ABSTRACT
Attenuated measles virus (MV) is one of the most effective and safe vaccines available, making it an attractive candidate vector for preventing other infectious diseases. Yet the great capacity of this vaccine still needs to be understood at the molecular level. MV vaccine strains have different type I interferon (IFN)-inducing abilities that partially depend on the presence of 5′ copy-back defective interfering genomes (DI-RNAs). DI-RNAs are pathogen-associated molecular patterns recognized by RIG-I-like receptors (RLRs) (RIG-I, MDA5, and LGP2) that activate innate immune signaling and shape the adaptive immune response. In this study, we characterized the DI-RNAs produced by various modified recombinant MVs (rMVs), including vaccine candidates, as well as wild-type MV. All tested rMVs produced 5′ copy-back DI-RNAs that were different in length and nucleotide sequence but still respected the so-called “rule of six.” We correlated the presence of DI-RNAs with a larger stimulation of the IFN-β pathway and compared their immunostimulatory potentials. Importantly, we revealed that encapsidation of DI-RNA molecules within the MV nucleocapsid abolished their immunoactive properties. Furthermore, we identified specific interactions of DI-RNAs with both RIG-I and LGP2 but not MDA5. Our results suggest that DI-RNAs produced by rMV vaccine candidates may indeed strengthen their efficiency by triggering RLR signaling.
IMPORTANCE Having been administered to hundreds of millions of children, the live attenuated measles virus (MV) vaccine is the safest and most widely used human vaccine, providing high protection with long-term memory. Additionally, recombinant MVs carrying heterologous antigens are promising vectors for new vaccines. The great capacity of this vaccine still needs to be elucidated at the molecular level. Here we document that recombinant MVs produce defective interfering genomes that have high immunostimulatory properties via their binding to RIG-I and LGP2 proteins, both of which are cytosolic nonself RNA sensors of innate immunity. Defective interfering genome production during viral replication should be considered of great importance due to the immunostimulatory properties of these genomes as intrinsic adjuvants produced by the vector that increase recognition by the innate immune system.
KEYWORDS: measles virus, defective interfering RNA, type I interferon, RIG-I-like receptors, innate immunity
INTRODUCTION
Defective interfering (DI) genomes are truncated forms of the viral genome generated by most viruses during viral replication. DI and viral genomes share the minimum essential characteristics for replication: a competent initiation site at the 3′ end, its complementary sequence at the 5′ end, and a structure or sequence required for encapsidation into a nucleocapsid (1). However, DI genomes are defective for replication in the absence of a complete functional virus genome that provides the missing functions. DI genomes, and more precisely the 5′ copy-back DI genomes (DI-RNAs), are well described for paramyxoviruses. Specific DI-RNAs are produced when the viral polymerase, due to a yet unknown mechanism, detaches from the template and reattaches itself to copy back the 5′ end of the genome (reviewed in references 1 to 3).
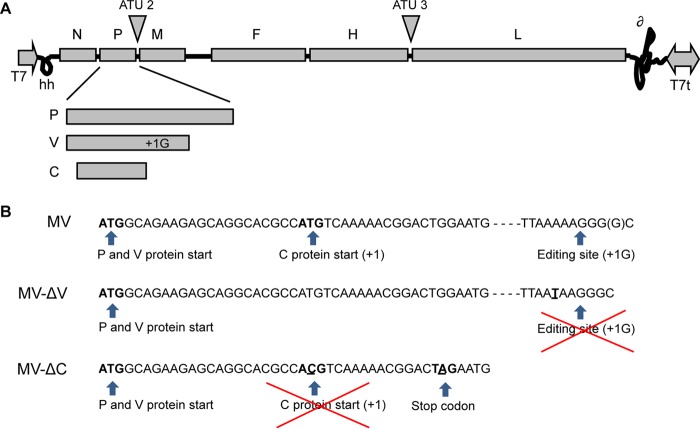
Paramyxoviruses, like other members of the order Mononegavirales, possess nonsegmented single-stranded negative-sense RNA genomes, with virus replication set in the cytoplasm. The RNA genomes of viruses of the Paramyxoviridae encode six structural proteins. The hemagglutinin (H) and fusion (F) proteins are transmembrane glycoproteins that are part of the viral envelope. The nucleoprotein (N), the phosphoprotein (P), and the viral RNA-dependent RNA polymerase (RdRp, or L) assemble into ribonucleoprotein complexes (RNPs) that ensure viral genome transcription and replication in the cytoplasm of infected cells. Replication of these viruses is coupled to encapsidation so that full-length antigenomes and genomes are found only inside assembled nucleocapsids (4). The nonglycosylated matrix protein (M) forms the inner layer of the viral envelope, recruiting assembled RNPs underneath the cellular membrane, and is involved in the budding of viral particles. In addition to the structural proteins, the P genes of paramyxoviruses encode nonstructural, genus-specific virulence factors. For measles virus (MV), these are the C and V proteins (Fig. 1A), whose expression is achieved by site-specific editing of the P mRNA by the viral RdRp to produce V protein and by the initiation of translation from the overlapping open reading frame (ORF) on the P mRNA to produce the C protein (5, 6).
FIG 1.
Representation of rMV-ΔV and rMV-ΔC cloning. (A) Schematic diagram of the MV genome and gene order showing that the P gene encodes the P, V, and C proteins. Sites used for insertion of additional transcription units (ATUs) within the MV genome are represented. (B) Portion of the nucleotide sequence of the P gene from MV (Schwarz strain; GenBank accession no. AF266291.1). For MV-ΔV construction, the editing site was mutated by replacing the adenine at position 2493 of the MV genome with uracil. For MV-ΔC, the start codon was mutated by replacing the uracil at position 1830 with cytosine, and a stop codon was introduced by replacing the guanine at position 1845 with adenine (the introduced nucleotides are underlined).
5′ copy-back DI-RNAs of paramyxoviruses have been shown to induce type I interferon (IFN-I) signaling due to their structural characteristics, i.e., a double-stranded stem with perfectly complementary blunt ends and a 5′ triphosphate extremity (7–10). These RNA molecules activate innate signaling through cytosolic RIG-I-like receptors (RLRs). Thus far, 5′ copy-back DI genomes have been described as pathogen-associated molecular patterns (PAMPs) for RIG-I (11–13) and MDA5 (12). Whether LGP2, which is the third known RLR, can also specifically interact with 5′ copy-back DI genomes is not known. Furthermore, RIG-I-dependent IFN-β signaling mediated by 5′ copy-back DI genomes is enhanced in the presence of IFN-inducible double-stranded RNA (dsRNA)-dependent protein kinase activator A (PACT), which also binds the DI-RNA (14). Additionally, accumulation of DI-RNAs triggers double-stranded RNA-dependent protein kinase (PKR) activation, which generates a cellular stress response resulting in translational arrest and formation of stress granules (15). However, the underlying mechanisms for how 5′ copy-back DI genomes provide their dsRNA the potential to activate type I IFN signaling remain to be elucidated, since their synthesis is coupled to their assembly into nucleocapsids.
DI genomes have also been identified in human infections with influenza virus (16), dengue virus (17), hepatitis C virus (18), and (more recently) respiratory syncytial virus (RSV), another paramyxovirus (19). Interestingly, the rate of accumulation of DI genomes during infection with RSV has been correlated with the quality of the antiviral response. Indeed, early accumulation of DI genomes is a good prognostic for viral clearance and full disease recovery (19). For MV, 5′ copy-back DI genomes have not yet been documented in vivo during natural infection (20). It is well known that viral stocks of standard live attenuated vaccines are variable producers of DI genomes (8, 20–22). Measles vaccine strains have shown different abilities to induce type I IFN, depending on the presence of 5′ copy-back DI-RNAs (8). The mechanism of viral attenuation and the resultant efficacy of live attenuated vaccines are not yet fully understood, but it is now clear that a robust adaptive immunity is induced by triggering innate immune system signaling (23). The attenuated MV-Schwarz vaccine strain that is routinely used is a poor producer of DI genomes. In cell cultures, production of DI-RNAs depends on the multiplicity of infection (MOI) and the number of cell passages (8, 10, 24). However, several modifications of the MV genome lead to efficient DI-RNA production by modified strains, giving a recombinant MV (rMV) expressing an additional copy of MV-N (rMV-N) (10), as well as rMV-ΔC (24) and rMV-ΔV (13), lacking the expression of two virulence factors, i.e., the C and V proteins, respectively (Fig. 1B).
Recombinant MV is a promising vaccine vector for delivery of heterologous antigens (25). Indeed, a recent human trial of an rMV-chikungunya vaccine candidate demonstrated that measles preimmunity in volunteers did not interfere with acquisition of strong functional antibodies to the chikungunya virus heterologous pathogen (26). It now needs to be evaluated if such a capacity of rMV to provide strong functional immunity might be explained by an increased production of DI-RNAs compared to that of the backbone MV-Schwarz strain. This would suggest an additional intrinsic immunostimulatory advantage for rMV-based vaccines. In this study, we characterized the 5′ copy-back DI-RNAs produced by recombinant MV strains, including rMV-based vaccines, and by wild-type MV (wt-MV), and we compared the production efficiencies of these DI-RNAs in different cell types. For the first time, the specific binding of 5′ copy-back DI-RNAs to RIG-I, MDA5, and LGP2 was assessed and linked to type I IFN signaling of encapsidated or nonencapsidated DI genomes.
RESULTS
Various 5′ copy-back DI-RNAs are produced by modified recombinant MVs.
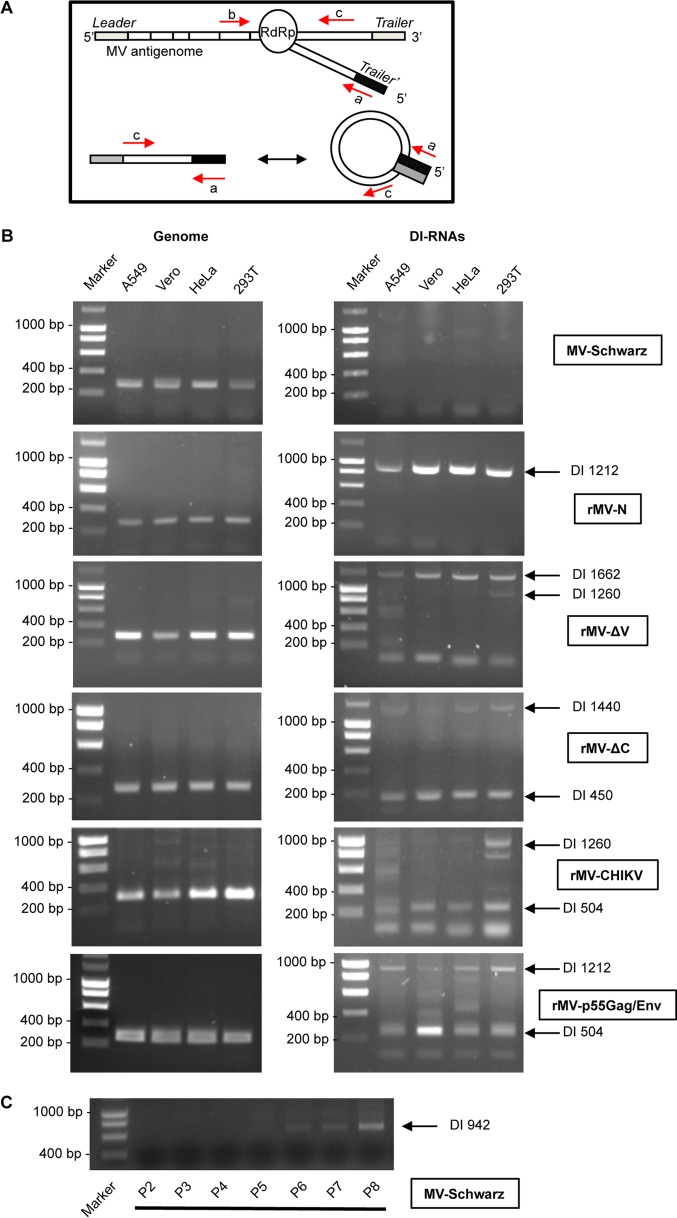
Production of 5′ copy-back DI genomes has previously been shown in separate independent studies for three modified rMVs: rMV-N (10), rMV-ΔV (13), and rMV-ΔC (24). To investigate the differential composition of 5′ copy-back DI-RNAs produced by rMV-N, rMV-ΔV, and rMV-ΔC in parallel under the same experimental conditions, we applied a unique reverse transcription-PCR (RT-PCR) protocol to molecularly characterize these RNAs in four different cell types. rMVs were generated using the same reverse genetics system as that used for MV-Schwarz, from which rMVs are derived (27). DI genomes were detected by RT-PCR by use of two primers, both of negative polarity in accordance with the full-length MV genome, so that only 5′ copy-back DI-RNAs could be amplified (Fig. 2A; Table 1). The presence of DI genomes was assessed in the total RNAs purified at 24 h postinfection for Vero, A549, 293T, and HeLa cells infected at an MOI of 1. As expected, we observed that various DI-RNAs were produced upon infection with modified rMVs but not with the control MV-Schwarz strain (Fig. 2B). Nevertheless, after several passages (P) on Vero cells at an MOI of 0.1, we observed an amplified production of DI-RNAs starting from P5 (Fig. 2C). Importantly, no mutation was detected after sequencing of the full-length viral genome of the MV-Schwarz P8 viral stock.
FIG 2.
Detection of DI-RNAs produced by rMVs. (A) Schematic representation of 5′ copy-back DI-RNA generation by the viral polymerase (RdRp). Primers used for PCR are represented for the genome (primers a and b) and DI-RNAs (primers a and c). (B) Total RNAs were extracted 24 h after infection with MV-Schwarz, rMV-N, rMV-ΔV, rMV-ΔC, rMV-CHIKV, and rMVp55Gag/Env at an MOI of 1. DI-RNAs and genomes in four different cell types (A549, Vero, HeLa, and 293T) were detected by RT-PCR. (C) DI-RNAs were detected by RT-PCR on total RNAs extracted 24 h after infection with MV-Schwarz strains that were obtained after 2 to 8 passages on Vero cells at an MOI of 0.1.
TABLE 1.
Primers and probes used in this study
| Primer or probe | Targeted RNA | Sequence (5′–3′)a | Reference |
|---|---|---|---|
| RT-PCR primers | |||
| A2 (a) | AAAGCTGGGAATAGAAACTTCG | 15 | |
| JM 402 (b) | TTTATCCAGAATCTCAATCCGG | 8 | |
| JM 403 (c) | CGAAGATATTCTGGTGTAAGTCTAGTA | 8 | |
| RT-qPCR primers and probes | |||
| Forward primer | Measles virus genome | TCAGGCATACCCACTAGTGTGAA | |
| Reverse primer | TGACAGATAGCGAGTCCATAACG | ||
| TaqMan probe (reporter dye) | CATCAGAATTAAGAAAAACGTAG (VIC) | ||
| Forward primer | β-Actin | ACCGAGCGCGGCTACAG | |
| Reverse primer | CTTAATGTCACGCACGATTTCC | ||
| TaqMan probe (reporter dye) | CACCACCACGGCCGA (FAM) | ||
| Forward primer | 504-nt DI-RNA | CGGAGTTCAACCAATTAGTCCTTAA | |
| Reverse primer | TGTGCCCCCAGAATTTGC | ||
| TaqMan probe (reporter dye) | CAGGGCACTATCTAGG (FAM) | ||
| Forward primer | 1,212-nt DI-RNA | TTGCAAATAATGCCTAACCACCTA | |
| Reverse primer | ACACTGCCTACCCACGTGACT | ||
| TaqMan probe (reporter dye) | CAGGATTAGGGTTCCGGG (FAM) | ||
| Forward primer | 1,662-nt DI-RNA | TTACCTTAAAAACCCACTCACGTTT | |
| Reverse primer | AGATTGCCCCCTGAAATGG | ||
| TaqMan probe (reporter dye) | AACACAAGCAAGCACA (FAM) | ||
| Forward primer | 450-nt DI-RNA | TTATCAACTTTTTGTTCCCGGAGTA | |
| Reverse primer | ACCACCTAGGGCAGGATTAGG | ||
| TaqMan probe (reporter dye) | AGATAATTGGTTGAACTCCGGAA (FAM) | ||
| Forward primer | 942-nt DI-RNA | GAGATGAAGTTGCTGTATCTAGGG | |
| Reverse primer | AATGCCTAACCACCTAGGGCAGG | ||
| TaqMan probe (reporter dye) | TGAACTCCGGAACCC (FAM) |
VIC, 4,7,2′-trichloro-7-phenyl-6-carboxyfluorescein; FAM, 6-carboxyfluorescein.
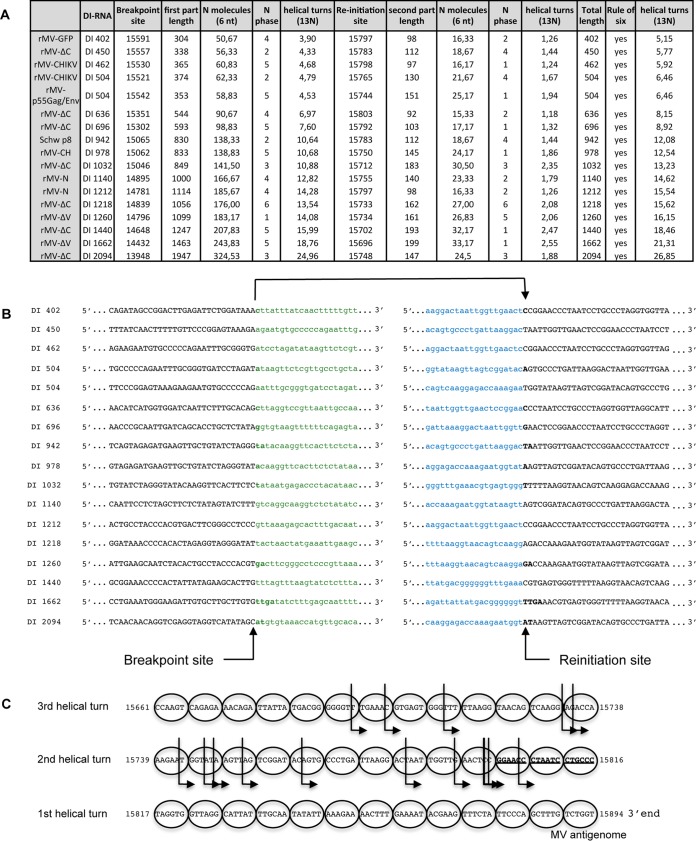
We further performed molecular characterization of various 5′ copy-back DI-RNAs produced upon infection with rMV-N, rMV-ΔV, and rMV-ΔC. The cDNA molecules encompassing the corresponding DI genome fragments were extracted from agarose gels, cloned, amplified, and sequenced, and the sequences were aligned with the MV-Schwarz sequence. The known structure of 5′ copy-back DI genomes allowed deduction of the entire sequence of the DI-RNA from the amplified cDNA. Table 2 summarizes important points that define each 5′ copy-back DI-RNA: the breakpoint site corresponds to the last nucleotide incorporated by the RdRp before it leaves the template antigenome strand, and the reinitiation site corresponds to the first nucleotide where the polymerase begins the synthesis of a cRNA strand of opposite polarity. We validated that rMV-N produced a single population of 1,212-nucleotide (nt) DI-RNAs (10). rMV-ΔV produced two DI-RNAs, of 1,662 and 1,260 nt, and rMV-ΔC also produced two DI-RNAs, of 450 and 1,440 nt. We observed that all rMVs produced a predominant DI-RNA that was found in the four different cell types (Fig. 2B) but could also produce other minor DI-RNAs depending on the cell type (for instance, a 1,260-nt DI-RNA in 293T cells for rMV-ΔV).
TABLE 2.
Characterization of DI-RNAs produced by different rMVs
| Virus | Rescue | DI-RNA length (nt) | Breakpoint sitea | Reinitiation sitea | ssRNA loop length (nt) | dsRNA stem length (nt) | Insertion/deletion | Mutation | Rule of six |
|---|---|---|---|---|---|---|---|---|---|
| MV-Schwarz | a | ||||||||
| MV-Schwarz P8b | a | 942 | 15,065 | 15,783 | 718 | 112 | 157 × 6 | ||
| rMV-N | a | 1,212 | 14,781 | 15,797 | 1,016 | 98 | 202 × 6 | ||
| b | 1,140 | 14,895 | 15,755 | 860 | 140 | 190 × 6 | |||
| rMV-ΔV | a | 1,662 | 14,428 | 15,696 | 1,264 | 199 | 277 × 6 | ||
| a | 1,260 | 14,793 | 15,737 | 938 | 161 | 210 × 6 | |||
| b (13) | 1,236 | 14,861 | 15,694 | 834 | 201 | +1 (A) | 206 × 6 | ||
| rMV-ΔC | a | 1,440 | 14,648 | 15,702 | 1,054 | 193 | 240 × 6 | ||
| a | 450 | 15,557 | 15,783 | 226 | 112 | 75 × 6 | |||
| b | 2,094 | 13,948 | 15,748 | 1,800 | 147 | 349 × 6 | |||
| b | 1,032 | 15,046 | 15,712 | 666 | 183 | A → G (36/286 A's [in loop]) | 172 × 6 | ||
| b | 696 | 15,302 | 15,792 | 490 | 103 | 116 × 6 | |||
| c | 1,218 | 14,839 | 15,733 | 894 | 162 | 203 × 6 | |||
| c | 636 | 15,351 | 15,803 | 452 | 92 | 106 × 6 | |||
| rMV-GFP | a | 402 | 15,591 | 15,797 | 206 | 98 | 67 × 6 | ||
| rMV-CH | a | 978 | 15,062 | 15,750 | 688 | 145 | 163 × 6 | ||
| rMV-CHIKV | a | 504 | 15,521 | 15,765 | 244 | 130 | 84 × 6 | ||
| a | 1,260 | 14,793 | 15,737 | 938 | 161 | 210 × 6 | |||
| b | 462 | 15,530 | 15,798 | 268 | 97 | 77 × 6 | |||
| rMV-p55Gag/Env | a | 504 | 15,542 | 15,744 | 202 | 151 | 84 × 6 | ||
| a | 1,212 | 14,781 | 15,797 | 1,016 | 98 | 202 × 6 | |||
| Wild type | 1,212 | 14,781 | 15,797 | 1,016 | 98 | 202 × 6 |
Nucleotide positions of breakpoints and reinitiation sites are aligned to the reference sequence for the Schwarz vaccine virus (GenBank accession no. AF266291.1).
MV-Schwarz P8 was obtained after 8 passages on Vero cells at an MOI of 0.1.
All identified 5′ copy-back DI-RNAs produced by the different tested rMVs differed in size for both the loop and stem, but they all respected the so-called “rule of six” postulating that, for the paramyxovirus subfamily Paramyxovirinae, only hexameric-length genomes are replicated efficiently (28).
Recombinant MVs expressing heterologous antigens are efficient producers of 5′ copy-back DI-RNAs.
We investigated whether addition of genetic material into the MV genome has an impact on the production of DI genomes. In a previous study, rMVs expressing the green fluorescent protein (GFP) gene from an additional transcription unit in the viral genome were poor producers of DI genomes and behaved similarly to the parental virus (15). In contrast, we demonstrated that our viral stocks of rMV-GFP and rMV-CH (expressing Cherry red fluorescent protein) contained DI-RNAs of 402 and 978 nt, respectively (Table 2).
We further examined whether rMVs expressing heterologous antigens could produce DI genomes in different cell lines. A549, Vero, HeLa, and 293T cells were infected with either rMV-CHIKV, expressing the structural proteins of chikungunya virus (E1, E2, E3, C, and 6K) (29), or rMV-p55Gag/Env, expressing the HIV-1 p55Gag polyprotein and EnvΔV1V2 envelope glycoprotein (30). Total RNAs were extracted, and DI-RNAs were detected by RT-PCR using two DI genome-specific primers (Table 1). The production of 5′ copy-back DI genomes was observed in cells infected by both recombinant viruses (Fig. 2B). We then performed molecular characterization of their DI genomes. While rMV-CHIKV produced two different 5′ copy-back DI genomes, of 1,260 and 504 nt, rMV-p55Gag/Env produced two different DI-RNAs, of 1,212 and 504 nt (Table 2; Fig. 2B). Interestingly, the 1,212-nt DI-RNA produced by rMV-p55Gag/Env corresponded perfectly to the unique DI-RNA detected in cells infected with rMV-N, whereas the 504-nt DI genome made by the same virus differed in sequence from the 504-nt DI-RNAs of rMV-CHIKV.
Therefore, we identified that all tested rMVs expressing heterologous genes produced various 5′ copy-back DI-RNAs. All these DI-RNAs respected the “rule of six” (Table 2).
Production of various DI genomes is an intrinsic feature of MV infection.
To investigate whether the difference in DI genome composition depends on the type of genetic modification introduced into the viral genome or on the rescue itself, we characterized DI genomes produced by different stocks of rMV-ΔC, rMV-N, and rMV-CHIKV viruses obtained from different rescues. We observed that the sequence pattern of DI genomes was different for each rescue (rescues a, b, and c) (Table 2). Indeed, previous studies showed that DI-RNAs were produced very early after the rescue and that this initial sequence pattern defined the DI genome pattern of each viral stock (24). To ensure that these DI-RNAs were not generated by the rescue system, we amplified a wt-MV isolate from a patient sample on Vero/hSLAM cells permissive to wt-MV. We next infected Vero/hSLAM cells with wt-MV at an MOI of 1 and assessed the presence of 5′ copy-back DI genomes by RT-PCR. As a result, we identified a 1,212-nt DI-RNA with the same breakpoint and reinitiation sites as those of DI-RNAs produced by rMV-N and rMV-dbp55Gag/Env but containing differences in nucleotide sequence corresponding to the wt-MV sequence (Table 2) (see Materials and Methods).
These results validate that DI genome production is an intrinsic feature of MV infection and that viruses with exactly the same genomic sequence but obtained from different rescues can produce distinct 5′ copy-back DI genomes.
RNP and RNA structure analysis of DI-RNA- and DI genome-forming regions.
To understand how DI genomes are generated by the viral polymerase, we determined the positions of breakpoint/reinitiation sites within the MV RNP, the primary and secondary RNA structures within these regions, and their GC compositions.
Within the MV RNP, each nucleoprotein encapsidates exactly 6 nucleotides, and each helical turn encompasses 13 nucleoproteins. We observed that all DI genomes found in this study respected the rule of six necessary for efficient encapsidation (Table 2). First, we analyzed the positions of breakpoint and reinitiation sites in accordance with the phase of N encapsidation or the phase of the RNP helical turn (Fig. 3). The breakpoint and reinitiation sites were observed in the six different positions of nucleotides covered by nucleoprotein encapsidation (Fig. 3A, N phase). No similarities for the positioning of breakpoint and reinitiation sites within the viral RNP complex were observed (Fig. 3A to C, helical turns).
FIG 3.
Analysis of DI-RNA sequences. (A) Characteristics of breakpoint sites and reinitiation sites for each DI-RNA. (B) Sequences encompassing breakpoint sites and reinitiation sites. (C) Graphic representation of reinitiation sites. Each nucleoprotein encapsidates 6 nucleotides and is represented by a circle. Each row corresponds to a helical turn encompassing 13 nucleoproteins. The B box sequence is represented in bold and underlined. The different reinitiation sites are represented by arrows. Reinitiation sites that are represented more than once are shown in bold.
By applying different bioinformatics tools, we studied RNA sequence motifs that could explain DI genome generation by the viral polymerase. We used weblogo and MUSCLE (multiple-sequence comparison by log expectation) software for multiple-sequence alignment analysis of 50-nt RNA fragments encompassing RNA sequences before or after breakpoint and reinitiation sites. However, we failed to find any conserved sequence or repeated motifs (data not shown). Further, using the mFold algorithm (mFold Web server), we performed in silico analysis of the potential to form RNA secondary structures within 50-nt RNA fragments corresponding to the sequences before and after breakpoint and reinitiation sites. In some DI genomes (402-, 504-, 1,140-, and 1,212-nt DI-RNAs), we found hairpin structures just after the breakpoint sites, but this was not observed for all DI-RNAs under study (data not shown). The measles virus GC ratio was also analyzed to see if a high GC content could slow down the polymerase and favor its detachment. However, we observed no significant and conserved change of GC content at breakpoint sites.
Thus, neither RNP folding nor specific RNA motifs or secondary structure within the breakpoint and reinitiation sites could explain DI genome generation by the viral polymerase.
Efficiency of DI genome production varies for different rMVs.
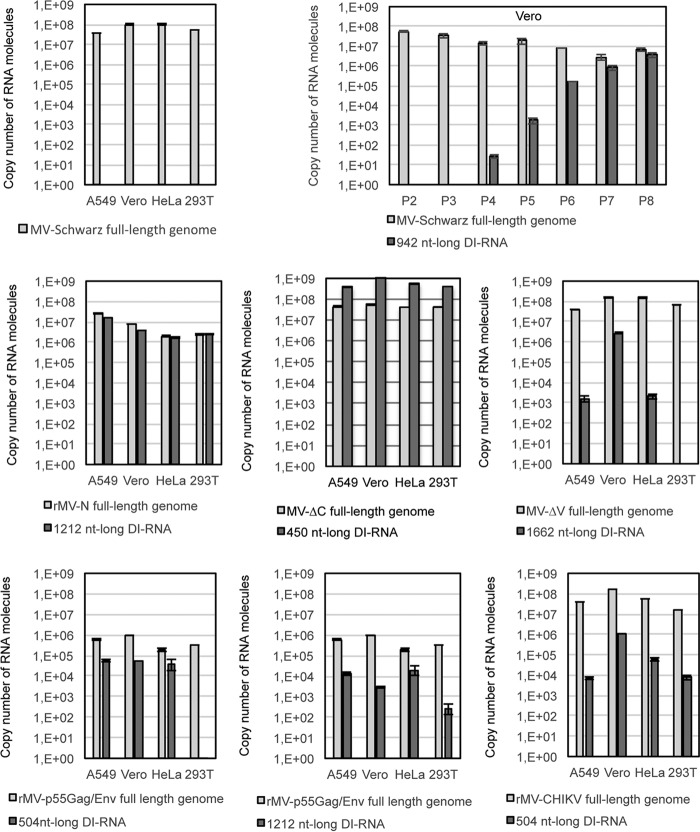
DI genomes are designated “defective interfering” because they interfere with infectious virus replication (31). To assess the interference capacity of the DI genomes, we compared the numbers of full-length and DI genomes produced by five recombinant viruses, namely, rMV-N, rMV-ΔV, rMV-ΔC, rMV-CHIKV, and rMV-dbp55Gag/Env, by using quantitative RT-PCR (RT-qPCR). A549, Vero, HeLa, and HEK-293T cells were infected at an MOI of 1, and total RNA was purified from infected cells at 24 h postinfection. Absolute quantification was performed using serial dilutions of in vitro-transcribed (IVT) specific DI-RNAs or an MV full-length genomic RNA fragment. MV-Schwarz was used as an “empty of DI-RNA MV” control. The absolute quantification of MV-Schwarz full-length genomes in the four different cell lines reached the same level, about 1 × 108 copies of RNA molecules, thus providing a “gold standard” for MV replication efficiency (Fig. 4). Interestingly, we observed that the level of full-length genomes produced by MV-Schwarz along 8 passages on Vero cells decreased with the amplification of the 942-nt DI-RNAs (Fig. 4). The ratio of DI genomes to full-length viral genomes varied greatly for each rMV (Fig. 4). We observed the following four situations: (i) there was a lack of detectable DI genome production (MV-Schwarz), (ii) DI genome production was as efficient as that of the full-length genome (rMV-N), (iii) DI genome production was more efficient than that of the full-length genome (rMV-ΔC), and (iv) DI genome production was less efficient than that of the full-length genome (rMV-ΔV, rMV-dbp55Gag/Env, and rMV-CHIKV). Furthermore, the level of DI genomes also differed in different cell types. In most cases, Vero cells, which are interferon deficient (32), were more permissive for DI-RNA production (Fig. 4). However, we failed to quantify some minor DI genomes by RT-qPCR (1,260-nt DI-RNAs of rMV-ΔV and rMV-CHIKV), probably due to their low levels of production. Only rMV-p55Gag/Env produced fewer full-length genomes than all other recombinant viruses and the “gold standard” MV-Schwarz strain.
FIG 4.
Absolute quantification of genomes and DI-RNAs for MV-Schwarz, rMV-N, rMV-ΔC, rMV-ΔV, rMV-p55Gag/Env, and rMV-CHIKV in four different cell lines and for MV-Schwarz after 2 to 8 passages on Vero cells (P2 to P8). Total RNAs (400 ng) purified from the different cell types (A549, Vero, HeLa, and 293T) at 24 h postinfection were analyzed by RT-qPCR. Absolute quantification was performed using serial dilutions of in vitro-transcribed MV DI-RNAs or an MV genome RNA fragment. Results are indicated in copy numbers of RNA molecules. Samples were tested in triplicate, and data represent means and standard deviations (SD).
Thus, the high levels of DI genomes produced by the different rMVs did not necessarily interfere with full-length genome replication.
rMVs are strong inducers of the type I IFN signaling pathway.
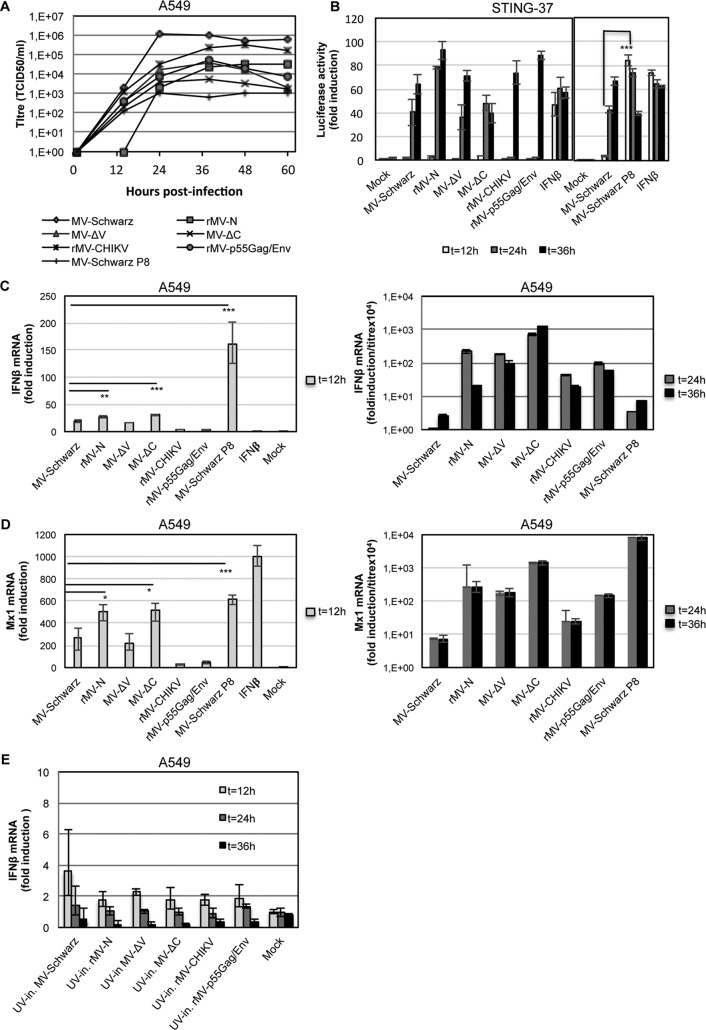
We evaluated the ability of rMVs to induce type I IFN signaling in comparison to that of the parental MV-Schwarz strain under the same experimental conditions. First, we assessed the growth kinetics of MV-Schwarz, rMV-N, rMV-ΔV, rMV-ΔC, rMV-CHIKV, rMV-p55Gag/Env, and MV-Schwarz P8. A549 cells were infected with the five rMVs and the MV-Schwarz control strain at an MOI of 1, and titration was performed at six different time points. We observed that all rMVs and MV-Schwarz P8 grew more slowly than the parental MV-Schwarz strain, with final viral titers varying from 1 × 103 to 1 × 106 50% tissue culture infective doses (TCID50)/ml (Fig. 5A). Then, to assess the immunostimulatory activity induced by rMVs, we used two different approaches: (i) measuring the type I IFN-stimulatory activity of rMVs at the protein level by using STING-37 cells, which stably express the luciferase gene under the control of a promoter sequence containing five IFN-stimulated response elements (ISRE) (33); and (ii) determining the transcription of IFN-β and Mx1, an interferon-stimulated gene (ISG).
FIG 5.
Evaluation of type I IFN response to rMVs. (A) Growth curves for MV-Schwarz, rMV-N, rMV-ΔV, rMV-ΔC, rMV-CHIKV, rMV-p55Gag/Env, and MV-Schwarz P8 for 60 h after infection of A549 cells at an MOI of 1. Titers are indicated in TCID50 per milliliter. (B) Luciferase reporter activities at 12, 24, and 36 h postinfection for STING-37 cells (33) infected with various rMVs at an MOI of 1. IFN-β at 300 IU/ml was used as a positive control, and noninfected cells (mock) were used as a negative control. Fold induction of IFN-β mRNA (C) and Mx1 mRNA (D) was determined 12, 24, and 36 h after infection of A549 cells at an MOI of 1. Quantification of mRNA was done using GAPDH as the reference gene and the relative qPCR gene expression protocol. t test results are represented as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Data obtained at 24 and 36 h postinfection were normalized to the viral titer, and results are presented as fold induction/(titer × 104). (E) Fold induction of IFN-β mRNA 12, 24, and 36 h after A549 cells were infected by UV-inactivated (UV-in.) rMVs at an MOI of 1. Experiments were performed two times, and data represent the upper and lower 95% confidence intervals for technical triplicates of the most representative experiment.
STING-37 cells were infected with the different rMVs and then lysed at 12, 24, and 36 h postinfection. All rMVs, including the parental MV-Schwarz strain, appeared to be strong IFN inducers starting from 24 h postinfection (Fig. 5B). Only MV-Schwarz P8 induced a fast and high ISRE response at 12 h postinfection (t test; P < 0.001). Interestingly, for rMV-CHIKV and rMV-p55Gag/Env, which are vaccine candidates, even if the ISRE activation by IFN-β was observed at a later time point of infection, it was almost as strong as that of rMV-N and rMV-ΔV.
To measure the potential to induce a type I IFN response, A549 cells were infected with the different rMVs and lysed at 12, 24, and 36 h postinfection, and total RNA was extracted. IFN-β and Mx1 mRNA levels were analyzed by RT-qPCR (Fig. 5C and D). We observed similar profiles for the IFN-β and Mx1 responses: (i) MV-Schwarz P8 (t test; P < 0.001 for IFN-β and Mx1), rMV-N (t test; P < 0.01 for IFN-β and P < 0.05 for Mx1), and rMV-ΔC (t test; P < 0.001 for IFN-β and P < 0.05 for Mx1), which produced high levels of DI-RNAs, induced early increases of IFN-β and Mx1 mRNA production at 12 h postinfection (Fig. 5C and D, left panels); and (ii) at 24 and 36 h postinfection, all infected cells produced high levels of IFN-β and Mx1 mRNAs compared to those in noninfected cells. To compare the immunostimulatory properties of the different viruses at 24 and 36 h postinfection, we normalized the results to the viral titers and demonstrated that all rMVs induced more production of IFN-β mRNA than that of the parental Schwarz virus (Fig. 5C and D, right panels). In parallel, the same protocol was applied to UV-inactivated viruses. UV treatment abrogates virus replication, preventing the accumulation of DI genomes within the cytoplasm of infected cells. As a result, we observed no induction of IFN-β mRNA production upon infection with UV-irradiated recombinant viruses (Fig. 5E).
Altogether, these results show that DI-RNAs produced by MV during replication have a strong capacity to stimulate the type I IFN signaling pathway and at least partly reduce viral growth.
Efficient type I IFN signaling is independent of DI-RNA size or sequence but is lost upon DI-RNA encapsidation into MV nucleocapsids.
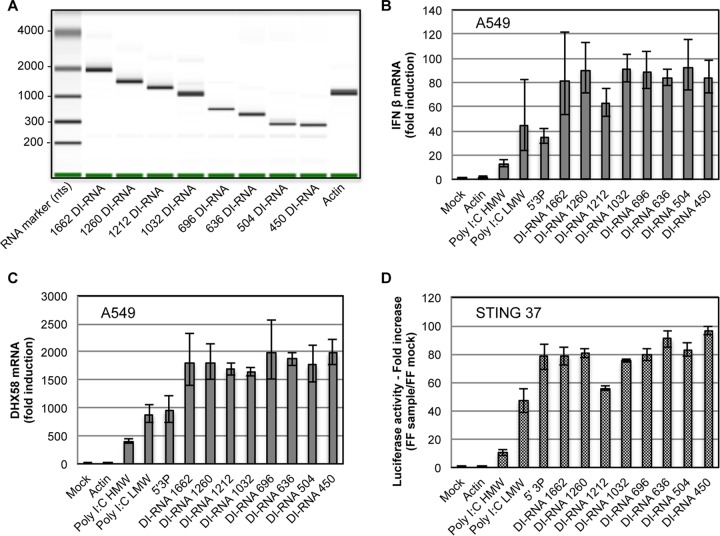
To compare levels of type I IFN activation by the different DI-RNAs identified in this study (Table 2), DI-RNAs were synthesized by in vitro transcription and transfected into STING-37 or A549 cells. We analyzed eight IVT DI-RNAs that were different in size and sequence, i.e., 1,662-, 1,260-, 1,212-, 1,032-, 696-, 636-, 504-, and 450-nt DI-RNAs (Fig. 6A; Table 2). β-Actin RNA was used as a negative control, and three classical RLR PAMPs were used as positive controls, namely, poly(I·C) of high molecular weight (HMW) or low molecular weight (LMW) and 5′ triphosphate RNA (5′3P). IFN transcription efficiency in A549 cells was assessed by RT-qPCR analysis of IFN-β mRNA (Fig. 6B) and DHX58 mRNA, a human ISG (Fig. 6C). In parallel, the efficiency of type I IFN signaling was assessed using an ISRE-luciferase reporter assay (Fig. 6D). We observed that all eight IVT DI-RNAs efficiently activated the type I IFN pathway, with few differences between them, independently of their size or sequence.
FIG 6.
Type I IFN response to DI-RNAs. (A) Results of IVT DI-RNA and actin RNA analysis on an Agilent bioanalyzer. (B to D) STING-37 and A549 cells were transfected with 10 ng of one of eight IVT DI-RNAs of different lengths, actin RNA, HMW poly(I·C), LMW poly(I·C), or 5′3P and lysed at 24 h posttransfection. The graphs show IFN-β mRNA (B) and DHX58 mRNA (C) expression profiles in A549 cells and luciferase reporter activities in STING-37 cells (D). Data were normalized to GAPDH and compared to those for mock-transfected cells. Experiments were performed two times, and the upper and lower 95% confidence intervals are shown as error bars for technical triplicates of the most representative experiment. FF, firefly luciferase activity.
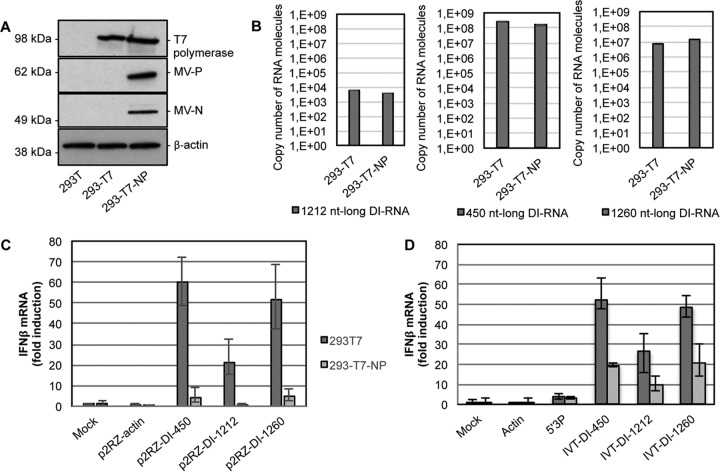
To investigate whether DI-RNAs encapsidated within MV nucleocapsids were as efficient at stimulating type I IFN as nonencapsidated DI-RNAs, we used the HEK293-T7 and HEK293-T7-NP cell lines, both of which stably expressed T7 RNA polymerase (Fig. 7A), but only HEK293-T7-NP cells also produced the MV N and P proteins. HEK293-T7 and HEK293-T7-NP cells were transfected with p2RZ vectors expressing the 1,212-nt DI-RNA (p2RZ-DI1212), the 450-nt DI-RNA (p2RZ-DI450), the 1,260-nt DI-RNA (p2RZ-DI1260), or a 1,212-nt fragment of actin mRNA (p2RZ-actin). The p2RZ plasmid allows the production of RNA transcripts under the control of the T7 polymerase promoter sequence and possesses a 3′-end cis-acting ribozyme of hepatitis delta virus (HDV) for direct transcriptional processing to precisely define the RNA transcript 3′ end. Upon transfection with the p2RZ-DI1212, p2RZ-DI450, and p2RZ-DI1260 vectors, both cell lines produced similar quantities of DI-RNAs (Fig. 7B). However, only HEK293-T7 cells showed a significantly increased level of IFN-β production, while HEK293-T7-NP cells did not (Fig. 7C). As expected, no type I IFN signaling activation was observed in both cell lines upon transfection with the negative-control p2RZ-actin vector. Furthermore, we analyzed the potential of DI-RNAs to stimulate type I IFN by transfecting the IVT DI-RNAs into HEK293-T7 and HEK293-T7-NP cells (Fig. 7D). As before, HEK293-T7 cells presented a higher type I IFN activation potential than that of HEK293-T7-NP cells upon transfection of the IVT DI-RNAs, whereas control stimulation levels by 5′3P RNA were identical in the two cell lines. The presence of MV N and P in HEK293-T7-NP cells decreased type I IFN activation by DI genomes but did not abrogate it. Importantly, we observed similar type I IFN activation levels by plasmid-derived and IVT DI-RNAs in HEK293-T7 cells, whereas in HEK293-T7-NP cells the plasmid-derived DI genomes were less potent. Therefore, the rapid encapsidation immediately after synthesis from p2RZ-DI-RNA vectors in HEK293-T7-NP cells hid the viral RNA and abolished its recognition and downstream activation of type I IFN signaling. By simultaneous transfection of a viral polymerase-encoding plasmid, we validated that type I IFN signaling upon DI-RNA encapsidation into MV nucleocapsids was independent of the presence of the MV L protein (data not shown).
FIG 7.
Variation in type I IFN response activation by DI-RNAs in the presence of the MV N and P proteins. (A) Western blot analysis of T7 polymerase, MV-N, MV-P, and β-actin in HEK293-T7 and HEK293-T7-NP cells. A total of 1 × 106 cells were plated, and lysis was performed 12 h later. (B) Absolute quantification of 1,212-, 450-, and 1,260-nt DI-RNAs produced by HEK293-T7 and HEK293-T7-NP cells transfected with the p2RZ-DI1212, p2RZ-DI450, and p2RZ-DI1260 vectors. Total RNA (400 ng) purified from cells at 24 h posttransfection was treated with RQ1 RNase-free DNase, and 100 ng was analyzed by RT-qPCR. Absolute quantification was performed using serial dilutions of in vitro-transcribed MV DI-RNAs and normalized with actin. Results are expressed in copy numbers of RNA molecules. Experiments were performed two times, and samples were tested in triplicate. (C and D) HEK293-T7 and HEK293-T7-NP cells were transfected with p2RZ vectors encompassing DI-RNAs or an actin sequence (C) or with in vitro-transcribed RNAs (actin, 5′3P, or 3 different IVT DI-RNAs) (D). Relative IFN-β mRNA expression levels were measured at 24 h posttransfection, normalized to GAPDH, and compared to those of mock-transfected cells. Experiments were performed two times, and the upper and lower 95% confidence intervals are shown as error bars for technical triplicates of the most representative experiment.
These results show that nonencapsidated DI-RNAs efficiently activated type I IFN signaling independently of their size or sequence and that their encapsidation abolished type I IFN stimulation.
MV 5′ copy-back DI-RNAs are specific agonists of RIG-I and LGP2 but not MDA5.
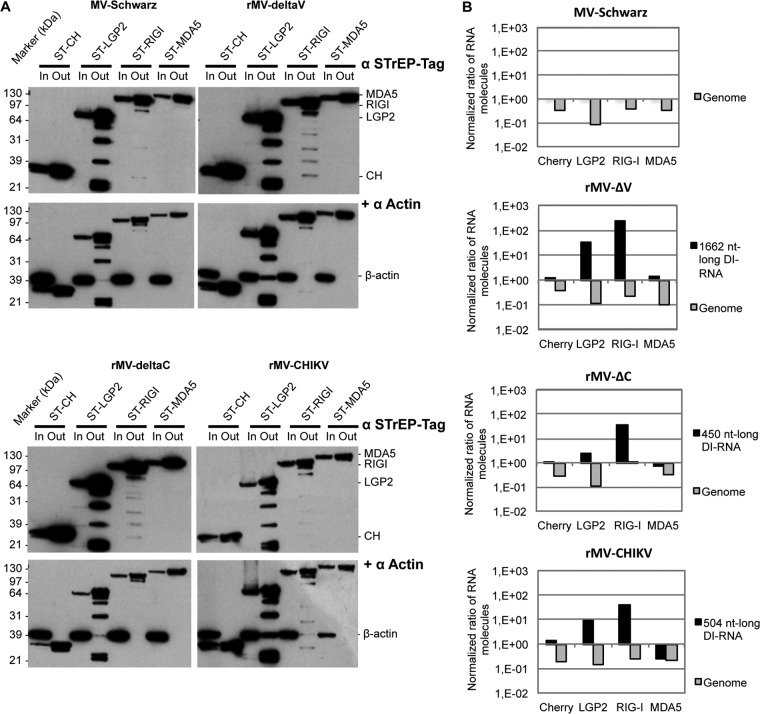
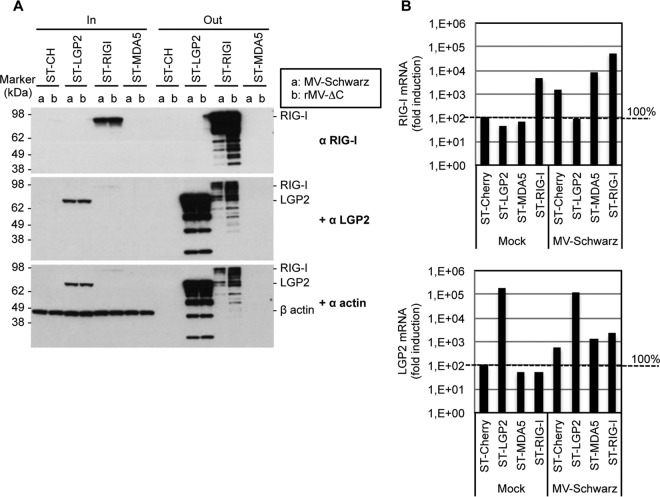
To determine which of the three RLRs are involved in DI genome sensing, we compared LGP2, MDA5, and RIG-I binding efficiencies with various 5′ copy-back DI genomes produced during infection. To this end, we used human HEK293 cell lines stably expressing tagged LGP2, MDA5, and RIG-I proteins (ST-RLR cells). We previously showed that these cells constitute an effective tool for purifying viral agonists of RLRs (13). ST-RLR cells were infected with MV-Schwarz, rMV-ΔV, rMV-ΔC, and rMV-CHIKV for 24 h at an MOI of 1. RLR-specific RNA ligands were purified as previously described (13). In addition, a stable cell line (ST-CH) expressing the Cherry protein instead of tagged RLRs was used as a negative control to allow subtraction of nonspecific RNA binding (13). The efficacy of purification was assessed by Western blotting to confirm the enrichment of RLRs and the absence of nonspecific binding (Fig. 8A). Absolute quantification of DI-RNAs, genomes, and β-actin mRNA was done using RT-qPCR. Ratios of full-length genomes to β-actin and of DI-RNAs to β-actin indicated that DI-RNAs were always enriched on RIG-I and, to a lesser extent, on LGP2 but never on MDA5 (Fig. 8B). Full-length genomes were equally depleted on the three RLRs for MV-Schwarz and any of the rMV infections.
FIG 8.
5′ copy-back DI-RNA-specific binding to RLRs. (A) Western blot of infected ST-RLR cells before (input [In]) and after (output [Out]) One-STrEP tag affinity purification. Western blot analysis of β-actin served as a control for nonspecific binding. (B) Ratios of genomes and DI-RNA molecules after to before One-STrEP tag affinity purification of ST-RLR cell lines. Ten nanograms of either total RNA or RNA obtained after affinity chromatography of STrEP-tagged Cherry (negative control), LGP2, RIG-I, or MDA5 was analyzed by RT-qPCR. Absolute quantification was performed as described in the legend to Fig. 4 and normalized with β-actin mRNA (nonspecific binding). Samples were analyzed in triplicate, and two biological replicates were performed.
To ensure that the enrichment of DI-RNAs on LGP2 was not due to a RIG-I–LGP2 protein-protein interaction (34), we performed Western blotting to confirm that endogenous RIG-I proteins were missing in the LGP2-specific protein complexes purified from MV-infected cells (Fig. 9A). The results were similar to previously published data in which RIG-I failed to be coprecipitated with LGP2 (35). Additionally, we quantified RIG-I and LGP2 mRNA levels in total RNA extracted from ST-RLR cells infected or not with MV-Schwarz. While endogenous RIG-I was not induced in the MV-infected ST-LGP2 cell line, these cells expressed 3 log more LGP2 transcripts due to overexpression (Fig. 9B). Thus, the bias for the presence of specific RNA partners on LGP2 due to the presence of endogenous RIG-I in our ST-LGP2 cells should be minimal, if present at all.
FIG 9.
Endogenous RIG-I is absent from the LGP2-specific protein complex. (A) Western blot analysis of infected ST-RLR cells before (total lysate [In]) and after (beads [Out]) One-STrEP tag affinity purification. Cells were infected with MV-Schwarz (a) or rMV-ΔC (b) at an MOI of 1. The presence of RIG-I and LGP2 was assessed, and β-actin served as a control for nonspecific binding. (B) Relative quantification of RIG-I and LGP2 mRNAs in mock- and MV-Schwarz-infected ST-RLR cell lines by qRT-PCR, normalized with GAPDH mRNA.
Thus, we validated that various DI-RNAs specifically interact with RIG-I and observed, for the first time, their specific binding to LGP2.
DISCUSSION
DI-RNAs are known to be produced by modified MVs (rMV-N [10], rMV-ΔV [13], and rMV-ΔC [24]) or by measles vaccine strains after cell passages at a high MOI (8, 20, 22). By comparing different stocks of rMVs that had different genetic modifications but were identically produced using the reverse genetics approach, we observed that any modification of the MV genome increased its capacity to produce DI-RNAs. Indeed, all rMVs tested in this study significantly produced one or more distinct DI-RNAs, while the parental MV-Schwarz stock did not, unless it was passaged at least five times in cell culture (Table 2; Fig. 2B and C). The genome modifications included an addition of genetic material (rMV-GFP, rMV-CH, rMV-N, rMV-CHIKV, and rMVp55Gag/Env) or a deletion of the MV C or V virulence factor (rMV-ΔC and rMV-ΔV) (Fig. 1). The MV C and V proteins are multifunctional proteins. One of their functions is to regulate viral RNA synthesis. Indeed, the C protein is described as an important factor that stabilizes the RNP-polymerase complex, as a lack of C protein results in more frequent chain termination occurring during both transcription and replication (15). The V protein helps to maintain the ratio of genome to antigenome synthesis during replication (36).
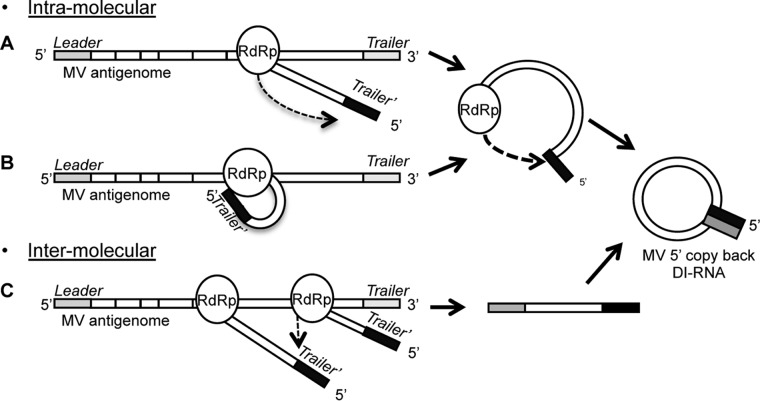
We observed that the production of DI genomes is an intrinsic quality of MV infection, not a side effect of the reverse genetics protocol, since DI-RNAs were also detected during infection with wt-MV isolated from a patient sample (Table 2). However, the exact mechanism by which the polymerase stops replicating and starts copying back remains unknown. We suggest two hypotheses for DI genome generation (Fig. 10). The first is an intramolecular model in which the nascent genome is the template for the polymerase to copy back the complementary ending. This could happen with a polymerase “jump” (Fig. 10A) or, more likely, by spatial proximity due to a temporal interaction of the 5′ end of the nascent genome with the polymerase during the replication process (Fig. 10B). The second hypothesis is an intermolecular model in which another nascent genome is a template for the polymerase due to spatial proximity (Fig. 10C). We found a different sequencing pattern of DI-RNAs for each viral stock and identified multiple breakpoint and reinitiation sites, especially for DI-RNAs of rMV-ΔC. This pattern was defined early after rescue, as shown by Pfaller et al. (24). Nevertheless, we found DI genomes of 1,212 nt with identical breakpoint and reinitiation sites in three different viral stocks (rMV-N, rMV-P55Gag/Env, and wt-MV), suggesting that some conserved motifs were involved. However, whatever the mechanism of DI genome generation, we could not find any correlation with specific RNA motifs, RNA secondary structure, or RNP folding (Fig. 3).
FIG 10.
Hypotheses on DI genome generation by the viral polymerase. (A and B) The intramolecular model suggests that the nascent genome is a template (black) for the polymerase (RdRp) to synthetize the complementary ending (gray) of the DI-RNA after a polymerase “jump” (A) or by spatial proximity due to a temporal interaction of the 5′ end of the nascent genome with the RdRp (B). (C) The intermolecular model suggests that another nascent genome is a template (black) for the RdRp due to spatial proximity.
DI-RNA lengths ranged from 402 nt to 2,094 bp. The loop and stem sizes varied from one DI-RNA to another, but all identified sequences respected the “rule of six” necessary for efficient encapsidation (21, 24, 28). The minimal size of the double-stranded stem region, 93 nt, included the B′ box of the trailer sequence (37). This is also the minimal size observed by Pfaller et al. (24) and is comparable to the minimal length of 96 nt observed in copy-back DI-RNAs of Sendai virus (SeV), another member of the paramyxovirus subfamily Paramyxovirinae (38). This minimal sequence is necessary for genome encapsidation by N and P proteins (39, 40). The MV genome is known to be encapsidated immediately after the beginning of replication (4), and the “rule of six” necessary for encapsidation may have advantaged replication, transmission of DI-RNAs from cell to cell, and stability within the cytoplasm. However, DI-RNAs with lengths that do not respect the “rule of six” have also been described (14, 24).
Defective viral genomes are named defective “interfering” RNA because DI-RNA particles were postulated to interfere with the replication of the infectious virus by competing for viral proteins in the infected cell (41). In our study, all rMVs had decreased growth compared to that of MV-Schwarz, meaning that the genetic modifications decreased the viral fitness (Fig. 5A). The lowest growth rates were observed for MV-Schwarz P8, rMV-N, and rMV-ΔC, which produced large amounts of DI-RNAs at levels similar to those of full-length genomes (Fig. 4). These DI-RNAs may indeed decrease viral growth not only by interfering directly with viral replication but also by enhancing the host response due to their detection as PAMPs by the innate immune system. Interestingly, for rMV-ΔC, we quantified as many full-length genomes as those of MV-Schwarz by RT-qPCR, whereas the viral titer was a hundred times lower (Fig. 4 and 5A). This could be explained by reduced virus protein translation being responsible for fewer complete viral particles. Indeed, the translation inhibition observed in A549 cells infected by rMV-ΔC has been correlated with phosphorylation of the alpha subunit of eukaryotic translation inhibition factor 2 (eIF2α) (42).
5′ copy-back DI-RNAs are known to be PAMPs to intracellular RLRs, specifically to RIG-I (11, 13) and also MDA5 (12). Here we demonstrated a direct interaction between DI-RNAs and the RIG-I and LGP2 receptors by infecting ST-RLR cell lines (Fig. 8B). These cells enabled isolation of RLR-specific interactors by chromatographic affinity purification (13). DI-RNAs were amplified among RNAs associated with RIG-I and LGP2 receptors but not MDA5. We described here, for the first time, an interaction between LGP2 and DI-RNA. LGP2 differs from RIG-I and MDA5 in the absence of a CARD domain necessary to activate the final IFN signaling cascade via MAVS activation (43). The role of LGP2 as a regulator of MDA5 and RIG-I responses is controversial and not well defined. It acts mainly as a negative regulator of RIG-I (34, 44) and a positive regulator of MDA5 (45). Common ligands between MDA5 and LGP2 have been suggested (13, 46, 47), and MDA5 may play a role in the induction of an antiviral state in cells infected by the measles vaccine by recognizing MV PAMPs other than DI-RNA (13, 48). Here we described the 5′ copy-back DI-RNA molecule as a common ligand between RIG-I and LGP2. Although rMVs shared high immunostimulatory properties for IFN and ISG expression (Fig. 5 and 6), both regulatory functions of LGP2, i.e., (i) an enhancement of the MDA5 response and (ii) a downregulation via RIG-I to regulate the activation and avoid exacerbation of the innate immune response, may still be involved.
DI-RNA sequences were highly preserved, with no mutations observed for all DI-RNAs except the 1,032-nt DI-RNA. This DI-RNA, produced by rMV-ΔC, accumulated clusters of A-to-G transitions (Table 2). This pattern was already observed by Pfaller et al. (24) for DI-RNAs produced by the Moraten vaccine strain and the wild-type IC-B strain lacking expression of the MV C protein. ADAR1 editing has been hypothesized to destabilize free DI-RNA molecules that have dsRNA structures (24). However, in our case, the A-to-G transitions, observed for 36 of 286 A nucleotides (12.7%), were located only in the loop sequence. These mutations did not affect the immunostimulatory properties, as the general structure was conserved. Indeed, cells transfected with eight in vitro-transcribed DI-RNAs differing in size and sequence showed similar high levels of IFN-β and ISG expression in STING-37 and A549 cells (Fig. 6). These experiments confirmed that the PAMPs recognized by RIG-I and LGP2 are structure dependent and do not rely on the sequence or the size of the single-stranded RNA (ssRNA) loop. Moreover, Ho et al. (14) recently confirmed that the 5′3P DI-RNA extremity plays a major role in RIG-I recognition.
MV vaccine strains are stronger inducers of the IFN pathway than wt-MV (20), even in the absence of DI-RNA. In this study, we showed that MV-Schwarz P8 and all tested rMVs possessed higher immunostimulatory properties than that of the parental strain MV-Schwarz due to the higher levels of DI-RNAs (Fig. 5). Viral replication is necessary for these immunostimulatory properties, as UV-inactivated virus did not induce type I IFN production even when the viral stock contained DI-RNAs (Fig. 5E). More importantly, we showed that nonencapsidated DI-RNAs induced a strong type I IFN response, whereas encapsidated DI-RNAs did not. These results seem to contradict an immediate encapsidation of each new strand of RNA. We suggest that modifications of the MV genome via gene deletion or insertion induce replication disturbances that might decrease efficient replication/encapsidation coupling. This allows a population of DI-RNAs to remain nonencapsidated in the cytoplasm by exceeding the capacity of MV-N encapsidation. Indeed, SeV DI-RNAs in extracts of infected cells, analyzed by use of a CsCl density gradient, were found to be mostly encapsidated, but a small amount (<5%) remained naked (7). Pfaller et al. also found more naked dsRNA in cells infected by rMV-ΔC (15). Because the C protein stabilizes the MV RdRp complex, its absence might enhance more production of DI-RNAs that cannot be encapsidated due to the inhibition of translation (40) leading to a lack of a sufficient amount of MV-N. Otherwise, the failure of encapsidated DI-RNAs to induce IFN-β (Fig. 7C and D) also contests the previously suggested ability of RLRs to use the ATP-driven receptor dynamics for remodeling viral RNPs to facilitate their accessibility to RNA ligands (49).
Our experiments demonstrating a difference in type I IFN responses to nonencapsidated and encapsidated DI-RNAs were obtained upon their transfection into either HEK293T-T7 or HEK293T-T7-NP cells. This protocol may involve a “passive” encapsidation by N and P that may be different from a bona fide cotranscriptional encapsidation occurring during regular viral replication. However, MV-N is the only viral protein that encapsidates the MV genome, thus forming nucleocapsids that constitute the functional template for viral RdRp. MV-P serves as an anchor that brings viral RdRp to nucleocapsids via protein-protein interaction (for a recent review, see reference 50). In addition, MV N-P protein interaction maintains nucleoproteins in a soluble monomeric form and prevents illegitimate encapsidation of cellular RNA (50, 51). In our transfection experiments, we used HEK293-T7-NP cells that express both MV-N and MV-P. The above-mentioned data show that the presence in the cytoplasm of both the N and P proteins together with defective viral genomes is sufficient to form functional nucleocapsids in HEK293-T7-NP cells upon transfection of DI-RNAs. Importantly, HEK293-T7-NP cells were the cells in which viral genome encapsidation was functionally validated, since these cells are routinely used to rescue recombinant MV by reverse genetics (27, 52).
Vaccine efficacy depends on the immunostimulatory properties of the vaccine. We suggest that 5′ copy-back DI-RNAs act as intrinsic adjuvants naturally produced by the measles virus vector during replication, as they enhance recognition and activation of the innate immune system. They certainly play a major role in MV effectiveness as a vector against heterologous antigens, and their presence should be considered to have great importance. Obviously, standardization of DI production by rMVs is required to produce homogeneous vaccine batches, and a modified vaccine production protocol should be developed in the future.
MATERIALS AND METHODS
Cells, viruses, plasmid vectors, and antibodies.
HEK293T (human embryonic kidney cells), Vero (African green monkey kidney cells), A549 (human alveolar basal epithelial adenocarcinoma cells), and HeLa cells were maintained in Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 10% heat-inactivated fetal calf serum (FCS; GE Healthcare) and 10,000 U/ml of penicillin-streptomycin (Life Technologies). Vero/hSLAM and HEK293 cell lines expressing One-STrEP-tagged RIG-I (ST-RIG-I), MDA5 (ST-MDA5), LGP2 (ST-LGP2), and Cherry (ST-CH) (13) were maintained in the same medium supplemented with G418 (Sigma) at 500 μg/ml. The STING-37 cell line corresponded to HEK293 cells stably transfected with an ISRE-luciferase reporter gene (33). HEK293-T7 (kindly provided by Yves Jacob, Unité de Génétique Moléculaire des Virus ARN, Institut Pasteur, and Pierre Charneau, Unité de Virologie Moléculaire et Vaccinologie, Institut Pasteur) and HEK293-T7-NP (27) cells expressed the T7 polymerase in the cytosol. HEK-293-T7-NP cells also stably expressed MV-N and MV-P.
The MV-Schwarz vaccine strain (GenBank accession no. AF266291.1) (27) and seven recombinant MVs were used: rMV-N, expressing an additional nucleoprotein in additional transcription unit 2 (ATU2) (Fig. 1A) (10); rMV-ΔC, lacking the MV C protein; rMV-ΔV, lacking the MV V protein (13); rMV-CHIKV, expressing the chikungunya virus structural proteins (E1, E2, E3, C, and 6K) in ATU2 (29); rMV-p55Gag/Env, expressing the HIV-1 p55Gag polyprotein and EnvΔV1V2 envelope glycoprotein in ATU2 (30); rMV-GFP, expressing green fluorescent protein in ATU2 (27); and rMV-CH, expressing Cherry red fluorescent protein in ATU3 (A. Meignié C. Combredet, A. Arbabian, F. Tangy, and A. V. Komarova, unpublished data). rMV-ΔC was generated by site-directed mutagenesis on a cassette vector covering the P gene sequence. The second translation initiation codon present in the N-terminal P ORF in the +1 frame was mutated (U → C), and then a substitution (G → A) was generated at nucleotide position 1845 of the MV genome to introduce a stop codon within the C ORF. Both mutations do not disturb the P and V ORFs (Fig. 1B). MV-Schwarz P8 was obtained after 8 passages of MV-Schwarz on Vero cells at an MOI of 0.1. wt-MV was obtained from a patient urine sample and assigned G2V wt-MV (GenBank accession no. MF449469). A stock of wt-MV was produced by virus multiplication on Vero/hSLAM cells at an MOI of 0.1. Virus titers were determined by TCID50 titration on Vero cells (or Vero-SLAM cells for wt-MV). Two different viral stocks from two technical replicates of rescues were obtained for rMV-ΔC, rMV-N, and rMV-CHIKV, using the protocol described by Radecke et al. (52) and modified by Parks et al. (53).
The plasmid vector pCRII-TOPO from a TOPO TA cloning kit (Invitrogen) was used to clone RT-PCR products before sequencing. The DI-RNA sequences were cloned into the in vitro transcription vector p2RZ (Addgene) by use of the HindIII and NheI restriction sites. In vitro transcription was carried out at 37°C by using T7 RiboMAX (Promega), and RNA products were purified using an RNeasy clean-up kit (Qiagen).
The following antibodies were used: anti-STrEP tag (34850; Qiagen), a monoclonal anti-β-actin antibody (A5441; Sigma), a polyclonal anti-T7 polymerase antibody (kindly provided by Yves Jacob), monoclonal anti-MV-N (clone 25; kindly provided by Chantal Rabourdin-Combe [54]), monoclonal anti-MV-P (9H4; Novus), anti-RIG-I (D14G6; Cell Signaling), and anti-LGP2 (HPA019570; Sigma).
Virus growth curves.
Monolayers of A549 cells in 24-mm-diameter dishes (6-well plates) were infected with MV-Schwarz, MV-Schwarz P8, or rMVs at an MOI of 1. At various times postinfection, cells were scraped into culture medium. After freeze-thawing of cells and medium and clarification of cell debris, virus titers were determined. For this purpose, Vero cells were seeded into 96-well plates (7,500 cells/well) and infected with serial 1:10 dilutions of virus sample in DMEM-5% FCS. After incubation for 7 days, cells were stained with crystal violet, and the TCID50 values were calculated by use of the Karber method.
Virus infection for RT-PCR detection of 5′ copy-back DI-RNAs.
Cells were seeded into T25 flasks 1 day before infection. Virus infections were carried out at an MOI of 1. Viruses were diluted with Opti-MEM to obtain a final inoculum volume of 2 ml. Cells were incubated with virus for 2 h at 37°C. Four milliliters of DMEM containing 10% fetal bovine serum (FBS) was added to each T25 flask, and cells were incubated at 37°C until infections were stopped by cell lysis 24 h later.
Total RNA was extracted by use of an RNeasy minikit (Qiagen). cDNA was generated from 300 ng of total RNA by using Superscript III (Thermo Fisher Scientific) and primer a (Fig. 2A and Table 1) in a total volume of 20 μl. A total of 2 μl of the resultant cDNA was then amplified with primers a and b for genome amplification (Fig. 2A and Table 1) and primers a and c for DI-RNA amplification (Fig. 2A and Table 1), using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific) in a total volume of 50 μl (95°C for 2 min; 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and 72°C for 10 min). At the end of the PCR, Taq DNA polymerase (Thermo Fisher Scientific) was added and incubated at 72°C for 10 min. The products were analyzed in a 1% agarose gel, with a Smartladder (MW-1700-10; Eurogentec) as the size standard, and gel purified with a QIAquick gel extraction kit (Qiagen). The PCR-amplified products were cloned into the pCRII-TOPO vector and sequenced.
TaqMan RT-qPCR analysis of DI-RNAs and full-length MV genomes.
Analysis was performed using Applied Biosystems StepOnePlus technology. Various MV DI-RNA primers and probes were designed using Primer Express software (Applied Biosystems) (Table 1). Reactions were performed on 400 ng of total RNA by use of TaqMan RNA-to-Ct 1-Step kits (Thermo Fisher Scientific) for one-step RT-qPCR analyses. Reactions were performed in a final volume of 20 μl in the presence of 100 nM TaqMan DI-RNA-specific probe and 100 nM (each) DI-RNA-specific forward and reverse primers. For absolute quantification of DI-RNAs, standard curves were established by using serially diluted RNAs obtained by in vitro transcription of p2RZ, encompassing DI-RNAs specific for each rMV. The protocol for absolute quantification of β-actin or MV genomes was described elsewhere (10). The standard curves were generated by the StepOnePlus software system by plotting the threshold cycle (CT) values against the logarithms of the calculated initial copy numbers. The unknown initial sample copy numbers were then automatically calculated from their CT values and compared with the RNA standard curves.
Affinity chromatography of RLR-RNP complexes and subsequent RNA purification.
ST-RLR cells (4 × 107) were infected at an MOI of 1 for 24 h with MV-Schwarz, rMV-ΔV, rMV-ΔC, or rMV-CHIKV. Cells were washed twice with cold phosphate-buffered saline (PBS) and lysed in 4 ml of lysis buffer (20 mM morpholinepropanesulfonic acid [MOPS]-KOH, pH 7.4, 120 mM KCl, 0.5% Igepal, 2 mM β-mercaptoethanol) supplemented with RNasin at 1/200 (Promega) and with Complete protease inhibitor cocktail (Roche). Cell lysates were incubated on ice for 20 min, with gentle mixing every 5 min, and then clarified by centrifugation at 16,000 × g for 15 min at 4°C. A 100-μl aliquot of each cell lysate was used to perform total RNA purification, using Tri reagent LS (Sigma). The rest of the cell lysate was incubated for 2 h on a spinning wheel at 4°C with 200 μl of StrepTactin Sepharose (high performance; GE Healthcare). Beads were collected by centrifugation (1,600 × g for 5 min at 4°C) and washed three times for 5 min each on a spinning wheel with 5 ml of washing buffer (20 mM MOPS-KOH, pH 7.4, 120 mM KCl, 2 mM β-mercaptoethanol) supplemented with RNasin at 1/400 and with Complete protease inhibitor cocktail. RNA purification was performed using Tri reagent LS. RNA was dissolved in 50 μl of DNase-free and RNase-free ultrapure water. Extracted RNAs were analyzed using Nanovue (GE Healthcare) and a Bioanalyser RNA nanokit (Agilent).
To validate the efficacy of RLR-ribonucleoprotein complex purification, protein extracts were resolved by SDS-polyacrylamide gel electrophoresis on 4 to 12% Criterion gels (Bio-Rad) with MOPS running buffer and transferred to cellulose membranes (GE Healthcare) by use of a Criterion blotter system (Bio-Rad).
Absolute quantification of DI-RNAs, genomes, and β-actin mRNA was done by RT-qPCR on total RNA and on RNA obtained after One-STrEP tag affinity purification. A β-actin mRNA fragment was used to normalize the quantities of RNA before and after affinity purification.
Analysis of rMV immunostimulatory activities.
STING-37 cells (33) were infected with MV-Schwarz, MV-Schwarz P8, or rMVs at an MOI of 1, left noninfected (mock), or transfected with 10 ng of in vitro-transcribed DI-RNAs by use of a TransIT-mRNA transfection kit (Mirus). Poly(I·C) of high molecular weight (HMW; Invivogen) and low molecular weight (LMW; Invivogen), short 5′3P RNA (synthetized in vitro with a T7 RiboMAX Express kit), and recombinant human IFN-β 1a (PBL) were used as positive controls. Cells were lysed at different time points postinfection. The firefly luciferase activity was measured using the Bright-Glo luciferase assay system (Promega) following the manufacturer's recommendations.
TaqMan RT-qPCR of IFN-β and ISG expression.
Gene expression qPCR analysis was performed by using Applied Biosystems StepOnePlus technology. Total RNA was extracted from A549 cells infected by different rMVs or from A549, HEK293-T7, or HEK293-T7-NP cells transfected with different in vitro-transcribed DI-RNAs at 24 h, using an RNeasy minikit. Total RNAs extracted from HEK293-T7 and HEK293-T7-NP cells transfected with p2RZ vector in the presence or absence of pEMC-La (a kind gift of M. A. Billeter) were treated with RQ1 RNase-free DNase (Promega). Expression levels of IFN-β or ISG (Mx1 and DHX58) were quantified by one-step real-time PCR, using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA expression as an internal control. One hundred nanograms of total RNA was amplified with 20× custom TaqMan gene expression assays (IFN-β, Hs01077958_s1; Mx1, Hs00895608_m1; DHX58, Hs00225561_m1; and GAPDH, Hs99999905_m1) (Life Technologies) by using a TaqMan RNA-to-Ct 1-Step kit in accordance with the manufacturer's instructions. All measurements were performed in triplicate and analyzed by use of the StepOnePlus software system.
Bioinformatics tools for DI-RNA analysis.
Weblogo and MUSCLE (multiple-sequence comparison by log expectation) software was used for multiple-sequence alignment analysis. The mFold Web server was used to predict RNA secondary structures. GC ratios were calculated using the GC Content Calculator (BiologicsCorp). All software consulted is available via free access online.
ACKNOWLEDGMENTS
We thank Martin Billeter, Yves Jacob, and Pierre Charneau for providing material, Phanramphoei Frantz (Unité de Génomique Virale et Vaccination, Institut Pasteur) for a critical reading of the manuscript, and Bernd Jagla (Hub Bioinformatique et Biostatistique, Institut Pasteur) for help with bioinformatics analysis of DI-RNAs. We thank all the members of the Viral Genomics and Vaccination Research Unit for their help and useful discussions.
This work was supported by the Institut Pasteur and the CNRS. M.M. was supported by the Institut de Recherche Biomédicale des Armées (IRBA), and R.Y.S.D. was supported by the Fondation pour la Recherche Médicale (grant FDT20140931129).
REFERENCES
- 1.Lazzarini RA, Keene JD, Schubert M. 1981. The origins of defective interfering particles of the negative-strand RNA viruses. Cell 26:145–154. doi: 10.1016/0092-8674(81)90298-1. [DOI] [PubMed] [Google Scholar]
- 2.Dimmock NJ, Easton AJ. 2014. Defective interfering influenza virus RNAs: time to reevaluate their clinical potential as broad-spectrum antivirals? J Virol 88:5217–5227. doi: 10.1128/JVI.03193-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez CB. 2014. Defective viral genomes: critical danger signals of viral infections. J Virol 88:8720–8723. doi: 10.1128/JVI.00707-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gubbay O, Curran J, Kolakofsky D. 2001. Sendai virus genome synthesis and assembly are coupled: a possible mechanism to promote viral RNA polymerase processivity. J Gen Virol 82:2895–2903. doi: 10.1099/0022-1317-82-12-2895. [DOI] [PubMed] [Google Scholar]
- 5.Bellini WJ, Englund G, Rozenblatt S, Arnheiter H, Richardson CD. 1985. Measles virus P gene codes for two proteins. J Virol 53:908–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattaneo R, Kaelin K, Baczko K, Billeter MA. 1989. Measles virus editing provides an additional cysteine-rich protein. Cell 56:759–764. doi: 10.1016/0092-8674(89)90679-X. [DOI] [PubMed] [Google Scholar]
- 7.Strahle L, Garcin D, Kolakofsky D. 2006. Sendai virus defective-interfering genomes and the activation of interferon-beta. Virology 351:101–111. doi: 10.1016/j.virol.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 8.Shingai M, Ebihara T, Begum NA, Kato A, Honma T, Matsumoto K, Saito H, Ogura H, Matsumoto M, Seya T. 2007. Differential type I IFN-inducing abilities of wild-type versus vaccine strains of measles virus. J Immunol 179:6123–6133. doi: 10.4049/jimmunol.179.9.6123. [DOI] [PubMed] [Google Scholar]
- 9.Mercado-Lopez X, Cotter CR, Kim WK, Sun Y, Munoz L, Tapia K, Lopez CB. 2013. Highly immunostimulatory RNA derived from a Sendai virus defective viral genome. Vaccine 31:5713–5721. doi: 10.1016/j.vaccine.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komarova AV, Combredet C, Sismeiro O, Dillies MA, Jagla B, Sanchez David RY, Vabret N, Coppee JY, Vidalain PO, Tangy F. 2013. Identification of RNA partners of viral proteins in infected cells. RNA Biol 10:944–956. doi: 10.4161/rna.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baum A, Sachidanandam R, Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci U S A 107:16303–16308. doi: 10.1073/pnas.1005077107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Runge S, Sparrer KM, Lassig C, Hembach K, Baum A, Garcia-Sastre A, Soding J, Conzelmann KK, Hopfner KP. 2014. In vivo ligands of MDA5 and RIG-I in measles virus-infected cells. PLoS Pathog 10:e1004081. doi: 10.1371/journal.ppat.1004081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez David RY, Combredet C, Sismeiro O, Dillies MA, Jagla B, Coppee JY, Mura M, Guerbois Galla M, Despres P, Tangy F, Komarova AV. 2016. Comparative analysis of viral RNA signatures on different RIG-I-like receptors. eLife 5:e11275. doi: 10.7554/eLife.11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ho TH, Kew C, Lui PY, Chan CP, Satoh T, Akira S, Jin DY, Kok KH. 2015. PACT- and RIG-I-dependent activation of type I interferon production by a defective interfering RNA derived from measles virus vaccine. J Virol 90:1557–1568. doi: 10.1128/JVI.02161-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaller CK, Radeke MJ, Cattaneo R, Samuel CE. 2014. Measles virus C protein impairs production of defective copyback double-stranded viral RNA and activation of protein kinase R. J Virol 88:456–468. doi: 10.1128/JVI.02572-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Saira K, Lin X, DePasse JV, Halpin R, Twaddle A, Stockwell T, Angus B, Cozzi-Lepri A, Delfino M, Dugan V, Dwyer DE, Freiberg M, Horban A, Losso M, Lynfield R, Wentworth DN, Holmes EC, Davey R, Wentworth DE, Ghedin E, INSIGHT FLU002 Study Group, INSIGHT FLU003 Study Group. 2013. Sequence analysis of in vivo defective interfering-like RNA of influenza A H1N1 pandemic virus. J Virol 87:8064–8074. doi: 10.1128/JVI.00240-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li D, Lott WB, Lowry K, Jones A, Thu HM, Aaskov J. 2011. Defective interfering viral particles in acute dengue infections. PLoS One 6:e19447. doi: 10.1371/journal.pone.0019447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prince AM, Huima-Byron T, Parker TS, Levine DM. 1996. Visualization of hepatitis C virions and putative defective interfering particles isolated from low-density lipoproteins. J Viral Hepat 3:11–17. doi: 10.1111/j.1365-2893.1996.tb00075.x. [DOI] [PubMed] [Google Scholar]
- 19.Sun Y, Jain D, Koziol-White CJ, Genoyer E, Gilbert M, Tapia K, Panettieri RA Jr, Hodinka RL, Lopez CB. 2015. Immunostimulatory defective viral genomes from respiratory syncytial virus promote a strong innate antiviral response during infection in mice and humans. PLoS Pathog 11:e1005122. doi: 10.1371/journal.ppat.1005122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kessler JR, Kremer JR, Muller CP. 2011. Interplay of measles virus with early induced cytokines reveals different wild type phenotypes. Virus Res 155:195–202. doi: 10.1016/j.virusres.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 21.Calain P, Roux L. 1988. Generation of measles virus defective interfering particles and their presence in a preparation of attenuated live-virus vaccine. J Virol 62:2859–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whistler T, Bellini WJ, Rota PA. 1996. Generation of defective interfering particles by two vaccine strains of measles virus. Virology 220:480–484. doi: 10.1006/viro.1996.0335. [DOI] [PubMed] [Google Scholar]
- 23.Pulendran B, Oh JZ, Nakaya HI, Ravindran R, Kazmin DA. 2013. Immunity to viruses: learning from successful human vaccines. Immunol Rev 255:243–255. doi: 10.1111/imr.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller CK, Mastorakos GM, Matchett WE, Ma X, Samuel CE, Cattaneo R. 2015. Measles virus defective interfering RNAs are generated frequently and early in the absence of C protein and can be destabilized by adenosine deaminase acting on RNA-1-like hypermutations. J Virol 89:7735–7747. doi: 10.1128/JVI.01017-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brandler S, Tangy F. 2008. Recombinant vector derived from live attenuated measles virus: potential for flavivirus vaccines. Comp Immunol Microbiol Infect Dis 31:271–291. doi: 10.1016/j.cimid.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Ramsauer K, Schwameis M, Firbas C, Mullner M, Putnak RJ, Thomas SJ, Despres P, Tauber E, Jilma B, Tangy F. 2015. Immunogenicity, safety, and tolerability of a recombinant measles-virus-based chikungunya vaccine: a randomised, double-blind, placebo-controlled, active-comparator, first-in-man trial. Lancet Infect Dis 15:519–527. doi: 10.1016/S1473-3099(15)70043-5. [DOI] [PubMed] [Google Scholar]
- 27.Combredet C, Labrousse V, Mollet L, Lorin C, Delebecque F, Hurtrel B, McClure H, Feinberg MB, Brahic M, Tangy F. 2003. A molecularly cloned Schwarz strain of measles virus vaccine induces strong immune responses in macaques and transgenic mice. J Virol 77:11546–11554. doi: 10.1128/JVI.77.21.11546-11554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolakofsky D, Roux L, Garcin D, Ruigrok RW. 2005. Paramyxovirus mRNA editing, the “rule of six” and error catastrophe: a hypothesis. J Gen Virol 86:1869–1877. doi: 10.1099/vir.0.80986-0. [DOI] [PubMed] [Google Scholar]
- 29.Brandler S, Ruffie C, Combredet C, Brault JB, Najburg V, Prevost MC, Habel A, Tauber E, Despres P, Tangy F. 2013. A recombinant measles vaccine expressing chikungunya virus-like particles is strongly immunogenic and protects mice from lethal challenge with chikungunya virus. Vaccine 31:3718–3725. doi: 10.1016/j.vaccine.2013.05.086. [DOI] [PubMed] [Google Scholar]
- 30.Guerbois M, Moris A, Combredet C, Najburg V, Ruffie C, Fevrier M, Cayet N, Brandler S, Schwartz O, Tangy F. 2009. Live attenuated measles vaccine expressing HIV-1 Gag virus like particles covered with gp160DeltaV1V2 is strongly immunogenic. Virology 388:191–203. doi: 10.1016/j.virol.2009.02.047. [DOI] [PubMed] [Google Scholar]
- 31.Pathak KB, Nagy PD. 2009. Defective interfering RNAs: foes of viruses and friends of virologists. Viruses 1:895–919. doi: 10.3390/v1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Desmyter J, Melnick JL, Rawls WE. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J Virol 2:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lucas-Hourani M, Munier-Lehmann H, Helynck O, Komarova A, Despres P, Tangy F, Vidalain PO. 2014. High-throughput screening for broad-spectrum chemical inhibitors of RNA viruses. J Vis Exp 2014:51222. doi: 10.3791/51222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saito T, Hirai R, Loo YM, Owen D, Johnson CL, Sinha SC, Akira S, Fujita T, Gale M Jr. 2007. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc Natl Acad Sci U S A 104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li S, Wang L, Berman M, Kong YY, Dorf ME. 2011. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parks CL, Witko SE, Kotash C, Lin SL, Sidhu MS, Udem SA. 2006. Role of V protein RNA binding in inhibition of measles virus minigenome replication. Virology 348:96–106. doi: 10.1016/j.virol.2005.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Parks CL, Lerch RA, Walpita P, Wang HP, Sidhu MS, Udem SA. 2001. Analysis of the noncoding regions of measles virus strains in the Edmonston vaccine lineage. J Virol 75:921–933. doi: 10.1128/JVI.75.2.921-933.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salinas Y, Roux L. 2005. Replication and packaging properties of short paramyxovirus defective RNAs. Virus Res 109:125–132. doi: 10.1016/j.virusres.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 39.Bhella D, Ralph A, Yeo RP. 2004. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J Mol Biol 340:319–331. doi: 10.1016/j.jmb.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 40.Tapparel C, Maurice D, Roux L. 1998. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol 72:3117–3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perrault J. 1981. Origin and replication of defective interfering particles. Curr Top Microbiol Immunol 93:151–207. [DOI] [PubMed] [Google Scholar]
- 42.Nakatsu Y, Takeda M, Ohno S, Koga R, Yanagi Y. 2006. Translational inhibition and increased interferon induction in cells infected with C protein-deficient measles virus. J Virol 80:11861–11867. doi: 10.1128/JVI.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, Taira K, Foy E, Loo YM, Gale M Jr, Akira S, Yonehara S, Kato A, Fujita T. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol 175:2851–2858. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 44.Rothenfusser S, Goutagny N, DiPerna G, Gong M, Monks BG, Schoenemeyer A, Yamamoto M, Akira S, Fitzgerald KA. 2005. The RNA helicase Lgp2 inhibits TLR-independent sensing of viral replication by retinoic acid-inducible gene-I. J Immunol 175:5260–5268. doi: 10.4049/jimmunol.175.8.5260. [DOI] [PubMed] [Google Scholar]
- 45.Bruns AM, Horvath CM. 2015. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine 74:198–206. doi: 10.1016/j.cyto.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bruns AM, Leser GP, Lamb RA, Horvath CM. 2014. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol Cell 55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Deddouche S, Goubau D, Rehwinkel J, Chakravarty P, Begum S, Maillard PV, Borg A, Matthews N, Feng Q, van Kuppeveld FJ, Reis e Sousa C. 2014. Identification of an LGP2-associated MDA5 agonist in picornavirus-infected cells. eLife 3:e01535. doi: 10.7554/eLife.01535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berghall H, Siren J, Sarkar D, Julkunen I, Fisher PB, Vainionpaa R, Matikainen S. 2006. The interferon-inducible RNA helicase, mda-5, is involved in measles virus-induced expression of antiviral cytokines. Microbes Infect 8:2138–2144. doi: 10.1016/j.micinf.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 49.Yao H, Dittmann M, Peisley A, Hoffmann HH, Gilmore RH, Schmidt T, Schmid-Burgk JL, Hornung V, Rice CM, Hur S. 2015. ATP-dependent effector-like functions of RIG-I-like receptors. Mol Cell 58:541–548. doi: 10.1016/j.molcel.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Longhi S, Bloyet LM, Gianni S, Gerlier D. 2017. How order and disorder within paramyxoviral nucleoproteins and phosphoproteins orchestrate the molecular interplay of transcription and replication. Cell Mol Life Sci 74:3091–3118. doi: 10.1007/s00018-017-2556-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Curran J, Marq JB, Kolakofsky D. 1995. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J Virol 69:849–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J 14:5773–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parks CL, Lerch RA, Walpita P, Sidhu MS, Udem SA. 1999. Enhanced measles virus cDNA rescue and gene expression after heat shock. J Virol 73:3560–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giraudon P, Wild TF. 1981. Monoclonal antibodies against measles virus. J Gen Virol 54:325–332. doi: 10.1099/0022-1317-54-2-325. [DOI] [PubMed] [Google Scholar]