and peptide pulldown. (A) Use of biotinylated eVP40 WT (MRRVILPTAPPEYMEAI[Lys-biotin]) peptide (50 μg) to screen a proline-rich reading array. The GST-WW domain fusion proteins are arrayed in duplicate and at different angles, as indicated in enlarged box C. Box C shows duplicate samples of all four WW domains from WWP1, WWP2, and ITCH as indicated. Additional positive interactions are indicated in the highlighted red boxes and ovals (A to H). The eVP40 mutant peptide (MRRVILPTAAAEAMEAI[Lys-biotin]) did not interact with any GST-WW domain fusion protein (data not shown). (B) Exogenously expressed FLAG-tagged WWP1-WT was pulled down with streptavidin beads bound to either eVP40 WT (WT) or PPXY mutant (mut) peptides and detected by Western blotting using anti-Flag antiserum (top). Expression controls for WWP1 and actin are shown (bottom).
To further evaluate the interaction between eVP40 PPXY and the host WWP1, we used a peptide pulldown assay of WWP1 transfected cell extracts (Fig. 1B). Briefly, HEK293T cells were transfected with FLAG-tagged WWP1-WT, and the cell extract was incubated with streptavidin beads bound with either eVP40 WT or PPXY mutant peptides (Fig. 1B). WWP1-WT was pulled down by the eVP40 WT peptide but not by the eVP40 PPXY mutant peptide, as determined by Western blotting (Fig. 1B). The selective interaction between the eVP40 WT peptide and the WW domains of WWP1 (Fig. 1A) in conjunction with the results of the peptide pulldown assay (Fig. 1B) suggest that an eVP40-WWP1 interaction is likely to occur in mammalian cells.
IP/Western analysis of eVP40 and WWP1.
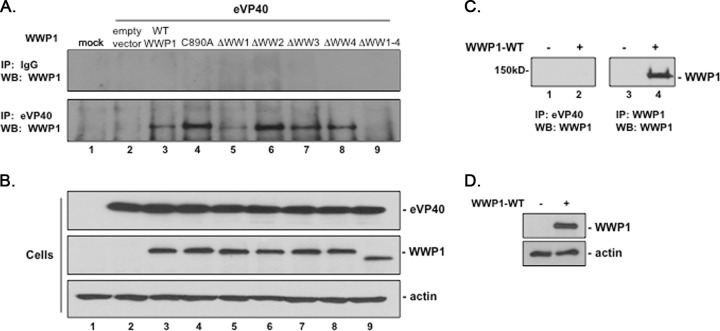
We used an immunoprecipitation (IP)/Western assay to validate the interaction of full-length eVP40 and WWP1 proteins in mammalian cells as predicted by the results described above. Briefly, HEK293T cells were mock transfected or transfected with eVP40 WT plus either pCAGGS vector alone, WWP1-WT, WWP1-C890A, or one of the WW-domain deletion mutants of WWP1, as indicated in Fig. 2A. Cell extracts were immunoprecipitated with either normal IgG (Fig. 2A, top panel) or polyclonal anti-eVP40 antiserum (Fig. 2A, bottom panel), and FLAG-tagged WWP1 was detected in precipitated samples by Western blotting using anti-FLAG antiserum (Fig. 2A). With the exception of mutant WWP1-ΔWW1–4 (Fig. 2A, lane 9), all other variations of WWP1 were detected, albeit at different levels, in eVP40 precipitates (Fig. 2A, lanes 3 to 8). Of note was the strong interaction detected between eVP40 and both the enzymatically inactive mutant WWP1-C890A and particularly WWP1-ΔWW2 (Fig. 2A, lane 4), and the weakest interaction was detected between the eVP40 and mutant WWP1-ΔWW1 (lane 5). One possibility for the robust interaction between VP40 and WWP1-ΔWW2 is that deletion of WW2 may alter the structure of WWP1 such that the most important interacting modular domain (WW1) now becomes more accessible or more efficient at binding to the PPXY motif of VP40. Expression controls for eVP40 and the various WWP1 proteins (Fig. 2B), as well as controls to confirm that anti-eVP40 antiserum does not pull down WWP1 (Fig. 2C), are shown. In sum, these results indicated the following: (i) eVP40 WT interacted with both WT and enzymatically inactive forms of WWP1, (ii) eVP40 WT interacted with all four individual WW-domain deletion mutants, (iii) eVP40 WT did not interact with mutant WWP1-ΔWW1–4 lacking all four WW domains, and (iv) WW domain 1 of WWP1 appears to be the most important modular domain for mediating eVP40-WWP1 binding.
FIG 2.
IP/Western analysis of WW-domain-dependent interactions between WWP1 and eVP40. (A) HEK293T cells were mock transfected or transfected with the indicated plasmids; extracts were first immunoprecipitated (IP) with either nonspecific (IgG) or polyclonal anti-eVP40 antiserum, and the indicated Flag-tagged WWP1 protein was detected in precipitated samples by Western blotting (WB) using anti-Flag antiserum (lanes 2 to 9). (B) Western blots of expression controls for eVP40, the indicated WWP1 protein, and actin. (C) HEK293T cells were mock transfected (lanes 1 and 3) or transfected with HA-tagged WWP1-WT (lanes 2 and 4). Cell extracts were first immunoprecipitated with either anti-eVP40 antiserum (lanes 1 and 2) or anti-HA (WWP1) antiserum (lanes 3 and 4), and HA-tagged WWP1 was detected in precipitated samples by Western blotting. (D) Western blots of expression controls for WWP1 and actin in cell extracts used in the experiment shown in panel C.
Colocalization of eVP40 and WWP1 visualized using confocal microscopy.
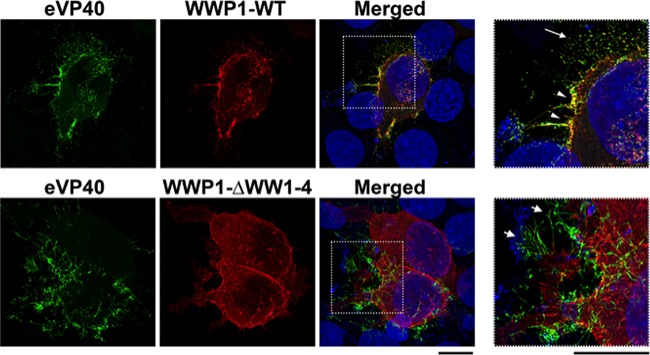
We next used confocal microscopy to visualize the intracellular localization patterns of eVP40, WWP1-WT, and mutant WWP1-ΔWW1–4 in HEK293T cells (Fig. 3). Transfected cells were fixed and permeabilized at 16 h posttransfection, and specific antisera were used to detect eVP40 (green) and Flag-tagged WWP1 (red) (Fig. 3). Clear evidence of eVP40 and WWP1-WT colocalization (yellow) was observed at the plasma membrane of cotransfected cells (Fig. 3, top row, white arrowheads), whereas little to no evidence of colocalization was observed in cells expressing eVP40 plus the mutant WWP1-ΔWW1–4 (Fig. 3, bottom row). In addition, efficient budding of eVP40 VLPs was observed from cells coexpressing WWP1-WT, as evidenced by released VLPs (Fig. 3, top row, white arrow), whereas budding of eVP40 VLPs appeared to be restricted in cells coexpressing mutant WWP1-ΔWW1–4, as evidenced by an abundance of long, thin VLP projections from the cell surface (Fig. 3, bottom row, white arrows). These data correlate well with those described above and indicate that WWP1 interacts with eVP40 and colocalizes with eVP40 at the site of VLP egress in a WW-domain-dependent manner to promote VLP budding.
FIG 3.
Confocal microscopy to visualize colocalization of eVP40 and WWP1. HEK293T cells were cotransfected with eVP40 plus WWP1-WT (top) or eVP40 plus mutant WWP1-ΔWW1–4 (bottom). Deconvolved confocal images are shown for eVP40 alone (green), WWP1-WT or WWP1-ΔWW1–4 alone (red), and merged panels, with the boxed insets enlarged. The white arrowheads highlight colocalization (yellow) of eVP40 and WWP1, the long white arrow highlights budding VLPs, and the short white arrows highlight long, thin projections typical of inhibited VLP budding.
Endogenous WWP1 is required for efficient egress of eVP40 VLPs.
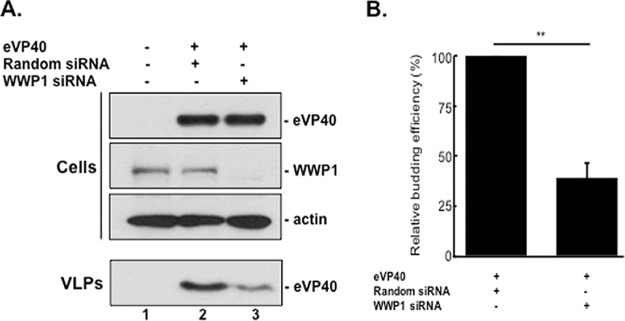
We next sought to confirm whether endogenously expressed WWP1 could influence eVP40 VLP budding. Toward this end, we used a small interfering RNA (siRNA) assay to specifically knock down expression of endogenous WWP1 in eVP40 transfected cells. Briefly, HEK293T cells were mock transfected or transfected with eVP40 WT plus random or WWP1-specific siRNAs, and cell extracts and VLPs were harvested and subjected to Western blot analysis (Fig. 4A). We observed a >90% knockdown of endogenous expression of WWP1 in siRNA-treated cells as confirmed by Western analysis (Fig. 4A, lane 3). Moreover, in multiple experiments, knockdown of endogenous WWP1 led to an approximately 3-fold reduction in eVP40 VLPs (Fig. 4B) compared to the level in random siRNA controls (Fig. 4A, compare lanes 2 and 3). These results indicate that expression of endogenous WWP1 is important for efficient egress of eVP40 VLPs.
FIG 4.
siRNA knockdown of endogenous WWP1 regulates eVP40 VLP egress. (A) Western blot of cell extracts and VLPs from HEK293T cells mock transfected (lane 1) or transfected with the indicated siRNAs and eVP40 (lanes 2 and 3). (B) Relative budding efficiency or mean relative VLP intensity (normalized to that of random siRNA) of eVP40 VLPs from cells treated with a WWP1-specific siRNA relative to that from a random siRNA control. Error bars represent the standard deviation of the mean from three independent experiments (n = 3). **, P = 0.002.
WWP1 regulates egress of eVP40 VLPs.
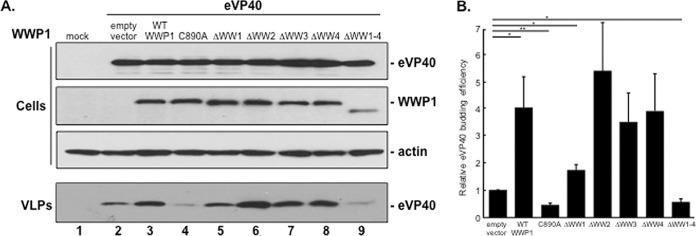
We next sought to evaluate how WT or mutant forms of WWP1 would influence egress of eVP40 VLPs in a functional budding assay. Briefly, HEK293T cells were mock transfected or transfected with eVP40 WT plus either the pCAGGS vector alone, WWP1-WT, WWP1-C890A, or one of the WW-domain deletion mutants of WWP1, as indicated in Fig. 5A. Cell extracts and VLPs were harvested at 16 h posttransfection, and the proteins were detected by Western analysis (Fig. 5A, cells and VLPs). Levels of the expression controls for eVP40, the various forms of WWP1, and actin were equivalent in all samples; however, significant differences were observed in the levels of eVP40 in VLPs (Fig. 5A). For example, we observed a reproducible enhancement of eVP40 VLP egress in the presence of exogenous WWP1-WT compared to the level with the eVP40-alone control (Fig. 5A, compare lanes 2 and 3). In contrast, we observed a consistent decrease of eVP40 VLP egress in the presence of exogenous WWP1-C890A mutant compared to the level in the eVP40-alone control (Fig. 5A, compare lanes 2 and 4). Budding of eVP40 VLPs was generally enhanced in the presence of all four of the individual WW-domain deletion mutants of WWP1 compared to that with eVP40 alone; however, the level of enhancement was repeatedly lowest in samples expressing WWP1-ΔWW1 (Fig. 5A, compare lane 2 and lanes 5 to 8). Last, as expected, budding of eVP40 VLPs was greatly reduced in samples expressing mutant WWP1-ΔWW1–4 lacking all four WW domains (Fig. 5A, compare lanes 2 and 9). Quantification of the relative budding efficiency of eVP40 alone or in combination with the plasmids indicated in Fig. 5B. In sum, results from this functional assay correlate well with the observed physical interactions between eVP40 and WWP1 and indicate that WWP1 expression regulates egress of eVP40 VLPs in a PPXY WW-domain-dependent manner.
FIG 5.
Enzymatically active WWP1 promotes enhanced eVP40 VLP egress in a WW-domain-dependent manner. (A) Western analysis of cell extracts and VLPs from HEK293T cells mock transfected (lane 1) or transfected with eVP40 plus the indicated plasmids (lanes 2 to 9). (B) Budding efficiency or mean relative VLP intensity (normalized to empty vector) and standard error are shown for n = 5 experiments. *, P < 0.05; **, P < 0.01.
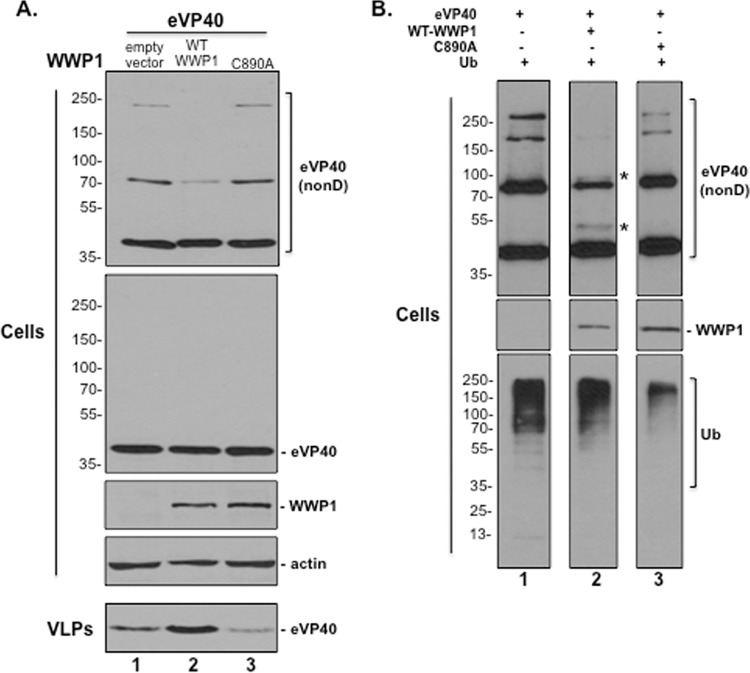
WWP1 ubiquitination of eVP40 leads to reduced expression of higher-MW forms of cellular eVP40 and increased VLP production.
WWP1 localizes in part to the plasma membrane (PM) (57), where eVP40 must self-assemble and homo-oligomerize properly to facilitate efficient VLP and virus egress (59, 61–64). Since expression of WWP1 enhances eVP40 VLP egress, we sought to determine whether enhanced egress correlated with WWP1-mediated ubiquitination of eVP40 and subsequent formation of eVP40 oligomers. Briefly, HEK293T cells were transfected with eVP40 plus an empty vector, WWP1-WT, or mutant WWP1-C890A, and both cells and VLPs were harvested and analyzed by SDS-PAGE under nondenaturing or denaturing conditions as indicated in Fig. 6A. In repeated experiments, we observed a marked decrease in expression levels of higher-MW forms of eVP40 migrating at approximately 80 and 240 kDa in the presence of WWP1-WT under nondenaturing conditions (Fig. 6A, top gel, lane 2), whereas the levels of these higher-MW species remained unaltered in the presence of empty vector (lane 1) and mutant WWP1-C890A (lane 3). The levels of monomeric eVP40 (40 kDa) remained essentially equivalent in all samples (Fig. 6A, top gel). The levels of eVP40, WWP1, and actin were equivalent in all samples under normal denaturing conditions (Fig. 6A, cells). As expected, the eVP40 level in VLPs was enhanced in the presence of WWP1-WT (Fig. 6A, VLPs, lane 2) and reduced in the presence of mutant WWP1-C890A (lane 3) compared to the level in the plasmid-alone control (lane 1). These results suggest that expression of WWP1-WT, but not WWP1-C890A, leads to a reduction in the levels of higher-MW oligomers of eVP40 in cell extracts, presumably due to the corresponding increase in the release of VLPs.
FIG 6.
WWP1 ubiquitinates eVP40 and reduces expression of higher-MW oligomers of eVP40. (A) Western analysis of cell extracts and VLPs from HEK293T cells transfected with eVP40 plus an empty vector (lane 1), WWP1-WT, or mutant WWP1-C890A. Cell extracts were harvested and analyzed under nondenaturing (nonD) conditions where indicated. Higher-molecular-mass forms of eVP40 were detected under nondenaturing conditions and migrated at approximately 80 and 240 kDa, compared to monomeric eVP40 detected at approximately 40 kDa. (B) Western analysis of cell extracts and VLPs from HEK293T cells transfected with eVP40 plus ubiquitin (Ub) alone (lane 1), WWP1-WT plus Ub (lane 2), or mutant WWP1-C890A plus Ub (lane 3). Cell extracts were harvested and analyzed under nondenaturing (nonD) conditions where indicated. The asterisks (*) represent potential ubiquitinated forms of eVP40 (lane 2).
Since the reduction of higher-MW oligomers of eVP40 occurred only in the presence of enzymatically active WWP1, we next examined the ubiquitination status of eVP40 in the presence of exogenously added ubiquitin (Fig. 6B). Briefly, HEK293T cells were transfected with eVP40 plus either ubiquitin (Ub) alone, WWP1-WT plus Ub, or mutant WWP1-C890A plus Ub, and cells were harvested and analyzed by SDS-PAGE under nondenaturing or denaturing conditions as indicated in Fig. 6B. As described above, higher-MW forms of eVP40 were reduced in the presence of Ub and WWP1-WT (Fig. 6B, lane 2) compared to those detected in samples expressing Ub alone (lane 1) or Ub and mutant WWP1-C890A (lane 3). In addition, protein bands corresponding in size to mono-ubiquitinated monomers and dimers of eVP40 were detected only in the WWP1-WT sample (Fig. 6B, lane 2, asterisks). Expression controls under denaturing conditions for WWP1 and the overall cellular ubiquitination profile in the cells are shown (Fig. 6B). Together, these results suggest that WWP1-WT ubiquitinates eVP40, which regulates budding efficiency, as evidenced by increased levels of eVP40 in budding VLPs and a corresponding decrease in levels of cellular eVP40 oligomers necessary for VLP assembly and maturation.
Effect of WWP1 expression on eVP40 oligomerization profiles.
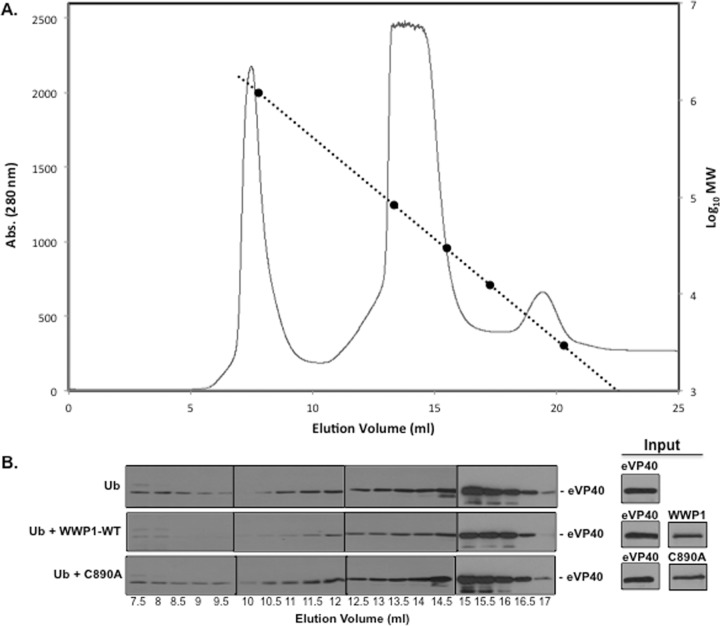
Next, we used gel filtration analysis to further examine the oligomerization profiles of eVP40 when it is coexpressed with WT or mutant WWP1. Briefly, HEK293T cells were transfected with eVP40 plus either Ub, Ub plus WWP1-WT, or Ub plus WWP1-C890A (Fig. 7). Cell extracts were clarified and filtered through a 0.22-μm-pore-size filter, and proteins were separated on a Superdex-200 10/30 high-resolution, fast protein liquid chromatography (FPLC) column, along with internal molecular weight standards (Fig. 7A). Samples from the eluted fractions were analyzed by SDS-PAGE and Western blotting using anti-eVP40 antiserum (Fig. 7B). The input levels of eVP40, WWP1-WT, and WWP1-C890A prior to gel filtration were detected by Western blotting (Fig. 7B). eVP40 WT eluted from the FPLC column in a major peak at 14.5 to 15.5 ml represented its monomeric form (Fig. 7B) while eVP40 WT eluted at 10.5 to 12 ml likely represents the hexameric form (∼240 kDa). Interestingly, eVP40 levels in the samples expressing Ub plus WWP1-WT were reduced slightly in the monomeric (14.5 to 15.5 ml) fractions and more significantly in the hexameric fractions (10.5 to 12 ml) compared to eVP40 levels in the same fractions of samples expressing Ub-alone and Ub plus WWP1-C890A (Fig. 7B). The observed reduction in hexameric forms of eVP40 correlated well with our results described in Fig. 6, and these findings further support the role for E3 ligase WWP1 and ubiquitination in facilitating and enhancing egress of eVP40 VLPs.
FIG 7.
Gel filtration analysis of eVP40 in the presence of WWP1-WT or mutant WWP1-C890A. HEK293T cells were transfected with eVP40 plus either Ub, Ub plus WWP1-WT, or Ub plus WWP1-C890A as indicated. Lysates were separated by size on a Superdex-200 10/30 high-resolution fast-performance liquid chromatography column. (A) The chromatogram for eVP40 elution is shown as absorbance (Abs) versus elution volume. Additionally, molecular weight standards were plotted along the dashed line as log values versus elution volume. (B) Western blot analysis for eVP40 in the indicated elution volumes. Input controls for eVP40, WWP1-WT, and WWP1-C890A are shown.
DISCUSSION
We have identified host WWP1, an HECT-type E3 ubiquitin ligase, as a physical and functional interactor with EBOV VP40. WWP1 contains an N-terminal C2 domain, a C-terminal HECT domain, and four tandem WW domains that mediate interactions with PPXY-containing substrates including, but not limited to, JunB, Smad2, p63, and ErbB4/HER4 (57). Indeed, WWP1 functions in multiple cellular processes, including protein degradation, signaling, and trafficking, and has been implicated previously in virus budding. As such, this ubiquitously expressed protein is a central player in pathways regulating transforming growth factor β (TGF-β) signaling, apoptosis, neurological diseases, epidermal growth factor (EGF) signaling, metastasis, and virus egress (24, 52–55, 57).
Here, we show that WWP1 expression enhances EBOV VLP egress in a PPXY/WW-domain-dependent manner (1, 6, 16–19, 21). Indeed, WWP1 contains four individual WW domains, and we show that all four WW domains can interact with the PPXY motif of eVP40, albeit WW domain 1 appears to be the most critical for mediating the eVP40-WWP1 interaction. In concurrence with coimmunoprecipitation analysis, results from confocal microscopy experiments demonstrated that WWP1 colocalizes with eVP40 at the site of budding at the plasma membrane in a WW-domain-dependent manner. The functional relevance of WWP1 expression to eVP40-mediated egress was demonstrated by quantifying VP40 VLP egress in the absence or presence of WT and mutant forms of WWP1, as well as by siRNA knockdown assays. The ability of WWP1 to enhance the efficiency of VLP maturation and egress was evident by the significant reduction in cellular expression of higher-MW oligomers of VP40 in the presence of WWP1-WT but not in the presence of inactive mutant WWP1-C890A, as shown by nondenaturing SDS-PAGE and gel filtration. Indeed, the detection of abundant VP40 oligomers in cells expressing WWP1 mutant C890A, but not WT WWP1, suggests that VP40 oligomers remain trapped at the plasma membrane in C890A-expressing cells due to a decreased level of VP40 ubiquitination, thereby decreasing the efficiency of VLP budding. Enhanced egress of VP40 VLPs was concomitant with WWP1-mediated ubiquitination of VP40.
The positive effect of host E3 ligase WWP1 on eVP40 VLP budding reported here further highlights the probudding function associated with the host ubiquitination process (1, 18, 21, 65). In contrast, we along with others identified a counteracting, “anti-budding” role for the interferon-induced, ubiquitin-like host protein ISG15 and the process of ISGylation (16, 17, 66). These mechanistically competing processes highlight the interplay between the virus and the host innate immune response in regulating eVP40-mediated egress during filovirus infection. A more comprehensive understanding of this virus-host interplay will be crucial for our understanding of filovirus pathogenesis and the development of novel antiviral strategies and treatments.
While the precise mechanism by which ubiquitin and E3 ubiquitin ligases facilitate efficient virus egress remains unclear, there does appear to be a general requirement for enzymatically active E3 ligases and subsequent mono-ubiquitination of viral matrix proteins. However, there is a plethora of host E3 ligases that can interact and ubiquitinate viral matrix proteins such as VP40. The seemingly global regulatory role of the various host E3 ubiquitin ligase interactors to enhance VLP egress may simply represent redundancy in the ability of eVP40 to hijack or recruit host E3 ligases to promote budding. Alternatively, eVP40 may interact with E3 ligases in a cell-type-specific manner during infection that is regulated by their levels of expression. As such, cell-type-specific expression levels of individual E3 ligases and the sequences/structures of their individual modular WW domains likely determine their ability to interact with eVP40 (67). Indeed, the observed selectivity and specificity of the eVP40 PPXY motif for binding to only a subset of mammalian WW domains in our WW- and SH3-domain array (Fig. 1) predict that these interacting host proteins are likely to be biologically relevant during the virus life cycle.
Due to the broad-spectrum nature of this virus-host interaction, the PPXY-WW domain interface represents an intriguing target for the identification and development of small-molecule inhibitors as potential broad-spectrum antiviral therapeutics (19, 68, 69). Indeed, we have identified lead candidate budding inhibitors that target and block the VP40/Nedd4 PPXY-WW domain interaction, resulting in inhibition of both VP40 VLPs and live virus egress (19, 20, 69). These host-oriented budding inhibitors would represent novel first-in-class therapeutics that could potentially target not only filoviruses but also other RNA viruses that utilize PPXY L-domain motifs to complete their budding and maturation processes. It will be of interest to determine whether our current lead PPXY inhibitors can also block VP40 interactions with WWP1 and Itch E3 ligases to similarly reduce virus budding and spread.
MATERIALS AND METHODS
Cell lines, plasmids, and reagents.
HEK293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), and penicillin (100 U/ml)-streptomycin (100 μg/ml) at 37°C in a humidified 5% CO2 incubator. The pCAGGs-based eVP40 WT expression plasmid has been described previously (1, 6, 17). Plasmids expressing FLAG-tagged WWP1-WT, WWP1-C890A, WWP1-ΔWW1, WWP1-ΔWW2, WWP1-ΔWW3, WWP1-ΔWW4, and WWP1-ΔWW1–4 were kindly provided by C. Prunier (INSERM UMR S 938, Université Pierre et Marie Curie, Paris, France), and a plasmid expressing hemagglutinin (HA)-tagged ubiquitin (Ub-HA) was kindly provided by H. Gottlinger (University of Massachusetts Medical School, Worcester, MA). Mouse anti-β-actin (A1978) antiserum was obtained from Sigma-Aldrich, mouse anti-Flag antiserum was obtained from Cell Biolabs, Inc., and polyclonal anti-eVP40 antiserum was obtained from ProSci, Inc.
Protein array experiments.
The proline-rich reading array containing WW and SH3 domains was codon optimized for bacterial expression and cloned into a pGex vector. All WW and SH3 domains were expressed as GST fusions in Escherichia coli and purified on glutathione-Sepharose beads. The recombinant domains were arrayed onto nitrocellulose-coated glass slides (OncyteAvid slides; Grace Bio-Labs, Bend, Oregon), using an Aushon 2470 arrayer with solid pins, as described previously (50). The fluorescent labeling of the biotinylated peptide probe and binding to slides have been described previously (50). Two EBOV VP40 peptides were tested on the array, the eVP40 WT (MRRVILPTAPPEYMEAI[Lys-biotin]) and eVP40 mutant (MRRVILPTAAAEAMEAI[Lys-biotin]) peptides. Fluorescence was detected using a GeneTAC LSIV scanner (Genomic Solutions).
GST peptide pulldown assays.
Streptavidin agarose beads (Millipore) were prewashed once with 1× mild buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.1% NP-40, 5 mM EDTA, 5 mM EGTA, 15 mM MgCl2), and 15 μg of the WT or PPXY mutant eVP40 peptide was incubated with the prewashed streptavidin beads in 500 μl of 1× mild buffer for 1 h at 4°C with rocking. The beads were washed three times with mild buffer and then incubated with cell extracts from WWP1-WT transfected HEK293T cells. The beads were then washed three times with 1× mild buffer and suspended in 30 μl of 2× loading buffer with boiling. Protein samples were analyzed by SDS-PAGE and Western blotting.
siRNA analysis.
HEK293T cells were plated in Opti-MEM in collagen-coated six-well plates and transfected two times with either a random control siRNA or WWP1-specific siRNA (Dharmacon, Inc.) at a final concentration of 200 nM using Lipofectamine (Invitrogen) at 2-day intervals. eVP40 WT plasmid DNA (0.5 μg) was transfected with the second round of siRNAs. Cell extracts and VLPs were harvested at 24 h posttransfection, and the proteins were detected in cell and VLP samples by Western blotting using specific antisera.
IP/Western analysis.
Human HEK293T cells were transfected with the plasmids indicated in Fig. 2A using Lipofectamine reagent (Invitrogen) and the protocol of the supplier. Cells were harvested and lysed in nondenaturing buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 1.0% Nonidet P-40 [NP-40], 2.0 mM EDTA, 2.0 mM EGTA, and 10% glycerol) at 18 to 20 h posttransfection. Cell lysates were clarified for 10 min at 3,000 rpm. Supernatants were incubated with anti-eVP40 or normal IgG (Cell Signaling) for 5 h at 4°C. Protein A agarose beads (Invitrogen) were added to the samples and incubated with agitation overnight at 4°C. The beads were washed five times in nondenaturing lysis buffer, suspended in loading buffer with boiling, and then fractionated by SDS-PAGE. Proteins were detected in precipitates by Western blotting using specific antisera.
VLP budding assays.
Filovirus VLP budding assays using HEK293T cells and eVP40 have been described previously (1, 6, 12, 19, 68). For nondenaturing conditions, cells were harvested and lysed in nondenaturing buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 1.0% Nonidet P-40 [NP-40], 2.0 mM EDTA, 2.0 mM EGTA, and 10% glycerol) at 20 h posttransfection. Cell lysates were clarified for 10 min at 3,000 rpm. Protein samples were prepared with NuPAGE lithium dodecyl sulfate (LDS) sample buffer without dithiothreitol (DTT) and boiling.
Gel filtration assay.
HEK293T cells were transfected with the plasmids indicated in Fig. 7B for 24 h, and cells were lysed with phosphate-buffered saline (PBS) containing 1% NP-40 plus protease inhibitors and incubated at 4°C for 30 min. Cleared lysates were filtered through a 0.22-μm-pore-size filter and separated on a Superdex-200 10/30 high-resolution, fast-performance liquid chromatography column (GE Healthcare) using an ÄKTA 10 purifier system (GE Healthcare). Eluted proteins were collected in 0.5-ml fractions and analyzed by SDS-PAGE and Western blotting with anti-eVP40 antiserum. The chromatogram plotting absorbance (280 nm) versus elution volume was generated with Unicorn software. Molecular mass standards depicted on the chromatogram had molecular masses of 670, 158, 44, 17, and 1.35 kDa (Bio-Rad).
Confocal microscopy.
HEK293T cells on glass coverslips were transfected with the plasmids indicated in Fig. 3 for 16 h. Cells were fixed and permeabilized with 4% paraformaldehyde (Affymetrix) and 0.1% Triton X-100 and then incubated in 1× PBS containing 5% dried milk. Cells were incubated with rabbit polyclonal anti-eVP40 antiserum followed by staining with Alexa 488-conjugated goat anti-rabbit secondary antibody (green) and with mouse anti-Flag antiserum to detect WWP1, followed by staining with Alexa 594-conjugated goat anti-mouse secondary antibody (red). Images were acquired on a Leica SP5 inverted confocal microscope with a 100× (numerical aperture [NA], 1.46) objective lens. The confocal images were subsequently deconvolved with Huygens Essential deconvolution software.
Statistical analysis.
Significance for all statistical tests was determined at P values of <0.05 and <0.01. Western blot protein intensities were quantified using ImageJ software, and intensities were normalized to the intensity of the empty vector within each experiment (n = 5) before values were compared across various constructs using Welch's t test.
ACKNOWLEDGMENTS
We thank C. Prunier for kindly providing reagents and members of the Freedman and Harty labs for helpful comments.
This work was supported in part by NIH grants AI102104, AI113952, and AI103785 to R.N.H., by CPRIT grant RP13042 to M.T.B. for the protein array analysis, and by seed grants from The National University of Singapore Medical School and Mechanobiology Institute in Singapore to M.S. Imaging experiments were performed in the PennVet Imaging Core Facility on instrumentation supported by NIH S10RR027128, the School of Veterinary Medicine, the University of Pennsylvania, and the Commonwealth of Pennsylvania.
REFERENCES
- 1.Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. 2000. A PPXY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci U S A 97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin-Serrano J, Zang T, Bieniasz PD. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med 7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 3.Timmins J, Scianimanico S, Schoehn G, Weissenhorn W. 2001. Vesicular release of ebola virus matrix protein VP40. Virology 283:1–6. doi: 10.1006/viro.2001.0860. [DOI] [PubMed] [Google Scholar]
- 4.Kolesnikova L, Bugany H, Klenk HD, Becker S. 2002. VP40, the matrix protein of Marburg virus, is associated with membranes of the late endosomal compartment. J Virol 76:1825–1838. doi: 10.1128/JVI.76.4.1825-1838.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noda T, Sagara H, Suzuki E, Takada A, Kida H, Kawaoka Y. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J Virol 76:4855–4865. doi: 10.1128/JVI.76.10.4855-4865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Licata JM, Simpson-Holley M, Wright NT, Han Z, Paragas J, Harty RN. 2003. Overlapping motifs (PTAP and PPEY) within the Ebola virus VP40 protein function independently as late budding domains: involvement of host proteins TSG101 and VPS-4. J Virol 77:1812–1819. doi: 10.1128/JVI.77.3.1812-1819.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jasenosky LD, Kawaoka Y. 2004. Filovirus budding. Virus Res 106:181–188. doi: 10.1016/j.virusres.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 8.Swenson DL, Warfield KL, Kuehl K, Larsen T, Hevey MC, Schmaljohn A, Bavari S, Aman MJ. 2004. Generation of Marburg virus-like particles by co-expression of glycoprotein and matrix protein. FEMS Immunol Med Microbiol 40:27–31. doi: 10.1016/S0928-8244(03)00273-6. [DOI] [PubMed] [Google Scholar]
- 9.Hartlieb B, Weissenhorn W. 2006. Filovirus assembly and budding. Virology 344:64–70. doi: 10.1016/j.virol.2005.09.018. [DOI] [PubMed] [Google Scholar]
- 10.Noda T, Ebihara H, Muramoto Y, Fujii K, Takada A, Sagara H, Kim JH, Kida H, Feldmann H, Kawaoka Y. 2006. Assembly and budding of Ebolavirus. PLoS Pathog 2:e99. doi: 10.1371/journal.ppat.0020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Z, Madara JJ, Liu Y, Liu W, Ruthel G, Freedman BD, Harty RN. 2015. ALIX rescues budding of a double PTAP/PPEY L-domain deletion mutant of Ebola VP40: a role for ALIX in Ebola virus egress. J Infect Dis 212(Suppl 2):S138–S145. doi: 10.1093/infdis/jiu838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Qu Y, Liu Y, Jambusaria R, Han Z, Ruthel G, Freedman BD, Harty RN. 2013. Host IQGAP1 and Ebola virus VP40 interactions facilitate virus-like particle egress. J Virol 87:7777–7780. doi: 10.1128/JVI.00470-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Lee MS, Olson MA, Harty RN. 2011. Bimolecular complementation to visualize filovirus VP40-host complexes in live mammalian cells: toward the identification of budding inhibitors. Adv Virol 2011:341816. doi: 10.1155/2011/341816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yasuda J, Nakao M, Kawaoka Y, Shida H. 2003. Nedd4 regulates egress of Ebola virus-like particles from host cells. J Virol 77:9987–9992. doi: 10.1128/JVI.77.18.9987-9992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Timmins J, Schoehn G, Ricard-Blum S, Scianimanico S, Vernet T, Ruigrok RW, Weissenhorn W. 2003. Ebola virus matrix protein VP40 interaction with human cellular factors Tsg101 and Nedd4. J Mol Biol 326:493–502. doi: 10.1016/S0022-2836(02)01406-7. [DOI] [PubMed] [Google Scholar]
- 16.Malakhova OA, Zhang DE. 2008. ISG15 inhibits Nedd4 ubiquitin E3 activity and enhances the innate antiviral response. J Biol Chem 283:8783–8787. doi: 10.1074/jbc.C800030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okumura A, Pitha PM, Harty RN. 2008. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A 105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Urata S, Yasuda J. 2010. Regulation of Marburg virus (MARV) budding by Nedd4.1: a different WW domain of Nedd4.1 is critical for binding to MARV and Ebola virus VP40. J Gen Virol 91:228–234. doi: 10.1099/vir.0.015495-0. [DOI] [PubMed] [Google Scholar]
- 19.Han Z, Lu J, Liu Y, Davis B, Lee MS, Olson MA, Ruthel G, Freedman BD, Schnell MJ, Wrobel JE, Reitz AB, Harty RN. 2014. Small-molecule probes targeting the viral PPXY-host Nedd4 interface block egress of a broad range of RNA viruses. J Virol 88:7294–7306. doi: 10.1128/JVI.00591-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loughran HM, Han Z, Wrobel JE, Decker SE, Ruthel G, Freedman BD, Harty RN, Reitz AB. 2016. Quinoxaline-based inhibitors of Ebola and Marburg VP40 egress. Bioorg Med Chem Lett 26:3429–3435. doi: 10.1016/j.bmcl.2016.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Z, Sagum CA, Bedford MT, Sidhu SS, Sudol M, Harty RN. 2016. ITCH E3 ubiquitin ligase interacts with Ebola virus VP40 to regulate budding. J Virol 90:9163–9171. doi: 10.1128/JVI.01078-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lewis B, Whitney S, Hudacik L, Galmin L, Huaman MC, Cristillo AD. 2014. Nedd4-mediated increase in HIV-1 Gag and Env proteins and immunity following DNA-vaccination of BALB/c mice. PLoS One 9:e91267. doi: 10.1371/journal.pone.0091267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sette P, Nagashima K, Piper RC, Bouamr F. 2013. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology 10:79. doi: 10.1186/1742-4690-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhadina M, Bieniasz PD. 2010. Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog 6:e1001153. doi: 10.1371/journal.ppat.1001153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss ER, Popova E, Yamanaka H, Kim HC, Huibregtse JM, Gottlinger H. 2010. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog 6:e1001107. doi: 10.1371/journal.ppat.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. 2010. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4-1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol 84:8181–8192. doi: 10.1128/JVI.00634-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Usami Y, Popov S, Popova E, Inoue M, Weissenhorn W, Gottlinger HG. 2009. The ESCRT pathway and HIV-1 budding. Biochem Soc Trans 37:181–184. doi: 10.1042/BST0370181. [DOI] [PubMed] [Google Scholar]
- 28.Calistri A, Del Vecchio C, Salata C, Celestino M, Celegato M, Gottlinger H, Palu G, Parolin C. 2009. Role of the feline immunodeficiency virus L-domain in the presence or absence of Gag processing: involvement of ubiquitin and Nedd4-2s ligase in viral egress. J Cell Physiol 218:175–182. doi: 10.1002/jcp.21587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pincetic A, Medina G, Carter C, Leis J. 2008. Avian sarcoma virus and human immunodeficiency virus, type 1 use different subsets of ESCRT proteins to facilitate the budding process. J Biol Chem 283:29822–29830. doi: 10.1074/jbc.M804157200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chung HY, Morita E, von Schwedler U, Muller B, Krausslich HG, Sundquist WI. 2008. NEDD4L overexpression rescues the release and infectivity of human immunodeficiency virus type 1 constructs lacking PTAP and YPXL late domains. J Virol 82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhadina M, McClure MO, Johnson MC, Bieniasz PD. 2007. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc Natl Acad Sci U S A 104:20031–20036. doi: 10.1073/pnas.0708002104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urata S, Noda T, Kawaoka Y, Yokosawa H, Yasuda J. 2006. Cellular factors required for Lassa virus budding. J Virol 80:4191–4195. doi: 10.1128/JVI.80.8.4191-4195.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klinger PP, Schubert U. 2005. The ubiquitin-proteasome system in HIV replication: potential targets for antiretroviral therapy. Expert Rev Anti Infect Ther 3:61–79. doi: 10.1586/14787210.3.1.61. [DOI] [PubMed] [Google Scholar]
- 34.Vana ML, Tang Y, Chen A, Medina G, Carter C, Leis J. 2004. Role of Nedd4 and ubiquitination of Rous sarcoma virus Gag in budding of virus-like particles from cells. J Virol 78:13943–13953. doi: 10.1128/JVI.78.24.13943-13953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakurai A, Yasuda J, Takano H, Tanaka Y, Hatakeyama M, Shida H. 2004. Regulation of human T-cell leukemia virus type 1 (HTLV-1) budding by ubiquitin ligase Nedd4. Microbes Infect 6:150–156. doi: 10.1016/j.micinf.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 36.Yasuda J, Hunter E, Nakao M, Shida H. 2002. Functional involvement of a novel Nedd4-like ubiquitin ligase on retrovirus budding. EMBO Rep 3:636–640. doi: 10.1093/embo-reports/kvf132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harty RN, Brown ME, McGettigan JP, Wang G, Jayakar HR, Huibregtse JM, Whitt MA, Schnell MJ. 2001. Rhabdoviruses and the cellular ubiquitin-proteasome system: a budding interaction. J Virol 75:10623–10629. doi: 10.1128/JVI.75.22.10623-10629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for Gag budding from cells. Proc Natl Acad Sci U S A 98:11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bork P, Sudol M. 1994. The WW domain: a signalling site in dystrophin? Trends Biochem Sci 19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 40.Chen HI, Sudol M. 1995. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci U S A 92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. 1995. Characterization of a novel protein-binding module—the WW domain. FEBS Lett 369:67–71. doi: 10.1016/0014-5793(95)00550-S. [DOI] [PubMed] [Google Scholar]
- 42.Sudol M. 1996. The WW module competes with the SH3 domain? Trends Biochem Sci 21:161–163. doi: 10.1016/S0968-0004(96)30018-2. [DOI] [PubMed] [Google Scholar]
- 43.Sudol M. 1996. Structure and function of the WW domain. Prog Biophys Mol Biol 65:113–132. doi: 10.1016/S0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 44.Sudol M, Sliwa K, Russo T. 2001. Functions of WW domains in the nucleus. FEBS Lett 490:190–195. doi: 10.1016/S0014-5793(01)02122-6. [DOI] [PubMed] [Google Scholar]
- 45.Ilsley JL, Sudol M, Winder SJ. 2002. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell Signal 14:183–189. doi: 10.1016/S0898-6568(01)00236-4. [DOI] [PubMed] [Google Scholar]
- 46.Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L, James M, Davis R, Sudol M, Rodwell J, Herrero JJ. 2004. A map of WW domain family interactions. Proteomics 4:643–655. doi: 10.1002/pmic.200300632. [DOI] [PubMed] [Google Scholar]
- 47.Sudol M, Hunter T. 2000. NeW wrinkles for an old domain. Cell 103:1001–1004. doi: 10.1016/S0092-8674(00)00203-8. [DOI] [PubMed] [Google Scholar]
- 48.Einbond A, Sudol M. 1996. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Lett 384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- 49.Liang J, Sagum CA, Bedford MT, Sidhu SS, Sudol M, Han Z, Harty RN. 2017. Chaperone-mediated autophagy protein BAG3 negatively regulates Ebola and Marburg VP40-mediated egress. PLoS Pathog 13:e1006132. doi: 10.1371/journal.ppat.1006132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espejo A, Cote J, Bednarek A, Richard S, Bedford MT. 2002. A protein-domain microarray identifies novel protein-protein interactions. Biochem J 367:697–702. doi: 10.1042/bj20020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia ML, Reynolds TD, Mothes W, Robek MD. 2013. Functional characterization of the putative hepatitis B virus core protein late domain using retrovirus chimeras. PLoS One 8:e72845. doi: 10.1371/journal.pone.0072845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rauch S, Martin-Serrano J. 2011. Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J Virol 85:3546–3556. doi: 10.1128/JVI.02045-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heidecker G, Lloyd PA, Soheilian F, Nagashima K, Derse D. 2007. The role of WWP1-Gag interaction and Gag ubiquitination in assembly and release of human T-cell leukemia virus type 1. J Virol 81:9769–9777. doi: 10.1128/JVI.00642-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. 2005. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol 168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heidecker G, Lloyd PA, Fox K, Nagashima K, Derse D. 2004. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J Virol 78:6636–6648. doi: 10.1128/JVI.78.12.6636-6648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Galinier R, Gout E, Lortat-Jacob H, Wood J, Chroboczek J. 2002. Adenovirus protein involved in virus internalization recruits ubiquitin-protein ligases. Biochemistry 41:14299–14305. doi: 10.1021/bi020125b. [DOI] [PubMed] [Google Scholar]
- 57.Zhi X, Chen C. 2012. WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol Life Sci 69:1425–1434. doi: 10.1007/s00018-011-0871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bornholdt ZA, Noda T, Abelson DM, Halfmann P, Wood MR, Kawaoka Y, Saphire EO. 2013. Structural rearrangement of Ebola virus VP40 begets multiple functions in the virus life cycle. Cell 154:763–774. doi: 10.1016/j.cell.2013.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gc JB, Gerstman BS, Stahelin RV, Chapagain PP. 2016. The Ebola virus protein VP40 hexamer enhances the clustering of PI(4,5)P2 lipids in the plasma membrane. Phys Chem Chem Phys 18:28409–28417. doi: 10.1039/C6CP03776C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gc JB, Johnson KA, Husby ML, Frick CT, Gerstman BS, Stahelin RV, Chapagain PP. 2016. Interdomain salt-bridges in the Ebola virus protein VP40 and their role in domain association and plasma membrane localization. Protein Sci 25:1648–1658. doi: 10.1002/pro.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson KA, Taghon GJ, Scott JL, Stahelin RV. 2016. The Ebola virus matrix protein, VP40, requires phosphatidylinositol 4,5-bisphosphate (PI(4,5)P2) for extensive oligomerization at the plasma membrane and viral egress. Sci Rep 6:19125. doi: 10.1038/srep19125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adu-Gyamfi E, Digman MA, Gratton E, Stahelin RV. 2012. Investigation of Ebola VP40 assembly and oligomerization in live cells using number and brightness analysis. Biophys J 102:2517–2525. doi: 10.1016/j.bpj.2012.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adu-Gyamfi E, Soni SP, Xue Y, Digman MA, Gratton E, Stahelin RV. 2013. The Ebola virus matrix protein penetrates into the plasma membrane: a key step in viral protein 40 (VP40) oligomerization and viral egress. J Biol Chem 288:5779–5789. doi: 10.1074/jbc.M112.443960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stahelin RV. 2014. Membrane binding and bending in Ebola VP40 assembly and egress. Front Microbiol 5:300. doi: 10.3389/fmicb.2014.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu Y, Harty RN. 2010. Viral and host proteins that modulate filovirus budding. Future Virol 5:481–491. doi: 10.2217/fvl.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Harty RN, Pitha PM, Okumura A. 2009. Antiviral activity of innate immune protein ISG15. J Innate Immun 1:397–404. doi: 10.1159/000226245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garcia M, Cooper A, Shi W, Bornmann W, Carrion R, Kalman D, Nabel GJ. 2012. Productive replication of Ebola virus is regulated by the c-Abl1 tyrosine kinase. Sci Transl Med 4:123ra124. doi: 10.1126/scitranslmed.3003500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu J, Han Z, Liu Y, Liu W, Lee MS, Olson MA, Ruthel G, Freedman BD, Harty RN. 2014. A host-oriented inhibitor of Junin Argentine hemorrhagic fever virus egress. J Virol 88:4736–4743. doi: 10.1128/JVI.03757-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Madara JJ, Han Z, Ruthel G, Freedman BD, Harty RN. 2015. The multifunctional Ebola virus VP40 matrix protein is a promising therapeutic target. Future Virol 10:537–546. doi: 10.2217/fvl.15.6. [DOI] [PMC free article] [PubMed] [Google Scholar]