ABSTRACT
Adenovirus (Ad)-based immunization is a popular approach in vaccine development, and Ad-based vectors are renowned for their potential to induce strong CD8+ T cell responses to the encoded transgene. Surprisingly, we previously found in the mouse Friend retrovirus (FV) model that Ad-based immunization did not induce CD8+ T cell responses to the FV Leader-Gag-derived immunodominant epitope GagL85–93. We show now that induction of GagL85–93-specific CD8+ T cells was highly effective when leader-Gag was delivered by plasmid DNA immunization, implying a role for Ad-derived epitopes in mediating unresponsiveness. By immunizing with DNA constructs encoding strings of GagL85–93 and the two Ad-derived epitopes DNA-binding protein418–426 (DBP418–426) and hexon486–494, we confirmed that Ad epitopes prevent induction of GagL85–93-specific CD8+ T cells. Interestingly, while DBP418–426 did not interfere with GagL85–93-specific CD8+ T cell induction, the H-2Dd-restricted hexon486–494 suppressed the CD8+ T cell response to the H-2Db-restricted GagL85–93 strongly in H-2b/d mice but not in H-2b/b mice. This finding indicates that competition occurs at the level of responding CD8+ T cells, and we could indeed demonstrate that coimmunization with an interleukin 2 (IL-2)-encoding plasmid restored GagL85–93-specific CD8+ T cell responses to epitope strings in the presence of hexon486–494. IL-2 codelivery did not restore GagL85–93 responsiveness in Ad-based immunization, however, likely due to the presence of further epitopes in the Ad vector. Our findings show that seemingly immunodominant transgene epitopes can be dominated by Ad-derived epitopes. These findings underline the importance of thorough characterization of vaccine vectors, and modifications of vectors or immunogens may be required to prevent impaired transgene-specific immune responses.
IMPORTANCE Ad-based vectors are widely used in experimental preclinical and clinical immunization studies against numerous infectious agents, such as human immunodeficiency virus, Ebola virus, Plasmodium falciparum, or Mycobacterium tuberculosis. Preexisting immunity to Ad-based vectors is widely recognized as a hindrance to the widespread use of Ad-based vectors for immunizations in humans; however, our data show that an immune response to Ad-derived T cell epitopes can also result in loss or impairment of transgene-specific immune responses in prenaive vaccinees due to immune competition. Our results highlight that seemingly immunodominant epitopes may be affected by dominance of vector-derived epitopes, and modifications of the vector design or the immunogens employed in immunization may lead to more effective vaccines.
KEYWORDS: Friend virus, adenovirus, immunization, immunodominance, retrovirus, vaccine, vector
INTRODUCTION
Adenovirus (Ad)-based vectors are popular tools for the development of new immunization strategies and have been employed in experimental immunization studies against a large variety of pathogens. Clinical trials in humans have already been advanced into phase IIb for HIV immunization and phase II for Ebola virus immunization (1, 2). The high immunogenicity of Ad-based vectors is widely considered to be an advantage for immunization, as components of the Ad particle induce a significant range of innate immune responses (3, 4), and the subsequent induction of cytokines can have an adjuvant effect on the immune responses to the delivered immunogen. Ad-based vectors are renowned for their capacity to induce strong CD8+ T cell responses (5), which likely contribute to vaccine protection in many infections.
Friend virus (FV) is an immunosuppressive complex of two viruses, Friend murine leukemia virus (F-MuLV) and spleen focus-forming virus (SFFV), that causes severe splenomegaly and erythroleukemia in susceptible adult mice, whereas mice that are genetically resistant to the FV-induced disease develop a persistent infection (6). The FV infection is an excellent model for the study of mechanisms underlying immune protection and immune suppression in retrovirus infection (7–10) and has also been employed to elucidate mechanisms underlying vaccine-mediated protection from retrovirus infection (11–13). In the acute phase of FV infection, the CD8+ T cell response is crucial for immune control, and a large fraction of the CD8+ T cells that show an activated profile are reactive to the leader-Gag-derived epitope GagL85–93 (9, 14, 15). The GagL85–93 epitope is considered to be the immunodominant CD8+ T cell epitope in FV infection, and many efforts to identify further CD8+ T cell epitopes in FV-encoded proteins have failed to yield positive results (reference 16 and unpublished observation).
In vaccine experiments, it was shown in the past that attenuated F-MuLV was the most potent vaccine against FV, as it conferred very strong protection even to highly susceptible mice (17, 18), and transfer experiments showed that complex immune responses comprising CD8+ T cells as well as CD4+ T cells and antibody responses were required for complete protection (12). More recently we could show that Ad-based vaccines could also induce very strong protection from FV infection to highly susceptible mice by mechanisms that apparently differed from those for attenuated F-MuLV as shown in side-by-side experiments, as the protection mediated by the Ad-based vaccine clearly depended on very strong GagL85–93-specific CD8+ T cell responses (13). Similarly, we could demonstrate high, albeit incomplete, prophylactic vaccine efficacy for calcium phosphate nanoparticles, which relies exclusively on the induction of cellular immune responses (19). These results highlight the important role that CD8+ T cells play in vaccine-mediated protection from retrovirus infection.
Interestingly, we previously showed that the induction of GagL85–93-specific CD8+ T cells by immunization with a leader-Gag-encoding Ad vector required the modification of the immunogen, as the immunization with the native sequence did not result in any detectable induction of GagL85–93-specific CD8+ T cells (20). The modification of the epitope-flanking amino acid that was introduced to improve proteasomal degradation led to improved stimulation of GagL85–93-specific CD8+ T cells in vitro and to their improved induction in vivo in immunization experiments. The finding that the immunogen sequence had to be modified to allow for any induction of GagL85–93-specific CD8+ T cells was surprising because of the immunodominance of the GagL85–93 epitope in the native setting of an FV infection. We assumed that the more competitive environment created by Ad-derived epitopes hindered the induction of GagL85–93-specific CD8+ T cells and that the improved proteasomal degradation of the modified leader-Gag partially counterregulated this hindrance.
We have now investigated this hypothesis further, analyzing the influence of Ad-derived epitopes on GagL85–93 in epitope string constructs, and have evaluated the potential of cytokines as genetic adjuvants to alleviate immunocompetition as a more widely applicable solution to vector immunodominance.
RESULTS
GagL85–93-specific CD8+ T cells are efficiently induced by DNA immunization with a plasmid encoding the native leader-Gag sequence.
In previous work, we were surprised to find that the native leader-Gag sequence did not induce any GagL85–93-specific CD8+ T cell responses in mice if it was delivered by an Ad-based vector (20); this was an unexpected finding because of the apparent immunodominance of the GagL85–93 epitope in the native FV infection. We hypothesized that either other FV-derived proteins, such as F-MuLV protease, may be necessary for efficient processing of the GagL85–93 epitope or the GagL85–93-specific response is dominated by Ad-derived T cell epitopes.
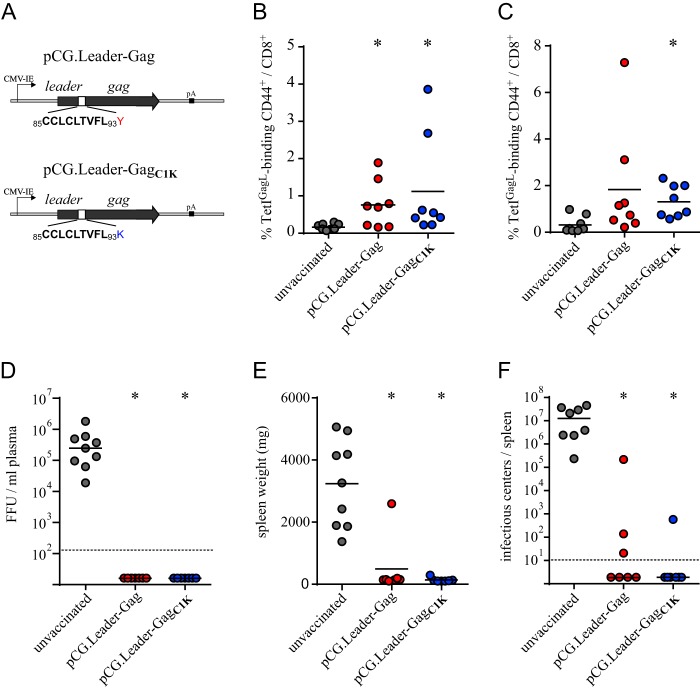
To address the first hypothesis, we evaluated whether the native leader-Gag alone can induce GagL85–93-specific CD8+ T cell responses in the context of DNA-based immunization. For this, we constructed DNA expression plasmids encoding either the native leader-Gag sequence or the modified leader-GagC1K sequence where the amino acid flanking the C terminus of the GagL85–93 epitope was changed from tyrosine to lysine as described before (Fig. 1A). We immunized highly susceptible CB6F1 mice with either plasmid by intramuscular injection followed by electroporation in a prime-boost immunization schedule and then analyzed the GagL85–93-specific CD8+ T cell response by major histocompatibility complex (MHC) I tetramer staining 2 weeks after each immunization. Both groups of mice were able to mount a CD8+ T cell response against the GagL85–93 epitope that was significantly higher than the background response detected in unvaccinated mice after the first immunization (Fig. 1B). Importantly, there was no difference in the response to either leader-Gag sequence; similar results were obtained after the second immunization (Fig. 1C). Three weeks after the second immunization, mice were challenged with 5,000 spleen focus-forming units (SFFU) FV; when the plasma viral load was analyzed 10 days after the infection, unvaccinated mice had high viremia levels, while mice immunized with either plasmid had undetectable viral loads in plasma (Fig. 1D). Three weeks after FV challenge infection, mice were sacrificed and the spleen weights and spleen viral loads were determined. While all unvaccinated mice had gravely enlarged spleens, spleens of immunized mice were significantly smaller, with most of them showing no increased weight (Fig. 1E); similarly, the viral load in spleens was significantly reduced in immunized mice compared to the unvaccinated control mice, and many mice did not exhibit any detectable viral loads (Fig. 1F). These findings confirm our previous observation that a vaccine that induces GagL85–93-specific CD8+ T cells, even at low levels, confers very strong protection from FV challenge infection to susceptible mice (13). Furthermore, these results indicate that processing of the leader-Gag protein resulting in the GagL85–93 peptide does not require the presence of any other FV-derived proteins.
FIG 1.
Efficient induction of GagL85–93-specific CD8+ T cell responses by plasmids encoding the native or modified leader-Gag sequence. (A) CB6F1 mice were immunized twice with DNA plasmids encoding F-MuLV leader-Gag either as the native sequence or as the Y94K variant leader-GagC1K. Mice received 30 μg plasmid by intramuscular injection, followed by in vivo electroporation. (B to F) The GagL85–93-specific CD8+ T cell response was analyzed 2 weeks after the first (B) and second (C) immunizations by MHC I tetramer staining. Three weeks after the second immunization, mice were infected with 5,000 SFFU FV, and viral loads in plasma were determined 10 days later (D). Three weeks after FV infection, the spleens were removed and weighed (E), and viral loads in spleen cells were determined (F). Data were acquired in two independent experiments, using four mice per group per experiment. Each dot indicates an individual mouse, lines indicate mean (B, C, and E) or median (D and F) values, and dashed lines indicate the detection limits. Data were analyzed by Kruskal-Wallis one-way analysis of variance on ranks and Dunn's multiple-comparison procedure for statistical significance. *, statistically significant differences (P < 0.05) compared to unvaccinated mice.
The presence of Ad-derived epitopes in the DNA vaccine abolishes the induction of GagL85–93-specific CD8+ T cells.
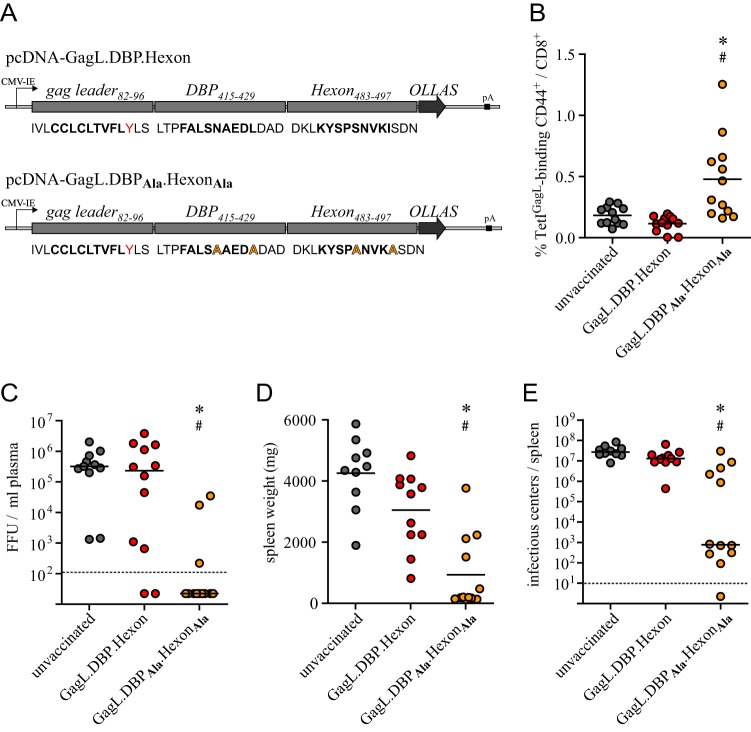
As the induction of GagL85–93-specific CD8+ T cells by the native and the modified leader-Gag sequences was equally effective when they were delivered by DNA plasmids, we turned our focus on our second hypothesis, suggesting that GagL85–93-specific CD8+ T cell induction may be hampered in Ad-based immunization because of immunodominance of Ad-derived T cell epitopes. To address this hypothesis, we constructed new DNA plasmids encoding epitope strings of the GagL85–93 epitope and two epitopes derived from the Ad proteins hexon (hexon486–494) and DNA-binding protein (DBP) (DBP418–426) that were previously described in C57BL/6 mice (DBP418–426; H-2Db) or BALB/c mice (hexon486–494; H-2Dd) (21). In order to allow for correct proteasomal processing, each epitope was flanked on either side by three amino acids derived from their native sequences, and we used the epitope prediction tool NetMHC 3.2 (22–24) to verify that merging the different sequences did not give rise to new T cell epitopes. To assess the induction of GagL85–93-specific CD8+ T cells by the epitope strings in the presence or absence of the Ad-derived epitopes, we used either the native Ad epitope sequences (epitope presence) or alanine-modified sequences where the epitope anchoring positions 5 and 9 were changed to alanine to prevent MHC I binding (epitope absence [25]). As we had not observed any differences between the immune responses to the native and the modified leader-Gag in DNA-based immunization, we used the native leader-Gag-derived sequence in the epitope strings (Fig. 2A shows a schematic of the epitope strings).
FIG 2.
The presence of adenovirus-derived epitopes abrogates the induction of GagL85–93-specific CD8+ T cell responses. (A) CB6F1 mice were immunized once with DNA plasmids encoding strings of epitope-comprising sequences of F-MuLV leader-Gag, Ad5 DNA-binding protein (DBP), and Ad5 hexon. Two different constructs were employed, comprising the native leader-Gag-derived sequence and the native or alanine-modified DBP and hexon-derived sequences; an OLLAS tag was fused to the C terminus for detection of the fusion peptide. (B) Mice were immunized by intramuscular injection of 30 μg DNA, followed by in vivo electroporation. The GagL85–93-specific CD8+ T cell response was analyzed 2 weeks after immunization by MHC I tetramer staining. (C) Three weeks after immunization, mice were challenged with 5,000 SFFU FV, and the viral load in plasma was analyzed 10 days later. (D and E) Three weeks after FV challenge infection, the spleens were removed and weighed (D), and viral loads in spleen cells were determined (E). Data were acquired in three independent experiments, using four mice per group per experiment. Each dot indicates an individual mouse, lines indicate mean (B and D) or median (C and E) values, and dashed lines indicate the detection limits. Data were analyzed by Kruskal-Wallis one-way analysis of variance on ranks and Dunn's multiple-comparison procedure for statistical significance. * and #, statistically significant differences (P < 0.05) compared to unvaccinated mice and compared to mice immunized with the respective plasmid comprising the native Ad-derived epitopes, respectively.
When we immunized CB6F1 mice (BALB/c × C57BL/6; H-2Db/d) with the epitope string-encoding plasmids, we found no induction of GagL85–93-specific CD8+ T cells in mice that received the epitope string comprising the native Ad-derived epitopes. In contrast, mice that were immunized with the epitope string comprising the alanine-modified Ad-derived epitopes were able to mount a GagL85–93-specific CD8+ T cell response that was significantly higher than that in unvaccinated mice or mice that were immunized with the respective epitope strings comprising the native Ad-derived epitopes (Fig. 2B).
As mice had mounted an adequate GagL85–93-specific CD8+ T cell response after one immunization and we had seen in the experiment described before that a second immunization did not have an appreciable boosting effect, mice were infected with FV after this single immunization to assess the protection from challenge infection. As before, we infected the mice with 5,000 SFFU FV and analyzed viral loads in plasma on day 10 after FV infection. While we found some variation in plasma viremia in mice that had been immunized with epitope strings comprising the native Ad-derived epitopes, there was no significant reduction in viral loads in these groups of mice compared to those in unvaccinated control mice. In contrast, mice that were immunized with epitope strings comprising alanine-modified Ad-derived epitopes mostly showed reduced virus loads in plasma; the plasma viral load in 9 of 12 mice of this group was below the detection limit and thus was significantly reduced in comparison to those in unvaccinated mice and mice immunized with the respective epitope string comprising the native Ad-derived epitopes (Fig. 2C).
Three weeks after FV infection, we removed the spleens and determined spleen weights and spleen virus loads. Mice that had been immunized with the epitope string comprising the intact Ad-derived epitopes had slightly reduced spleen weights compared to those of unvaccinated mice, but the difference was not statistically significant. On the other hand, mice that were immunized with the epitope string comprising the alanine-modified Ad-derived epitopes had spleens significantly smaller than those of both unvaccinated mice and mice that were immunized with the respective epitope strings comprising the native Ad-derived epitopes (Fig. 2D). Similarly, the viral loads in spleens of mice that were immunized with the epitope string comprising the native Ad-derived epitopes were not significantly reduced compared to those in unvaccinated control mice, whereas mice that were immunized with the epitope string comprising the alanine-modified Ad-derived epitopes had significantly reduced viral loads (Fig. 2E). The protective effect observed after alanine-modified epitope string-based immunization was slightly weaker than that after immunization with leader-Gag-encoding plasmids, which is probably attributable to the lack of CD4+ T cell epitopes in the epitope string constructs.
Our results demonstrate that the presence of Ad-derived epitopes gravely impairs the induction of GagL85–93-specific CD8+ T cells, reflecting the situation observed after Ad-based immunization.
The inhibition of GagL85–93-specific CD8+ T cell responses by hexon486–494 is not mediated by competition for MHC I presentation.
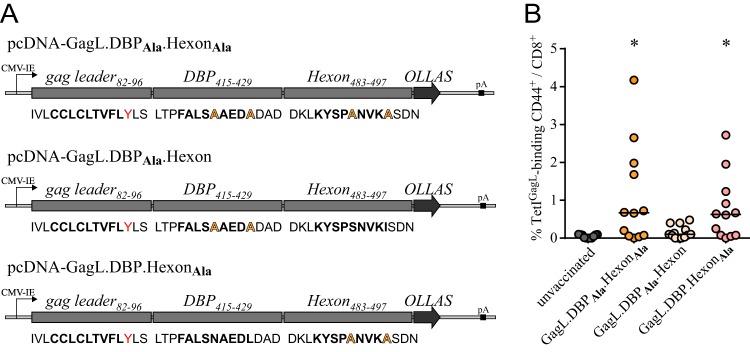
To gain further insight into the mechanism underlying the suppression by the Ad-derived epitopes, we constructed epitope strings that comprised one native (epitope present) and one alanine-modified (epitope absent) Ad-derived epitope (Fig. 3A shows a schematic of the epitope string constructs) and immunized mice with these new constructs to assess the immunodominant potential of each Ad-derived epitope.
FIG 3.
Hexon486–494, but not DBP418–426, impairs the induction of GagL85–93-specific CD8+ T cell responses. CB6F1 mice were immunized once with DNA plasmids encoding strings of epitope-comprising sequences of F-MuLV leader-Gag, Ad5 DNA-binding protein (DBP), and Ad5 hexon. Three different constructs were employed, comprising the native leader-Gag-derived sequence and two alanine-modified Ad-derived sequences or one native sequence and one alanine-modified Ad-derived sequence. (A) Mice were immunized by intramuscular injection of 30 μg DNA, followed by in vivo electroporation. The GagL85–93-specific CD8+ T cell response was analyzed 2 weeks after immunization by MHC I tetramer staining. (B) Data were acquired in three independent experiments, using four mice per group per experiment. Each dot indicates an individual mouse, and lines indicate mean values. Data were analyzed by Kruskal-Wallis one-way analysis of variance on ranks and Dunn's multiple-comparison procedure for statistical significance. *, statistically significant differences (P < 0.05) compared to unvaccinated mice.
We immunized CB6F1 mice (H-2Db/d) with the epitope string expression plasmids and analyzed the GagL85–93-specific CD8+ T cell response 2 weeks after immunization. As observed before, most mice that were immunized with the epitope string comprising two alanine-modified Ad-derived epitopes mounted a robust GagL85–93-specific CD8+ T cell response that was significantly higher than that in unvaccinated mice. Also, mice that received the epitope string comprising the native DBP418–426 and the alanine-modified hexon486–494 epitope were able to mount a GagL85–93-specific CD8+ T cell response. However, this response was potently suppressed after immunization with the epitope string comprising the alanine-modified DBP418–426 and the native hexon486–494 epitope (Fig. 3B). These results clearly indicate that hexon486–494 exerts a stronger immunodominance over GagL85–93 than DBP418–426. Interestingly, while GagL85–93 and DBP418–426 are both presented on H-2Db molecules, the hexon486–494 epitope is H-2Dd restricted; therefore, hexon486–494 and GagL85–93 do not compete for binding to the same MHC I allele.
To assess whether the dominance of hexon486–494 over GagL85–93 relies on higher efficiency of intracellular antigen processing or rather on direct competition of the responding T cells, we repeated the immunization experiment in C57BL/6 mice, which are H-2Db/b homozygous and therefore cannot present the hexon486–494 epitope; while intracellular processing of the epitope strings should be unaffected, there can be no presentation of, and CD8+ T cell response to, the hexon486–494 epitope in these mice. As expected, mice that were immunized with the epitope string comprising two alanine-modified Ad-derived epitopes were able to mount a GagL85–93-specific CD8+ T cell response, and importantly, mice that were immunized with the epitope string comprising the native hexon-derived sequence mounted a comparable GagL85–93-specific CD8+ T cell response (Fig. 4A). These results indicate that in the absence of a CD8+ T cell response to hexon486–494, the epitope loses its immunodominance.
FIG 4.
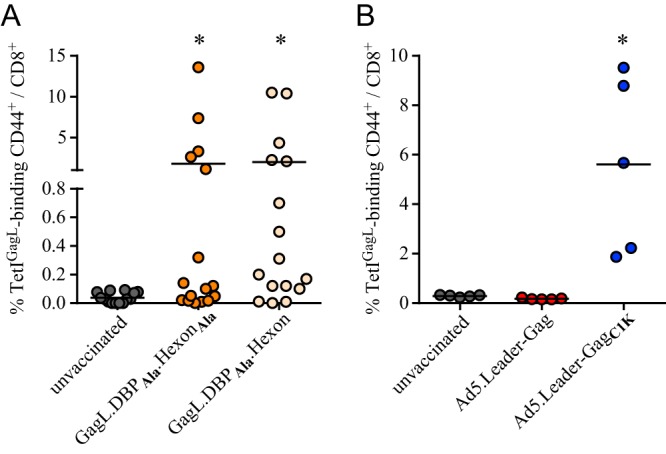
No impairment of GagL85–93-specific CD8+ T cell responses by hexon486–494 in H-2b/b C57BL/6 mice. (A) C57BL/6 mice were immunized once with DNA plasmids encoding strings of epitope-comprising sequences of F-MuLV leader-Gag, Ad5 DNA-binding protein (DBP), and Ad5 hexon. Two different constructs were employed, comprising the native leader-Gag-derived sequence, the alanine-modified DBP-derived sequence, and either the native or the alanine-modified hexon-derived sequence. Mice were immunized by intramuscular injection of 30 μg DNA, followed by in vivo electroporation. The GagL85–93-specific CD8+ T cell response was analyzed 2 weeks after immunization by MHC I tetramer staining. (B) For Ad-based immunization, C57BL/6 mice were immunized with 109 viral particles of Ad5.Leader-Gag or Ad5.Leader-GagC1K by intramuscular injection. The GagL85–93-specific CD8+ T cell response was analyzed 2 weeks after immunization by MHC I tetramer staining. Data were acquired in four independent experiments (A) or one experiment (B), using four (A) or five (B) mice per group per experiment. Each dot indicates an individual mouse, and lines indicate mean values. Data were analyzed by Kruskal-Wallis one-way analysis of variance on ranks and Dunn's multiple-comparison procedure for statistical significance. *, statistically significant differences (P < 0.05) compared to unvaccinated mice.
As DBP418–426 did not have a dominance effect over GagL85–93 and the dominance of hexon486–494 was lost in the C57BL/6 H-2Db/b background, we enquired next whether C57BL/6 mice would actually show responsiveness toward an Ad-delivered native leader-Gag. While we had performed immunization experiments in C57BL/6 mice in the past, in those experiments we had employed a mixture of the leader-Gag vector with an envelope-encoding vector (11), which we later showed to interfere with CD8+ T cell induction (11, 26). We immunized C57BL/6 mice once with Ad5.Leader-Gag, or with Ad5.Leader-GagC1K as a positive control, and analyzed the GagL85–93-specific CD8+ T cell response 2 weeks later by MHC I tetramer staining. Unfortunately, in mice that were immunized with Ad5.Leader-Gag, no GagL85–93-specific CD8+ T cell response was detectable, whereas we found a robust response in mice that were immunized with the modified Ad5.Leader-GagC1K (Fig. 4B), reflecting our previous results.
Ad immunization of mice induces a broad Ad-specific CD8+ T cell response.
The lack of a GagL85–93-specific CD8+ T cell response after Ad5.Leader-Gag immunization in C57BL/6 mice strongly indicates that while the two known epitopes DPB418–426 and hexon486–494 do not exhibit immunodominance in C57BL/6 mice, other epitopes that impede the induction of transgene-specific CD8+ T cell responses are present in the Ad vector. We used the epitope prediction tools of the Immune Epitope Database and Analysis Resource (IEDB) (23, 27–29) to evaluate the presence of further epitopes in the hexon, penton, and fiber proteins, which are three major constituents of the Ad capsid. We selected 20 H-2Db-or H-2Kb-restricted epitopes with the highest prediction scores (Table 1) and tested the reactivity of Ad-specific CD8+ T cells against these epitopes by enzyme-linked immunosorbent spot (ELISpot) assay. CD8+ T cells from Ad5-immunized CB6F1 mice showed reactivity not only to the two known epitopes DBP418–426 and hexon486–494 but also to the H-2Db- and H-2Kb-restricted peptide pools, indicating that a broad CD8+ T cell response against multiple Ad-derived epitopes is induced in mice by Ad-based vectors (Fig. 5A and B).
TABLE 1.
Ad-derived epitope pools
| Pool | Peptide | Sequence | IEDB scorea |
|---|---|---|---|
| Hexon H-2Db | Hexon567–575 | FAIKNLLLL | 0.84 |
| Hexon418–426 | GGVINTETL | −0.05 | |
| Hexon318–327 | QSMPNRPNYI | −0.91 | |
| Hexon H-2Kb | Hexon790–798 | RMYSFFRNF | 1.58 |
| Hexon676–683 | RGWAFTRL | 0.66 | |
| Hexon673–683 | AAFRGWAFTRL | 0.37 | |
| Hexon78–86 | YSYKARFTL | 0.36 | |
| Hexon938–945 | TVYLRTPF | 0.33 | |
| Hexon840–850 | QAYPANFPYPL | −0.71 | |
| Hexon699–708 | YTYSGSIPYL | −0.96 | |
| Penton H-2Db | Penton455–463 | RQISNFPVV | −0.06 |
| Penton137–148 | FMFTNKFKARVM | −1.27 | |
| Penton H-2Kb | Penton53–60 | NSIRYSEL | 0.42 |
| Penton485–492 | LIRQFTSL | 0.21 | |
| Penton398–408 | STFTQYRSWYL | 0.16 | |
| Fiber H-2Db | Fiber568–578 | FATSSYTFSYI | −1.93 |
| Fiber117–129 | AAPLMVAGNTLTM | −2.32 | |
| Fiber H-2Kb | Fiber571–578 | SSYTFSYI | 0.94 |
| Fiber272–282 | VSYPFDAQNQL | 0.85 | |
| Fiber301–309 | INYNKGLYL | 0.65 |
The IEDB score combines the predictions for proteasomal cleavage, TAP transport, and MHC binding; a higher score indicates a higher predicted efficiency.
FIG 5.
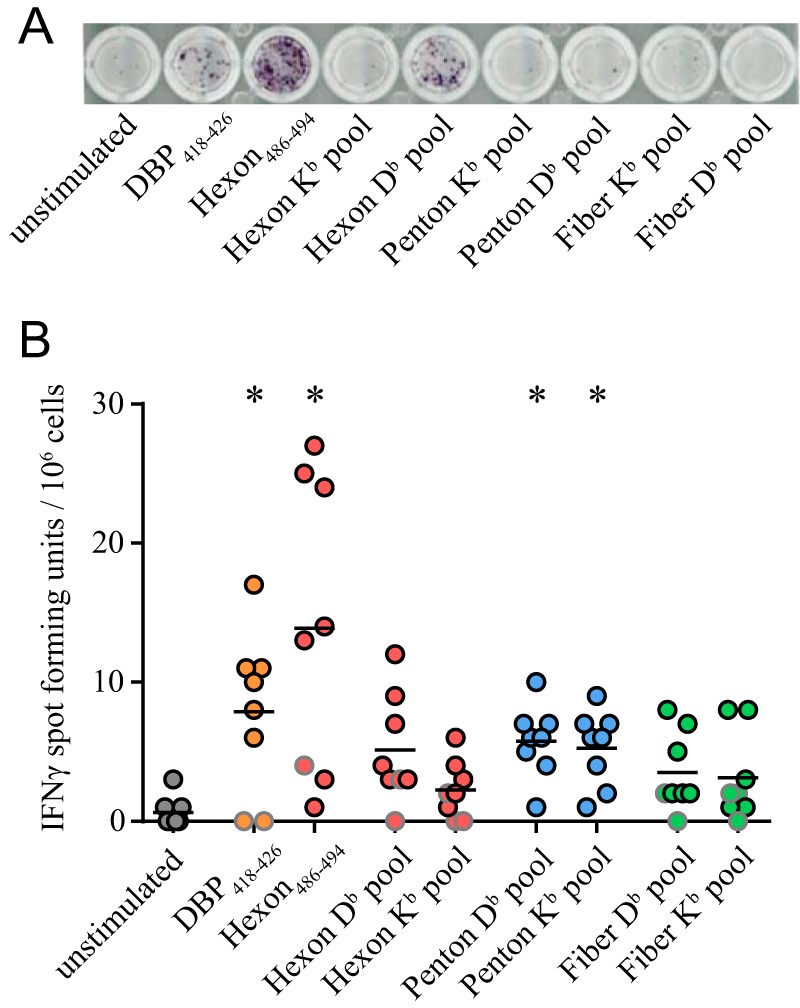
Ad-based immunization induces a broad CD8+ T cell response. CB6F1 mice were immunized twice with 109 viral particles of Ad5.empty, and 2 weeks after the second immunization, spleens were removed and spleen cells were stimulated with the Ad peptides DBP418–426 and hexon486–494 or with hexon-, penton-, and fiber-derived peptides that were pooled according to their predicted MHC I restriction. IFN-γ-producing cells were detected by ELISpot assay. (A) Exemplary sample; (B) data for eight mice. Data are representative of two experiments. Each dot indicates an individual mouse; dots with a gray border indicate values that did not reach the threshold of a 2-fold increase compared to the unstimulated control. Lines indicate mean values. Data were analyzed by Kruskal-Wallis one-way analysis of variance on ranks and Dunn's multiple-comparison procedure for statistical significance. *, statistically significant differences (P < 0.05) compared to the unstimulated control.
Inhibition of GagL85–93-specific CD8+ T cell responses is partly relieved by coimmunization with cytokine-encoding plasmids.
The immunization experiments in C57BL/6 mice clearly demonstrated that the native leader-Gag sequence delivered by an Ad vector is not able to induce a GagL85–93-specific CD8+ T cell response, even though the immunodominance of hexon486–494 is lost in the H-2Db background, and the stimulation with predicted epitope pools confirmed the induction of a broad Ad-specific CD8+ T cell response with further specificities. Therefore, the construction of modified Ad vectors with alanine-substituted epitopes does not seem to be a feasible approach to circumvent Ad epitope immunodominance. Our data indicate that not all Ad epitopes are restricted by H-2Db and therefore do not compete with GagL85–93 directly for MHC binding. While competition on the level of intracellular processes such as transport into the endoplasmic reticulum cannot be excluded for all epitopes, we could demonstrate that the competition by hexon486–494 is lost in the H-2Db background and therefore seems to be mediated by the responding CD8+ T cells rather than by intracellular processes. A possible circumvention strategy for this immunodominance might be the manipulation of the cytokine milieu to provide transgene-specific CD8+ T cells with stronger stimulation. Therefore, we evaluated the potential of cytokine coadministration for relief of Ad immunodominance.
We immunized CB6F1 mice with the epitope string comprising the native leader-Gag-, DBP-, and hexon-derived sequences alone or in combination with expression plasmids encoding interleukin 2 (IL-2), IL-15, IL-21, or IL-23; these cytokines were selected because they play an important role in CD8+ T cell stimulation (30, 31). While no GagL85–93-specific CD8+ T cell response was induced in mice immunized with the epitope string alone or in combination with IL-15- or IL-21-encoding plasmids, mice that were coimmunized with an IL-2-encoding plasmid showed a significantly improved induction of GagL85–93-specific CD8+ T cells, as did some of the mice coimmunized with the IL-23-encoding plasmid (Fig. 6A). We then performed a similar experiment using Ad-based immunization, immunizing CB6F1 mice with Ad5.Leader-Gag alone or in combination with Ad5.IL-2. However, the coadministration of the IL-2-encoding Ad vector did not lead to a significant rescue of GagL85–93-specific CD8+ T cell induction (Fig. 6B).
FIG 6.
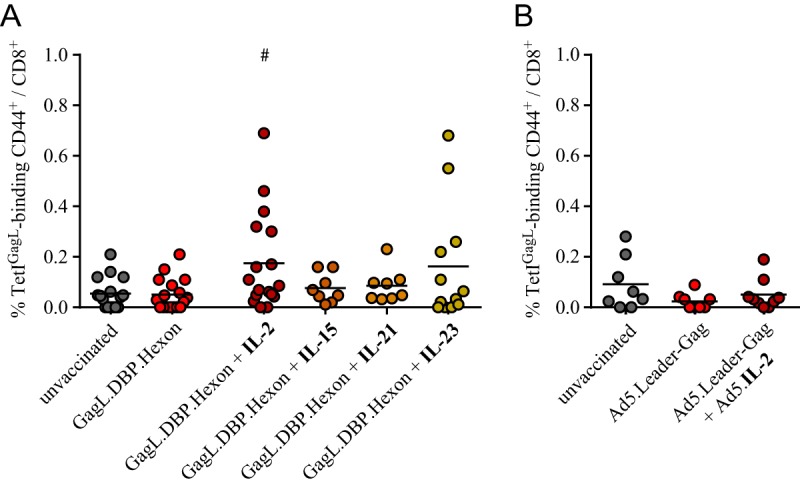
Coadministration of cytokine-encoding plasmids. (A) CB6F1 mice were immunized once with a DNA plasmid encoding the string of native epitope-comprising sequences of F-MuLV leader-Gag, Ad5 DNA-binding protein (DBP), and Ad5 hexon, either alone or in combination with DNA plasmids encoding interleukin 2 (IL-2), IL-15, IL-21, or IL-23. Mice were immunized by intramuscular injection of 30 μg of each DNA plasmid, followed by in vivo electroporation. The GagL85–93-specific CD8+ T cell response was analyzed 2 weeks after immunization by MHC I tetramer staining. (B) For Ad-based immunization, CB6F1 mice were immunized with 109 viral particles of Ad5.Leader-Gag alone or in combination with Ad5.IL-2. The GagL85–93-specific CD8+ T cell response was analyzed 2 weeks after immunization by MHC I tetramer staining. Data were acquired in two to four independent experiments, using four mice per group per experiment. Each dot indicates an individual mouse, and lines indicate mean values. Data were analyzed by Kruskal-Wallis one-way analysis of variance on ranks and Dunn's multiple-comparison procedure for statistical significance. #, statistically significant differences (P < 0.05) compared to mice immunized with the epitope string plasmid alone.
These results confirm that the Ad-derived hexon486–494 exerts immunodominance by a mechanism that can be compensated for by cytokine coadministration; however, the cytokine coadministration was not sufficient to relieve dominance of all Ad-derived epitopes.
DISCUSSION
Preexisting Ad-specific immune responses are widely acknowledged to pose a significant problem for the use of Ad-based vectors for immunization, but the role of Ad-specific immunity in Ad-seronegative individuals, on the other hand, is rarely appreciated. Our results clearly demonstrate that the presence of relatively strong epitopes in Ad proteins can lead to loss of reactivity against transgene-derived epitopes also in prenaive vaccine recipients, and they emphasize the importance of considering the relative strength of an epitope in a vector-modified environment.
The GagL85–93 epitope presents as the immunodominant epitope of FV, and in acutely FV infected mice, a high frequency of effector CD8+ T cells specific for GagL85–93 is crucial for the control of the infection (14); the identification of other epitopes has remained elusive in spite of numerous attempts, and this was also attributed to the dominance of GagL85–93. However, once the leader-Gag protein is placed in an environment conditioned by a viral vector, the GagL85–93 epitope seems to lose its dominance. Our work with Ad-based vectors (20) and with the epitope strings presented here clearly demonstrates an impaired induction of GagL85–93-specific CD8+ T cells in the presence of Ad-derived epitopes, and results from earlier studies using vaccinia virus-based vectors to deliver leader-Gag indicate a similar situation (32); while those authors did not analyze the CD8+ T cell response to the GagL85–93 epitope directly, which was described only some years later (15), their experiments indicated that the matrix protein contained the most protective determinant, not the Gag-leader, suggesting that GagL85–93-specific CD8+ T cells were not induced by the vaccinia virus-based vaccine.
Ad-based vectors are notorious for their capacity to induce strong transgene-specific CD8+ T cell responses; therefore, finding that CD8+ T cell responses to a seemingly immunodominant epitope were lost after delivery by an Ad-based vector was surprising. In experimental immunization studies, CD8+ T cell responses are often tested by stimulation with peptide pools, thereby masking individual responses and possibly losing the sensitivity to detect the loss of individual specificities. In Ad-based immunization against hepatitis B virus, it was shown that CD8+ T cell responses to a subdominant HBsAg epitope were lost after delivery of HBsAg by an Ad-based vector, whereas the responsiveness to an immunodominant HBsAg epitope was maintained (33), indicating that weakly immunogenic epitopes are dominated by Ad-derived epitopes. The loss of CD8+ T cell reactivity to a seemingly dominant epitope that we describe here clearly shows that immunodominance is a relative characteristic, and relative epitope strength has to be considered in vector-based vaccine design. The GagL85–93 epitope, while dominant in FV, appears to be comparatively weak when placed into a vector context, which may actually be a result of FV host adaption.
There are multiple steps preceding epitope recognition by CD8+ T cells that are prone to promoting dominance, such as the rate of protein degradation by the proteasome, the efficiency of epitope trimming, the affinity for the transporter associated with presentation (TAP), and the affinity for MHC I binding; furthermore, when multiple epitopes are presented on the same cell, the responding CD8+ T cells may compete for available cytokines (34), and higher-avidity CD8+ T cells may deplete lower-avidity CD8+ T cells of cytokines, leaving them without the appropriate stimuli and preventing their priming or survival. While not proving that competition for cytokines underlies the Ad-mediated suppression observed in our model, our experiments show that the immunodominance of some Ad-derived epitopes is dependent on a CD8+ T cell response and can be relieved using cytokine-encoding vectors as genetic adjuvants. Cytokine coadministration did not relieve immunodominance in the Ad vector-based immunization, indicating that further mechanisms of immunodominance affect transgene-specific CD8+ T cell responses. The epitope screen yielded new peptides that are able to induce interferon gamma (IFN-γ) production by Ad-induced CD8+ T cells, some of them with the same MHC I restriction as the GagL85–93 epitope and thus direct competitors for MHC I binding. The high number of epitopes in the Ad proteins that we already see in mice and that is an even more important problem for use in humans renders the development of epitope-modified Ad vectors hardly feasible. On the other hand, the approach that we employed in the past, i.e., a modification of the epitope-flanking region (20), to increase the efficiency of proteasomal processing and thereby improve the overall presentation rate is a feasible and widely applicable approach that should be considered in vector-based vaccine development.
The Ad-derived epitopes may originate either from components of the viral particles administered as the vaccine or from newly synthesized proteins that are expressed due to the leaky late gene expression of E1-deleted Ad-based vectors, such as the E1- and E3-deleted Ad vectors used in our studies. Interestingly, it was shown before that immunization with gutless Ad-based vectors, i.e., vectors that do not contain any Ad-derived coding sequences in the vector genome, allowed for the induction of CD8+ T cell responses against subdominant transgene epitopes, indicating that particle-derived protein has little impact on transgene-specific CD8+ T cell induction (35). Gutless Ad vectors are not very popular gene transfer vectors, and most Ad-based vectors in preclinical and clinical immunization studies are first-generation E1- and optionally E3-deleted vectors. However, strategies that reduce late gene expression by first-generation Ad vectors have been described, such as the additional deletion of early Ad genes E2A and E4 (reviewed in reference 36) or the replacement of Ad promoters (37), which may help to reduce Ad-specific immune responses. These strategies should be examined in greater detail for vaccine development, also with regard to Ad immunodominance.
Our data demonstrate the importance of Ad immunocompetition for the use of Ad-based vaccine vectors, as the CD8+ T cell response to seemingly immunodominant epitopes can be gravely affected. It would be interesting and provide important insight to investigate Ad immunocompetition in other immunization models, such as Ad-based immunization of nonhuman primates against simian immunodeficiency virus. Our data strongly suggest that optimization of the immunogens as well as the vectors may contribute to more potent Ad-based vaccines and may have an impact on current efforts in vaccine development against pathogens such as HIV or Ebola virus.
MATERIALS AND METHODS
Cells and cell culture.
The human embryonic kidney cell lines 293 (Microbix Biosystems, Toronto, ON, Canada) and 293T (CRL-11268; American Type Culture Collection, Manassas, VA) were propagated in Dulbecco's modified Eagle medium with high glucose. A murine fibroblast cell line from Mus dunni (38) was maintained in RPMI medium (Invitrogen/Gibco, Karlsruhe, Germany). Cell culture media were supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen/Gibco) and either 1% penicillin-streptomycin (PAA, Pasching, Austria) (for M. dunni) or 50 μg/ml gentamicin (for 293). Cell lines were maintained in a humidified 5% CO2 atmosphere at 37°C.
DNA plasmids.
The plasmid pCG.Leader-Gag encodes the full-length F-MuLV leader-Gag protein and has been described before (39). The plasmid pCG.Leader-GagC1K encodes F-MuLV leader-Gag with a Y94K mutation as described before (20). To generate epitope string constructs, synthetic DNA fragments encoding a string of the F-MuLV-derived GagL85–93 epitope and the Ad5-derived DBP418–426 and hexon486–494 epitopes, where each epitope was flanked by 3 additional amino acids on either side, were cloned into the standard cloning plasmid pcDNA3.1 (Thermo Fisher Scientific, Schwerte, Germany). We used the native sequences of all epitopes (GagL.DBP.Hexon) or the GagL sequence with one or two alanine-modified Ad5 sequences (GagL.DBPAla.HexonAla, GagL.DBP.HexonAla, and GagL.DBPAla.Hexon); an OLLAS tag was fused to the epitope strings for detection in Western blotting (40). The identity of the plasmids was confirmed by sequencing, and expression of the vectors was verified by Western blotting.
Plasmids encoding the murine cytokines IL-2, IL-15, IL-21, and IL-23 have been described before as pShuttle expression plasmids (11, 41).
All plasmids were purified by cesium chloride gradient centrifugation, and absence of endotoxin contamination was verified by Limulus amebocyte lysate assay (detection limit, 0.1 endotoxin units [EU]/ml; Thermo Fisher Scientific, Schwerte, Germany).
Adenoviral vectors.
The following adenoviral vectors have been described before: Ad5.Leader-Gag encodes the leader-Gag protein of F-MuLV FB29 (11), Ad5.IL-2 encodes murine interleukin 2 (11), and Ad5.empty does not encode any transgene sequence (13). All vectors are replication-deficient, E1- and E3-deleted Ad5-based vectors.
All adenoviral vectors were purified with the Vivapure AdenoPACK 100 kit (Vivascience, Hannover, Germany). The adenovirus particle concentrations were determined by spectrophotometry as described previously (42) and expressed as viral particles (VP)/ml; infectivity was verified in a 50% tissue culture infective dose (TCID50) assay.
Mice.
Female CB6F1 hybrid mice (BALB/c × C57BL/6 F1; H-2b/d Fv1b/b Fv2r/s Rfv3r/s) and female BALB/c mice were purchased from Charles River Laboratories (Sulzfeld, Germany). C57BL/6 mice (H-2b/b Fv1b/b Fv2r/r Rfv3r/r) were purchased from Envigo (Rossdorf, Germany). All mice were used when they were between 8 and 9 weeks of age. Mice were treated in accordance with the national law and the institutional guidelines of the University Hospital Essen, Germany. The study was approved by the Northrhine-Westphalia State Office for Nature, Environment and Consumer Protection (LANUV NRW).
Immunization.
For DNA immunization, mice were injected intramuscularly into the gastrocnemius muscle with 30 μg plasmid DNA diluted to 30 μl in phosphate-buffered saline (PBS), which was immediately followed by in vivo electroporation using the BTX AgilePulse system (BTX Molecular Delivery Systems, Holliston, MA) or the Ichor electroporation system (Ichor Medical Systems, San Diego, CA) using electroporation protocols described before (43, 44). Electroporation with both devices gave similar results.
For adenovirus-based immunization, 1 × 109 VP of the adenoviral vector were injected intramuscularly into the gastrocnemius muscle diluted in 30 μl of PBS.
FV and challenge infection.
Uncloned, lactate dehydrogenase-elevating virus (LDV)-free FV stock was obtained from BALB/c mouse spleen cell homogenate (10%, wt/vol) 14 days after infection with a B-tropic, polycythemia-inducing FV complex (45). CB6F1 mice were infected with 5,000 spleen focus-forming units (SFFU) and C57BL/6 mice with 20,000 SFFU FV, by intravenous injection into the tail vein.
The course of infection of CB6F1 mice was determined twice a week by palpation of the spleen of each animal under general anesthesia. Spleen sizes were rated on a scale ranging from 1 (normal spleen size) to 4 (severe splenomegaly) as described previously (46).
Viremia assay.
At 10 days postchallenge (p.c.), plasma samples from CB6F1 mice were obtained, and viremia was determined in a focal infectivity assay (47). Serial dilutions of plasma were incubated with M. dunni cells for 3 days under standard tissue culture conditions. When cells reached ∼100% confluence, they were fixed with ethanol and labeled first with F-MuLV Env-specific monoclonal antibody (MAb) 720 (48) and then with a horseradish peroxidase (HRP)-conjugated rabbit anti-mouse Ig antibody (Dako, Hamburg, Germany), followed by aminoethylcarbazole (Sigma-Aldrich, Deisenhofen, Germany) as the substrate to detect foci. Foci were counted, and focus-forming units (FFU)/ml plasma were calculated.
IC assay.
FV-infected animals were sacrificed by cervical dislocation, the spleens were removed and weighed, and single-cell suspensions were prepared. Serial dilutions of isolated spleen cells were seeded onto M. dunni cells and incubated under standard tissue culture conditions for 3 days. When they reached 100% confluence, they were fixed with ethanol and stained as described for the viremia assay. The resulting foci were counted, and infectious centers (IC)/spleen were calculated.
MHC I tetramer staining of GagL85–93-specific CD8+ T cells.
GagL85–93-specific CD8+ T cells were analyzed in peripheral blood at 2 weeks after immunization. After lysis of erythrocytes, blood cells were stained with phycoerythrin (PE)-coupled MHC I tetramer (containing the H-2Db-restricted F-MuLV Gag-leader epitope AbuAbuLAbuLTVFL, in which cysteine residues of the original amino acid sequence GagL85–93 [CCLCLTVFL] were replaced by amino-butyric acid [Abu] to prevent disulfide bonding [15] [iTAg MHC Tetramer; Biozol Diagnostica, Eching, Germany]), eFluor450-anti-CD8 and BV510-anti-CD44 antibodies (Becton Dickinson, Heidelberg, Germany), and Fixable Viability Dye eFluor 780 (eBioscience, Frankfurt, Germany). Data were acquired on an LSR II flow cytometer (Becton Dickinson, Mountain View, CA) and analyzed using FlowJo software (Tree Star, Ashton, OR).
Detection of Ad-specific CD8+ T cells by ELISpot assay.
For the detection of Ad-specific interferon gamma (IFN-γ)-producing CD8+ T cells, 106 spleen cells from Ad5.empty-immunized mice were subjected to ELISpot assay using the mouse IFN-γ ELISpotPlus kit (Mabtech, Nacka Strand, Sweden); cells were stimulated for 42 h in vitro with DBP418–426 peptide (FALSNAEDL), hexon486–494 peptide (KYSPSNVKI), or pools of predicted H-2Db- or H-2Kb-restricted hexon-, penton-, or fiber-derived peptides (Table 1) (all peptides were synthesized by peptides&elephants, Henningsdorf, Germany), using 10 μg/ml of each peptide. Detection of IFN-γ secretion was carried out according to the manufacturer's instructions.
Statistical analyses.
Statistical analyses were performed using the software SigmaStat 3.1 (Systat Software GmbH, Erkrath, Germany), testing with Kruskal-Wallis one-way analysis of variance on ranks and Dunn's multiple-comparison procedure.
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft to W.B. (DFG grant BA 4432/1-1) and M.T. (RTG 1949/1). C.P.H. and D.L. were supported by the DFG-funded research training group RTG 1949/1.
REFERENCES
- 1.Zhu FC, Wurie AH, Hou LH, Liang Q, Li YH, Russell JB, Wu SP, Li JX, Hu YM, Guo Q, Xu WB, Wurie AR, Wang WJ, Zhang Z, Yin WJ, Ghazzawi M, Zhang X, Duan L, Wang JZ, Chen W. 2017. Safety and immunogenicity of a recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in Sierra Leone: a single-centre, randomised, double-blind, placebo-controlled, phase 2 trial. Lancet 389:621–628. doi: 10.1016/S0140-6736(16)32617-4. [DOI] [PubMed] [Google Scholar]
- 2.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del RC, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartman ZC, Kiang A, Everett RS, Serra D, Yang XY, Clay TM, Amalfitano A. 2007. Adenovirus infection triggers a rapid, MyD88-regulated transcriptome response critical to acute-phase and adaptive immune responses in vivo. J Virol 81:1796–1812. doi: 10.1128/JVI.01936-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCaffrey AP, Fawcett P, Nakai H, McCaffrey RL, Ehrhardt A, Pham TT, Pandey K, Xu H, Feuss S, Storm TA, Kay MA. 2008. The host response to adenovirus, helper-dependent adenovirus, and adeno-associated virus in mouse liver. Mol Ther 16:931–941. doi: 10.1038/mt.2008.37. [DOI] [PubMed] [Google Scholar]
- 5.Fougeroux C, Holst PJ. 2017. Future prospects for the development of cost-effective adenovirus vaccines. Int J Mol Sci doi: 10.3390/ijms18030686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friend C. 1957. Cell-free transmission in adult Swiss mice of a disease having the character of a leukemia. J Exp Med 105:307–318. doi: 10.1084/jem.105.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manzke N, Akhmetzyanova I, Hasenkrug KJ, Trilling M, Zelinskyy G, Dittmer U. 2013. CD4+ T cells develop antiretroviral cytotoxic activity in the absence of regulatory T cells and CD8+ T cells. J Virol 87:6306–6313. doi: 10.1128/JVI.00432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair SR, Zelinskyy G, Schimmer S, Gerlach N, Kassiotis G, Dittmer U. 2010. Mechanisms of control of acute Friend virus infection by CD4+ T helper cells and their functional impairment by regulatory T cells. J Gen Virol 91:440–451. doi: 10.1099/vir.0.015834-0. [DOI] [PubMed] [Google Scholar]
- 9.Zelinskyy G, Balkow S, Schimmer S, Schepers K, Simon MM, Dittmer U. 2004. Independent roles of perforin, granzymes, and Fas in the control of Friend retrovirus infection. Virology 330:365–374. doi: 10.1016/j.virol.2004.08.040. [DOI] [PubMed] [Google Scholar]
- 10.Zelinskyy G, Dietze K, Sparwasser T, Dittmer U. 2009. Regulatory T cells suppress antiviral immune responses and increase viral loads during acute infection with a lymphotropic retrovirus. PLoS Pathog 5:e1000406. doi: 10.1371/journal.ppat.1000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bayer W, Schimmer S, Hoffmann D, Dittmer U, Wildner O. 2008. Evaluation of the Friend virus model for the development of improved adenovirus-vectored anti-retroviral vaccination strategies. Vaccine 26:716–726. doi: 10.1016/j.vaccine.2007.11.050. [DOI] [PubMed] [Google Scholar]
- 12.Dittmer U, Brooks DM, Hasenkrug KJ. 1999. Requirement for multiple lymphocyte subsets in protection by a live attenuated vaccine against retroviral infection. Nat Med 5:189–193. doi: 10.1038/5550. [DOI] [PubMed] [Google Scholar]
- 13.Kaulfuss M, Wensing I, Windmann S, Hrycak CP, Bayer W. 2017. Induction of complex immune responses and strong protection against retrovirus challenge by adenovirus-based immunization depends on the order of vaccine delivery. Retrovirology 14:8. doi: 10.1186/s12977-017-0336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zelinskyy G, Kraft AR, Schimmer S, Arndt T, Dittmer U. 2006. Kinetics of CD8+ effector T cell responses and induced CD4+ regulatory T cell responses during Friend retrovirus infection. Eur J Immunol 36:2658–2670. doi: 10.1002/eji.200636059. [DOI] [PubMed] [Google Scholar]
- 15.Chen W, Qin H, Chesebro B, Cheever MA. 1996. Identification of a gag-encoded cytotoxic T-lymphocyte epitope from FBL-3 leukemia shared by Friend, Moloney, and Rauscher murine leukemia virus-induced tumors. J Virol 70:7773–7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Messer RJ, Lavender KJ, Hasenkrug KJ. 2014. Mice of the resistant H-2(b) haplotype mount broad CD4(+) T cell responses against 9 distinct Friend virus epitopes. Virology 456-457:139–144. doi: 10.1016/j.virol.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dittmer U, Brooks DM, Hasenkrug KJ. 1999. Protection against establishment of retroviral persistence by vaccination with a live attenuated virus. J Virol 73:3753–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dittmer U, Race B, Hasenkrug KJ. 1999. Kinetics of the development of protective immunity in mice vaccinated with a live attenuated retrovirus. J Virol 73:8435–8440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knuschke T, Bayer W, Rotan O, Sokolova V, Wadwa M, Kirschning CJ, Hansen W, Dittmer U, Epple M, Buer J, Westendorf AM. 2014. Prophylactic and therapeutic vaccination with a nanoparticle-based peptide vaccine induces efficient protective immunity during acute and chronic retroviral infection. Nanomedicine (Lond) 10:1787–1798. doi: 10.1016/j.nano.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Godel P, Windmann S, Dietze KK, Dittmer U, Bayer W. 2012. Modification of one epitope-flanking amino acid allows for the induction of Friend retrovirus-specific CD8+ T cells by adenovirus-based immunization. J Virol 86:12422–12425. doi: 10.1128/JVI.01607-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKelvey T, Tang A, Bett AJ, Casimiro DR, Chastain M. 2004. T-cell response to adenovirus hexon and DNA-binding protein in mice. Gene Ther 11:791–796. doi: 10.1038/sj.gt.3302232. [DOI] [PubMed] [Google Scholar]
- 22.Buus S, Lauemoller SL, Worning P, Kesmir C, Frimurer T, Corbet S, Fomsgaard A, Hilden J, Holm A, Brunak S. 2003. Sensitive quantitative predictions of peptide-MHC binding by a ‘Query by Committee’ artificial neural network approach. Tissue Antigens 62:378–384. doi: 10.1034/j.1399-0039.2003.00112.x. [DOI] [PubMed] [Google Scholar]
- 23.Lundegaard C, Lund O, Nielsen M. 2008. Accurate approximation method for prediction of class I MHC affinities for peptides of length 8, 10 and 11 using prediction tools trained on 9mers. Bioinformatics 24:1397–1398. doi: 10.1093/bioinformatics/btn128. [DOI] [PubMed] [Google Scholar]
- 24.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O. 2003. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci 12:1007–1017. doi: 10.1110/ps.0239403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hudrisier D, Riond J, Burlet-Schiltz O, von Herrath MG, Lewicki H, Monsarrat B, Oldstone MB, Gairin JE. 2001. Structural and functional identification of major histocompatibility complex class I-restricted self-peptides as naturally occurring molecular mimics of viral antigens. Possible role in CD8+ T cell-mediated, virus-induced autoimmune disease. J Biol Chem 276:19396–19403. [DOI] [PubMed] [Google Scholar]
- 26.Bongard N, Lapuente D, Windmann S, Dittmer U, Tenbusch M, Bayer W. 2017. Interference of retroviral envelope with vaccine-induced CD8+ T cell responses is relieved by co-administration of cytokine-encoding vectors. Retrovirology 14:28. doi: 10.1186/s12977-017-0352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carmichael A, Bateman N, Nayagam M. 1991. Examination of induced sputum in the diagnosis of Pneumocystis carinii pneumonia. Cytopathology 2:61–66. doi: 10.1111/j.1365-2303.1991.tb00388.x. [DOI] [PubMed] [Google Scholar]
- 28.Lundegaard C, Lamberth K, Harndahl M, Buus S, Lund O, Nielsen M. 2008. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8-11. Nucleic Acids Res 36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tenzer S, Peters B, Bulik S, Schoor O, Lemmel C, Schatz MM, Kloetzel PM, Rammensee HG, Schild H, Holzhutter HG. 2005. Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci 62:1025–1037. doi: 10.1007/s00018-005-4528-2. [DOI] [PubMed] [Google Scholar]
- 30.Kaech SM, Wherry EJ, Ahmed R. 2002. Effector and memory T-cell differentiation: implications for vaccine development. Nat Rev Immunol 2:251–262. doi: 10.1038/nri778. [DOI] [PubMed] [Google Scholar]
- 31.Schluns KS, Lefrancois L. 2003. Cytokine control of memory T-cell development and survival. Nat Rev Immunol 3:269–279. doi: 10.1038/nri1052. [DOI] [PubMed] [Google Scholar]
- 32.Miyazawa M, Nishio J, Chesebro B. 1992. Protection against Friend retrovirus-induced leukemia by recombinant vaccinia viruses expressing the gag gene. J Virol 66:4497–4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schirmbeck R, Reimann J, Kochanek S, Kreppel F. 2008. The immunogenicity of adenovirus vectors limits the multispecificity of CD8 T-cell responses to vector-encoded transgenic antigens. Mol Ther 16:1609–1616. doi: 10.1038/mt.2008.141. [DOI] [PubMed] [Google Scholar]
- 34.Chen W, Pang K, Masterman KA, Kennedy G, Basta S, Dimopoulos N, Hornung F, Smyth M, Bennink JR, Yewdell JW. 2004. Reversal in the immunodominance hierarchy in secondary CD8+ T cell responses to influenza A virus: roles for cross-presentation and lysis-independent immunodomination. J Immunol 173:5021–5027. doi: 10.4049/jimmunol.173.8.5021. [DOI] [PubMed] [Google Scholar]
- 35.Kron MW, Engler T, Schmidt E, Schirmbeck R, Kochanek S, Kreppel F. 2011. High-capacity adenoviral vectors circumvent the limitations of dE1 and dE1/dE3 adenovirus vectors to induce multispecific transgene product-directed CD8 T-cell responses. J Gene Med 13:648–657. doi: 10.1002/jgm.1629. [DOI] [PubMed] [Google Scholar]
- 36.Schagen FH, Ossevoort M, Toes RE, Hoeben RC. 2004. Immune responses against adenoviral vectors and their transgene products: a review of strategies for evasion. Crit Rev Oncol Hematol 50:51–70. doi: 10.1016/S1040-8428(03)00172-0. [DOI] [PubMed] [Google Scholar]
- 37.Fang B, Koch P, Roth JA. 1997. Diminishing adenovirus gene expression and viral replication by promoter replacement. J Virol 71:4798–4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lander MR, Chattopadhyay SK. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ectropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J Virol 52:695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dittmer U, Brooks DM, Hasenkrug KJ. 1998. Characterization of a live-attenuated retroviral vaccine demonstrates protection via immune mechanisms. J Virol 72:6554–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park SH, Cheong C, Idoyaga J, Kim JY, Choi JH, Do Y, Lee H, Jo JH, Oh YS, Im W, Steinman RM, Park CG. 2008. Generation and application of new rat monoclonal antibodies against synthetic FLAG and OLLAS tags for improved immunodetection. J Immunol Methods 331:27–38. doi: 10.1016/j.jim.2007.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohs I, Windmann S, Wildner O, Dittmer U, Bayer W. 2013. Interleukin-encoding adenoviral vectors as genetic adjuvant for vaccination against retroviral infection. PLoSOne 8:e82528. doi: 10.1371/journal.pone.0082528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mittereder N, March KL, Trapnell BC. 1996. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol 70:7498–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roos AK, Eriksson F, Walters DC, Pisa P, King AD. 2009. Optimization of skin electroporation in mice to increase tolerability of DNA vaccine delivery to patients. Mol Ther 17:1637–1642. doi: 10.1038/mt.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Storcksdieck genannt Bonsmann M, Niezold T, Temchura V, Pissani F, Ehrhardt K, Brown EP, Osei-Owusu NY, Hannaman D, Hengel H, Ackerman ME, Streeck H, Nabi G, Tenbusch M, Uberla K. 2015. Enhancing the quality of antibodies to HIV-1 envelope by GagPol-specific Th cells. J Immunol 195:4861–4872. doi: 10.4049/jimmunol.1501377. [DOI] [PubMed] [Google Scholar]
- 45.Chesebro B, Wehrly K, Stimpfling J. 1974. Host genetic control of recovery from Friend leukemia virus-induced splenomegaly: mapping of a gene within the major histocompatability complex. J Exp Med 140:1457–1467. doi: 10.1084/jem.140.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hasenkrug KJ, Brooks DM, Robertson MN, Srinivas RV, Chesebro B. 1998. Immunoprotective determinants in friend murine leukemia virus envelope protein. Virology 248:66–73. doi: 10.1006/viro.1998.9264. [DOI] [PubMed] [Google Scholar]
- 47.Sitbon M, Nishio J, Wehrly K, Lodmell D, Chesebro B. 1985. Use of a focal immunofluorescence assay on live cells for quantitation of retroviruses: distinction of host range classes in virus mixtures and biological cloning of dual-tropic murine leukemia viruses. Virology 141:110–118. doi: 10.1016/0042-6822(85)90187-4. [DOI] [PubMed] [Google Scholar]
- 48.Robertson MN, Miyazawa M, Mori S, Caughey B, Evans LH, Hayes SF, Chesebro B. 1991. Production of monoclonal antibodies reactive with a denatured form of the Friend murine leukemia virus gp70 envelope protein: use in a focal infectivity assay, immunohistochemical studies, electron microscopy and western blotting. J Virol Methods 34:255–271. doi: 10.1016/0166-0934(91)90105-9. [DOI] [PubMed] [Google Scholar]