ABSTRACT
In the human hepatoma cell line Huh7, the coexpression of the coactivators peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α), cyclic AMP-responsive element binding protein binding protein (CBP), steroid receptor coactivator 1 (SRC1), and protein arginine methyltransferase 1 (PRMT1) only modestly increase hepatitis B virus (HBV) biosynthesis. However, by utilizing the human embryonic kidney cell line HEK293T, it was possible to demonstrate that PGC1α alone can support viral biosynthesis independently of the expression of additional coactivators or transcription factors. In contrast, additional coactivators failed to support robust HBV replication in the absence of PGC1α. These observations indicate that PGC1α represents a novel adaptor molecule capable of recruiting the necessary transcriptional machinery to the HBV nucleocapsid promoter to modestly enhance viral pregenomic 3.5-kb RNA synthesis. Although this change in transcription is associated with a similar modest change in hepatitis B virus core antigen polypeptide (HBcAg/p21) synthesis, it mediates a dramatic increase in viral capsid production and robust viral replication. Therefore, it is apparent that the synthesis of cytoplasmic HBcAg/p21 above a critical threshold level is required for the efficient assembly of HBV replication-competent viral capsids.
IMPORTANCE Hepatitis B virus (HBV) is a major human pathogen, and novel targets for the development of additional therapeutic agents are urgently needed. Here we demonstrate that the coactivator peroxisome proliferator-activated receptor γ coactivator 1α (PGC1α) serves as a unique adaptor molecule for the recruitment of additional coactivator proteins, which can finely regulate HBV transcription. The consequence of this precise regulation of viral RNA levels by PGC1α is a subtle increase in cytoplasmic HBcAg/p21 polypeptide translation, which shifts the equilibrium from dimer formation dramatically in favor of viral capsid assembly. These findings suggest that both PGC1α and capsid assembly may represent attractive targets for the development of antiviral agents against chronic HBV infection.
KEYWORDS: PGC1α, capsid assembly, hepatitis B virus, transcriptional coactivators
INTRODUCTION
Hepatitis B virus (HBV) replicates by the reverse transcription of the viral pregenomic 3.5-kb RNA (1, 2). As the transcription of viral genomic DNA is essentially limited to hepatocytes (3, 4), it appears that HBV biosynthesis is restricted to the liver by the action of liver-enriched transcription factors, especially nuclear receptors such as hepatocyte nuclear factor 4 (HNF4), retinoid X receptor (RXR), peroxisome proliferator-activated receptor (PPAR), farnesoid X receptor (FXR), and liver receptor homolog 1 (LRH1), which also play a major role in governing the metabolic function of the liver (3, 5, 6). However, the molecular mechanisms leading to the formation of a functional transcription preinitiation complex on viral promoter sequences are poorly defined, and the potential roles of specific coactivators in governing tissue-specific viral biosynthesis have not been extensively investigated (7–10). Furthermore, it is apparent that HBV transcription in vivo occurs in the context of chromatin, which must be modified by transcriptional coactivators to permit efficient viral transcription (11).
Transcriptional coactivators are generally defined as proteins that directly or indirectly interact with the transcription factors bound to the enhancer and promoter DNA sequences and regulate gene expression levels by recruiting the basic transcription machinery, including the general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH), mediator, and RNA polymerase II. Transcriptional coactivators often have enzymatic activities that can covalently modify DNA-bound transcription factors, transcriptional coactivators, and chromatin-associated histones to alter their regulatory functions (12–16). Histone acetyltransferases (HATs) acetylate histones, altering the conformation of chromatin so that enhancer and promoter sequences are more accessible for the recruitment of the transcriptional machinery necessary to mediate RNA synthesis. In particular, the cyclic AMP-responsive element binding protein binding protein (CBP)/p300 and steroid receptor coactivator 1 to 3 (SRC1-3)/p160 classes of HATs have been shown to increase the levels of transcription of genes when they have been recruited to their associated proximal promoter sequences (17). Furthermore, acetylation of DNA binding transcription factors by HATs can also influence their effect on transcriptional activity (13). Similarly, protein arginine methyltransferases (PRMTs) such as PRMT1 can modulate promoter activities by methylating arginine residues present in transcription factors, transcriptional coactivators, and histones, hence altering preinitiation complex formation and activity at the RNA start sites for targeted genes (12, 14–22).
In contrast to coactivators that display enzymatic functions, some transcriptional coactivators, such as the peroxisome proliferator-activated receptor γ coactivator 1 (PGC1) family members PGC1α, PGC1β, and peroxisome proliferator-activated receptor γ coactivator-related 1 (PRC), appear to lack such properties but serve as adaptor molecules governing the subsequent recruitment of additional distinct coactivators to specific promoters (23). Furthermore, it is clear that many of these coactivators possess peptide domains that promote their interactions not only with the transcription factors that recruit them to enhancer and promoter sequences but also with other coactivators (15, 17, 22–26). In this manner, it appears that coactivators of various classes may exist within cells in various coactivator complexes, permitting their simultaneous recruitment to specific promoters as a consequence of these interactions (23). In this study, we demonstrate that PGC1α can serve as an essential adaptor molecule for the recruitment of additional coactivators to the HBV nucleocapsid promoter to modestly enhance the expression of the viral pregenomic 3.5-kb RNA while dramatically stimulating HBV biosynthesis. This pathway may be distinct from the previously described activation of HBV transcription and replication by nuclear receptors (HNF4, RXR, PPAR, FXR, and LRH1), which can activate viral biosynthesis in the absence of PGC1 coactivators (3, 5, 6). These observations suggest that there are multiple independent transcriptional pathways capable of supporting HBV biosynthesis. Furthermore, it has been shown that modest increases in HBcAg/p21 protein synthesis can be associated with dramatic increases in viral capsid formation and the associated cytoplasmic HBV replication intermediate DNA. These observations indicate that HBV biosynthesis is subject to posttranscriptional regulation mediated by a threshold concentration of HBcAg/p21 above which efficient subunit oligomerization leads to efficient capsid formation and viral replication.
RESULTS
Transcriptional coactivators enhance HBV biosynthesis in human hepatoma Huh7 cells.
Transfection of the HBV DNA (4.1-kbp) construct into Huh7 cells supports HBV transcription and replication (Fig. 1A and B, lane 1). Furthermore, cotransfection of transcriptional coactivators enhances the synthesis of HBV RNA and DNA to a modest extent (Fig. 1). Various combinations of coactivators of different classes, including PGC1α, CBP (histone lysine acetyltransferase, p300 class), SRC1 (histone lysine acetyltransferase, p160 class), and PRMT1 (protein arginine methyltransferase), were more effective, in general, than individual coactivators alone, which were relatively ineffective at modulating viral biosynthesis. As the effects on Huh7 cells were modest (Fig. 1C), it is not possible to determine the potential roles of the individual coactivators in HBV biosynthesis without examining their effects in a more responsive system.
FIG 1.
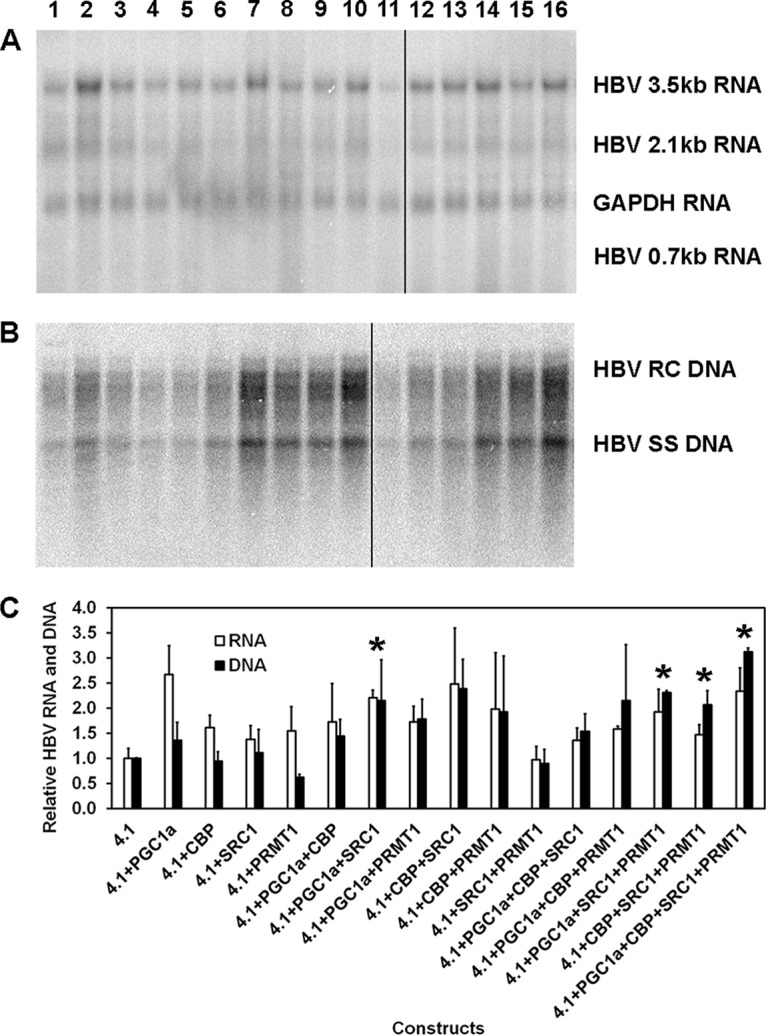
Effects of transcriptional coactivators on HBV biosynthesis in the human hepatoma cell line Huh7. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcript was used as an internal control for RNA loading per lane. The black line indicates noncontiguous lanes from a single filter hybridization analysis. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. HBV RC DNA, HBV relaxed circular DNA; HBV SS DNA, HBV single-stranded DNA. Cells were transfected with the HBV DNA (4.1-kbp) construct (lanes 1 to 16) plus PGC1α (lanes 2, 6 to 8, 12 to 14, and 16), CBP (lanes 3, 6, 9, 10, 12, 13, 15, and 16), SRC1 (lanes 4, 7, 9, 11, 12, and 14 to 16), and PRMT1 (lanes 5, 8, 10, 11, and 13 to 16) expression vectors, as indicated. The black line indicates noncontiguous lanes from a single filter hybridization analysis. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. The levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates are reported relative to the value for the HBV DNA (4.1-kbp) construct (lane 1). The mean RNA and DNA levels plus standard deviations from two independent analyses are shown. Levels of the transcripts (lane 7) and replication intermediates (lanes 14 to 16) in coactivator-expressing cells that are statistically significantly higher than the levels in cells transfected with the HBV DNA (4.1-kbp) construct only (lane 1), as determined by Student's t test (P < 0.05), are indicated with an asterisk.
Transcriptional coactivators support HBV biosynthesis in human embryonic kidney HEK293T cells.
Transfection of the HBV DNA (4.1-kbp) construct into HEK293T cells resulted in detectable levels of HBV 3.5-kb RNA but no detectable viral replication intermediates (Fig. 2A and B, lane 1). However, cotransfection of the HBV DNA (4.1-kbp) construct with the PGC1α expression vector increased HBV 3.5-kb RNA levels approximately 2-fold, while HBV replication intermediate DNA synthesis became readily apparent (Fig. 2A and B, lane 2). This observation suggested that a modest change in HBV transcription can be associated with a large change in HBV replication in this system. Furthermore, PGC1α was the only coactivator tested alone with the ability to support readily detectable levels of HBV DNA synthesis (Fig. 2B, lanes 2 to 5). Combinations of the coactivators CBP, SRC1, and PRMT1 enhanced PGC1α-mediated HBV transcription and replication further only if at least two additional coactivators were present (Fig. 2A and B, lanes 12 to 14 and 16). In the absence of PGC1α expression, HBV RNA levels were essentially unchanged by the expression of additional coactivators, and viral biosynthesis was not readily detectable (Fig. 2A and B, lanes 9 to 11 and 15). These observations suggested that PGC1α acted as an adaptor molecule for the recruitment of the other coactivators CBP, SRC1, and PRMT1 to enhance the synthesis of HBV RNA and DNA.
FIG 2.
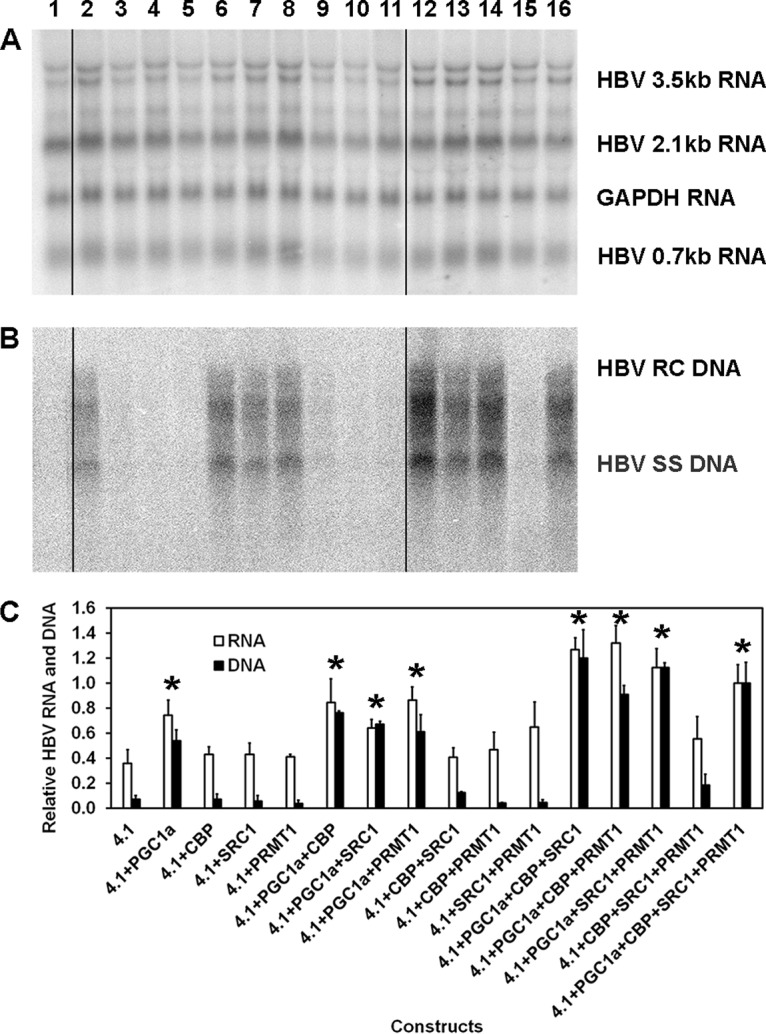
Effects of transcriptional coactivators on HBV biosynthesis in the human embryonic kidney cell line HEK293T. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The 3.9-kb transcript observed above the HBV 3.5-kb RNA probably represents the previously reported HBV long xRNA that initiates from the X promoter region (55). The GAPDH transcript was used as an internal control for RNA loading per lane. The black lines indicate noncontiguous lanes from a single filter hybridization analysis. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. Cells were transfected with the HBV DNA (4.1-kbp) construct (lanes 1 to 16) plus PGC1α (lanes 2, 6 to 8, 12 to 14, and 16), CBP (lanes 3, 6, 9, 10, 12, 13, 15, and 16), SRC1 (lanes 4, 7, 9, 11, 12, and 14 to 16), and PRMT1 (lanes 5, 8, 10, 11, and 13 to 16) expression vectors, as indicated. The black lines indicate noncontiguous lanes from a single filter hybridization analysis. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. The levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates are reported relative to the values for the HBV DNA (4.1-kbp) construct in the presence of the expression of the four coactivators (lane 16). The mean RNA and DNA levels plus standard deviations from three independent analyses are shown. Levels of the transcripts (lanes 2, 7, 8, 12 to 14, and 16) and replication intermediates (lanes 2, 6 to 8, 12 to 14, and 16) in coactivator-expressing cells that are statistically significantly higher than the levels in cells transfected with the HBV DNA (4.1-kbp) construct only (lane 1), as determined by Student's t test (P < 0.05), are indicated with an asterisk.
Role of the proximal nuclear receptor binding site in transcriptional coactivator-mediated activation of HBV biosynthesis in HEK293T cells.
A major transcriptional regulatory element within the HBV nucleocapsid promoter governing HBV biosynthesis is the proximal nuclear receptor binding site located approximately 60 nucleotides upstream of the viral pregenomic 3.5-kb RNA transcription site (6, 27). Furthermore, it is known that PGC1α can interact with various nuclear receptors to activate HBV transcription and replication (9, 10). Therefore, the role of this binding site in coactivator-mediated HBV biosynthesis was investigated (Fig. 3). As noted previously, HNF4-mediated HBV transcription and replication were highly sensitive to mutation of the HBV nucleocapsid proximal nuclear receptor binding site (Fig. 3, lanes 2, 6, 8, and 12) (6, 27). Similarly, coactivator-mediated HBV biosynthesis was also mediated, although to a lesser extent, through the HBV nucleocapsid proximal nuclear receptor binding site (Fig. 3, lanes 3 to 5 and 9 to 11). These observations indicate that PGC1α-mediated coactivator-enhanced HBV biosynthesis is governed, in part, by transcription factors binding to the proximal nuclear receptor binding site and, presumably, additional viral regulatory sequence elements.
FIG 3.
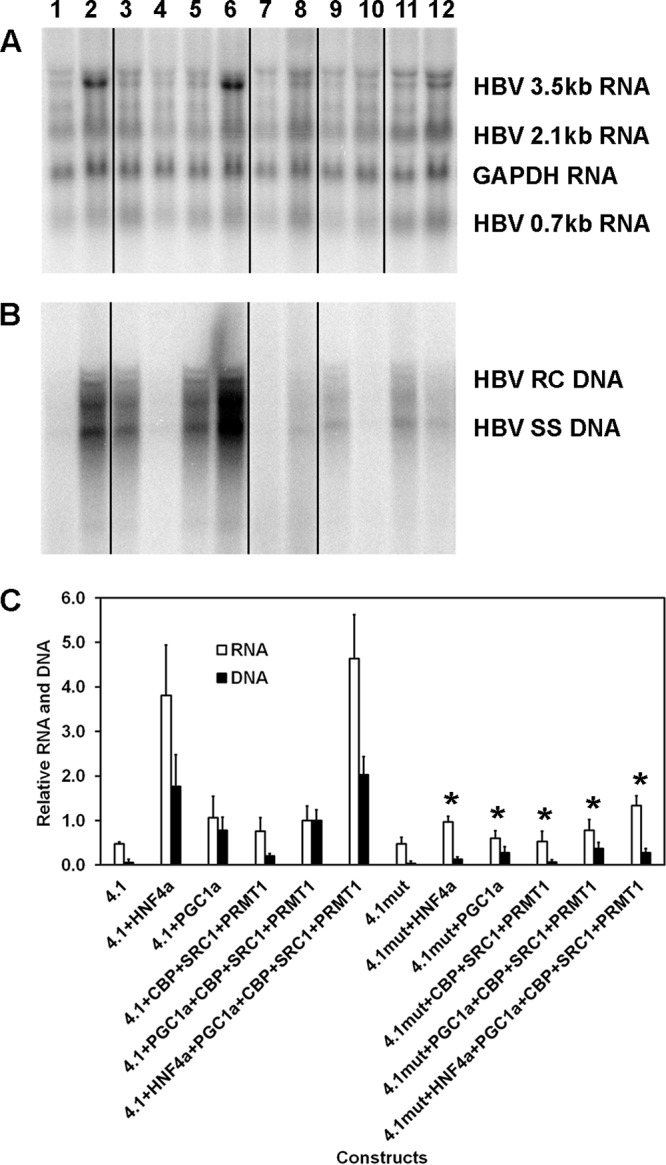
Role of the proximal nuclear receptor binding site in transcriptional coactivator-mediated activation of HBV biosynthesis in the human embryonic kidney cell line HEK293T. (A) RNA (Northern) filter hybridization analysis of HBV transcripts. The 3.9-kb transcript observed above the HBV 3.5-kb RNA probably represents the previously reported HBV long xRNA that initiates from the X promoter region (55). The GAPDH transcript was used as an internal control for RNA loading per lane. The black lines indicate noncontiguous lanes from a single filter hybridization analysis. (B) DNA (Southern) filter hybridization analysis of HBV replication intermediates. Cells were transfected with the HBV DNA (4.1-kbp) or HBVHNF4mut DNA (4.1-kbp) constructs (lanes 1 to 6 and 7 to 12, respectively) plus the HNF4α (lanes 2 and 8), PGC1α (lanes 3, 5, 6, 9, 11, and 12), CBP, SRC1, and PRMT1 (lanes 4 to 6 and 10 to 12) expression vectors. The black lines indicate noncontiguous lanes from a single filter hybridization analysis. (C) Quantitative analysis of the HBV 3.5-kb RNA and HBV DNA replication intermediates. The levels of the HBV 3.5-kb RNA and total HBV DNA replication intermediates are reported relative to the value for HBV DNA (4.1-kbp) construct in the presence of the expression of the four coactivators (lane 5). Mean RNA and DNA levels plus standard deviations from five independent analyses are shown. Levels of the transcripts (lanes 8 and 12) and replication intermediates (lanes 8 to 12) in HBVHNF4mut DNA (4.1-kbp) construct-transfected cells (lanes 7 to 12) that are statistically significantly lower than the levels in HBV DNA (4.1-kbp) construct-transfected cells expressing the same transcription factors and/or coactivators (lanes 1 to 6), as determined by Student's t test (P < 0.05), are indicated with an asterisk.
Mechanism of regulation of HBV biosynthesis by transcriptional coactivators in HEK293T cells.
Analysis of the effects of coactivators on HBV biosynthesis in HEK293T cells indicated that very modest changes in viral transcription can be associated with the induction of viral replication (Fig. 2). However, the mechanism responsible for this observation is not apparent from this analysis. Consequently, this issue was investigated further by initially determining the relative HBV precore and pregenomic 3.5-kb RNA levels by primer extension analysis (Fig. 4A). The ratio of HBV precore to pregenomic 3.5-kb RNAs did not change greatly (mean, 1.32; standard deviation [SD], 0.39) as a result of coactivator expression and correlated with the level of HBV 3.5-kb RNA detected by RNA filter hybridization analysis (Fig. 2). Furthermore, the level of the HBcAg/p21 polypeptide was consistent with the abundance of the HBV pregenomic 3.5-kb RNA (Fig. 4B). Remarkably, HBcAg/capsids and HBV replication intermediate DNA were readily detectable only in HEK293T cells that were expressing PGC1α (Fig. 4C and D). These observations support the contention that small increases in the levels of HBV pregenomic 3.5-kb RNA mediated by the adaptor function of the PGC1α coactivator lead to small increases in HBcAg/p21 polypeptide synthesis, which subsequently supports HBcAg/capsid formation, and, hence, viral replication, when the concentration of the HBcAg/p21 polypeptide reaches a critical threshold level. Quantitative analysis of these parameters supports this contention (Fig. 4E). The maximum relative level of HBV pregenomic 3.5-kb RNA that fails to support HBV capsid formation is approximately 0.3, whereas the minimum relative level of this transcript that supports robust HBV capsid formation is approximately 0.4 (Fig. 4E). Similarly, the maximum relative level of HBcAg/p21 that fails to support HBV capsid formation and viral replication is approximately 0.4, whereas the minimum relative level of this polypeptide that supports robust HBV capsid formation is approximately 0.7 (Fig. 4E). These observations indicate that an ∼2-fold increase in the levels of HBV pregenomic 3.5-kb RNA and HBcAg/p21 polypeptide is sufficient for the induction of efficient HBV biosynthesis in HEK293T cells (Fig. 2 and 4).
FIG 4.
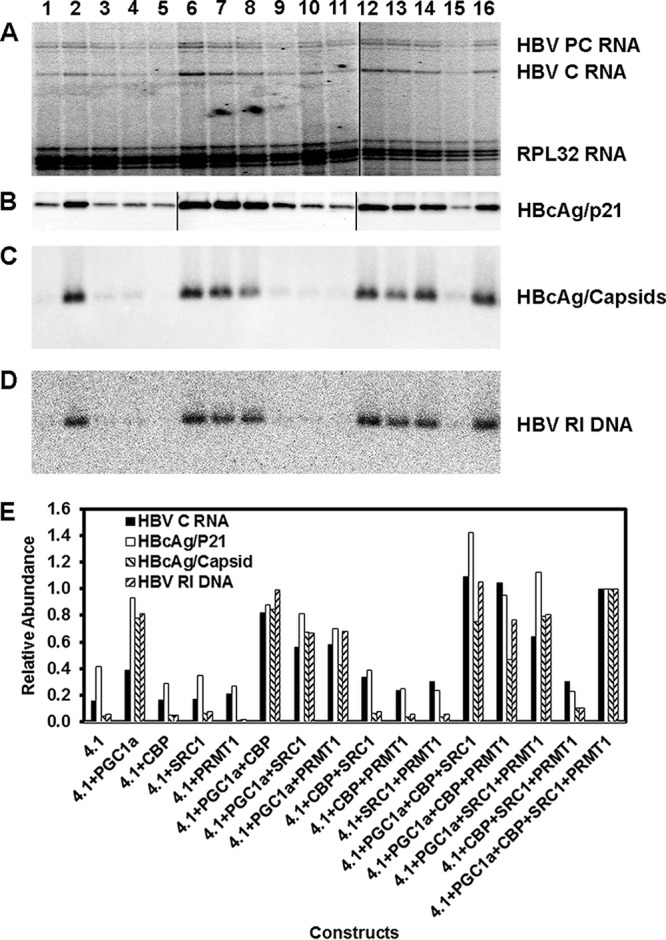
Effects of transcriptional coactivators on HBV 3.5-kb RNA, HBcAg/p21, HBcAg/capsid, and capsid-associated HBV replication intermediate levels in the human embryonic kidney cell line HEK293T. (A) Primer extension analysis of HBV precore (PC) and pregenomic or core (C) 3.5-kb RNAs. The 32-kDa large ribosomal protein subunit (RPL32) transcript was used as an internal control for RNA loading per lane. The black line indicates noncontiguous lanes derived from a single primer extension analysis resolved on two individual sequencing gels. (B) Protein (Western) filter immunodetection analysis of immunoprecipitated HBcAg/p21 protein present in cytoplasmic cell extracts. The black lines indicate noncontiguous lanes derived from the same immunodetection analysis of three individual membranes. (C) Protein (Western) filter immunodetection analysis of HBcAg/capsids present in cytoplasmic cell extracts. (D) DNA (Southern) filter hybridization analysis of HBV replication intermediate (RI) DNA present within HBcAg/capsids. Cells were transfected with the HBV DNA (4.1-kbp) construct (lanes 1 to 16) plus PGC1α (lanes 2, 6 to 8, 12 to 14, and 16), CBP (lanes 3, 6, 9, 10, 12, 13, 15, and 16), SRC1 (lanes 4, 7, 9, 11, 12, and 14 to 16), and PRMT1 (lanes 5, 8, 10, 11, and 13 to 16) expression vectors, as indicated. (E) Quantitative analysis of the HBV pregenomic/core (C) 3.5-kb RNA, HBcAg/p21 protein, HBcAg/capsid protein, and HBV replication intermediate (RI) DNA. The levels of HBV RNA, DNA, and protein are reported relative to the values for the HBV DNA (4.1-kbp) construct in the presence of the expression of the four coactivators (lane 16).
DISCUSSION
Transcriptional coactivators represent an additional level of regulation of gene expression (28, 29). Previously, it was shown that the transcriptional coactivator PGC1α was capable of activating the synthesis of HBV RNA and DNA in human hepatoma cell lines and differentially regulating HBV biosynthesis through its interactions with several nuclear receptors in HEK293T cells (9, 10). However, it was unclear to which extent PGC1α could interact functionally with the endogenous transcription factors present within HEK293T cells and whether or not it could generate a productive preinitiation complex capable of supporting HBV 3.5-kb RNA synthesis (9, 10). Data from RNA filter hybridization analyses suggested that HBV 3.5-kb RNA was transcribed from the viral genome in HEK293T cells at a modest level in the absence of exogenously expressed factors (Fig. 2A, lane 1). Primer extension analysis confirmed that the HBV 3.5-kb RNA transcribed from the viral genome represented approximately equal amounts of precore and pregenomic RNAs (Fig. 4A, lane 1). Therefore, it was unclear why no viral DNA synthesis was readily apparent (Fig. 2B, lane 1). However, these observations indicated that the transcription factors and coactivators present in HEK293T cells were capable of supporting limited transcription of HBV 3.5-kb RNA but they were not sufficient to support HBV replication (Fig. 5, steps 1 and 2).
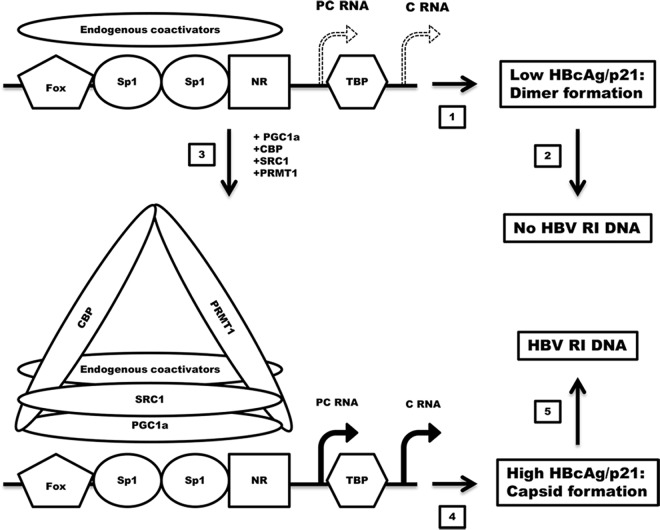
FIG 5.
Diagrammatic representation of the coactivators assembled on the HBV nucleocapsid promoter and their effects on HBV transcription, HBcAg/p21, HBcAg/capsid, and capsid-associated HBV replication intermediate (RI) DNA. PC RNA, HBV precore 3.5-kb RNA; C RNA, HBV pregenomic or core 3.5-kb RNA; Fox, fork head box transcription factor (56); Sp1, specificity protein 1 transcription factor (57); NR, nuclear receptor transcription factor (i.e., HNF4, RXR, PPAR, FXR, LRH1, and estrogen-related receptor [ERR]) (3, 6, 48); TBP, TATA-binding protein. (1 and 2) Endogenous coactivators support limited HBV 3.5-kb RNA expression (1) and relatively low levels of HBcAg/p21 polypeptide synthesis (2), which are not sufficient to support capsid formation, and hence, there is no viral DNA replication. (3) Recruitment of CBP, SRC1, and PRMT1 plus potentially additional endogenous coactivators by PGC1α to the HBV nucleocapsid promoter. (4) PGC1α, CBP, SRC1, and PRMT1 plus endogenous coactivators support modestly increased levels of HBV 3.5-kb RNA expression. (5) The translation of modestly increased levels of HBcAg/p21 polypeptide synthesis crosses a critical threshold that is required to support capsid formation and associated HBV DNA replication.
The expression of PGC1α alone was sufficient to induce robust HBV replication (Fig. 2B, lane 2). Viral replication was associated with only a modest increase in HBV 3.5-kb RNA synthesis (Fig. 2A, lane 2). This observation suggested that small changes in viral transcription can be associated with dramatic increases in HBV DNA synthesis (Fig. 5, steps 3 to 5). The expression of additional individual coactivators of various classes, including CBP, SRC1, and PRMT1, was not able to support robust viral biosynthesis, indicating that PGC1α displayed a distinct functional activity that was not associated with other coactivators. Furthermore, HBV biosynthesis could be enhanced further by combinations of the CBP, SRC1, and PRMT1 coactivators only in the presence of PGC1α but not in its absence (Fig. 2). These observations indicate that PGC1α serves as an adaptor molecule for the recruitment and/or activation of the other coactivators as they assemble at the HBV nucleocapsid proximal promoter region (23) (Fig. 5, step 3). Consistent with this suggestion is the finding that PGC1α, CBP, and SRC1 are known to associate and form a larger coactivator complex, which might permit their simultaneous recruitment to the transcriptional elements that are subject to their regulation (23). In the case of the HBV nucleocapsid promoter, it appears that these coactivators are recruited, either individually or as a complex, through their interaction with PGC1α, which, acting as a bridging adaptor molecule, must recognize the assembled transcription factors associated with the regulatory elements of this promoter. Consistent with this suggestion is the observation that mutation of the nuclear receptor binding site within the HBV nucleocapsid promoter leads to reductions in coactivator-mediated viral 3.5-kb RNA transcription and viral replication (Fig. 3).
Analysis of coactivator-mediated HBV biosynthesis indicated that the level of the HBcAg/p21 polypeptide synthesized was consistent with the observed level of HBV pregenomic 3.5-kb RNA expressed within HEK293T cells (Fig. 4A and B). However, HBcAg/capsid formation and, hence, capsid-associated viral replication intermediate DNA were observed only in cells expressing HBV 3.5-kb RNA and the HBcAg/p21 polypeptide at levels above a relatively low threshold limit (Fig. 4 and 5, step 4). Indeed, increases in HBV 3.5-kb RNA and HBcAg/p21 polypeptide levels of <2-fold appear to be sufficient to promote the robust induction of HBV replication intermediate DNA synthesis (Fig. 4 and 5, step 5). This observation is consistent with previous observations that indicated that the conversion of HBcAg dimers into capsids occurred by a highly cooperative process at a concentration of approximately 1 μM (30, 31). The observation that a similar process for the assembly of HBV capsids can occur in cell culture may help to explain various previous in vivo findings where limited alterations in HBV 3.5-kb RNA were associated with dramatic changes in viral replication (32–34). Indeed, the loss of capsids mediated by cytokine signaling, possibly governed by HBcAg phosphorylation (35–40), might be due to alterations in the equilibrium between HBcAg/dimers and capsids, preventing capsid assembly and consequently eliminating HBV replication intermediate DNA synthesis without affecting viral transcription or translation to a great extent (41, 42).
MATERIALS AND METHODS
Plasmid construction.
The HBV DNA (4.1-kbp) construct that contains 1.3 copies of the HBV genome includes the viral sequence from nucleotides 1072 to 3182 plus nucleotides 1 to 1990 (3). The pCMVHNF4, pcDNA-HA-hPGC1α, pSG5-HA-CBP, pSG5-HA-SRC1e, and pIRESneoPRMT1 vectors express HNF4α, PGC1α, CBP, SRC1, and PRMT1 from the corresponding cDNAs, respectively, using the cytomegalovirus (CMV) immediate early promoter (pCMV, pcDNA3, and pIRESneo) or the simian virus 40 early promoter (pSG5) (25, 43–46).
Cells and transfections.
The human hepatoma cell line Huh7 and the human embryonic kidney cell line HEK293T were grown in RPMI 1640 medium with 10% fetal bovine serum at 37°C in 5% CO2–air. Transfections for viral RNA and DNA analyses were performed as previously described (47), using 10-cm plates containing approximately 1 × 106 cells. Isolation of DNA and RNA was performed at 3 days posttransfection. The transfected DNA mixture was composed of 10 μg of HBV DNA (4.1 kbp) plus 1 μg of the nuclear receptor expression vector pCMVHNF4 or the transcriptional coactivator expression vectors pcDNA-HA-hPGC1α, pSG5-HA-CBP, pSG5-HA-SRC1e, and pIRESneoPRMT1 (3, 25, 43–46), as indicated. Controls were derived from cells transfected with HBV DNA and the expression vectors lacking a nuclear receptor or transcriptional coactivator cDNA insert (48).
Characterization of HBV transcripts and viral replication intermediates.
Transfected cells from a single plate were divided equally and used for the preparation of total cellular RNA and viral DNA replication intermediates as described previously (49), with minor modifications. RNA (Northern) and DNA (Southern) filter hybridization analyses were performed by using 10 μg of total cellular RNA and 30 μl of viral DNA replication intermediates, respectively, as described previously (50). Transcription initiation sites for the HBV 3.5-kb transcripts were examined by primer extension analysis using 5 U of avian myeloblastosis virus reverse transcriptase (Promega), 12 ng 32P-labeled HBV oligonucleotide probe 5′-GGAAAGAAGTCAGAAGGCAAAAACGAGAGTAACTCC-3′ (HBV positions 1976 to 1941), 9 ng 32P-labeled human 32-kDa large ribosomal protein subunit (RPL32) oligonucleotide probe 5′-CTCTTTTTGACGATCTTGGGCTTCAC-3′ (nucleotide positions +98 to +73), and 10 μg of total cellular RNA, as described previously (27). Data from filter hybridization and primer extension analyses were quantitated by phosphorimaging using a Molecular Dynamics Storm 5000 PhosphorImager system.
Analysis of HBcAg/p21 protein and capsids.
Transfected cells from a single plate were lysed in 200 μl of a solution containing 25 mM Tris hydrochloride (pH 7.4), 150 mM NaCl, 1 mM EDTA, 5% glycerol, and 1% (vol/vol) NP-40 and used for the preparation of cytoplasmic protein extracts as described previously (51). HBcAg was immunoprecipitated with 1 μl rabbit anti-HBc antibody (from Adam Zlotnick, Indiana University) plus protein G-agarose (Santa Cruz Biotechnology) according to the manufacturer's instructions (Abcam), resolved by SDS-PAGE, transferred onto polyvinylidene difluoride (PVDF) membranes, and identified by using rabbit anti-HBc antibody (1:1,000 dilution; Dako), followed by incubation with horseradish peroxidase-labeled goat anti-rabbit IgG (1:2,000; Cell Signaling Technology). HBcAg/p21 was detected by using enhanced chemiluminescence according to the manufacturer's instructions (Thermo Fisher Scientific) and quantitated by using the ChemiDoc MP imaging system (Bio-Rad).
HBcAg/capsids present in 20 μl of the cytoplasmic extract were resolved by 1% agarose gel electrophoresis as described previously (52–54). The HBcAg/capsid protein was detected by using rabbit anti-HBc antibody (1:1,000 dilution; Dako) as described previously for HBcAg/p21 (52–54). Capsid-associated HBV replication intermediate DNA was detected by DNA filter hybridization analysis as described previously (52, 54).
ACKNOWLEDGMENTS
We are grateful to Eric F. Johnson (The Scripps Research Institute, La Jolla, CA) for plasmid pCMVHNF4, Anastasia Kralli (The Scripps Research Institute, La Jolla, CA) for plasmids pcDNA3-HA-hPGC1α and pSG5-HA-CBP, Malcolm G. Parker (Imperial Cancer Research Fund, London, UK) for plasmid pSG5-HA-SRC1e, and Robert G. Roeder (The Rockefeller University, New York, NY) for plasmid pIRESneoPRMT1. We thank Adam Zlotnick (Indiana University, Bloomington, IN) and Haitao Guo (Indiana University School of Medicine, Indianapolis, IN) for the generous gifts of antibodies against HBcAg.
This work was supported by Public Health Service grant AI125401 from the National Institutes of Health and an Egypt Fulbright Missions Program (EFMP) graduate fellowship.
REFERENCES
- 1.Will H, Reiser W, Weimer T, Pfaff E, Buscher M, Sprengle R, Cattaneo R, Schaller H. 1987. Replication strategy of human hepatitis B virus. J Virol 61:904–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summers J, Mason WS. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403–415. doi: 10.1016/0092-8674(82)90157-X. [DOI] [PubMed] [Google Scholar]
- 3.Tang H, McLachlan A. 2001. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc Natl Acad Sci U S A 98:1841–1846. doi: 10.1073/pnas.98.4.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guidotti LG, Matzke B, Schaller H, Chisari FV. 1995. High-level hepatitis B virus replication in transgenic mice. J Virol 69:6158–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reese VC, Oropeza CE, McLachlan A. 2013. Independent activation of hepatitis B virus biosynthesis by retinoids, peroxisome proliferators, and bile acids. J Virol 87:991–997. doi: 10.1128/JVI.01562-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reese V, Ondracek C, Rushing C, Li L, Oropeza CE, McLachlan A. 2011. Multiple nuclear receptors may regulate hepatitis B virus biosynthesis during development. Int J Biochem Cell Biol 43:230–237. doi: 10.1016/j.biocel.2009.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shlomai A, Paran N, Shaul Y. 2006. PGC-1α controls hepatitis B virus through nutritional signals. Proc Natl Acad Sci U S A 103:16003–16008. doi: 10.1073/pnas.0607837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oropeza CE, Li L, McLachlan A. 2008. Differential inhibition of nuclear hormone receptor-dependent hepatitis B virus replication by the small heterodimer partner. J Virol 82:3814–3821. doi: 10.1128/JVI.02507-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ondracek CR, Reese VC, Rushing CN, Oropeza CE, McLachlan A. 2009. Distinct regulation of hepatitis B virus biosynthesis by peroxisome proliferator-activated receptor γ coactivator 1α and small heterodimer partner in human hepatoma cell lines. J Virol 83:12545–12551. doi: 10.1128/JVI.01624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ondracek CR, Rushing CN, Reese VC, Oropeza CE, McLachlan A. 2009. Peroxisome proliferator-activated receptor γ coactivator 1α and small heterodimer partner differentially regulate nuclear receptor-dependent hepatitis B virus biosynthesis. J Virol 83:12535–12544. doi: 10.1128/JVI.01623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. 1995. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol 69:3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teyssier C, Ma H, Emter R, Kralli A, Stallcup MR. 2005. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev 19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Bieker JJ. 1998. Acetylation and modulation of erythroid Krüppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc Natl Acad Sci U S A 95:9855–9860. doi: 10.1073/pnas.95.17.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao W, Xiao R, Peng B, Xu H, Shen H, Huang M, Shi T, Yi J, Zhang W, Wu X, Gao X, Lin X, Dorrestein PC, Rosenfeld MG, Liu W. 2015. Arginine methylation of HSP70 regulates retinoid acid-mediated RARβ2 gene activation. Proc Natl Acad Sci U S A 112:E3327–E3336. doi: 10.1073/pnas.1509658112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, Ma H, Hong H, Koh SS, Huang S-M, Schurter BT, Aswad DW, Stallcup MR. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 16.Ceschin DG, Walia M, Wenk SS, Duboé C, Gaudon C, Xiao Y, Fauquier L, Sankar M, Vandel L, Gronemeyer H. 2011. Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev 25:1132–1146. doi: 10.1101/gad.619211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D, Huang S-M, Stallcup MR. 2000. Synergistic, p160 coactivator-dependent enhancement of estrogen receptor function by CARM1 and p300. J Biol Chem 275:40810–40816. doi: 10.1074/jbc.M005459200. [DOI] [PubMed] [Google Scholar]
- 18.Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. 2001. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr Biol 11:1981–1985. doi: 10.1016/S0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 19.Feng Q, Yi P, Wong J, O'Malley BW. 2006. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol Cell Biol 26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao F, Li S, Chavez V, Lanting L, Natarajan R. 2006. Coactivator-associated arginine methyltransferase-1 enhances nuclear factor-kappaB-mediated gene transcription through methylation of histone H3 at arginine 17. Mol Endocrinol 20:1562–1573. doi: 10.1210/me.2005-0365. [DOI] [PubMed] [Google Scholar]
- 21.Zika E, Fauquier L, Vandel L, Ting JPY. 2005. Interplay among coactivator-associated arginine methyltransferase 1, CBP, and CIITA in IFN-γ-inducible MHC-II gene expression. Proc Natl Acad Sci U S A 102:16321–16326. doi: 10.1073/pnas.0505045102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y-H, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. 2005. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc Natl Acad Sci U S A 102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puigserver P, Adelmant C, Wu ZD, Fan M, Xu JM, O'Malley B, Spiegelman BM. 1999. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 24.Ma H, Hong H, Huang S-M, Irvine RA, Webb P, Kushner PJ, Coetzee GA, Stallcup MR. 1999. Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol Cell Biol 19:6164–6173. doi: 10.1128/MCB.19.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang S-M, Cheng Y-S. 2004. Analysis of two CBP (cAMP-response-element-binding protein-binding protein) interacting sites in GRIP1 (glucocorticoid-receptor-interacting protein), and their importance for the function of GRIP1. Biochem J 382:111–119. doi: 10.1042/BJ20040206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee Y-H, Koh SS, Zhang X, Cheng X, Stallcup MR. 2002. Synergy among nuclear receptor coactivators: selective requirement for protein methyltransferase and acetyltransferase activities. Mol Cell Biol 22:3621–3632. doi: 10.1128/MCB.22.11.3621-3632.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tang H, McLachlan A. 2002. A pregenomic RNA sequence adjacent to DR1 and complementary to epsilon influences hepatitis B virus replication efficiency. Virology 303:199–210. doi: 10.1006/viro.2002.1645. [DOI] [PubMed] [Google Scholar]
- 28.Spiegelman BM, Heinrich R. 2004. Biological control through regulated transcriptional coactivators. Cell 119:157–167. doi: 10.1016/j.cell.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 29.Rosenfeld MG, Lunyak VV, Glass CK. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 30.Seifer M, Zhou S, Standring DN. 1993. A micromolar pool of antigenically distinct precursors is required to initiate cooperative assembly of hepatitis B virus capsids in Xenopus oocytes. J Virol 67:249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Porterfield JZ, Dhason MS, Loeb DD, Nassal M, Stray SJ, Zlotnick A. 2010. Full-length hepatitis B virus core protein packages viral and heterologous RNA with similarly high levels of cooperativity. J Virol 84:7174–7184. doi: 10.1128/JVI.00586-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guidotti LG, Eggers CM, Raney AK, Chi SY, Peters JM, Gonzalez FJ, McLachlan A. 1999. In vivo regulation of hepatitis B virus replication by peroxisome proliferators. J Virol 73:10377–10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raney AK, Eggers CM, Kline EF, Guidotti LG, Pontoglio M, Yaniv M, McLachlan A. 2001. Nuclear covalently closed circular viral genomic DNA in the liver of hepatocyte nuclear factor 1α-null hepatitis B virus transgenic mice. J Virol 75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banks KE, Anderson AL, Tang H, Hughes DE, Costa RH, McLachlan A. 2002. Hepatocyte nuclear factor 3β inhibits hepatitis B virus replication in vivo. J Virol 76:12974–12980. doi: 10.1128/JVI.76.24.12974-12980.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ning X, Basagoudanavar SH, Liu K, Luckenbaugh L, Wei D, Wang C, Wei B, Zhao Y, Yan T, Delaney W, Hu J. 2017. Capsid phosphorylation state and hepadnavirus virion secretion. J Virol 91:e00092-17. doi: 10.1128/JVI.00092-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su P-Y, Yang C-J, Chu T-H, Chang C-H, Chiang C, Tang F-M, Lee C-Y, Shih C. 2016. HBV maintains electrostatic homeostasis by modulating negative charges from phosphoserine and encapsidated nucleic acids. Sci Rep 6:38959. doi: 10.1038/srep38959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selzer L, Kant R, Wang JC-Y, Bothner B, Zlotnick A. 2015. Hepatitis B virus core protein phosphorylation sites affect capsid stability and transient exposure of the C-terminal domain. J Biol Chem 290:28584–28593. doi: 10.1074/jbc.M115.678441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ludgate L, Liu K, Luckenbaugh L, Streck N, Eng S, Voitenleitner C, Delaney WE, Hu J. 2016. Cell-free hepatitis B virus capsid assembly dependent on the core protein C-terminal domain and regulated by phosphorylation. J Virol 90:5830–5844. doi: 10.1128/JVI.00394-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lan YT, Li J, Liao WY, Ou JH. 1999. Roles of the three major phosphorylation sites of hepatitis B virus core protein in viral replication. Virology 259:342–348. doi: 10.1006/viro.1999.9798. [DOI] [PubMed] [Google Scholar]
- 40.Roossinck MJ, Siddiqui A. 1987. In vivo phosphorylation and protein analysis of hepatitis B virus core antigen. J Virol 61:955–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieland SF, Eustaquio A, Whitten-Bauer C, Boyd B, Chisari FV. 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc Natl Acad Sci U S A 102:9913–9917. doi: 10.1073/pnas.0504273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieland SF, Guidotti LG, Chisari FV. 2000. Intrahepatic induction of alpha/beta interferon eliminates viral RNA-containing capsids in hepatitis B virus transgenic mice. J Virol 74:4165–4173. doi: 10.1128/JVI.74.9.4165-4173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen D, Lepar G, Kemper B. 1994. A transcriptional regulatory element common to a large family of hepatic cytochrome P450 genes is a functional binding site of the orphan receptor HNF-4. J Biol Chem 269:5420–5427. [PubMed] [Google Scholar]
- 44.Knutti D, Kaul A, Kralli A. 2000. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol Cell Biol 20:2411–2422. doi: 10.1128/MCB.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalkhoven E, Valentine JE, Heery DM, Parker MG. 1998. Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J 17:232–243. doi: 10.1093/emboj/17.1.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.An W, Kim J, Roeder RG. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 47.McLachlan A, Milich DR, Raney AK, Riggs MG, Hughes JL, Sorge J, Chisari FV. 1987. Expression of hepatitis B virus surface and core antigens: influences of pre-S and precore sequences. J Virol 61:683–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raney AK, Johnson JL, Palmer CNA, McLachlan A. 1997. Members of the nuclear receptor superfamily regulate transcription from the hepatitis B virus nucleocapsid promoter. J Virol 71:1058–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Summers J, Smith PM, Huang M, Yu M. 1991. Morphogenetic and regulatory effects of mutations in the envelope proteins of an avian hepadnavirus. J Virol 65:1310–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 51.Ondracek CR, McLachlan A. 2011. Role of peroxisome proliferator-activated receptor gamma coactivator 1α in AKT/PKB-mediated inhibition of hepatitis B virus biosynthesis. J Virol 85:11891–11900. doi: 10.1128/JVI.00832-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yu M, Summers J. 1994. Multiple functions of capsid protein phosphorylation in duck hepatitis B virus replication. J Virol 68:4341–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung J, Hwang SG, Chwae Y-J, Park S, Shin H-J, Kim K. 2014. Phosphoacceptors threonine 162 and serines 170 and 178 within the carboxyl-terminal RRRS/T motif of the hepatitis B virus core protein make multiple contributions to hepatitis B virus replication. J Virol 88:8754–8767. doi: 10.1128/JVI.01343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan R, Zhao X, Cai D, Liu Y, Block TM, Guo J-T, Guo H. 2015. The interferon-inducible protein tetherin inhibits hepatitis B virus virion secretion. J Virol 89:9200–9212. doi: 10.1128/JVI.00933-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Doitsh G, Shaul Y. 2003. A long HBV transcript encoding pX is inefficiently exported from the nucleus. Virol 309:339–349. doi: 10.1016/S0042-6822(03)00156-9. [DOI] [PubMed] [Google Scholar]
- 56.Johnson JL, Raney AK, McLachlan A. 1995. Characterization of a functional hepatocyte nuclear factor 3 binding site in the hepatitis B virus nucleocapsid promoter. Virol 208:147–158. doi: 10.1006/viro.1995.1138. [DOI] [PubMed] [Google Scholar]
- 57.Zhang P, Raney AK, McLachlan A. 1993. Characterization of functional Sp1 transcription factor binding sites in the hepatitis B virus nucleocapsid promoter. J Virol 67:1472–1481. [DOI] [PMC free article] [PubMed] [Google Scholar]