ABSTRACT
Classically, natural killer (NK) cells have been defined by nonspecific innate killing of virus-infected and tumor cells. However, burgeoning evidence suggests that the functional repertoire of NK cells is far more diverse than has been previously appreciated, thus raising the possibility that there may be unexpected functional specialization and even adaptive capabilities among NK cell subpopulations. Some of the first evidence that NK cells respond in an antigen-specific fashion came from experiments revealing that subpopulations of murine NK cells were able to respond to a specific murine cytomegalovirus (MCMV) protein and that in the absence of T and B cells, murine NK cells also mediated adaptive immune responses to a secondary challenge with specific haptens. These data have been followed by demonstrations of NK cell memory of viruses and viral antigens in mice and primates. Herein, we discuss different forms of NK cell antigen specificity and how these responses may be tuned to specific viral pathogens, and we provide assessment of the current literature that may explain molecular mechanisms of the novel phenomenon of NK cell memory.
KEYWORDS: immune memory, innate immunity, natural killer cells
INTRODUCTION
Cellular components of the innate immune system are typically characterized as using a finite number of germ line-encoded pattern recognition receptors to sense pathogens, neoplastic cells, and other tissue damage (1–3). In contrast, the adaptive immune system, which includes T and B cells and their effector functions, relies on recombinase-activating gene (RAG)-dependent nonhomologous end-joining of chromosomal DNA and its recombination to generate a substantial T and B cell receptor repertoire capable of antigen-specific recognition (4). Activation of T and B cells by their cognate antigen leads to activation, proliferation, and the selection of high-affinity effector and memory cells and results in accelerated and enhanced recall responses by memory T and B cells upon reexposure (1–3). Medically, we exploit the ability to elicit memory immune cells via vaccination, as optimal vaccine strategies are designed to induce long-lived, antigen-specific memory T and B cells that mediate rapid, high-affinity recall responses upon encounter of the actual pathogen. In contrast, innate cells have been thought of solely as a nonspecific first line of defense against pathogens that may also serve to augment or tune adaptive responses but do not generate memory in their own right.
Natural killer (NK) cells are primary effector cells of the innate immune system that can rapidly eliminate tumor and virus-infected cells. Although NK cells have not traditionally been thought to carry adaptive capabilities or require antigen priming, they do encode a complex array of receptors to recognize specific ligands on target cells, and the tuned integration of these signals results in cytokine secretion and cytolysis or, alternatively, tolerance (5). In humans, the largest group of NK cell receptors belong to the killer cell immunoglobulin (Ig)-like receptor (KIR) family, which consists of type I integral membrane proteins that form a polymorphic family within the Ig superfamily (6, 7). In mice, a similar group of NK cell receptors are type II integral membrane, C-type lectin-like molecules belonging to the Ly49 family (8). Both KIRs and Ly49s are germ line encoded, highly polymorphic receptors and selectively expressed on most naive NK cells, but NK cells can express one or more receptors (6, 7). KIRs and Ly49 receptors recognize host-derived major histocompatibility complex class I (MHC-I) molecules, contribute to the processes of “licensing” and “education” which occur during NK cell development, and ensure that only NK cells capable of engaging self-MHC with their inhibitory receptors are allowed to become functionally competent but are also restrained from killing healthy cells (9–13). Other receptor families in both mice and humans are leukocyte immunoglobulin-like receptors (LIRs), C-type lectin-like receptors (LLRs), tumor necrosis factor (TNF) superfamily receptors, and natural cytotoxicity receptors (NCRs), including members of the NKG2 family, and the common NK cell receptors NKp30, NKp46, and NKp80 (7, 14–20). Interestingly, NK cells and T cells share a progenitor. Both express NCRs, and many effector functions, such as gamma interferon (IFN-γ) release and perforin/granzyme-mediated killing, overlap significantly between NK cells and cytotoxic T lymphocytes (CTLs) (21). Indeed multiple cellular and noncellular components of the innate and adaptive immune system have been conserved in vertebrates for hundreds of millions of years, making it tempting to speculate that evolutionary pressures may have led to the development of an adaptive immune system from its innate counterpart in higher-order vertebrates (22). Over the past 10 years, a multitude of independent studies have revealed that subsets of murine and primate NK cells are capable of antigen-dependent expansion and long-lived immunological memory. Together, these data suggest that NK cell function may traverse both innate and adaptive immune systems, thus representing a third lineage of lymphocytes capable of antigen specificity. Here, we summarize and discuss current knowledge of NK cell-mediated adaptive immunity, its origins, and potential clinical applications (Fig. 1; Table 1).
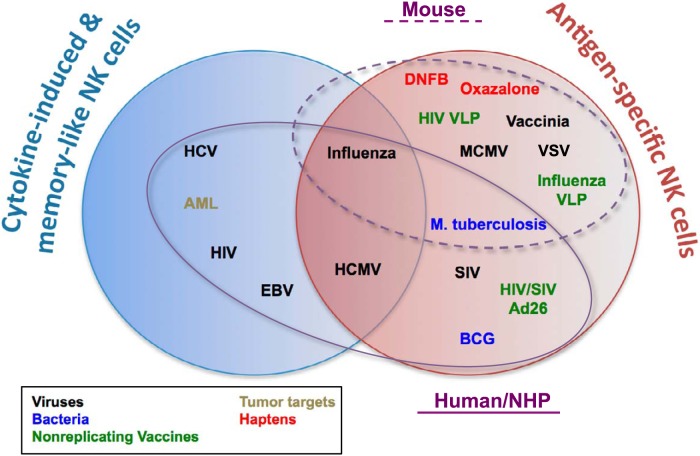
FIG 1.
Examples of antigen-specific, cytokine-induced, and memory-like NK cells in mice and humans/nonhuman primates (NHP). Venn diagrams represent examples of evidence for adaptive NK cells and whether these have been demonstrated as being truly antigen specific, cytokine induced, and memory-like or a combination of these. Microbial pathogens or other agents for which memory NK cells have been demonstrated are color coded. Ellipses indicated those agents that have been demonstrated in mice (dashed line) or humans/NHP (solid line). AML, acute myeloid leukemia; VLP, virus-like particles.
TABLE 1.
NK cell memory and memory-like responsesa
| Species | Type | Pathogen(s), antigen(s), or disease | Receptor(s) and/or cell phenotype | Ligand(s) or stimulus(i) | Reference(s) |
|---|---|---|---|---|---|
| Mice | Antigen specific | Haptens (DNFB, oxazalone) | NK1.1+ or DX5+ (CD49b) and CXCR6+ NKG2D+ CD90+ CD11b+ CD27− DX5− CD49a+ | ? | 26, 31, 32, 34 |
| HIV VLP (Gag, Env) | NK1.1+ or DX5+ and CXCR6+ | ? | 31 | ||
| Influenza VLP (M1 or HA/M1) | NK1.1+ or DX5+ and CXCR6+ | ? | 31 | ||
| Influenza virus | NK1.1+ or DX5+ and CXCR6+ DX5− CD49a+ | ? | 31 | ||
| 35 | |||||
| Vesicular stomatitis virus | NK1.1+ or DX5+ and CXCR6+ | ? | 31 | ||
| Vaccinia virus | NK1.1+ or DX5+ and CD90+ | ? | 33 | ||
| Murine cytomegalovirus | Ly49H+, Ly49P+ | m157, m04 | 43–45 | ||
| BCG, M. tuberculosis | ? | CD27, IL-21 | 85 | ||
| Cytokine induced | Ly49H+, NK1.1+ | IL-12, IL-15, IL-15 | 87 | ||
| Humans and NHP | Antigen specific | Human cytomegalovirus | NKG2C+ CD57+ | ||
| SIV, SHIV | NKG2A+, NKG2C+ | Gag, Env | 78 | ||
| HIV, SIV antigens by adenovirus vectors | NKG2A+, NKG2C+ | Gag, Env | 78 | ||
| BCG | CD56+ CD16lo | ? | 86 | ||
| Cytokine induced | Acute myeloid leukemia | NKG2D+ DNAM-1+ | IL-12, IL-15, IL-15 | 89 | |
| Inactivated influenza virus | IFN-αβR2+ | IL-2 | 88 | ||
| Memory-like | Human cytomegalovirus | NKG2C+, γ-chain− | Antibody | 90, 91 | |
| HIV | NKG2C+, γ-chain− | Antibody | 92 | ||
| HCV | NKG2C+, γ-chain− | Antibody | 93 | ||
| Epstein-Barr virus | CD56bright NKG2A+ CD56dim NKG2A+ NKG2A+ 2B4+ NKG2D+ | ? | 51, 57–60 |
VLP, virus-like particles; NHP, nonhuman primates.
ADAPTIVE NK CELL-MEDIATED IMMUNE RESPONSES TO ALTERED SELF-ANTIGENS
Exposure to haptens, self-proteins altered by the addition of a chemical moiety (23), induces a classical contact hypersensitivity (CHS) response, and typically the first exposure to hapten results only in sensitization. A second exposure to the same hapten generates an immune reaction resulting in a characteristic itchy rash, fluid-filled blisters, and hives. Human examples of hypersensitivity disease include asthma, rhinoconjunctivitis, otitis, rhinosinusitis, urticaria, angioedema, eczema, food allergy, drug allergy, insect allergy, occupational allergic diseases, and anaphylaxis (24). The CHS response is commonly used to investigate sensitization and antigen recall and was thought to be mediated primarily by T, NKT, and/or B cell activation (25). However, in 2006, O'Leary et al. reported that a novel subset of murine-liver-resident Thy1+ (CD90+) Ly49C+ (in C57BL/6 mice) DX5+ and NKG2D+ NK cells can mediate antigen-specific long-lived immunological recall responses to haptens in a RAG-independent manner (26). These findings were highly surprising, as they demonstrated that certain subsets of NK cells are capable of adaptive CHS responses, and flow cytometric analyses and confocal microscopy further confirmed that NK cells are recruited to sites of CHS upon hapten challenge (26). Sensitization of NK cells may occur in lymph nodes, since antibody-mediated blockade of P-, E-, and L-selectin or genetic deficiency in L-selectin (27–29) blocked CHS responses in sensitized mice. Interestingly, only sensitized liver-resident Thy1+ NK cells transferred CHS responses into naive lymphopenic hosts, while naive, sensitized hepatic Thy1− or splenic NK cells did not (26). In 2010, these findings were further clarified by Paust et al. in experiments demonstrating a requirement for adaptive NK cells to express CXCR6, which is expressed on about half of all murine-liver NK cells and for which the ligand, CXCL16, is constitutively expressed on liver sinusoidal endothelial cells and upregulated at sites of inflammation (30, 31). Indeed, CXCR6-deficient mice have reduced numbers of NK cells, exhibit poor NK cell survival upon adoptive transfer, and significantly reduced memory responses. Interestingly, administration of a blocking antibody specific to NKG2D also diminishes the CHS response (26), suggesting that NKG2D may somehow be important in NK cell-adaptive responses. Further phenotyping of hapten-sensitized NK cells via adoptive transfer and CHS identified adaptive murine NK cells as liver-resident NK cells that are positive for CD45, CD90, CD11b, Ly49C/I, CXCR6, and NK1.1 (C57BL/6) or DX5 (C57BL/6 or BALB/c) but negative for CD27 (26, 31–33). Of note, while four laboratories successfully transferred NK cell memory to non-cytomegalovirus (non-CMV) antigens using DX5+ as a selection marker for their antigen-sensitized liver NK cells (26, 31–33), one laboratory was unable to do so and suggested instead that CD49a-expressing NK cells mediate antigen-specific memory (34, 35).
NK CELL-MEDIATED ADAPTIVE IMMUNE RESPONSES TO NON-CMV PATHOGENS IN MICE
As discussed above, initial findings of antigen-dependent NK cell memory in mice against haptens have been described. Subsequent experiments expanded these findings of clinically relevant NK cell memory to human immunodeficiency virus (HIV) group antigen (gag)- or envelope (env)-containing virus-like particles (VLP) and those containing influenza A virus-derived matrix protein 1 (M1) (31). NK cell memory of M1 VLP was transferred to naive lymphopenic recipients of M1-sensitized lung NK cells, as demonstrated by delayed-type hypersensitivity (DTH) and prolonged survival of recipient lymphocyte-deficient mice upon challenge with influenza A PR8 virus, suggesting that liver NK cells may not be the only memory NK subset in mice. Even exposure to inactivated viruses, such as vaccination with UV-inactivated vesicular stomatitis virus (VSV), elicited adaptive immune responses in murine-liver-resident NK cells. These vaccine-induced memory NK cells were pathogen specific, were developed in the absence of RAG and T and B cells, and protected T and B cell-deficient mice from lethal viral challenge (26, 31). Independent verification of NK cell-mediated innate immune memory to vaccinia virus was published by Gillard et al., who demonstrated the ability of Thy1+ liver-derived NK cells to mediate adaptive immunity to this pathogen (33). In addition, Paust, et al. demonstrated clearly that NK cell memory of non-CMV viral antigens, like haptens, is confined to NK cells that express the chemokine receptor CXCR6 (31).
NK CELL-MEDIATED ADAPTIVE IMMUNE RESPONSES TO HERPESVIRUSES IN MICE AND HUMANS
Murine CMV (MCMV) is a commonly studied example of NK cell-mediated antiviral surveillance, as this virus has evolved elaborate mechanisms to evade NK cell responses and has also provided strong evidence in support of adaptive functions by NK cells (36–39). NK cells express on their cell surface activating receptors that specifically recognize MCMV-derived proteins, including Ly49H, which recognizes m157 (36–38, 40–42), and Ly49P, which recognizes m04 (43). However, this interaction is unique to B6 mice, as neither outbred mice nor inbred mice on a non-C57BL/6 background (BALB/c) express Ly49H and as such are highly susceptible to MCMV infection (44). NK cell-mediated long-term survival and memory responses to m157 MCMV antigen have recently been shown in B6 bone marrow chimera mice (45), in which Ly49H lymphopenia was induced through Dap12 deficiency. In this lymphopenic environment, Ly49H+ NK cells proliferated upon MCMV infection, contracted subsequently, and persisted in lymphoid and nonlymphoid organs for several weeks (45). These self-renewing memory NK cells rapidly degranulated and produced cytokines upon reactivation, and adoptive transfer of 10-fold-fewer memory NK cells was protective upon MCMV challenge compared to what occurred with naive NK cells. Since Ly49H is expressed on splenic and hepatic NK cells, both subsets responded to m157, although liver NK cells proliferated more vigorously than those derived from spleen. In contrast, there was no correlation between Ly49-activating receptors on hepatic NK cells and CHS activity, and splenic NK cells were unable to mediate CHS responses (26). Hence, the requirements of Ly49-activating receptors during NK cell-meditated memory responses may vary depending on the antigen and MHC haplotype, and their precise requirement to NK cell-mediated memory requires further study. Interestingly, while both splenic and hepatic NK cells respond to MCMV infection, they do so in an organ-specific manner (46, 47). In the spleen, perforin mediates viral clearance, while IFN-γ mediates protection in the liver. It would, therefore, be most informative for future studies to directly compare splenic and hepatic memory NK cell responses to MCMV challenge using adoptive transfers. While the full mechanisms mediating establishment of a long-lived memory NK cell pool of MCMV following infection are unclear, the proapoptotic factor Bim has been implicated in shaping the size and functional profile of long-lived memory NK cells in a mechanism analogous to that of memory CD8+ T cells (48). Also, as with autophagy-dependent formation of CD8+ T cell pools, surviving NK cells that undergo mitophagy during the contraction phase depend on BNIP3 to select survival of memory NK cells (49).
Although adaptive NK cell responses to CMV have been delineated most clearly for MCMV, empirical evidence suggests that a similar phenomenon may occur in humans. First identified as an expansion of NKG2C+ NK cells in response to human CMV (HCMV)-infected fibroblasts (50), it was later clarified that CD57+ NKG2Chi NK cells expand early after HCMV infection in vivo and are highly specific to the virus (51, 52). NK cells are typically the first lymphocytes to reconstitute after hematopoietic stem cell (HSC) transplantation and during reconstitution are reciprocally modulated by reactivated CMV (52, 53). Inhibition of CMV replication is modulated, at least in part, by NKG2C, which binds to HLA-E, which in the steady state presents signal peptides derived from other MHC-I proteins (54). It is currently unknown if a CMV-encoded ligand for NKG2C exists, but 5% of humans are NKG2C null and 20% are NKG2C heterozygous, and these genetic attributes carry significant implications in transplant immunology (55). Indeed, cord blood (CB) grafts expressing an NKG2C deletion allele possessed a high risk of CMV reactivation after CB transplantation and a reduced risk with the presence of the wild-type allele (56). Further evidence that NKG2C-mediated NK cell activation has a profound effect on the NK cell repertoire and CMV-specific NK memory in humans comes from a comparison of NK cells from CMV-seronegative and CMV-seropositive recipients of CB-derived HSCs (55). While NKG2C expression remained unchanged in patients without CMV viremia, in patients who reactivated CMV, NKG2C expression increased significantly during the acute phase of CMV infection, similar to what occurred with NK cells in other patients with CMV reactivation (53). Newly formed NK cells from patients who reactivated CMV after HSC transplantation also had a more mature phenotype and produced significantly more IFN-γ both before and after detection of CMV viremia and anti-CMV therapy. Interestingly, the NKG2C+ CD57+ NK cells that expand in CMV infection are not responsive to Epstein-Barr virus (EBV)-infected cells, suggesting that this phenotype is not a universal response to herpesvirus infections (51). However, several independent studies have indicated that specific subsets of NK cells are also enhanced in their responses to EBV infection (57–60), including a CD56bright NKG2A+ CD94+ CD45+ CD62L− population that accumulates in the tonsils of infected individuals (57). These NK cells secrete IFN-γ in response to EBV-infected cells and can restrict the transformation of EBV-infected B cells in vitro (57, 59). In a second study of pediatric patients, a subset of CD56dim NKG2A+ NK cells expands for several months following acute mononucleosis (caused by EBV) and preferentially responds to EBV-expressing B cells displaying lytic antigens, suggesting that this subset may play a key role in the control of primary EBV infection (60). Finally, an NKG2A+ 2B4+ CD16– CD57– NKG2D+ NK cell subset was recently shown to mediate a specific response to lymphoblastoid cell lines latently infected with EBV (58).
NK CELLS IN HUMAN IMMUNODEFICIENCY VIRUS INFECTION
Multiple studies have demonstrated an association between NK cells and control of HIV replication, as well as simian immunodeficiency virus (SIV) in macaque models. NK cells expand during primary infection (61, 62) prior to the development of CD8+ T lymphocytes and have evolved multiple mechanisms to recognize, lyse, or otherwise inhibit HIV- and SIV-infected cells via virus-induced downmodulation of MHC class I molecules (63) or upregulation of NKG2D ligands on target cells (64, 65) and secretion of the infection-blocking β-chemokines CCL3, CCL4, and CCL5 (66). Although these functions are innate in nature, burgeoning evidence has suggested that the NK cell response to HIV/SIV is robust and may not be entirely nonspecific. Indeed, NK cells have been linked to controlled viremia in HIV type 1 (HIV-1) elite controllers and long-term nonprogressors and reduced acquisition in HIV-1-exposed seronegative individuals, and peptide-specific NK cell responses have been shown to block HIV mother-to-child transmission (67–73). Longitudinal studies suggest that NK cells may be associated with preventing disease progression in SIV-infected macaques (74, 75), and experimental NK cell depletion results in increased virus replication (76, 77). Importantly, Reeves et al. also recently described evidence of antigen-specific NK cell memory responses mounted against SIV/simian-human immunodeficiency virus (SHIV) infections as well as against adenovirus 26 (Ad26) vaccine antigens in rhesus macaques (78). Responses were dependent on NKG2 molecules, but delineating the full mechanisms of primate NK cell memory will require further study. Many detailed analyses of NK cells in HIV and SIV have focused on KIR interactions with cognate HLA ligands that do not represent memory per se but demonstrate the potential for antigen-specific modulation of the NK cell response. One example is the coexpression of the KIR3DS1 allele in conjunction with HLA class I alleles from the HLA-Bw4 family being associated with delayed AIDS progression and greater suppression of virus replication in autologous CD4+ T cells (79–82). NK cells may also exert selective pressure on virus replication, as evidenced by HIV-1 polymorphisms associated with KIR2DL2 that can confer resistance to NK cells (69). Similarly, SIV peptides can modulate recognition of rhesus KIR; in one example, Mamu-KIR3DL05 is stabilized by certain peptides, but not by others, and the NK cell response can even be suppressed in this manner (83). A highly conserved HIV peptide that binds to HLA-E has also recently been shown to contribute to the sensitivity of HIV-infected cells to NKG2A-expressing NK cells (84).
NK CELL MEMORY OF MYCOBATERIUM TUBERCULOSIS
A nonviral form of memory NK cells has recently been described by Venkatasubramanian et al. and is present in spleens and draining lymph nodes of mice infected with Mycobacterium tuberculosis (85). Using a mouse model of tuberculosis (TB) infection, the authors were able to induce IFN-γ-producing CD27+ memory-like NK cells upon bacillus Calmette-Guerin (BCG) vaccination (the antigen used in the tuberculin test). Memory NK cells provided protection against subsequent TB challenge but not against challenge with other bacterial pathogens. Interestingly, murine TB-specific memory NK cells are distinct from CXCR6+ memory NK cells found in liver and are RAG dependent, although they do not require RAG expression but rather T cell-produced interleukin 21 (IL-21) for their induction and/or survival (85). Recently, BCG-specific memory NK cells were also identified in vaccinated humans and were found to be both long-lived and rapidly expanded upon BCG revaccination (86). Although the mechanisms and full phenotypic and functional profiles remain unclear, BCG-specific responses were found primarily among CD56+ CD16lo NK cells. All together, these exciting findings suggest that multiple subsets of distinct memory NK cells coexist and may protect their host using distinct mechanisms of induction, maintenance, and action.
OTHER FORMS OF TRAINED NK CELL-MEDIATED IMMUNITY
Cytokine-induced memory NK cells.
Another form of NK cell memory comes from initial studies by Cooper and colleagues, who demonstrated that cytokine-activated NK cells persist in naive hosts 7 to 22 days after adoptive transfer (87). Restimulation of these NK cells results in significantly elevated IFN-γ production, while granzyme B expression and killing ability are similar to those in naive NK cells. It is unlikely that this type of NK cell memory is entirely dependent on cytokine exposure after sensitization, since cytokine-mediated NK cell activation cannot fully explain antigen-specific responses. Nonetheless, NK cells retained an intrinsic memory of prior activation, a function until now attributed only to antigen-specific adaptive immune cells. Interestingly, recent similar studies have shown that influenza vaccination can also generate cytokine-induced memory NK cells (88). Hence, NK cell-mediated effector functions during antitumor responses and allergic and infectious diseases may clinically be more important than initially appreciated, and cytokine-induced memory NK cells may be attractive therapeutic targets for disease treatment (89).
Memory-like NK cells.
In addition to describing true antigen-specific NK cells, a recent study has identified a subpopulation of “memory-like” NK cells, which are exquisite effector cells when granted specificity through antibody binding. These cells, described in humans in 2013 by Zhang et al. (90), express high levels of FcγR (including CD16) but lack the intracellular γ-signaling chain. So-called (g–) or FcγRΔg NK cells are found at low frequencies in all individuals but expand in HCMV-seropositive persons. Following initial antibody binding, these cells become epigenetically modified and long-lived and are capable of significantly enhanced antibody-dependent functions and numerical expansion upon new antibody binding (91). FcγRΔg NK cells, partially identifiable by NKG2C and NKp30 expression, have been shown to exhibit potent antiviral functions against HCMV, herpes simplex virus (HSV), and influenza virus in the presence of their respective antiviral antibodies, regardless of previous antigen exposure. Recently, FcγRΔg NK cells with enhanced antibody-dependent cellular cytotoxicity (ADCC) have been shown to be increased 7-fold in HIV-infected persons and are also associated with protection from progressive liver disease in hepatitis C virus (HCV) infection (92, 93). Thus, this memory-like NK cell subpopulation has become an attractive target for antibody-based vaccines and immunotherapeutics.
NK CELL DIVERSITY IN VIRAL INFECTIONS AS A MECHANISM FOR MEMORY NK CELL GENERATION
It is unclear how NK cell memory of viral pathogens affects the diversity of the human NK cell repertoire. In the adaptive immune system, immune memory decreases repertoire diversity by increasing the frequency of cells expressing a single receptor specific for pathogens that have been encountered before. Among NK cells, this relationship is less clear, because with a few exceptions (such as the Ly49H-mediated recognition of m157 of murine CMV [45]), a specific receptor that mediates NK cell recognition and memory has not been identified. Instead, there are numerous associations between certain NK cell receptors and different viral infections in human, yet there are only a few situations in which there is a mechanistic understanding of how these receptors contribute to viral recognition (reviewed in reference 94). It is possible that distinct receptor combinations are required to respond to different viruses, making the elucidation of the requirements for a specific virus more challenging in light of the diversity receptor expression profiles within the NK cell repertoire. NK cell diversity may be best defined based on combinatorial expression patterns of activating and inhibitory receptors, whose signals are integrated to control NK cell function (95). Recent work has revealed that these receptors assort on the cell surface to generate a vast diversity of distinct phenotypic subsets, with 6,000 to 30,000 unique NK cell subsets per individual (95). This raises the possibility that specific subsets might be enhanced in their ability to recognize distinct pathogens, and a memory response may result in a shift in the NK cell repertoire to increase the frequency of these subsets. However, to date, only human CMV infection is associated with dramatic imprinting on the NK cell repertoire (96).
Interestingly, in vitro experiments suggest that short-term exposure to virus-infected cells actually increases human NK cell diversity (95). This shift in diversity may represent a short-term accommodation to “tune” the NK cell response to detect a specific pathogen. Consistently with this idea, a human immune repertoire with increased expression of maturity markers, such as CD57, is associated with shifts in the expression patterns of activating and inhibitory receptors in vivo (97). Such shifts, which generally favor greater expression of activating receptors, might decrease the threshold for NK cell activation upon a secondary exposure. Consistently with this idea, more mature CD57+ NK cells display enhanced cytokine secretion but diminished cytotoxic activity in response to autologous HIV-infected T cells in vitro (98), suggesting that the viral exposure modulates the quality of the NK cell response to subsequent infections. Consistently with this idea, exposure to many pathogens is associated with shifts in the expression patterns of a variety of natural killer cell receptors (94, 99).
Collectively, these findings raise the question of what effects chronic exposure to different viruses has on the NK cell repertoire. On one hand, exposure to viruses might give rise to adaptive and pathogen-specific cells, but on the other hand, it might lead to a more mature repertoire that favors cytokine secretion over direct killing. In a small study of women at risk of HIV infection, higher NK cell diversity was associated with increased HIV acquisition risk (98). Together, these data support a model in which exposure to a given pathogen might give rise to rare populations of memory cells that are difficult to detect among the overall increase in diversity. However, the cells that have diversified in response to one pathogen may be less “flexible” in their ability to respond to a de novo pathogen.
SPECULATION ON EVOLUTIONARILY CONSERVED ADAPTIVE IMMUNE MECHANISMS
The phenomenon of antigen-specific NK memory is entirely unprecedented and suggests an alternative pathway to generate immunological memory that is fundamentally distinct from all known cellular and molecular mechanisms of adaptive immunity. Based on the findings presented herein, we hypothesize that vertebrates have evolutionarily conserved the ability to generate a diverse antigen receptor family in a RAG-independent manner, resulting in NK cell-mediated adaptive immunity. Interestingly, evidence for a RAG-independent generation of a clonal repertoire of lymphocytes has been described in the only two surviving jawless vertebrates, lampreys and hagfish, which use recombinatorial assembly of leucine-rich-repeat genetic segments to generate diversified variable lymphocyte receptors (VLRs) (22, 100). Lamprey-expressed VLRs allow adaptive, clonal immune responses to a variety of antigens, rejection of secondary-skin allographs, and DTH not unlike DTH responses mediated by murine NK cell subsets. While lampreys express several genes or gene homologues that are important for adaptive immune responses (100–102), the numbers of immune gene homologs are comparatively low relative to that of jawed vertebrates. That said, lampreys are considered the most phylogenetically primitive species that may have an adaptive immune system. We are tempted to speculate that their ability to develop an adaptive immune system may have been key in their evolutionary survival. Further evidence that NK cell memory may not be restricted to higher-order vertebrates and may be highly evolutionarily conserved can be found in a recent report from Garcia-Valtanen et al. (103), who demonstrated that RAG-deficient zebrafish are also capable of antiviral innate immune memory. The responsible cell type could not be identified in this species, as regents to distinguish between innate immune cells of zebrafish are currently lacking; however, gene expression profiling did uncover an enhanced cytotoxic response. Whether mouse (or human) NK cells utilize similar or distinct mechanisms to generate a diverse antigen-receptor repertoire in a RAG-independent manner is under intense investigation. Indeed, data presented by Paust et al. outlined the development of a sorting strategy to identify and isolate 2,4-dinitrofluorobenzene (DNFB)-specific NK cells from livers of DNFB-sensitized RAG-knocked-out mice, whereby the nuclei were then transplanted into enucleated oocytes for the generation of embryonic stem cell lines that were used to clone mice (104). NK cells from cloned animals and about 50% of the F2 offspring instantly responded to DNFB without requiring prior sensitization but could not be sensitized to other haptens. These data strongly suggest that the nuclear information for hapten specificity of memory NK cells persists even in a donor nucleus whose epigenetic state was reset by nuclear reprogramming and subsequent breeding. This apparent genetic fixation would not be explained by epigenetic regulation of conventional “hard-wired” NK receptors but is expected if antigen receptor specificity is encoded at the level of genomic DNA and may suggest an entirely novel mechanism to generate receptor diversity (104).
CONCLUDING REMARKS
All together, the findings discussed herein challenge the notions that innate cells are incapable of innate immune memory or that adaptive immune memory is somehow strictly RAG dependent. Significant data also suggest that it is unique populations of NK cells or receptors that mediate adaptive immunity and that these functions might be conserved among species. Mechanistic evaluations remain ongoing, but the concept of NK cell memory has now evolved from immunologic heresy to a broad field of study, and it will be exciting to see how NK cell memory responses can be harnessed for improved vaccines and novel immunotherapies.
ACKNOWLEDGMENTS
Our efforts on this work were supported by NIH grants PO1 AI120756, RO1 DE026014 (R.K.R.), RO1 AI116282 (S.P.), R56 AI124788 (S.P. and C.A.B.), and DP2 AI112193 (C.A.B.). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Janeway CA Jr, Medzhitov R. 2002. Innate immune recognition. Annu Rev Immunol 20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Saxena M, Yeretssian G. 2014. NOD-like receptors: master regulators of inflammation and cancer. Front Immunol 5:327. doi: 10.3389/fimmu.2014.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee BL, Barton GM. 2014. Trafficking of endosomal Toll-like receptors. Trends Cell Biol 24:360–369. doi: 10.1016/j.tcb.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassing CH, Swat W, Alt FW. 2002. The mechanism and regulation of chromosomal V(D)J recombination. Cell 109(Suppl):S45–S55. doi: 10.1016/S0092-8674(02)00675-X. [DOI] [PubMed] [Google Scholar]
- 5.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manser AR, Weinhold S, Uhrberg M. 2015. Human KIR repertoires: shaped by genetic diversity and evolution. Immunol Rev 267:178–196. doi: 10.1111/imr.12316. [DOI] [PubMed] [Google Scholar]
- 7.Moretta L, Montaldo E, Vacca P, Del Zotto G, Moretta F, Merli P, Locatelli F, Mingari MC. 2014. Human natural killer cells: origin, receptors, function, and clinical applications. Int Arch Allergy Immunol 164:253–264. doi: 10.1159/000365632. [DOI] [PubMed] [Google Scholar]
- 8.Jonsson AH, Yokoyama WM. 2009. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol 101:27–79. doi: 10.1016/S0065-2776(08)01002-X. [DOI] [PubMed] [Google Scholar]
- 9.Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M, Mohanakumar T, Hsu KC, Dupont B, Yokoyama WM. 2008. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A 105:3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yokoyama WM, Kim S. 2006. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol Rev 214:143–154. doi: 10.1111/j.1600-065X.2006.00458.x. [DOI] [PubMed] [Google Scholar]
- 11.Yokoyama WM, Kim S. 2006. How do natural killer cells find self to achieve tolerance? Immunity 24:249–257. doi: 10.1016/j.immuni.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Orr MT, Lanier LL. 2010. Natural killer cell education and tolerance. Cell 142:847–856. doi: 10.1016/j.cell.2010.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shifrin N, Raulet DH, Ardolino M. 2014. NK cell self tolerance, responsiveness and missing self recognition. Semin Immunol 26:138–144. doi: 10.1016/j.smim.2014.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahim MM, Tu MM, Mahmoud AB, Wight A, Abou-Samra E, Lima PD, Makrigiannis AP. 2014. Ly49 receptors: innate and adaptive immune paradigms. Front Immunol 5:145. doi: 10.3389/fimmu.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. 2013. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol 31:413–441. doi: 10.1146/annurev-immunol-032712-095951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie J. 2012. The C-type lectin-like receptors of Dectin-1 cluster in natural killer gene complex. Glycoconj J 29:273–284. doi: 10.1007/s10719-012-9419-9. [DOI] [PubMed] [Google Scholar]
- 17.Orr MT, Lanier LL. 2011. Inhibitory Ly49 receptors on mouse natural killer cells. Curr Top Microbiol Immunol 350:67–87. doi: 10.1007/82_2010_85. [DOI] [PubMed] [Google Scholar]
- 18.Yokoyama WM, Riley JK. 2008. NK cells and their receptors. Reprod Biomed Online 16:173–191. doi: 10.1016/S1472-6483(10)60573-1. [DOI] [PubMed] [Google Scholar]
- 19.Joncker NT, Raulet DH. 2008. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol Rev 224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Culley FJ. 2009. Natural killer cells in infection and inflammation of the lung. Immunology 128:151–163. doi: 10.1111/j.1365-2567.2009.03167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanier LL. 2005. NK cell recognition. Annu Rev Immunol 23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 22.Cooper MD, Alder MN. 2006. The evolution of adaptive immune systems. Cell 124:815–822. doi: 10.1016/j.cell.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Gober MD, Gaspari AA. 2008. Allergic contact dermatitis. Curr Dir Autoimmun 10:1–26. doi: 10.1159/000131410. [DOI] [PubMed] [Google Scholar]
- 24.Sicherer SH, Leung DY. 2008. Advances in allergic skin disease, anaphylaxis, and hypersensitivity reactions to foods, drugs, and insects in 2007. J Allergy Clin Immunol 121:1351–1358. doi: 10.1016/j.jaci.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 25.Askenase PW. 2001. Yes T cells, but three different T cells (alphabeta, gammadelta and NK T cells), and also B-1 cells mediate contact sensitivity. Clin Exp Immunol 125:345–350. doi: 10.1046/j.1365-2249.2001.01619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Leary JG, Goodarzi M, Drayton DL, von Andrian UH. 2006. T cell- and B cell-independent adaptive immunity mediated by natural killer cells. Nat Immunol 7:507–516. doi: 10.1038/ni1332. [DOI] [PubMed] [Google Scholar]
- 27.Catalina MD, Carroll MC, Arizpe H, Takashima A, Estess P, Siegelman MH. 1996. The route of antigen entry determines the requirement for L-selectin during immune responses. J Exp Med 184:2341–2351. doi: 10.1084/jem.184.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Catalina MD, Estess P, Siegelman MH. 1999. Selective requirements for leukocyte adhesion molecules in models of acute and chronic cutaneous inflammation: participation of E- and P- but not L-selectin. Blood 93:580–589. [PubMed] [Google Scholar]
- 29.Diacovo TG, Catalina MD, Siegelman MH, von Andrian UH. 1998. Circulating activated platelets reconstitute lymphocyte homing and immunity in L-selectin-deficient mice. J Exp Med 187:197–204. doi: 10.1084/jem.187.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paust S, von Andrian UH. 2011. Natural killer cell memory. Nat Immunol 12:500–508. doi: 10.1038/ni.2032. [DOI] [PubMed] [Google Scholar]
- 31.Paust S, Gill HS, Wang BZ, Flynn MP, Moseman EA, Senman B, Szczepanik M, Telenti A, Askenase PW, Compans RW, von Andrian UH. 2010. Critical role for the chemokine receptor CXCR6 in NK cell-mediated antigen-specific memory of haptens and viruses. Nat Immunol 11:1127–1135. doi: 10.1038/ni.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majewska-Szczepanik M, Paust S, von Andrian UH, Askenase PW, Szczepanik M. 2013. Natural killer cell-mediated contact sensitivity develops rapidly and depends on interferon-alpha, interferon-gamma and interleukin-12. Immunology 140:98–110. doi: 10.1111/imm.12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gillard GO, Bivas-Benita M, Hovav AH, Grandpre LE, Panas MW, Seaman MS, Haynes BF, Letvin NL. 2011. Thy1+ NK [corrected] cells from vaccinia virus-primed mice confer protection against vaccinia virus challenge in the absence of adaptive lymphocytes. PLoS Pathog 7:e1002141. doi: 10.1371/journal.ppat.1002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng H, Jiang X, Chen Y, Sojka DK, Wei H, Gao X, Sun R, Yokoyama WM, Tian Z. 2013. Liver-resident NK cells confer adaptive immunity in skin-contact inflammation. J Clin Invest 123:1444–1456. doi: 10.1172/JCI66381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li T, Wang J, Wang Y, Chen Y, Wei H, Sun R, Tian Z. 2017. Respiratory influenza virus infection induces memory-like liver NK cells in mice. J Immunol 198:1242–1252. doi: 10.4049/jimmunol.1502186. [DOI] [PubMed] [Google Scholar]
- 36.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, Scalzo AA, Fremont DH, Yokoyama WM. 2002. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A 99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bubic I, Wagner M, Krmpotic A, Saulig T, Kim S, Yokoyama WM, Jonjic S, Koszinowski UH. 2004. Gain of virulence caused by loss of a gene in murine cytomegalovirus. J Virol 78:7536–7544. doi: 10.1128/JVI.78.14.7536-7544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voigt V, Forbes CA, Tonkin JN, Degli-Esposti MA, Smith HR, Yokoyama WM, Scalzo AA. 2003. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci U S A 100:13483–13488. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.French AR, Pingel JT, Wagner M, Bubic I, Yang L, Kim S, Koszinowski U, Jonjic S, Yokoyama WM. 2004. Escape of mutant double-stranded DNA virus from innate immune control. Immunity 20:747–756. doi: 10.1016/j.immuni.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Daniels KA, Devora G, Lai WC, O'Donnell CL, Bennett M, Welsh RM. 2001. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med 194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. 2001. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat Genet 28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 42.Brown MG, Dokun AO, Heusel JW, Smith HR, Beckman DL, Blattenberger EA, Dubbelde CE, Stone LR, Scalzo AA, Yokoyama WM. 2001. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science 292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 43.Kielczewska A, Pyzik M, Sun T, Krmpotic A, Lodoen MB, Munks MW, Babic M, Hill AB, Koszinowski UH, Jonjic S, Lanier LL, Vidal SM. 2009. Ly49P recognition of cytomegalovirus-infected cells expressing H2-Dk and CMV-encoded m04 correlates with the NK cell antiviral response. J Exp Med 206:515–523. doi: 10.1084/jem.20080954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scalzo AA, Manzur M, Forbes CA, Brown MG, Shellam GR. 2005. NK gene complex haplotype variability and host resistance alleles to murine cytomegalovirus in wild mouse populations. Immunol Cell Biol 83:144–149. doi: 10.1111/j.1440-1711.2005.01311.x. [DOI] [PubMed] [Google Scholar]
- 45.Sun JC, Beilke JN, Lanier LL. 2009. Adaptive immune features of natural killer cells. Nature 457:557–561. doi: 10.1038/nature07665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tay CH, Welsh RM. 1997. Distinct organ-dependent mechanisms for the control of murine cytomegalovirus infection by natural killer cells. J Virol 71:267–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Loh J, Chu DT, O'Guin AK, Yokoyama WM, Virgin HW IV. 2005. Natural killer cells utilize both perforin and gamma interferon to regulate murine cytomegalovirus infection in the spleen and liver. J Virol 79:661–667. doi: 10.1128/JVI.79.1.661-667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Min-Oo G, Bezman NA, Madera S, Sun JC, Lanier LL. 2014. Proapoptotic Bim regulates antigen-specific NK cell contraction and the generation of the memory NK cell pool after cytomegalovirus infection. J Exp Med 211:1289–1296. doi: 10.1084/jem.20132459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O'Sullivan TE, Johnson LR, Kang HH, Sun JC. 2015. BNIP3- and BNIP3L-mediated mitophagy promotes the generation of natural killer cell memory. Immunity 43:331–342. doi: 10.1016/j.immuni.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guma M, Budt M, Saez A, Brckalo T, Hengel H, Angulo A, Lopez-Botet M. 2006. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 107:3624–3631. doi: 10.1182/blood-2005-09-3682. [DOI] [PubMed] [Google Scholar]
- 51.Hendricks DW, Balfour HH Jr, Dunmire SK, Schmeling DO, Hogquist KA, Lanier LL. 2014. Cutting edge: NKG2ChiCD57+ NK cells respond specifically to acute infection with cytomegalovirus and not Epstein-Barr virus. J Immunol 192:4492–4496. doi: 10.4049/jimmunol.1303211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lopez-Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA, Houchins JP, Miller S, Kang SM, Norris PJ, Nixon DF, Lanier LL. 2011. Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A 108:14725–14732. doi: 10.1073/pnas.1110900108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Foley B, Cooley S, Verneris MR, Curtsinger J, Luo X, Waller EK, Anasetti C, Weisdorf D, Miller JS. 2012. Human cytomegalovirus (CMV)-induced memory-like NKG2C+ NK cells are transplantable and expand in vivo in response to recipient CMV antigen. J Immunol 189:5082–5088. doi: 10.4049/jimmunol.1201964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nash WT, Teoh J, Wei H, Gamache A, Brown MG. 2014. Know thyself: NK-cell inhibitory receptors prompt self-tolerance, education, and viral control. Front Immunol 5:175. doi: 10.3389/fimmu.2014.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Della Chiesa M, Falco M, Bertaina A, Muccio L, Alicata C, Frassoni F, Locatelli F, Moretta L, Moretta A. 2014. Human cytomegalovirus infection promotes rapid maturation of NK cells expressing activating killer Ig-like receptor in patients transplanted with NKG2C−/− umbilical cord blood. J Immunol 192:1471–1479. doi: 10.4049/jimmunol.1302053. [DOI] [PubMed] [Google Scholar]
- 56.Mehta RS, Shpall EJ, Rezvani K. 2015. Cord blood as a source of natural killer cells. Front Med (Lausanne) 2:93. doi: 10.3389/fmed.2015.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lunemann A, Vanoaica LD, Azzi T, Nadal D, Munz C. 2013. A distinct subpopulation of human NK cells restricts B cell transformation by EBV. J Immunol 191:4989–4995. doi: 10.4049/jimmunol.1301046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hatton O, Strauss-Albee DM, Zhao NQ, Haggadone MD, Pelpola JS, Krams SM, Martinez OM, Blish CA. 2016. NKG2A-expressing natural killer cells dominate the response to autologous lymphoblastoid cells infected with Epstein-Barr virus. Front Immunol 7:607. doi: 10.3389/fimmu.2016.00607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jud A, Kotur M, Berger C, Gysin C, Nadal D, Lunemann A. 2016. Tonsillar CD56brightNKG2A+ NK cells restrict primary Epstein-Barr virus infection in B cells via IFN-gamma. Oncotarget doi: 10.18632/oncotarget.14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Azzi T, Lunemann A, Murer A, Ueda S, Beziat V, Malmberg KJ, Staubli G, Gysin C, Berger C, Munz C, Chijioke O, Nadal D. 2014. Role for early-differentiated natural killer cells in infectious mononucleosis. Blood 124:2533–2543. doi: 10.1182/blood-2014-01-553024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alter G, Teigen N, Ahern R, Streeck H, Meier A, Rosenberg ES, Altfeld M. 2007. Evolution of innate and adaptive effector cell functions during acute HIV-1 infection. J Infect Dis 195:1452–1460. doi: 10.1086/513878. [DOI] [PubMed] [Google Scholar]
- 62.Alter G, Teigen N, Davis BT, Addo MM, Suscovich TJ, Waring MT, Streeck H, Johnston MN, Staller KD, Zaman MT, Yu XG, Lichterfeld M, Basgoz N, Rosenberg ES, Altfeld M. 2005. Sequential deregulation of NK cell subset distribution and function starting in acute HIV-1 infection. Blood 106:3366–3369. doi: 10.1182/blood-2005-03-1100. [DOI] [PubMed] [Google Scholar]
- 63.Bonaparte MI, Barker E. 2004. Killing of human immunodeficiency virus-infected primary T-cell blasts by autologous natural killer cells is dependent on the ability of the virus to alter the expression of major histocompatibility complex class I molecules. Blood 104:2087–2094. doi: 10.1182/blood-2004-02-0696. [DOI] [PubMed] [Google Scholar]
- 64.Fogli M, Mavilio D, Brunetta E, Varchetta S, Ata K, Roby G, Kovacs C, Follmann D, Pende D, Ward J, Barker E, Marcenaro E, Moretta A, Fauci AS. 2008. Lysis of endogenously infected CD4+ T cell blasts by rIL-2 activated autologous natural killer cells from HIV-infected viremic individuals. PLoS Pathog 4:e1000101. doi: 10.1371/journal.ppat.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ward J, Bonaparte M, Sacks J, Guterman J, Fogli M, Mavilio D, Barker E. 2007. HIV modulates the expression of ligands important in triggering natural killer cell cytotoxic responses on infected primary T-cell blasts. Blood 110:1207–1214. doi: 10.1182/blood-2006-06-028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fehniger TA, Herbein G, Yu H, Para MI, Bernstein ZP, O'Brien WA, Caligiuri MA. 1998. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J Immunol 161:6433–6438. [PubMed] [Google Scholar]
- 67.Thomas R, Low HZ, Kniesch K, Jacobs R, Schmidt RE, Witte T. 2012. NKG2C deletion is a risk factor of HIV infection. AIDS Res Hum Retroviruses 28:844–851. doi: 10.1089/aid.2011.0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Marras F, Nicco E, Bozzano F, Di Biagio A, Dentone C, Pontali E, Boni S, Setti M, Orofino G, Mantia E, Bartolacci V, Bisio F, Riva A, Biassoni R, Moretta L, De Maria A. 2013. Natural killer cells in HIV controller patients express an activated effector phenotype and do not up-regulate NKp44 on IL-2 stimulation. Proc Natl Acad Sci U S A 110:11970–11975. doi: 10.1073/pnas.1302090110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, Oniangue-Ndza C, Martin M, Li B, Khakoo SI, Carrington M, Allen TM, Altfeld M. 2011. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Scott-Algara D, Truong LX, Versmisse P, David A, Luong TT, Nguyen NV, Theodorou I, Barre-Sinoussi F, Pancino G. 2003. Cutting edge: increased NK cell activity in HIV-1-exposed but uninfected Vietnamese intravascular drug users. J Immunol 171:5663–5667. doi: 10.4049/jimmunol.171.11.5663. [DOI] [PubMed] [Google Scholar]
- 71.Ravet S, Scott-Algara D, Bonnet E, Tran HK, Tran T, Nguyen N, Truong LX, Theodorou I, Barre-Sinoussi F, Pancino G, Paul P. 2007. Distinctive NK-cell receptor repertoires sustain high-level constitutive NK-cell activation in HIV-exposed uninfected individuals. Blood 109:4296–4305. doi: 10.1182/blood-2006-08-040238. [DOI] [PubMed] [Google Scholar]
- 72.O'Connor GM, Holmes A, Mulcahy F, Gardiner CM. 2007. Natural killer cells from long-term non-progressor HIV patients are characterized by altered phenotype and function. Clin Immunol 124:277–283. doi: 10.1016/j.clim.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 73.Tiemessen CT, Shalekoff S, Meddows-Taylor S, Schramm DB, Papathanasopoulos MA, Gray GE, Sherman GG, Coovadia AH, Kuhn L. 2009. Cutting edge: unusual NK cell responses to HIV-1 peptides are associated with protection against maternal-infant transmission of HIV-1. J Immunol 182:5914–5918. doi: 10.4049/jimmunol.0900419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pereira LE, Johnson RP, Ansari AA. 2008. Sooty mangabeys and rhesus macaques exhibit significant divergent natural killer cell responses during both acute and chronic phases of SIV infection. Cell Immunol 254:10–19. doi: 10.1016/j.cellimm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 75.Bostik P, Kobkitjaroen J, Tang W, Villinger F, Pereira LE, Little DM, Stephenson ST, Bouzyk M, Ansari AA. 2009. Decreased NK cell frequency and function is associated with increased risk of KIR3DL allele polymorphism in simian immunodeficiency virus-infected rhesus macaques with high viral loads. J Immunol 182:3638–3649. doi: 10.4049/jimmunol.0803580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Takahashi Y, Mayne AE, Khowawisetsut L, Pattanapanyasat K, Little D, Villinger F, Ansari AA. 2013. In vivo administration of a JAK3 inhibitor to chronically SIV infected rhesus macaques leads to NK cell depletion associated with transient modest increase in viral loads. PLoS One 8:e70992. doi: 10.1371/journal.pone.0070992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takahashi Y, Byrareddy SN, Albrecht C, Brameier M, Walter L, Mayne AE, Dunbar P, Russo R, Little DM, Villinger T, Khowawisetsut L, Pattanapanyasat K, Villinger F, Ansari AA. 2014. In vivo administration of a JAK3 inhibitor during acute SIV infection leads to significant increases in viral load during chronic infection. PLoS Pathog 10:e1003929. doi: 10.1371/journal.ppat.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Reeves RK, Li H, Jost S, Blass E, Li H, Schafer JL, Varner V, Manickam C, Eslamizar L, Altfeld M, von Andrian UH, Barouch DH. 2015. Antigen-specific NK cell memory in rhesus macaques. Nat Immunol 16:927–932. doi: 10.1038/ni.3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, O'Brien SJ, Carrington M. 2002. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 80.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, Buchbinder S, Kirk GD, Telenti A, Connors M, O'Brien SJ, Walker BD, Parham P, Deeks SG, McVicar DW, Carrington M. 2007. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Carrington M, Martin MP, van Bergen J. 2008. KIR-HLA intercourse in HIV disease. Trends Microbiol 16:620–627. doi: 10.1016/j.tim.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song R, Lisovsky I, Lebouche B, Routy JP, Bruneau J, Bernard NF. 2014. HIV protective KIR3DL1/S1-HLA-B genotypes influence NK cell-mediated inhibition of HIV replication in autologous CD4 targets. PLoS Pathog 10:e1003867. doi: 10.1371/journal.ppat.1003867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Colantonio AD, Bimber BN, Neidermyer WJ, Reeves RK, Alter G, Altfeld M, Johnson RP, Carrington M, O'Connor DH, Evans DT. 2011. KIR polymorphisms modulate peptide-dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog 7:e1001316. doi: 10.1371/journal.ppat.1001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Davis ZB, Cogswell A, Scott H, Mertsching A, Boucau J, Wambua D, Le Gall S, Planelles V, Campbell KS, Barker E. 2016. A conserved HIV-1-derived peptide presented by HLA-E renders infected T-cells highly susceptible to attack by NKG2A/CD94-bearing natural killer cells. PLoS Pathog 12:e1005421. doi: 10.1371/journal.ppat.1005421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Venkatasubramanian S, Cheekatla S, Paidipally P, Tripathi D, Welch E, Tvinnereim AR, Nurieva R, Vankayalapati R. 2016. IL-21-dependent expansion of memory-like NK cells enhances protective immune responses against Mycobacterium tuberculosis. Mucosal Immunol doi: 10.1038/mi.2016.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suliman S, Geldenhuys H, Johnson JL, Hughes JE, Smit E, Murphy M, Toefy A, Lerumo L, Hopley C, Pienaar B, Chheng P, Nemes E, Hoft DF, Hanekom WA, Boom WH, Hatherill M, Scriba TJ. 2016. Bacillus Calmette-Guerin (BCG) revaccination of adults with latent Mycobacterium tuberculosis infection induces long-lived BCG-reactive NK cell responses. J Immunol 197:1100–1110. doi: 10.4049/jimmunol.1501996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM. 2009. Cytokine-induced memory-like natural killer cells. Proc Natl Acad Sci U S A 106:1915–1919. doi: 10.1073/pnas.0813192106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goodier MR, Rodriguez-Galan A, Lusa C, Nielsen CM, Darboe A, Moldoveanu AL, White MJ, Behrens R, Riley EM. 2016. Influenza vaccination generates cytokine-induced memory-like NK cells: impact of human cytomegalovirus infection. J Immunol 197:313–325. doi: 10.4049/jimmunol.1502049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Romee R, Rosario M, Berrien-Elliott MM, Wagner JA, Jewell BA, Schappe T, Leong JW, Abdel-Latif S, Schneider SE, Willey S, Neal CC, Yu L, Oh ST, Lee YS, Mulder A, Claas F, Cooper MA, Fehniger TA. 2016. Cytokine-induced memory-like natural killer cells exhibit enhanced responses against myeloid leukemia. Sci Transl Med 8:357ra123. doi: 10.1126/scitranslmed.aaf2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang T, Scott JM, Hwang I, Kim S. 2013. Cutting edge: antibody-dependent memory-like NK cells distinguished by FcRgamma deficiency. J Immunol 190:1402–1406. doi: 10.4049/jimmunol.1203034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee J, Zhang T, Hwang I, Kim A, Nitschke L, Kim M, Scott JM, Kamimura Y, Lanier LL, Kim S. 2015. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42:431–442. doi: 10.1016/j.immuni.2015.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhou J, Amran FS, Kramski M, Angelovich TA, Elliott J, Hearps AC, Price P, Jaworowski A. 2015. An NK cell population lacking FcRgamma is expanded in chronically infected HIV patients. J Immunol 194:4688–4697. doi: 10.4049/jimmunol.1402448. [DOI] [PubMed] [Google Scholar]
- 93.Oh JS, Ali AK, Kim S, Corsi DJ, Cooper CL, Lee SH. 2016. NK cells lacking FcepsilonRIgamma are associated with reduced liver damage in chronic hepatitis C virus infection. Eur J Immunol 46:1020–1029. doi: 10.1002/eji.201546009. [DOI] [PubMed] [Google Scholar]
- 94.Blish CA. 2016. Natural killer cell diversity in viral infection: why and how much? Pathog Immun 1:165–192. doi: 10.20411/pai.v1i1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Horowitz A, Strauss-Albee DM, Leipold M, Kubo J, Nemat-Gorgani N, Dogan OC, Dekker CL, Mackey S, Maecker H, Swan GE, Davis MM, Norman PJ, Guethlein LA, Desai M, Parham P, Blish CA. 2013. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 5:208ra145. doi: 10.1126/scitranslmed.3006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beziat V, Liu LL, Malmberg JA, Ivarsson MA, Sohlberg E, Bjorklund AT, Retiere C, Sverremark-Ekstrom E, Traherne J, Ljungman P, Schaffer M, Price DA, Trowsdale J, Michaelsson J, Ljunggren HG, Malmberg KJ. 2013. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121:2678–2688. doi: 10.1182/blood-2012-10-459545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Strauss-Albee DM, Horowitz A, Parham P, Blish CA. 2014. Coordinated regulation of NK receptor expression in the maturing human immune system. J Immunol 193:4871–4879. doi: 10.4049/jimmunol.1401821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Strauss-Albee DM, Fukuyama J, Liang EC, Yao Y, Jarrell JA, Drake AL, Kinuthia J, Montgomery RR, John-Stewart G, Holmes S, Blish CA. 2015. Human NK cell repertoire diversity reflects immune experience and correlates with viral susceptibility. Sci Transl Med 7:297ra115. doi: 10.1126/scitranslmed.aac5722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Strauss-Albee DM, Blish CA. 2016. Human NK cell diversity in viral infection: ramifications of ramification. Front Immunol 7:66. doi: 10.3389/fimmu.2016.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Amemiya CT, Saha NR, Zapata A. 2007. Evolution and development of immunological structures in the lamprey. Curr Opin Immunol 19:535–541. doi: 10.1016/j.coi.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tsutsui S, Nakamura O, Watanabe T. 2007. Lamprey (Lethenteron japonicum) IL-17 upregulated by LPS-stimulation in the skin cells. Immunogenetics 59:873–882. doi: 10.1007/s00251-007-0254-2. [DOI] [PubMed] [Google Scholar]
- 102.Uinuk-Ool T, Mayer WE, Sato A, Dongak R, Cooper MD, Klein J. 2002. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc Natl Acad Sci U S A 99:14356–14361. doi: 10.1073/pnas.212527699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Garcia-Valtanen P, Martinez-Lopez A, Lopez-Munoz A, Bello-Perez M, Medina-Gali RM, Ortega-Villaizan MD, Varela M, Figueras A, Mulero V, Novoa B, Estepa A, Coll J. 2017. Zebra fish lacking adaptive immunity acquire an antiviral alert state characterized by upregulated gene expression of apoptosis, multigene families, and interferon-related genes. Front Immunol 8:121. doi: 10.3389/fimmu.2017.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Paust S, Flynn M, Wang B, Dougan S, Ploegh H, Telenti A, Compans R, von Andrian U. 2012. Adaptive immunity mediated by natural killer cells. J Immunol 188(Suppl 1):45.20. [Google Scholar]