ABSTRACT
The neurotropic herpesvirus varicella-zoster virus (VZV) establishes a lifelong latent infection in humans following primary infection. The low abundance of VZV nucleic acids in human neurons has hindered an understanding of the mechanisms that regulate viral gene transcription during latency. To overcome this critical barrier, we optimized a targeted capture protocol to enrich VZV DNA and cDNA prior to whole-genome/transcriptome sequence analysis. Since the VZV genome is remarkably stable, it was surprising to detect that VZV32, a VZV laboratory strain with no discernible growth defect in tissue culture, contained a 2,158-bp deletion in open reading frame (ORF) 12. Consequently, ORF 12 and 13 protein expression was abolished and Akt phosphorylation was inhibited. The discovery of the ORF 12 deletion, revealed through targeted genome sequencing analysis, points to the need to authenticate the VZV genome when the virus is propagated in tissue culture.
IMPORTANCE Viruses isolated from clinical samples often undergo genetic modifications when cultured in the laboratory. Historically, VZV is among the most genetically stable herpesviruses, a notion supported by more than 60 complete genome sequences from multiple isolates and following multiple in vitro passages. However, application of enrichment protocols to targeted genome sequencing revealed the unexpected deletion of a significant portion of VZV ORF 12 following propagation in cultured human fibroblast cells. While the enrichment protocol did not introduce bias in either the virus genome or transcriptome, the findings indicate the need for authentication of VZV by sequencing when the virus is propagated in tissue culture.
KEYWORDS: ORF 12, VZV, deletion, genome, transcriptome
INTRODUCTION
Varicella-zoster virus (VZV) is a ubiquitous neurotropic human alphaherpesvirus. Serological studies show that over 95% of the world's population is exposed to the virus (1). VZV is typically acquired early in life, thereafter establishing latency in multiple cranial and dorsal root ganglia (2) as well as thoracic sympathetic (3) and enteric (4) ganglia. With declining cell-mediated immunity to VZV due to aging, disease, or immunosuppressive therapy, the virus can reactivate to produce zoster (5), which is often complicated by persistent, dermatomal distribution pain (postherpetic neuralgia), the leading cause of pain-related suicide in elderly patients (6). Zoster increases the risk of stroke 2.4-fold and of myocardial infarction 1.7-fold within 2 weeks of onset (7), and the risk remains elevated for the subsequent 3 months (8). Furthermore, VZV reactivation is associated with giant cell arteritis, the most common systemic vasculitis in the elderly (9). The successful treatment of an immunocompetent patient with 6 years of chronic progressive neurological disease attributed to VZV infection of the brain (10) highlights the protean nature of VZV infections (11).
A critical barrier to the study of VZV latency is the low virus burden in naturally infected human ganglia removed at autopsy (12) and in experimentally infected human neurons maintained in tissue culture (13–22). While protocols to enrich VZV DNA (23) and cDNA (24) relative to host nucleic acids have been developed, the sensitivity and specificity provided by PCR reaffirms the benefit of this technology for investigations into latent VZV gene transcription and its regulation (25–28). However, extending PCR technologies to virus whole-genome analysis requires multiple optimizations and recalibrations to maintain primer efficiency in multiplex reactions involving >20 individual amplicons (29).
The VZV genome is remarkably stable. The complete genome sequence of 66 VZV isolates is publically available (http://www.ncbi.nlm.nih.gov/nuccore/?term=varicella+zoster+virus+complete+genome), and the 124,947- ± 210-bp DNA can be partitioned into 5 defined clades (30), with the possible emergence of a 6th clade (31). While new VZV isolates can be genotyped by single-nucleotide polymorphism (SNP) analysis within VZV open reading frames (ORFs) 21 and 50 (32), VZV clades are genetic variations that are informative for epidemiology (33–36) and evolutionary studies (37–40) but do not describe different virus phenotypes in the human population. Naturally circulating VZV can undergo limited recombination (31, 41), but the only phenotypic mutation identified to date in clinical isolates is the D150N SNP in glycoprotein E, which results in increased virus spread in cell culture (42).
VZV DNA is stable, with no detectable mutation during 16 passages in vitro after clinical isolation (33). A total of 72 in vitro passages of the VZV32 strain introduced 31 SNPs, resulting in one codon change in ORF 33 (viral protease) and 6 codon changes in the major immediate-early transcriptional transactivating protein (IE62) encoded by VZV ORF 62, but these mutations did not hinder virus growth in culture (43). The 834-bp VZV ORF 63 encoding immediate-early protein 63 (IE63) is stable, and only 3 SNPs arose in ORF 63 between in vitro passages 1,060 and 1,206, none of which affected IE63 size, immunoreactivity on Western blots, or virus growth characteristics in culture (44). Based on the documented stability of VZV DNA in the human population and in tissue culture, we developed methods to enrich VZV sequences from a background of host nucleic acid and found an unexpected 2,158-bp deletion in ORF 12 of a VZV laboratory strain with no discernible growth defect in tissue culture. This novel feature was discovered only through targeted genome sequencing analysis, underscoring the need to authenticate the VZV genome when the virus is propagated in tissue culture.
RESULTS
Library construction and efficiency of virus nucleic acid enrichment.
Illumina sequencing libraries were constructed from total DNA or RNA extracted from biological replicates of VZV-DEN09-infected human fetal lung fibroblast (HFL) cells and sequenced pre- and postenrichment for virus nucleic acids. Infected cells were harvested 2 days after low-multiplicity infection to ensure the presence of the full virus transcriptome, as the virus propagated asynchronously (45). FastQC analysis to assess DNA sequence quality for each library (data not shown) identified each nucleotide with a high degree of confidence (average per-base sequence quality score of >36) and no significant bias in GC content (absence of peaks in average per-base sequence content). Consequently, no end trimming of DNA sequences was required, and the entire 51-nucleotide sequence from 2.17 × 108 individual sequencing events was analyzed.
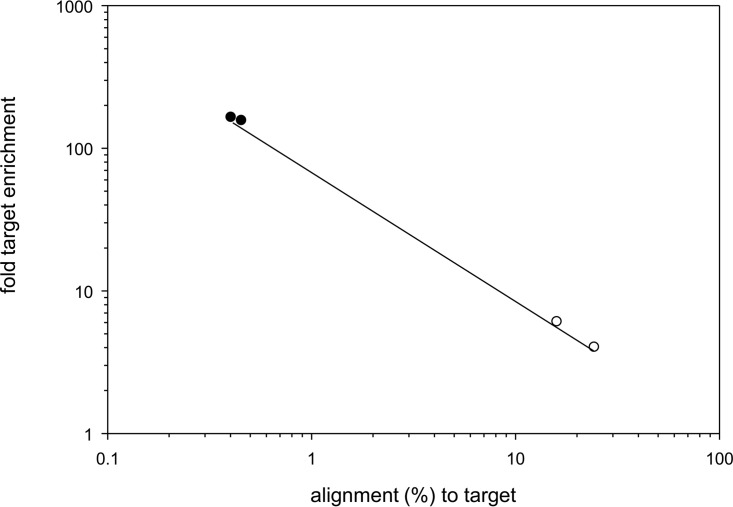
To determine virus nucleic acid enrichment efficiency, sequenced reads were aligned to the reference VZV32 passage 72 (DQ479963.1) and human genome assembly 19 (https://genome.ucsc.edu/cgi-bin/hgGateway) (Table 1). On average, 97.6% of total reads mapped to either virus or human sequences. In unenriched libraries, 20.2% ± 5.9% of DNA reads and 0.44% ± 0.02% of cDNA reads mapped to the VZV sequence. After targeted enrichment, the proportion of reads mapping to VZV increased to 97.7% ± 0.9% for DNA and 68.8% ± 3.3% for cDNA. An inverse linear relationship existed between the initial abundance of virus nucleic acid present and the amount of target DNA enrichment (Fig. 1), i.e., enrichment was greater with less virus nucleic acid. For example, 0.4% of total cDNA sequences mapped to the VZV genome in unenriched samples from VZV-infected HFL cells, while the amount of VZV cDNA sequences increased 162- ± 5-fold after enrichment. On the other hand, the overall enrichment of virus DNA was low when initial target DNA was abundant. For example, 24.1% of total DNA mapped to the VZV genome in unenriched samples, while the amount of VZV DNA sequences increased only 4% after enrichment. Nonetheless, the vast majority of DNA sequences (>83%) obtained after targeted enrichment of VZV DNA and cDNA mapped to the virus.
TABLE 1.
Alignment of DNA sequences to VZV and human genomes
| Parameter | Result for sample: |
|||||||
|---|---|---|---|---|---|---|---|---|
| A | A | B | B | C | C | D | D | |
| Nucleic acid used for library prepn | DNA | DNA | DNA | DNA | RNA | RNA | RNA | RNA |
| Targeted enrichment | No | Yes | No | Yes | No | Yes | No | Yes |
| Reads | ||||||||
| Total no. | 1,494,870 | 57,782,614 | 99,799,072 | 9,818,920 | 27,321,531 | 1,878,742 | 18,062,627 | 1,293,139 |
| No. mapping to hg19 | 1,082,286 | 196,461 | 80,408,112 | 159,729 | 26,567,457 | 492,794 | 17,116,145 | 390,011 |
| No. mapping to VZV strain 32 passage 72 | 361,609 | 56,834,979 | 15,808,173 | 9,531,226 | 122,947 | 1,338,040 | 72,251 | 860,067 |
| No. of unaligned reads | 50,975 | 751,174 | 3,582,787 | 60,877 | 631,127 | 47,908 | 874,231 | 43,062 |
| Alignment (%) to hg19 | 72.40 | 0.34 | 80.57 | 0.62 | 97.24 | 26.23 | 94.76 | 30.16 |
| Alignment (%) to VZV strain 32 passage 72 | 24.19 | 98.36 | 15.84 | 97.07 | 0.45 | 71.22 | 0.40 | 66.51 |
| Unaligned (%) reads | 3.41 | 1.30 | 3.59 | 2.31 | 2.31 | 2.55 | 4.84 | 3.33 |
| Fold enrichment (%) of hg19 | 0.00 | 0.01 | 0.27 | 0.32 | ||||
| Fold enrichment (%) of VZV strain 32 passage 72 | 4.07 | 6.13 | 158.27 | 166.28 | ||||
FIG 1.
VZV nucleic acid enrichment dependency on initial target abundance. The fold target enrichment (y axis) in VZV DNA (open circles) or cDNA sequences (closed circles) was plotted as a function of percent input VZV nucleic acid in each sample. The efficiency of virus DNA or cDNA enrichment spanned almost 2 orders of magnitude.
Comparison of VZV-DEN09 DNA to reference VZV32 passage 72 DNA.
VZV-DEN09 DNA sequences obtained with and without enrichment were independently mapped to VZV32 passage 72, and the number of times that each nucleotide was sequenced was recorded (counts per nucleotide, or cpn) (see Table S1 in the supplemental material). Combining the four VZV genomic sequencing events, a total of 4,175,583,047 cpn were obtained from 82,535,987 individual DNA reads, confirming the 51-nucleotide read length (4,175,583,047 cpn/82,535,987 reads = 50.6 nucleotides/read). The four VZV-DEN09 DNA sequences were combined into a single database that averaged 33,436 cpn. Differences between the reference sequence and VZV-DEN09 were identified only if the difference occurred ≥10,000 times at an individual site and only if ≥98% of nucleotide reads at the site were identical. A total of 41 SNPs were detected between the reference VZV DNA and VZV-DEN09, reflecting 0.04% of the 125,169 nucleotides in VZV32 passage 72 (Table S2). Fourteen mutations occurred in ORF 62 and were also present in ORF 71 by virtue of the gene's location in the repeat region of the unique short DNA segment. There were 27 unique SNPs in VZV-DEN09, 23 of which were within unique VZV ORFs and 4 SNPs in unannotated regions of the virus genome. The 23 SNPs included 14 synonymous mutations that would not affect protein sequence and 9 nonsynonymous mutations resulting in predicted amino acid changes to proteins encoded by VZV ORFs 2, 17, 51, 35, 51, and 62.
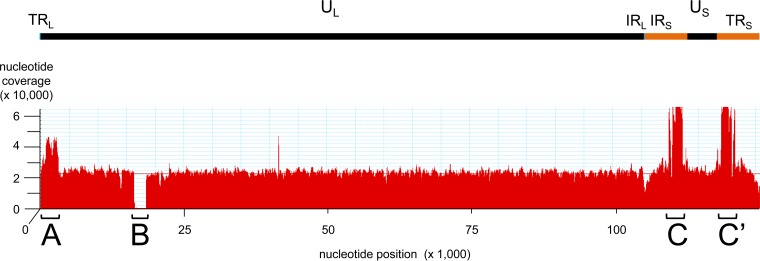
Alignment of the pooled VZV-DEN09 DNA data set to the reference genome showed remarkably consistent read depths, with the exception of three areas (Fig. 2): area A showed an ∼2-fold increase in read depth between nucleotides 800 and 3100 compared to the genome average, area B indicated an ∼1.2-kbp DNA deletion in the VZV-DEN09 genome, and area C indicated a 3- to 4-fold increase in read depth compared to the genome average between nucleotides 110000 and 111700. Since area C was located in the internal repeat of the unique short segment (IRS), the identical sequence (area C′) is located in the inverted terminal repeat of the unique short DNA segment (TRS) between nucleotides 120100 and 118600. Importantly, neither area C nor C′ extended throughout the internal or terminal invert repeat of the unique short segment of the virus genome, suggesting these areas were not the result of the mapping algorithm.
FIG 2.
Alignment of DNA sequence from VZV-DEN09 to VZV32 passage 72. The VZV genome contains a long unique (UL) and short unique (US) segment joined through the internal repeat of the long (IRL) and short (IRS) segments, which are inverted at the genome end as terminal repeats of the long (TRL) and short (TRS) segments, respectively. The 51-nucleotide VZV-DEN09 reads, mapped to the archived reference strain (VZV32 passage 72) genome, showed overrepresented DNA segments at A, C, and C′ along with a deletion at region B. The average read depth per nucleotide was 24,000 (red line).
Accumulation of short VZV DNA repeats in areas A and C/C′.
The overrepresented VZV DNA in areas A and C/C′ was confirmed by quantitative PCR (qPCR). Six sets of qPCR primers (Table 2) spanning most of the virus genome at ∼20-kbp intervals were selected to establish a baseline for comparison of qPCR results obtained with primers located within A and C/C′. Primer sets were specific for VZV DNA and produced single 40- to 46-bp amplicons with 93.03% ± 3.53% efficiency (Table 3, columns 2 to 4). Use of each of the eight PCR primer sets to quantify 2-fold dilutions of VZV bacterial artificial chromosome (BAC) DNA revealed a 2.09- ± 0.91-fold dilution of target DNA (Table 3, columns 5 to 7), indicating that qPCR could detect the predicted 2- to 4-fold increase of VZV DNA in areas A and C/C′ over baseline VZV DNA. Each of the 8 primer sets amplified 6,000 copies of BAC-derived VZV DNA extracted from Escherichia coli to the same extent (threshold cycle [CT] of 25.62 ± 1.31, n = 60) (Table 3, columns 8 to 11). VZV-DEN09 DNA extracted from infected cells was diluted to ∼6,000 copies per PCR and amplified with the 8 sets of PCR primers. Results of 4 to 6 separate assays showed that areas A and C/C′ contained 2.22- ± 0.79- and 4.32- ± 0.70-fold more DNA, respectively, than that detected using the six baseline primer sets (Table 3, columns 12 to 15), confirming the increased presence of VZV DNA in these areas indicated by whole-genome sequencing.
TABLE 2.
Oligonucleotide primers
| Name | Sequence (5′–3′) | Location of 5′ enda | Application |
|---|---|---|---|
| 5k-FOR | GGATTATTGCCTGCACCTCTAA | 8746 | PCR |
| 5k-REV | CAGAAGCGACTACGTCTTCTAAC | 8791 | PCR |
| 20k-FOR | AATGCTCAGGGCACGTAAA | 24248 | PCR |
| 20k-REV | GCCGGAGTGTCCAGATATAAAC | 24290 | PCR |
| 40k-FOR | CCGTTCGGCGTATCGTATTT | 40775 | PCR |
| 40k-REV | TGCACGCGCTAACCATAAT | 40815 | PCR |
| 60k-FOR | CCCGAAGAGAGTTTGGATTACC | 64941 | PCR |
| 60k-REV | CGTGTGGGCCATGGATAAA | 64988 | PCR |
| 80k-FOR | GTTGTGAAGCGTTCCAATTCC | 80249 | PCR |
| 80k-REV | ACGTCATCAGGCCGTTATTC | 80291 | PCR |
| 100k-FOR | CCTTGGTTGCATCTCCAAGTA | 100574 | PCR |
| 100k-REV | CCTCTACGGGAGCAAACTAAAG | 100619 | PCR |
| Area A For | CCCTTACTGCTGTCGTGGTCGCC | 495 | PCR |
| Area A Rev | GGACTCACGTATTGCCCGCGCG | 3344 | PCR |
| Area A-For first seq | GTTGCCGTTTTTCCCGAGGA | 501 | Sequence |
| Area A-Rev first seq | CGTGAGAGAGTCATACGTG | 3319 | Sequence |
| Area A-For second seq | CTCTGTTTATGGCGGGGTAA | 1213 | Sequence |
| Area A-Rev second seq | TGTTAGGGTTGGGAGAGTGG | 2383 | Sequence |
| Area B For | AGCGCGGAGTTCTTTGG | 18476 | PCR |
| Area B seq For | CGCAATGTTTTCTCGGTTTGCG | 16012 | Sequence |
| Area B seq Rev | CTGAAGTTCGCCGGTTAACG | 18707 | Sequence |
| Area B Rev | GCGTAGCCATGCATTTACTAC | 16198 | PCR |
| Area C/C′-FOR | GGAAGTGCGAACGGGTTTA | 117892 | PCR |
| Area C/C′-REV | CGAAGAACTTCATATCTGAGGCA | 117935 | PCR |
Location based on archive VZV strain 32 passage 72 DNA sequence (accession no. DQ479963.1).
TABLE 3.
Analysis of overrepresented DNA regions in VZV
| Primer set ID | PCR standardization |
Analysis of 2-fold diluted VZV DNAa |
CT value for 6,000 copies of VZV DNAa |
ΔΔ analysis of area: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ab |
C/C′c |
|||||||||||||
| Efficiency | R2 | n | Δd | SD | n | Trial 1 | Trial 2 | Trial 3 | Trial 4 | Avg | n | Avg | n | |
| 5k | 93.13 | 1.00 | 3 | 1.02 | 0.19 | 2 | 25.96 | 26.70 | 26.93 | −0.79 | 4 | −1.51 | 4 | |
| 25.87 | 26.53 | 26.92 | ||||||||||||
| 20k | 90.54 | 1.00 | 3 | 1.19 | 0.06 | 2 | 26.88 | 26.38 | 26.16 | −1.04 | 4 | −1.76 | 4 | |
| 26.80 | 26.58 | 26.36 | ||||||||||||
| 40k | 91.29 | 1.00 | 3 | 1.02 | 0.08 | 3 | 26.30 | 25.20 | 26.59 | 26.88 | −0.77 | 6 | −2.20 | 6 |
| 26.28 | 25.75 | 26.82 | 26.60 | |||||||||||
| 26.89 | ||||||||||||||
| 60k | 94.61 | 0.98 | 3 | 1.00 | 0.06 | 3 | 26.38 | 26.46 | 25.54 | 25.98 | −1.58 | 6 | −3.01 | 6 |
| 26.10 | 26.50 | 26.27 | 24.46 | |||||||||||
| 22.67 | ||||||||||||||
| 80k | 98.39 | 1.00 | 3 | 0.99 | 0.14 | 2 | 25.41 | 25.24 | 26.25 | −1.44 | 4 | −2.16 | 4 | |
| 25.20 | 24.92 | 26.02 | ||||||||||||
| 100k | 93.61 | 0.99 | 3 | 0.97 | 0.06 | 2 | 25.00 | 26.41 | 26.00 | −1.29 | 4 | −2.01 | 4 | |
| 24.34 | 26.39 | 25.76 | ||||||||||||
| Area A | 86.82 | 1.00 | 3 | 0.97 | 0.31 | 3 | 23.21 | 26.32 | 26.64 | 25.13 | ||||
| 22.90 | 26.11 | 26.98 | 24.52 | 0 | 6 | |||||||||
| 26.08 | ||||||||||||||
| Area C | 95.84 | 1.00 | 3 | 1.34 | 0.65 | 3 | 26.92 | 24.47 | 21.66 | 23.82 | 0 | 6 | ||
| 26.51 | 24.66 | 22.55 | 23.92 | |||||||||||
| 22.43 | ||||||||||||||
| Avg | 93.03 | 0.99 | 3 | 1.06 | 25.62 | −1.15 | −2.11 | |||||||
| SD | 3.53 | 0.01 | 0 | 0.13 | 1.31 | 0.34 | 0.51 | |||||||
| n | 8 | 8 | 8 | 8 | 60 | 6 | 6 | |||||||
| Fold differencee (avg) | 2.09 | 2.22 | 4.32 | |||||||||||
| Fold difference (SD) | 0.91 | 0.79 | 0.70 | |||||||||||
| N | 8 | 6 | 6 | |||||||||||
VZV DNA extracted from VZV BAC grown in E. coli.
[CT(area A) − CT(6,000 VZV DNA copies)] − [CT(background primer set) − CT(6,000 VZV DNA copies)].
[CT(area C/C′) − CT(6,000 VZV DNA copies)] − [CT(background primer set) −CT(6,000 VZV DNA copies)].
Difference in CT values from 2-fold dilutions of VZV DNA.
Two raised to the power of the absolute value Δ or ΔΔ.
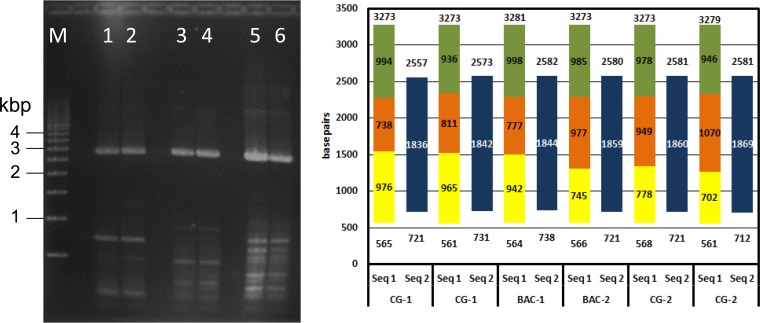
To determine whether the overrepresented virus DNA in region A (Fig. 2) resulted from unanticipated DNA repeats present in VZV-DEN09 but not in VZV32, two independent samples of VZV-DEN09 DNA were PCR amplified in duplicate and compared to fragments from PCR of VZV BAC DNA extracted from E. coli (Fig. 3, left). A predicted 2.7-kbp fragment was detected in all samples. To determine if new repeat regions were contained within the 2.7-kbp segment, the entire segment was Sanger sequenced using sets of internal primers (Fig. 3, right). The DNA sequence for each PCR product showed that the 2,711 ± 6 nucleotide fragment contained no repeated segment.
FIG 3.
VZV DNA region A sequence determination. (Left) Two independent samples of VZV-DEN09 DNA (sample 1, lanes 1 and 2; sample 2, lanes 5 and 6) along with duplicate samples of VZV BAC (lanes 3 and 4) were PCR amplified with primers flanking region A. All PCR products showed similar 2.8-kbp bands. M, 500-bp marker. (Right) The complete DNA sequence of the 2.8-kbp PCR product was assembled in two steps. Seq 1 obtained 702 to 976 nucleotides beginning at approximately nucleotide 565 (yellow) and 936 to 998 nucleotides ending at approximately nucleotide 3275 (green). Seq 2 completed the 738 to 1070 internal sequence (orange), with 1842 to 1869 nucleotides extending from nucleotides 712 to 738 to nucleotides 2557 to 2582 (blue).
The deletion in area B results in the absence of ORF 12.
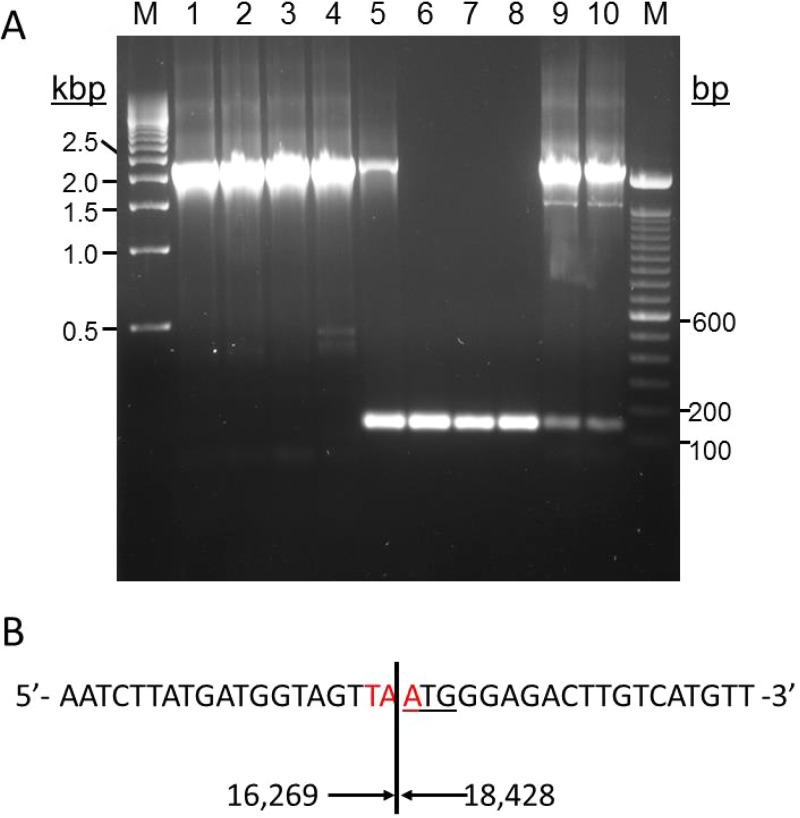
The DNA deletion (area B) indicated by whole-genome sequencing was confirmed by PCR using primers located 79 and 49 nucleotides before and after the expected deletion site, respectively (Fig. 4). An ∼2.3-kbp PCR band was present in VZV-DG, VZV-NASA, VZV Web A (RIT), VZV-DEN09, and VZV-DEN13 DNA samples (Fig. 4A). While the 2.3-kbp DNA fragment was predicted in all complete VZV genome sequences publically available and was present in VZV-DEN09 at passage 7, albeit to a lesser extent, in HFL cells (lane 5), it was undetectable in VZV-DEN09 after 20 passages in HFL cells (lanes 6 to 8). Instead, an ∼140-bp fragment was present in these samples and, to a lesser degree, in VZV-DEN13 after 2 passages in MRC-5 cells (lanes 9 and 10). The disappearance of the 2.3-kbp band and appearance of the ∼140-bp PCR product was consistent with the targeted genome sequencing data for VZV-DEN09 after 20 passages in HFL cells (Fig. 2) and suggested that a large deletion in VZV32 had occurred. DNA sequence analysis of the 140-bp PCR product in lane 6 confirmed the deletion of 2,160 bp from nucleotides 16269 to 18428 with respect to the VZV32 passage 72 genome (Fig. 4B).
FIG 4.
Deletion of 2,160 bp in VZV-DEN09 ORF 12. (A) Agarose gel of PCR products generated using primers spanning ORF 12 and VZV DNA from VZV-DG after 21 and 48 passages in MeWo cells (lanes 1 and 2, respectively), VZV-NASA after <10 passages in HFL cells (lane 3), VZV Web A (RIT) after >50 passages in BSC-1 cells (lane 4), VZV-DEN09 after 7 passages in HFL cells (lane 5), three independent DNA extractions of VZV-DEN09 in HFL cells after >20 passages (lanes 6 to 8), and two independent DNA extractions of VZV-DEN13 after 2 passages in MRC-5 cells (lanes 9 and 10). M, 100-bp marker (left) and 500-bp marker (right). (B) Sequence of the ORF 12 deletion junction determined from analysis of the PCR product from lane 6. Red nucleotides indicate newly formed ORF 12 termination codons, underlined nucleotides indicate ORF 13 initiation codons, and numbers indicate nucleotide location of deletion relative to archive VZV32 passage 72.
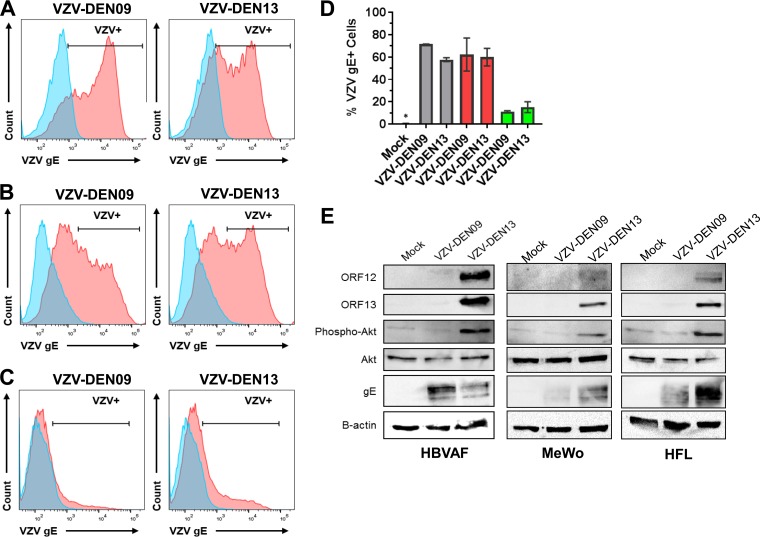
The outcome of this deletion preserved the coding sequence for the first 22 amino acids of ORF 12 but removed the coding sequence for the remaining C-terminal 211 amino acids. Interestingly, the TAA termination codon that truncated ORF 12 was generated from a fusion of TAC (tyrosine codon) of ORF 12 with the ATG (initiation codon) of ORF 13 (Fig. 4B). Therefore, we asked if the DNA deletion in VZV-DEN09 affected the protein expression of ORFs 12 and 13. MeWo, HFL, and quiescent human brain vascular adventitia fibroblast (HBVAF) cultures were infected with VZV-DEN09 and VZV-DEN13 for 72 h, and infection efficiency was quantitated by glycoprotein E (gE) expression (Fig. 5A to D). Expression of ORFs 12 and 13 then was examined by immunoblotting. In all three cell types, only infection with VZV-DEN13 supported expression of ORFs 12 and 13 (Fig. 5E). Since ORF 12 protein triggers phosphorylation of Akt (46), we also examined whether infection with VZV-DEN09 would phosphorylate Akt through a possible ORF 12-independent, redundant pathway. HBVAFs were included as a control because quiescence has previously been established in these cells (47), which allows background levels of phosphorylated Akt to be kept at a minimum (46, 48). Total Akt protein expression was similar in uninfected and infected cells. However, while VZV-DEN13 infection induced Akt phosphorylation, no phosphorylated Akt was detected following VZV-DEN09 infection (Fig. 5E).
FIG 5.
VZV-DEN09 does not express ORFs 12 or 13 and does not induce Akt phosphorylation. (A to C) Flow cytometry analyses of VZV gE expression in mock (blue)- and VZV (red)-infected cells. Human brain vascular adventitia fibroblasts (HBVAFs) (A), HFL (B), or MeWo (C) cultures were left uninfected or infected with VZV-DEN09 or VZV-DEN13 for 72 h. (D) Graphical representation of percent VZV gE+ cells from panels A to C. HBVAFs, gray; HFLs, red; MeWos, green. Error bars represent average percent VZV gE+ cells ± standard deviations from duplicates for HBVAFs. An asterisk denotes 0% of mock-infected cells expressed VZV gE+. (E) Cell lysates were prepared for immunoblotting and resolved on 12.5% SDS-PAGE gels. Membranes were probed for ORFs 12 and 13 and total and phospho-Akt. gE was included as an infection control, and β-actin was included as a loading control.
The ORF 12 deletion has minimal influence on VZV replication.
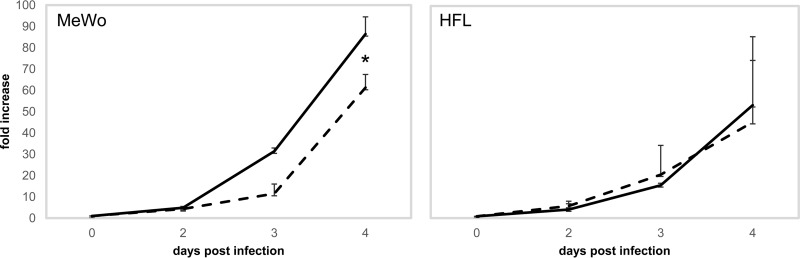
We examined if loss of ORF 12 and ORF 13 proteins could influence virus replication in the cell types in which the viruses were grown. While a minor, yet significant, decrease in replication was observed with VZV-DEN09 infection of MeWo cells at 4 dpi, infection of HFL cells with VZV-DEN09 and VZV-DEN13 did not result in any discernible replication differences (Fig. 6).
FIG 6.
Growth analysis of VZV-DEN09 and VZV-DEN13 in MeWo and HFL cells. DNA was isolated from infected cells (VZV-DEN09 [dashed lines] and VZV-DEN13 [solid lines]) and quantified by TaqMan-based qPCR at the indicated days postinfection. Fold increase in VZV DNA was determined by the delta CT method, normalized to day 0 (2[CT(day 0) − CT(day X)]). Data represent duplicate growth curves. *, P < 0.05.
Analysis of DNA sequence bias in enriched virus samples.
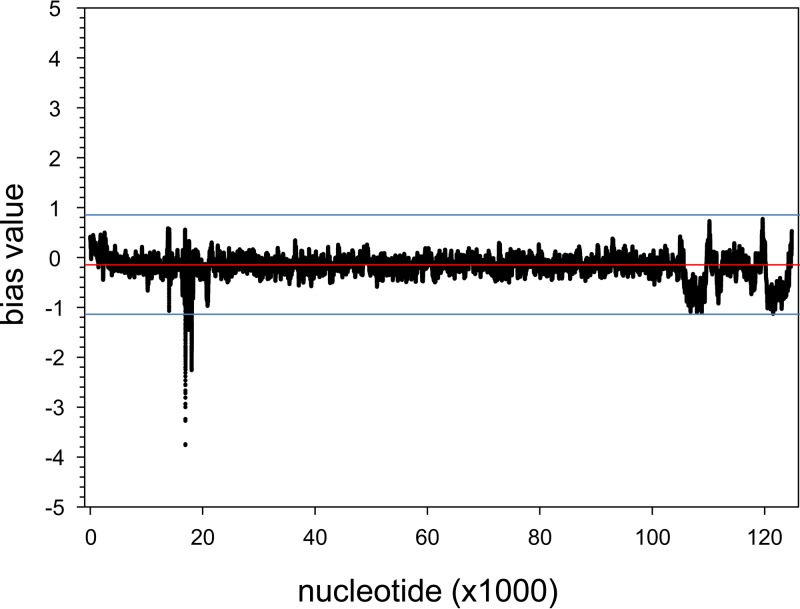
To determine whether DNA enrichment was uniform across the virus genome, the VZV-DEN09 cpn at each nucleotide location was normalized and compared before and after DNA enrichment. For each library, the cpn for the ith nucleotide was modified to reflect the read originated from a 51-bp DNA sequence by summing the cpn for each position 25 nucleotides before and after the ith position. For each nucleotide, the new cpn was divided by the maximum cpn for all positions, resulting in a normalized cpn for each position ranging from 0 to 1. The normalized cpn for each enriched library was then divided by the normalized cpn for each unenriched library, converted to log2 values, and averaged for each position across all library combinations. The resulting bias value reflected whether specific DNA sequences were overrepresented due to the postenrichment 14 PCR cycles (bias value, >1) or to the loss of specific DNA sequences during target enrichment (bias value, <1). The overall average bias value was −0.18 ± 0.23, suggesting a slight, but insignificant, negative bias during enrichment. Display of the bias value for each nucleotide (Fig. 7) indicated that the only area of VZV DNA showing a bias value of <1 was the deletion identified as region B in Fig. 2. No bias of >2-fold was encountered during enrichment of VZV DNA across the VZV genome.
FIG 7.
Bias value plot for VZV-DEN09 DNA. The bias value, an index of enrichment efficiency, was determined for each nucleotide of VZV-DEN09. Bias values of <0 indicate less-than-expected enrichment, while a bias value of >0 indicates more-than-expected enrichment. A single region of VZV-DEN09 DNA that was deleted from VZV32 was outside a 2-fold deviation (blue lines) from the average (red line).
VZV-DEN09 transcriptome.
Directional cDNA libraries were constructed from total RNA extracted from biological replicates of VZV-infected HFL cells (Table 1, samples C and D). After sequencing, reads for each library (with or without enrichment) were aligned to specific strands of the 123,025-bp VZV-DEN09 genome assembled from DNA sequence data obtained as described above (Table S3). For each library, the total number of reads divided by total cpn for each strand indicated a read length of 50.9 ± 0.2 nucleotides, verifying the integrity of alignment and mapping data.
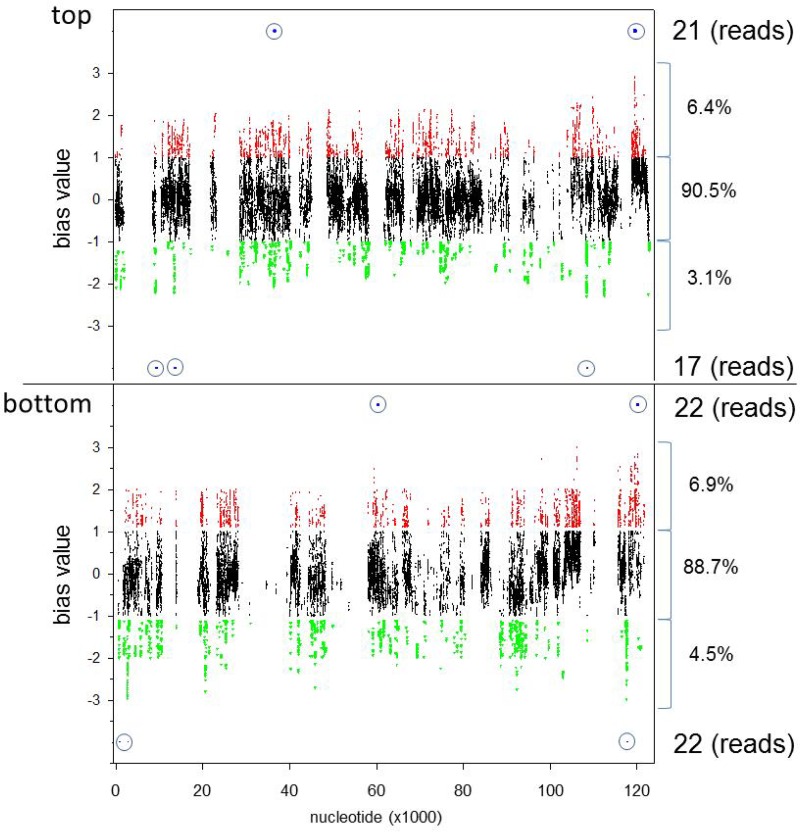
To determine whether cDNA enrichment was uniform for all nucleotides across the VZV genome, a bias value similar to that described above was determined. A total of 1.22 × 108 cpn were obtained, but each library was trimmed to omit <10 cpn for unenriched libraries and <100 cpn for enriched libraries, yielding 1.10 × 108 cpn used for further calculations. For each VZV DNA nucleotide, the trimmed cpn was normalized to the maximum cpn of the related DNA strand. The bias value plot (quotient of normalized enriched samples to normalized unenriched samples) showed the deviation from expectation (bias value, 0) for each nucleotide (Fig. 8). A total of 73,867,869 cpn mapped to the top strand of the virus genome, while 47,668,602 cpn mapped to the bottom DNA strand, showing 1.55:1 top-to-bottom virus strand coding usage. For both DNA strands, the majority of cpn (90.5% [top] and 88.7% [bottom]) showed bias values between −1 and +1, indicating a <2-fold relative difference between unenriched and enriched VZV cDNA. Bias values of <−1 were obtained for 3.1% of nucleotides on the top strand and 4.5% of nucleotides on the bottom strand of the virus genome, indicating a >2-fold relative enrichment of cDNA at these locations. Bias values of >1 were obtained for 6.4% of nucleotides on the top strand and 6.9% of nucleotides on the bottom strand, indicating a >2-fold enrichment of cDNA at these locations. Since cpn reflects alignment of 51-bp sequencing reads, a 51-bp sliding window was generated to map individual cpn to reads. Of the 9,435 nucleotides showing >2-fold (absolute value) bias values, only 82 (0.9%) mapped to potential sequencing reads. To determine whether these 82 reads were located within coding regions, the VZV-DEN09 genome was annotated based on the archived VZV32 passage 72 genome (accession no. DQ479963). While coding frames were shifted for some ORFs due to intergenic indels, only the ORF 12 truncation showed a length difference compared to the archived genome (Fig. 9). Based on the VZV-DEN09 annotation, the 5′ end of the outlying transcriptome sequencing (RNA-seq) reads were located at nucleotides 36283 (ORF 22), 60098 to 60095 (ORF 33), 119227 to 119246, and 119876 to 119894 (ORF 71). Analysis of transcripts per million mapped reads, calculated by normalizing the sum of cpn for each VZV-DEN09 ORF to ORF length followed by renormalization to total cpn, all divided by 106, revealed no significant difference (P = 0.11 to 0.99 by t test) between the two independent unenriched libraries and enriched libraries at any of the 74 VZV ORFs (Table S4), indicating that targeted enrichment of VZV cDNA did not alter the transcriptional profile of the annotated VZV genes.
FIG 8.
Bias value plot for VZV-DEN09 transcripts. Bias values were determined from transcriptome analysis for all nucleotide locations on both top and bottom DNA strands of VZV-DEN09. Bias values of >1 (red dots) indicate nucleotide locations with >2-fold the expected counts per nucleotide (cpn) in enriched samples. Bias values of <−1 (green dots) indicate nucleotide locations with <2-fold less than the expected cpn in enriched samples. Bias values between −1 and 1 (black dots) indicate nucleotide locations with cpn within 2-fold of the expected value. Percentages of total cpn that reside within each area are listed on the right. Total numbers and locations of cpn from 51-nucleotide reads residing outside the 2-fold expected value are listed on the right, with relative location shown by dots within blue circles.
FIG 9.
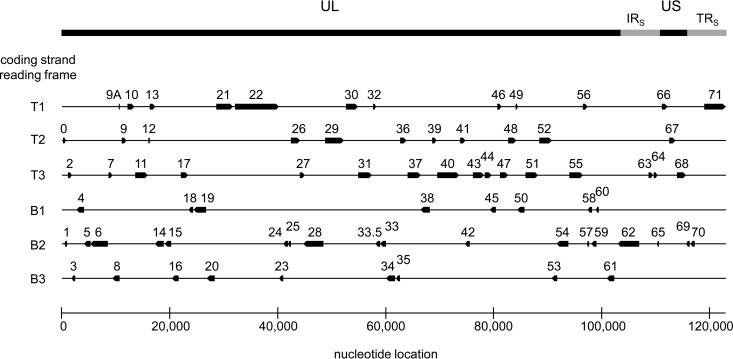
VZV-DEN09 annotated genome. Open reading frames as annotated in reference genome VZV32 passage 72 were mapped to 1st, 2nd, or 3rd reading frames of top (T) or bottom (B) DNA strands of VZV-DEN09.
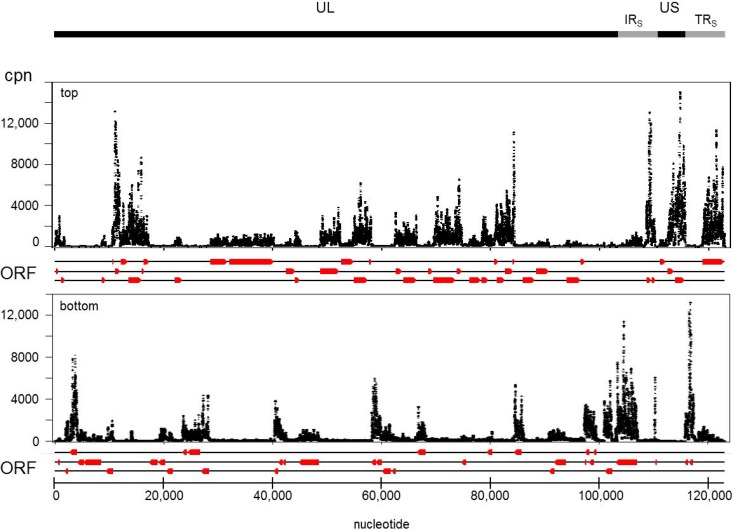
Summation of strand-specific cpn for all libraries of VZV-DEN09 was used to develop a transcriptome map (Fig. 10). All annotated VZV ORFs were transcribed in HFL cells, and their steady-state abundance was ranked in order of decreasing amounts (Table 4). To better determine relationships between transcript abundance, VZV genes were grouped into similar temporal and functional groups based on established or predicted homology to herpes simplex virus 1 (HSV-1). The average rank for VZV genes sorted into temporal classes was immediate early (8.3), early (47.7), late (38.4), and unknown class (36.8); the average rank for VZV genes sorted into functional classes was DNA replication (51.8), glycoproteins (33.0), nucleocapsid proteins (28.2), and regulation of transcription (8.3). These results indicate that steady-state amounts of immediate-early VZV gene transcripts involved in regulating virus gene transcription exceeded those of transcripts encoded by early or late virus genes involved in DNA replication or the structure of the virus.
FIG 10.
VZV-DEN09 transcriptome. cDNA reads were converted to counts per nucleotide (cpn) and plotted against the top or bottom strand of the annotated VZV-DEN09 genome. Direction and location of ORFs (red arrows) are as shown in Fig. 8.
TABLE 4.
VZV-DEN09 average transcripts per million mapped reads compared to HSV-1
| ORF | TPMa (avg) | TPM (SD) | Rank | HSV-1 homologue | ORF | Temporal classb | Rank | Avg rank | Functional class | ORF | Rank | Avg rank |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 6.946 | 1.389 | 32 | None | 70 | IE | 3 | 8.3 | DNA replication | 16 | 60 | 51.8 |
| 1 | 1.220 | 0.308 | 69 | None | 63 | IE | 4 | DNA replication | 28 | 55 | ||
| 2 | 2.563 | 0.343 | 57 | None | 4 | IE | 6 | DNA replication | 29 | 33 | ||
| 3 | 8.790 | 0.795 | 28 | UL55 | 62 | IE | 9 | DNA replication | 36 | 40 | ||
| 4 | 22.000 | 2.256 | 6 | UL54 | 71 | IE | 11 | DNA replication | 51 | 71 | ||
| 5 | 2.751 | 0.219 | 54 | UL53 | 61 | IE | 17 | Glycoproteins | 5 | 54 | 33.0 | |
| 6 | 1.417 | 0.057 | 67 | UL52 | 48 | E | 14 | 47.7 | Glycoproteins | 14 | 74 | |
| 7 | 2.922 | 0.359 | 53 | UL1 | 59 | E | 24 | Glycoproteins | 31 | 22 | ||
| 8 | 3.755 | 0.453 | 46 | UL50 | 29 | E | 33 | Glycoproteins | 37 | 37 | ||
| 9A | 6.529 | 0.576 | 35 | UL49.5 | 18 | E | 36 | Glycoproteins | 50 | 23 | ||
| 9 | 42.232 | 2.692 | 1 | UL49 | 36 | E | 40 | Glycoproteins | 60 | 42 | ||
| 10 | 6.921 | 0.659 | 34 | UL48 | 19 | E | 41 | Glycoproteins | 67 | 7 | ||
| 11 | 16.528 | 2.434 | 10 | UL47 | 8 | E | 46 | Glycoproteins | 68 | 5 | ||
| 12 | 3.847 | 0.569 | 44 | UL4 | 28 | E | 55 | Nucleocapsid | 21 | 51 | 28.2 | |
| 13 | 8.310 | 0.891 | 30 | None | 16 | E | 60 | Nucleocapsid | 23 | 19 | ||
| 14 | 0.321 | 0.039 | 74 | UL44 | 52 | E | 66 | Nucleocapsid | 33 | 16 | ||
| 15 | 3.426 | 0.070 | 49 | UL43 | 6 | E | 67 | Nucleocapsid | 40 | 27 | ||
| 16 | 1.977 | 0.241 | 60 | UL42 | 55 | E | 68 | Regulate transcription | 4 | 6 | 8.3 | |
| 17 | 2.457 | 0.104 | 58 | UL41 | 51 | E | 71 | Regulate transcription | 61 | 17 | ||
| 18 | 6.390 | 0.545 | 36 | UL40 | 9 | L | 1 | 38.4 | Regulate transcription | 62 | 9 | |
| 19 | 4.806 | 0.383 | 41 | UL39 | 49 | L | 2 | Regulate transcription | 63 | 4 | ||
| 20 | 8.588 | 0.669 | 29 | UL38 | 68 | L | 5 | Regulate transcription | 70 | 3 | ||
| 21 | 3.234 | 0.320 | 51 | UL37 | 67 | L | 7 | Regulate transcription | 71 | 11 | ||
| 22 | 3.234 | 0.422 | 50 | UL36 | 33.5 | L | 8 | 0 | 32 | |||
| 23 | 13.117 | 1.352 | 19 | UL35 | 11 | L | 10 | 1 | 69 | |||
| 24 | 5.945 | 0.748 | 38 | UL34 | 41 | L | 13 | 2 | 57 | |||
| 25 | 2.696 | 0.392 | 56 | UL33 | 58 | L | 15 | 3 | 28 | |||
| 26 | 1.675 | 0.058 | 61 | UL32 | 33 | L | 16 | 6 | 67 | |||
| 27 | 3.532 | 0.218 | 48 | UL31 | 47 | L | 18 | 7 | 53 | |||
| 28 | 2.720 | 0.234 | 55 | UL30 | 23 | L | 19 | 8 | 46 | |||
| 29 | 6.937 | 0.567 | 33 | UL29 | 46 | L | 20 | 9A | 35 | |||
| 30 | 1.674 | 0.200 | 62 | UL28 | 31 | L | 22 | 9 | 1 | |||
| 31 | 11.910 | 0.666 | 22 | UL27 | 50 | L | 23 | 10 | 34 | |||
| 32 | 12.010 | 1.344 | 21 | None | 64 | L | 25 | 11 | 10 | |||
| 33 | 13.614 | 1.374 | 16 | UL26 | 69 | L | 26 | 12 | 44 | |||
| 33.5 | 18.037 | 2.056 | 8 | UL26.5 | 40 | L | 27 | 13 | 30 | |||
| 34 | 4.319 | 0.427 | 43 | UL25 | 3 | L | 28 | 15 | 49 | |||
| 35 | 1.660 | 0.254 | 65 | UL24 | 20 | L | 29 | 17 | 58 | |||
| 36 | 5.406 | 0.308 | 40 | UL23 | 44 | L | 31 | 18 | 36 | |||
| 37 | 6.330 | 0.326 | 37 | UL22 | 10 | L | 34 | 19 | 41 | |||
| 38 | 5.626 | 0.242 | 39 | UL21 | 9A | L | 35 | 20 | 29 | |||
| 39 | 1.115 | 0.077 | 70 | UL20 | 37 | L | 37 | 22 | 50 | |||
| 40 | 8.839 | 1.045 | 27 | UL19 | 24 | L | 38 | 24 | 38 | |||
| 41 | 15.740 | 0.793 | 13 | UL18 | 38 | L | 39 | 25 | 56 | |||
| 42 | 1.669 | 0.260 | 64 | UL15 | 60 | L | 42 | 26 | 61 | |||
| 43 | 3.012 | 0.324 | 52 | UL17 | 34 | L | 43 | 27 | 48 | |||
| 44 | 7.917 | 0.868 | 31 | UL16 | 12 | L | 44 | 30 | 62 | |||
| 45 | 1.673 | 0.078 | 63 | UL15 | 53 | L | 45 | 32 | 21 | |||
| 46 | 13.117 | 1.682 | 20 | UL14 | 54 | L | 47 | 33.5 | 8 | |||
| 47 | 13.191 | 1.594 | 18 | UL13 | 27 | L | 48 | 34 | 43 | |||
| 48 | 15.054 | 0.967 | 14 | UL12 | 15 | L | 49 | 35 | 65 | |||
| 49 | 38.327 | 1.554 | 2 | UL1 | 22 | L | 50 | 38 | 39 | |||
| 50 | 11.103 | 0.715 | 23 | UL10 | 21 | L | 51 | 39 | 70 | |||
| 51 | 0.852 | 0.080 | 71 | UL9 | 43 | L | 52 | 41 | 13 | |||
| 52 | 1.518 | 0.132 | 66 | UL8 | 7 | L | 53 | 42 | 64 | |||
| 53 | 3.839 | 0.553 | 45 | UL7 | 5 | L | 54 | 43 | 52 | |||
| 54 | 3.539 | 0.572 | 47 | UL6 | 25 | L | 56 | 44 | 31 | |||
| 55 | 1.415 | 0.082 | 68 | UL5 | 17 | L | 58 | 45 | 63 | |||
| 56 | 0.835 | 0.177 | 72 | UL4 | 66 | L | 59 | 46 | 20 | |||
| 57 | 16.342 | 1.311 | 12 | None | 26 | L | 61 | 47 | 18 | |||
| 58 | 14.830 | 0.975 | 15 | UL3 | 30 | L | 62 | 48 | 14 | |||
| 59 | 10.028 | 0.958 | 24 | UL2 | 45 | L | 63 | 49 | 2 | |||
| 60 | 4.456 | 0.162 | 42 | UL1 | 42 | L | 64 | 52 | 66 | |||
| 61 | 13.237 | 0.310 | 17 | ICP0 | 35 | L | 65 | 53 | 45 | |||
| 62 | 16.548 | 3.813 | 9 | ICP4 | 39 | L | 70 | 54 | 47 | |||
| 63 | 34.880 | 4.937 | 4 | us1.5 | 56 | L | 72 | 55 | 68 | |||
| 64 | 9.462 | 2.572 | 25 | us10 | 65 | L | 73 | 56 | 72 | |||
| 65 | 0.784 | 0.165 | 73 | us9 | 14 | L | 74 | 57 | 12 | |||
| 66 | 2.160 | 0.190 | 59 | us3 | 57 | U | 12 | 36.8 | 58 | 15 | ||
| 67 | 18.250 | 1.183 | 7 | us7 | 32 | U | 21 | 59 | 24 | |||
| 68 | 31.449 | 1.539 | 5 | us8 | 13 | U | 30 | 64 | 25 | |||
| 69 | 9.259 | 2.643 | 26 | us10 | 0 | U | 32 | 65 | 73 | |||
| 70 | 35.131 | 4.209 | 3 | us1.5 | 2 | U | 57 | 66 | 59 | |||
| 71 | 16.447 | 3.946 | 11 | ICP4 | 1 | U | 69 | 69 | 26 |
TPM, transcripts per million reads.
IE, immediate early; E, early; L, late; U, unknown.
DISCUSSION
Despite its global distribution and an extremely long evolutionary history, VZV DNA is remarkably homogeneous throughout the world. The VZV mutation rate of 1 × 10−5 to 6 × 10−5 substitutions/site/year translates into <1 mutation in the 125-kbp genome per year (35). The VZV phenotype is even more stable; the only reported clinical strain of VZV with an altered in vitro growth phenotype has a nonsynonymous substitution in gE, a mutation that results in increased virulence in tissue culture and skin graft cultures (49), and is also present in an independent VZV isolate from British Columbia (42). In tissue culture, VZV is also extremely stable. Sequencing of VZV32 after 5, 22, and 72 passages has revealed significant sequence modification at only 3 locations. Three regions show expansion of DNA repeats: (i) an 8-bp region of ORF 11 at passage 5 was duplicated at passage 22 and maintained at passage 72; (ii) a 4-copy 54-bp repeat between ORFs 62 and 63 as well as the inverse ORFs 70 and 71 at passage 5 expanded to five copies by passage 22 and was maintained at passage 72; and (iii) one copy of an 18-bp duplication in ORF 22 at passage 5 was deleted by passage 22 and returned in passage 72. Notably, all of these variations occur in regions of the virus DNA that contain known sequence repeats, and none are contained within the origin of VZV DNA replication. Thus, while extended in vitro propagation produces local extension or retraction of known repeated DNA, our deep-sequence analysis of VZV-DEN09 showed a large deletion significantly affecting ORF 12 and 13 protein expression. In addition, specific areas (A, C, and C′) of overrepresented DNA, confirmed by multiple qPCR assays, were detected that most likely originated from fragments of virus DNA present in the infected cell nucleus but not contained in the virus genome or in VZV DNA prepared from E. coli as a stable bacterial artificial chromosome.
The 2,160-bp deletion in ORF 12 revealed by deep-sequence analysis represents the largest VZV DNA deletion identified to date in virus propagated under nonselective conditions. The 84.6-kDa tegument protein encoded by ORF 12 induces phosphorylation of ERK1/2 and Akt, which activate AP-1 and Akt pathways, respectively (46, 48). AP-1 activation protects MeWo cells from staurosporine-induced apoptosis, and Akt activation increases levels of cyclin B1 and D3 to force cells from stationary phase and into M and S phases, respectively (46, 48). Sequence analysis of the ORF 12 deletion within VZV-DEN09 showed the first 22 codons of ORF 12 were retained but that a new termination codon is formed from the fusion between codon 22 of ORF 12 with the initiation codon of ORF 13. This interesting arrangement would preserve the N-terminal 22 amino acids of ORF 12 but lack the gene's 3′-untranslated regulatory sequences. Our results, in agreement with previous findings (48), indicated that ORF 12 was required for activation of the Akt pathway, since VZV-DEN09 infection failed to phosphorylate Akt. ORF 13 transcription also could be affected in VZV-DEN09-infected cells, since the 2,160-bp deletion removed the entire 241-bp ORF 12/13 intergenic region that presumably contains the regulatory sequences for ORF 13 transcription. Indeed, we found that while RNA mapping to ORF 13 was present (likely a result of a read-through event from fused ORF 12/13), no ORF 13 protein was made. The thymidylate synthetase encoded by VZV ORF 13 is dispensable for virus propagation (50) and is a prime site for targeted insertion of foreign genes (51, 52). The nonessential nature of ORF 12 and ORF 13 proteins is echoed by our growth analysis, in which we did not observe a replication defect in HFL cells infected with VZV-DEN09. However, a slight decrease in replication was apparent in infected MeWo cells at later times postinfection. Since MeWo cells were derived from a malignant melanoma, these findings suggest that the ORF 12 and ORF 13 proteins serve different functions in normal and cancer cell lines.
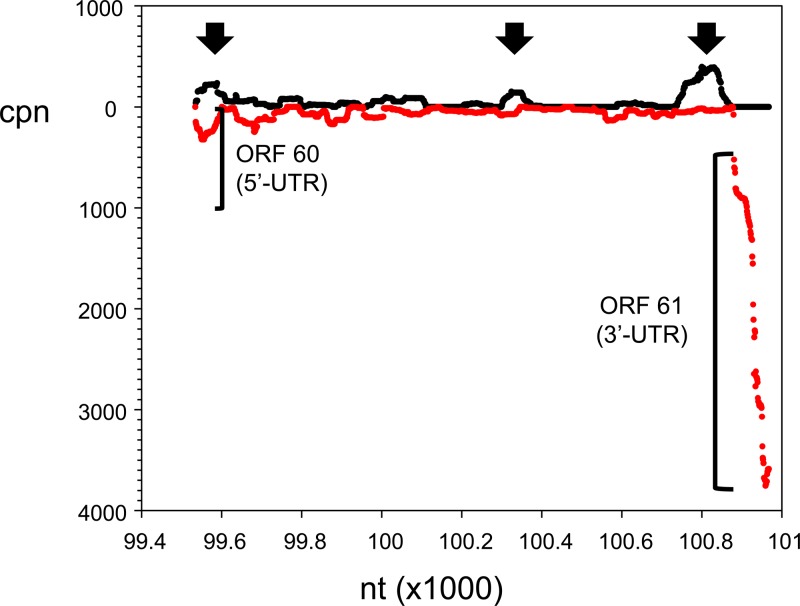
The VZV lytic transcriptome has been analyzed by PCR-based (53) and long oligonucleotide-based (54) arrays, by multiplex PCR (29), and by next-generation cDNA sequencing (55–57). While differences in transcript profiles are noted depending on cell type, time of harvest, and technique used, general themes emerge when the data are restricted to RNA-seq findings during lytic VZV infection. Combining published RNA-seq data derived from VZV-infected HFL cells or human neurons in culture with results in the present study showed individual differences in transcript abundance. For example, the five most abundant transcripts (in decreasing order) map to ORFs 57, 9, 68, 58, and 63 (55), ORFs 57, 49, 9, 68, and 67 (22), and ORFs 9, 49, 63, 68, and 4 (this study); however, when virus gene classes are analyzed, all studies identify immediate-early transcripts as the most abundant, followed by late and early transcripts. Similarly, transcripts that regulate virus gene transcription are the most abundant, followed by transcripts encoding glycoproteins, nucleocapsid proteins, and, finally, proteins involved in virus DNA replication (Table 5). In addition to determining steady-state amounts of previously mapped virus genes, RNA-seq analysis can indicate the presence of novel virus transcriptional units. For example, RNA mapping to the unannotated 1.4-kbp region between VZV ORFs 60 and 61 has been detected by Northern blot (58) and array (53) analyses, and preliminary cloning results suggest extensive splicing of short segments into single polyadenylated virus transcripts within this region (unpublished data). Mapping our current RNA-seq data to this region revealed multiple short and low-abundance RNA segments on the opposite coding strand of ORFs 60 and 61 (Fig. 11).
TABLE 5.
Comparison of RNA-seq data for VZV during lytic infection in culture
| Classd | Avg rank by cell type |
|||
|---|---|---|---|---|
| Fibroblasta | Fibroblastb | Neuronb | Neuronc | |
| Temporal | ||||
| Immediate early (4) | 7.3 | 13.0 | 12.5 | 20.5 |
| Early (13) | 42.0 | 45.2 | 42.8 | 43.4 |
| Late (44) | 33.4 | 32.0 | 32.7 | 32.1 |
| Functional | ||||
| DNA replication (5) | 45.8 | 52.2 | 48.4 | 49.2 |
| Glycoproteins (7) | 23.4 | 21.0 | 23.3 | 21.0 |
| Nucleocapsid (4) | 24.3 | 26.8 | 32.3 | 33.0 |
| Regulate transcription (4) | 7.3 | 13.0 | 12.5 | 20.5 |
FIG 11.
Location of candidate novel VZV transcripts. Enlargement of the 1.4-kbp ORF 60/61 intergenic segment of VZV-DEN09 shown in Fig. 9 revealed three locations of increased cpn (arrows) on the DNA strand (black) opposite that encoding ORFs 60 and 61 (red).
Overall, we have developed a technique to enrich low-abundance VZV DNA and cDNA in an unbiased manner; consequently, we identified a deletion in the coding region of a VZV gene which occurred through normal (no intentional selective pressure) in vitro propagation. Comparison of available RNA-seq data showed differences in individual gene transcript amounts but concordance at the level of gene groupings. Interestingly, transcripts mapping to immediate-early genes functionally identified as regulators of gene transcription were the most abundant transcriptional group detected during lytic infection in human fibroblasts or neurons harvested 2 days to 2 weeks postinfection, suggesting that the continuous presence of virus transactivators is required as the infection spreads throughout the culture.
The unique features of VZV propagated in vitro, as revealed by whole-genome analysis of VZV DNA and transcripts, raises several interesting questions. For example, how did the ORF 12 deletion develop, and what is its consequence on activation of cellular immediate-early response pathways? What mechanism leads to the overrepresented DNA that was identified by deep sequencing and confirmed by qPCR? Answers to these questions await further investigation.
In summary, the discovery of the ORF 12 deletion affecting ORF 12 and 13 protein production in the VZV-DEN09 strain of VZV32, revealed through whole-genome analysis, points to the need to authenticate the VZV genome when the virus is propagated in tissue culture. While this deletion appears to be restricted to VZV32, our data suggest that high passage can lead to changes in what is believed to be a stable VZV genome.
MATERIALS AND METHODS
Cells and virus.
Human fetal lung fibroblasts (HFL; ATCC-CCL-153), MeWo malignant melanoma cells (59), and MRC-5 human lung fibroblasts (ATCC CCL-171) were grown in Dulbecco's modified Eagle medium (Invitrogen, Grand Island, NY) supplemented with 10% fetal bovine serum (FBS; HyClone, Logan, UT). Primary human brain vascular adventitia fibroblasts (HBVAFs) were grown in 2% FBS, 1% fibroblast growth serum, and 1% 100× penicillin-streptomycin in basal fibroblast medium (Sciencell, Carlsbad, CA) as previously described (47). After 24 h, the medium was changed to 0.1% FBS for 6 to 7 days to establish quiescence.
VZV32 (provided to R. J. Cohrs in 2009 by Charles Grose, University of Iowa) was propagated in MeWo cells for >50 passages and transferred as Dounce-homogenized cell-free virus (60) into HFL cells. This virus was designated VZV-DEN09 to uniquely identify the virus DNA and cDNA sequences analyzed in the current study. VZV32 (provided to K. S. Lee in 2013 by Charles Grose) was also propagated in a separate laboratory for <3 passages in MRC-5 cells and designated VZV-DEN13. VZV-DG, a clinical isolate from a zoster lesion, was propagated in MeWo cells. The VZV Web A (RIT) strain was isolated from the vesicle of a patient with varicella and cultivated in human fibroblasts, WI-38 and MRC-5 cells (61), before the virus was transferred (62) and propagated in BSC-1 African green monkey kidney cells (63). VZV-NASA, isolated from saliva of an astronaut 1 day after spaceflight, was propagated for <20 passages in HFL cells (64). VZV BAC is the parental Oka virus strain containing the bacterial artificial chromosome initially propagated in MeWo cells (19) and subsequently transferred to HFL cells. All VZV isolates were propagated by cocultivation of infected and uninfected cells and never processed on the same day.
Virus nucleic acid extraction and library preparation.
Chromatin harvested from VZV-infected HFL cells at 2 days postinfection (dpi) was formaldehyde cross-linked (65) and sheered to an average size of 250 to 500 bp by isothermic sonication at 4°C for 16 min with 200 bursts/min at 140 W/burst (E220; Covaris, Inc., Woburn, MA, USA). DNA-containing virus nucleocapsids were removed by 30 min of centrifugation at 40,000 × g at 4°C (66). Formaldehyde cross-links were reversed at 65°C for 1.5 h in 0.2 M NaCl, and DNA was extracted by affinity chromatography (QIAquick PCR clean up kit; Qiagen, Valencia, CA, USA). Libraries for DNA sequence analysis were constructed using the KAPA Hyper Prep kit (Kapa Biosystems, Wilmington, MA, USA). For construction of cDNA libraries, total RNA was extracted (TriPure; Roche, San Francisco, CA, USA) at 2 dpi from biological replicates of VZV-DEN09-infected HFL cells and DNase treated (Turbo-DNase; Thermo-Fisher, Grand Island, NY, USA), and directional, rRNA-depleted cDNA libraries were constructed (NEBNext Ultra directional RNA library prep kit; New England BioLabs, Ipswich, MA, USA). Each sample from both the DNA and cDNA libraries was divided into two aliquots. One aliquot was used for virus-specific target enrichment (SeqCap EZ System; Roche, Madison, WI, USA) as described below, while the other aliquot was processed without enrichment.
Virus-specific nucleic acid enrichment.
Single-strand biotinylated probes covering 99.2% of the VZV genome (accession no. DQ452050.1) were designed with the NimbleGen SeqCap EZ Developer (Roche). Hybridization of VZV-infected HFL DNA or cDNA libraries to the biotinylated capture probes with subsequent washings and elution were performed according to the manufacturer's recommendations (SeqCap EZ SR User's Guide v5.0; Roche).
DNA and RNA sequence analysis.
All libraries were subjected to single-end 50-bp sequencing using an Illumina HiSeq4000 (San Diego, CA, USA). Resulting fastq files were mapped to VZV (strain 32 passage 72 [accession no. DQ479963] and VZV-DEN09) genomes using Bowtie2 (with default arguments) and SeqMan Pro (DNAStar, Madison, WI, USA).
PCR.
Oligonucleotide primers (Table 2) synthesized by Integrated DNA Technology (IDT; Coralville, Iowa, USA) were designed using VZV Dumas (accession no. X04370.1) as the template for standard and quantitative PCR (65).
Flow cytometry.
Mock- and VZV-infected MeWo, HFL, and HBVAF cultures at 72 h postinfection (hpi) were washed with fluorescence-activated cell sorter (FACS) buffer (phosphate-buffered saline [PBS] containing 1% FBS) and stained with mouse anti-VZV-gE R-phycoerythrin (Millipore) for 30 min at 4°C, washed with FACS buffer, and fixed with 1% paraformaldehyde. Isotype controls were used in staining. Cells were analyzed using an LSR-II flow cytometer (BD Immunocytometry Systems, San Jose, CA); more than 15,000 events were collected for all samples. Subsequent data were analyzed using Diva software (BD Biosciences) and FlowJo software (Tree Star, Ashland, OR).
Immunoblotting.
VZV-infected and control HBVAF, MeWo, and HFL cells were harvested at 72 hpi with 0.25% trypsin-EDTA (25200-056; Life Technologies). The harvested cells were washed once with PBS and centrifuged at 1,500 rpm for 5 min at 4°C, resuspended in E1A lysis buffer (50 mM HEPES, pH 7.2, 250 mM NaCl, 2 mM EDTA, 0.1% NP-40, 1 mM dithiothreitol [DTT], 1 μg/ml aprotinin, 1 μg/ml leupeptin, 50 μg/ml phenylmethylsulfonyl fluoride [PMSF], and 10 mM NaF), and kept on ice for 20 min. The lysates were clarified at 14,000 rpm for 10 min at 4°C. Total protein concentrations were determined using the DCTM protein assay (Bio-Rad Laboratories, Hercules, CA). Twenty-microgram aliquots of lysates were resolved on 12.5% SDS-PAGE gels, transferred onto polyvinylidene difluoride (PVDF) membranes, and probed for VZV ORF 12 (mouse anti-VZV ORF 12; 1:1000; Center for Proteomics, University of Rijeka, Croatia), VZV ORF13 (mouse anti-ORF 13; 1:1,000; Center for Proteomics, University of Rijeka, Croatia), Akt (rabbit anti-Akt; 1:1000; no. 4685; Cell Signaling Technology), phosphorylated Akt (rabbit anti-Akt [Thr308], 1:1,000; no. 13038; Cell Signaling Technology), VZV glycoprotein E (gE; mouse anti-gE; 1:500; sc-56995; Santa Cruz Biotechnology), and β-actin (mouse anti-β-actin; 1:10,000; no. 3700; Cell Signaling Technology), followed by appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (Santa Cruz Biotechnology). The blots were incubated with Amersham ECL Select Western blotting detection reagent (RPN2235; GE Healthcare) and visualized with the ChemiDoc XRS+ system (Bio-Rad).
Viral DNA replication.
HFL and MeWo cells were seeded in 6-well plates at 40% confluence. Both cell lines were infected (multiplicity of infection of 0.001) with VZV-DEN09 or VZV-DEN13 (propagated in their respective cell lineage) after lysis through a 28-gauge needle. Virus inoculum was adsorbed for 2.5 h at 37°C with occasional rocking, washed twice with PBS cultures, and replenished with growth medium. Cells were trypsinized at 0, 2, 3, and 4 dpi, and nucleic acid was extracted from the harvested cells (Zymo Duet DNA/RNA kit; Zymo Research, Irvine, CA). PCR was performed with primers and probes specific for VZV ORF68 (Table 2) using a 7500 fast-real-time PCR system (Applied Biosystems, Thermo Fisher Scientific, Grand Island, NY). Statistical significance was determined using a Student's paired t test.
Supplementary Material
ACKNOWLEDGMENTS
We thank Charles Grose for providing initial stocks of VZV32 to R.J.C. and to K.S.L. We also thank Hussain Badani for technical assistance, Alex Greninger for bioinformatics assistance, Joel Rovnak for insightful discussions, Marina Hoffman for editorial review, and Cathy Allen for manuscript preparation.
This work was supported by Public Health Service grants NS082228 (R.J.C.), AG032958 (R.J.C.), and NS093716 (N.L.B.) from the National Institutes of Health. Support for D.Y. was provided in part by NIH grant R25HL103286.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01141-17.
REFERENCES
- 1.Virgin HW, Wherry EJ, Ahmed R. 2009. Redefining chronic viral infection. Cell 138:30–50. doi: 10.1016/j.cell.2009.06.036. [DOI] [PubMed] [Google Scholar]
- 2.Eshleman E, Shahzad A, Cohrs RJ. 2011. Varicella zoster virus latency. Future Virol 6:341–355. doi: 10.2217/fvl.10.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagel MA, Rempel A, Huntington J, Kim F, Choe A, Gilden D. 2014. Frequency and abundance of alphaherpesvirus DNA in human thoracic sympathetic ganglia. J Virol 88:8189–8192. doi: 10.1128/JVI.01070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gershon AA, Chen J, Davis L, Krinsky C, Cowles R, Reichard R, Gershon M. 2012. Latency of varicella zoster virus in dorsal root, cranial, and enteric ganglia in vaccinated children. Trans Am Clin Climatol Assoc 123:17–33. [PMC free article] [PubMed] [Google Scholar]
- 5.Weinberg A, Levin MJ. 2010. VZV T cell-mediated immunity. Curr Top Microbiol Immunol 342:341–357. [DOI] [PubMed] [Google Scholar]
- 6.Schmader K. 1998. Postherpetic neuralgia in immunocompetent elderly people. Vaccine 16:1768–1770. doi: 10.1016/S0264-410X(98)00137-6. [DOI] [PubMed] [Google Scholar]
- 7.Minassian C, Thomas SL, Smeeth L, Douglas I, Brauer R, Langan SM. 2015. Acute cardiovascular events after herpes zoster: a self-controlled case series analysis in vaccinated and unvaccinated older residents of the United States. PLoS Med 12:e1001919. doi: 10.1371/journal.pmed.1001919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yawn BP, Wollan PC, Nagel MA, Gilden D. 2016. Risk of stroke and myocardial infarction after herpes zoster in older adults in a US community population. Mayo Clin Proc 91:33–44. doi: 10.1016/j.mayocp.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilden D, White T, Khmeleva N, Boyer PJ, Nagel MA. 2016. VZV in biopsy-positive and -negative giant cell arteritis: analysis of 100+ temporal arteries. Neurol Neuroimmunol Neuroinflamm 3:e216. doi: 10.1212/NXI.0000000000000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilden D, Grose C, White T, Nagae L, Hendricks RL, Cohrs RJ, Nagel MA. 2016. Successful antiviral treatment after 6 years of chronic progressive neurological disease attributed to VZV brain infection. J Neurol Sci 368:240–242. doi: 10.1016/j.jns.2016.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilden D. 2015. Varicella-zoster virus infections. Continuum (Minneap Minn) 21:1692–1703. [DOI] [PubMed] [Google Scholar]
- 12.Cohrs RJ, Randall J, Smith J, Gilden DH, Dabrowski C, van Der KH, Tal-Singer R. 2000. Analysis of individual human trigeminal ganglia for latent herpes simplex virus type 1 and varicella-zoster virus nucleic acids using real-time PCR. J Virol 74:11464–11471. doi: 10.1128/JVI.74.24.11464-11471.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pugazhenthi S, Nair S, Velmurugan K, Liang Q, Mahalingam R, Cohrs RJ, Nagel MA, Gilden D. 2011. Varicella-zoster virus infection of differentiated human neural stem cells. J Virol 85:6678–6686. doi: 10.1128/JVI.00445-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dukhovny A, Sloutskin A, Markus A, Yee MB, Kinchington PR, Goldstein RS. 2012. Varicella-zoster virus infects human embryonic stem cell-derived neurons and neurospheres but not pluripotent embryonic stem cells or early progenitors. J Virol 86:3211–3218. doi: 10.1128/JVI.06810-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee KS, Zhou W, Scott-McKean JJ, Emmerling KL, Cai GY, Krah DL, Costa AC, Freed CR, Levin MJ. 2012. Human sensory neurons derived from induced pluripotent stem cells support varicella-zoster virus infection. PLoS One 7:e53010. doi: 10.1371/journal.pone.0053010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baird NL, Yu X, Cohrs RJ, Gilden D. 2013. Varicella zoster virus (VZV)-human neuron interaction. Viruses 5:2106–2115. doi: 10.3390/v5092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baird NL, Bowlin JL, Yu X, Jonjic S, Haas J, Cohrs RJ, Gilden D. 2014. Varicella zoster virus DNA does not accumulate in infected human neurons. Virology 458-459:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baird NL, Bowlin JL, Hotz TJ, Cohrs RJ, Gilden D. 2015. Interferon gamma prolongs survival of varicella-zoster virus-infected human neurons in vitro. J Virol 89:7425–7427. doi: 10.1128/JVI.00594-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodwin TJ, McCarthy M, Osterrieder N, Cohrs RJ, Kaufer BB. 2013. Three-dimensional normal human neural progenitor tissue-like assemblies: a model of persistent varicella-zoster virus infection. PLoS Pathog 9:e1003512. doi: 10.1371/journal.ppat.1003512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu X, Seitz S, Pointon T, Bowlin JL, Cohrs RJ, Jonjic S, Haas J, Wellish M, Gilden D. 2013. Varicella zoster virus infection of highly pure terminally differentiated human neurons. J Neurovirol 19:75–81. doi: 10.1007/s13365-012-0142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markus A, Lebenthal-Loinger I, Yang IH, Kinchington PR, Goldstein RS. 2015. An in vitro model of latency and reactivation of varicella zoster virus in human stem cell-derived neurons. PLoS Pathog 11:e1004885. doi: 10.1371/journal.ppat.1004885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadaoka T, Depledge DP, Rajbhandari L, Venkatesan A, Breuer J, Cohen JI. 2016. In vitro system using human neurons demonstrates that varicella-zoster vaccine virus is impaired for reactivation, but not latency. Proc Natl Acad Sci U S A 113:E2403–E2412. doi: 10.1073/pnas.1522575113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Depledge DP, Palser AL, Watson SJ, Lai IY, Gray ER, Grant P, Kanda RK, Leproust E, Kellam P, Breuer J. 2011. Specific capture and whole-genome sequencing of viruses from clinical samples. PLoS One 6:e27805. doi: 10.1371/journal.pone.0027805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cohrs RJ, Barbour M, Gilden DH. 1996. Varicella-zoster virus (VZV) transcription during latency in human ganglia: detection of transcripts mapping to genes 21, 29, 62, and 63 in a cDNA library enriched for VZV RNA. J Virol 70:2789–2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ouwendijk WJ, Choe A, Nagel MA, Gilden D, Osterhaus AD, Cohrs RJ, Verjans GM. 2012. Restricted varicella-zoster virus transcription in human trigeminal ganglia obtained soon after death. J Virol 86:10203–10206. doi: 10.1128/JVI.01331-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azarkh Y, Bos N, Gilden D, Cohrs RJ. 2012. Human trigeminal ganglionic explants as a model to study alphaherpesvirus reactivation. J Neurovirol 18:456–461. doi: 10.1007/s13365-012-0123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagel MA, Choe A, Traktinskiy I, Cordery-Cotter R, Gilden D, Cohrs RJ. 2011. Varicella-zoster virus transcriptome in latently infected human ganglia. J Virol 85:2276–2287. doi: 10.1128/JVI.01862-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cohrs RJ, Gilden DH. 2007. Prevalence and abundance of latently transcribed varicella-zoster virus genes in human ganglia. J Virol 81:2950–2956. doi: 10.1128/JVI.02745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagel MA, Gilden D, Shade T, Gao G, Cohrs RJ. 2009. Rapid and sensitive detection of 68 unique varicella zoster virus gene transcripts in five multiplex reverse transcription-polymerase chain reactions. J Virol Methods 157:62–68. doi: 10.1016/j.jviromet.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Breuer J, Grose C, Norberg P, Tipples G, Schmid DS. 2010. A proposal for a common nomenclature for viral clades that form the species varicella-zoster virus: summary of VZV Nomenclature Meeting 2008, Barts and the London School of Medicine and Dentistry, 24-25 July 2008. J Gen Virol 91:821–828. doi: 10.1099/vir.0.017814-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norberg P, Depledge DP, Kundu S, Atkinson C, Brown J, Haque T, Hussaini Y, MacMahon E, Molyneaux P, Papaevangelou V, Sengupta N, Koay ES, Tang JW, Underhill GS, Grahn A, Studahl M, Breuer J, Bergstrom T. 2015. Recombination of globally circulating varicella-zoster virus. J Virol 89:7133–7146. doi: 10.1128/JVI.00437-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loparev VN, Rubtcova EN, Bostik V, Govil D, Birch CJ, Druce JD, Schmid DS, Croxson MC. 2007. Identification of five major and two minor genotypes of varicella-zoster virus strains: a practical two-amplicon approach used to genotype clinical isolates in Australia and New Zealand. J Virol 81:12758–12765. doi: 10.1128/JVI.01145-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sauerbrei A, Philipps A, Zell R, Wutzler P. 2007. Genotyping of varicella-zoster virus strains after serial passages in cell culture. J Virol Methods 145:80–83. doi: 10.1016/j.jviromet.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 34.Vaughan G, Rodriguez-Castillo A, Cruz-Rivera MY, Ruiz-Tovar K, Ramirez-Gonzalez JE, Rivera-Osorio P, Fonseca-Coronado S, Carpio-Pedroza JC, Cazares F, Vazquez-Pichardo M, Anaya L, Escobar-Gutierrez A. 2011. Is ultra-violet radiation the main force shaping molecular evolution of varicella-zoster virus? Virol J 8:370. doi: 10.1186/1743-422X-8-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Depledge DP, Gray ER, Kundu S, Cooray S, Poulsen A, Aaby P, Breuer J. 2014. Evolution of cocirculating varicella-zoster virus genotypes during a chickenpox outbreak in Guinea-Bissau. J Virol 88:13936–13946. doi: 10.1128/JVI.02337-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Depledge DP, Brown J, Macanovic J, Underhill G, Breuer J. 2016. Viral genome sequencing proves nosocomial transmission of fatal varicella. J Infect Dis 214:1399–1402. doi: 10.1093/infdis/jiw398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grose C. 2012. Pangaea and the out-of-Africa model of varicella-zoster virus evolution and phylogeography. J Virol 86:9558–9565. doi: 10.1128/JVI.00357-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolesnik M, Bonnekoh B, Tammer I, Gollnick H, Sauerbrei A. 2012. Varicella outbreak in an Indian couple living in Germany caused by VZV clade VI acquired during a trip to The Netherlands. Case Rep Med 2012:838241. doi: 10.1155/2012/838241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roycroft E, Rose L, Scallan MF, Crowley B. 2012. Molecular characterization of varicella-zoster virus clinical isolates from 2006 to 2008 in a tertiary care hospital, Dublin, Ireland, using different genotyping methods. J Med Virol 84:1672–1679. doi: 10.1002/jmv.23344. [DOI] [PubMed] [Google Scholar]
- 40.Weinert LA, Depledge DP, Kundu S, Gershon AA, Nichols RA, Balloux F, Welch JJ, Breuer J. 2015. Rates of vaccine evolution show strong effects of latency: implications for varicella zoster virus epidemiology. Mol Biol Evol 32:1020–1028. doi: 10.1093/molbev/msu406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun Z, Guo Y, Li M, Yao Z. 2016. Genotype analysis of varicella-zoster virus isolates from suburban Shanghai Municipal Province, China. J Med Microbiol 65:123–128. doi: 10.1099/jmm.0.000208. [DOI] [PubMed] [Google Scholar]
- 42.Grose C, Tyler S, Peters G, Hiebert J, Stephens GM, Ruyechan WT, Jackson W, Storlie J, Tipples GA. 2004. Complete DNA sequence analyses of the first two varicella-zoster virus glycoprotein E (D150N) mutant viruses found in North America: evolution of genotypes with an accelerated cell spread phenotype. J Virol 78:6799–6807. doi: 10.1128/JVI.78.13.6799-6807.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tyler SD, Peters GA, Grose C, Severini A, Gray MJ, Upton C, Tipples GA. 2007. Genomic cartography of varicella-zoster virus: a complete genome-based analysis of strain variability with implications for attenuation and phenotypic differences. Virology 359:447–458. doi: 10.1016/j.virol.2006.09.037. [DOI] [PubMed] [Google Scholar]
- 44.Liu M, Vafai N, Liu A, Hart J, Liu H, He J, Tang X, Wang D, Vafai A. 2008. Stability of varicella-zoster virus open reading frame 63. Arch Virol 153:1943–1947. doi: 10.1007/s00705-008-0197-4. [DOI] [PubMed] [Google Scholar]
- 45.Cohrs RJ, Hurley MP, Gilden DH. 2003. Array analysis of viral gene transcription during lytic infection of cells in tissue culture with varicella-zoster virus. J Virol 77:11718–11732. doi: 10.1128/JVI.77.21.11718-11732.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu X, Cohen JI. 2013. Varicella-zoster virus ORF12 protein activates the phosphatidylinositol 3-kinase/Akt pathway to regulate cell cycle progression. J Virol 87:1842–1848. doi: 10.1128/JVI.02395-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones D, Blackmon A, Neff CP, Palmer BE, Gilden D, Badani H, Nagel MA. 2016. Varicella-zoster virus downregulates programmed death ligand 1 and major histocompatibility complex class I in human brain vascular adventitial fibroblasts, perineural cells, and lung fibroblasts. J Virol 90:10527–10534. doi: 10.1128/JVI.01546-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu X, Li Q, Dowdell K, Fischer ER, Cohen JI. 2012. Varicella-zoster virus ORF12 protein triggers phosphorylation of ERK1/2 and inhibits apoptosis. J Virol 86:3143–3151. doi: 10.1128/JVI.06923-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Santos RA, Hatfield CC, Cole NL, Padilla JA, Moffat JF, Arvin AM, Ruyechan WT, Hay J, Grose C. 2000. Varicella-zoster virus gE escape mutant VZV-MSP exhibits an accelerated cell-to-cell spread phenotype in both infected cell cultures and SCID-hu mice. Virology 275:306–317. doi: 10.1006/viro.2000.0507. [DOI] [PubMed] [Google Scholar]
- 50.Thompson R, Honess RW, Taylor L, Morran J, Davison AJ. 1987. Varicella-zoster virus specifies a thymidylate synthetase. J Gen Virol 68:1449–1455. doi: 10.1099/0022-1317-68-5-1449. [DOI] [PubMed] [Google Scholar]
- 51.Cohen JI, Seidel KE. 1993. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc Natl Acad Sci U S A 90:7376–7380. doi: 10.1073/pnas.90.15.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Somboonthum P, Yoshii H, Okamoto S, Koike M, Gomi Y, Uchiyama Y, Takahashi M, Yamanishi K, Mori Y. 2007. Generation of a recombinant Oka varicella vaccine expressing mumps virus hemagglutinin-neuraminidase protein as a polyvalent live vaccine. Vaccine 25:8741–8755. doi: 10.1016/j.vaccine.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 53.Kaufer BB, Smejkal B, Osterrieder N. 2010. The varicella-zoster virus ORFS/L (ORF0) gene is required for efficient viral replication and contains an element involved in DNA cleavage. J Virol 84:11661–11669. doi: 10.1128/JVI.00878-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kennedy PG, Grinfeld E, Craigon M, Vierlinger K, Roy D, Forster T, Ghazal P. 2005. Transcriptomal analysis of varicella-zoster virus infection using long oligonucleotide-based microarrays. J Gen Virol 86:2673–2684. doi: 10.1099/vir.0.80946-0. [DOI] [PubMed] [Google Scholar]
- 55.Baird NL, Bowlin JL, Cohrs RJ, Gilden D, Jones KL. 2014. Comparison of varicella-zoster virus RNA sequences in human neurons and fibroblasts. J Virol 88:5877–5880. doi: 10.1128/JVI.00476-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones M, Dry IR, Frampton D, Singh M, Kanda RK, Yee MB, Kellam P, Hollinshead M, Kinchington PR, O'Toole EA, Breuer J. 2014. RNA-seq analysis of host and viral gene expression highlights interaction between varicella zoster virus and keratinocyte differentiation. PLoS Pathog 10:e1003896. doi: 10.1371/journal.ppat.1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Markus A, Waldman Ben-Asher H, Kinchington PR, Goldstein RS. 2014. Cellular transcriptome analysis reveals differential expression of pro- and antiapoptosis genes by varicella-zoster virus-infected neurons and fibroblasts. J Virol 88:7674–7677. doi: 10.1128/JVI.00500-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reinhold WC, Straus SE, Ostrove JM. 1988. Directionality and further mapping of varicella zoster virus transcripts. Virus Res 9:249–261. doi: 10.1016/0168-1702(88)90034-2. [DOI] [PubMed] [Google Scholar]
- 59.Grose C, Brunel PA. 1978. Varicella-zoster virus: isolation and propagation in human melanoma cells at 36 and 32 degrees C. Infect Immun 19:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harper DR, Mathieu N, Mullarkey J. 1998. High-titre, cryostable cell-free varicella zoster virus. Arch Virol 143:1163–1170. [DOI] [PubMed] [Google Scholar]
- 61.Gilden DH, Wroblewska Z, Kindt V, Warren KG, Wolinsky JS. 1978. Varicella-zoster virus infection of human brain cells and ganglion cells in tissue culture. Arch Virol 56:105–117. [DOI] [PubMed] [Google Scholar]
- 62.Gilden DH, Shtram Y, Friedmann A, Wellish M, Devlin M, Fraser N, Becker Y. 1982. The internal organization of the varicella-zoster virus genome. J Gen Virol 60:371–374. [DOI] [PubMed] [Google Scholar]
- 63.Hopps HE, Bernheim BC, Nisalak A, Tjio JH, Smadel JE. 1963. Biological characteristics of a continuous kidney cell line derived from the African green monkey. J Immunol 91:416–424. [PubMed] [Google Scholar]
- 64.Cohrs RJ, Mehta SK, Schmid DS, Gilden DH, Pierson DL. 2008. Asymptomatic reactivation and shed of infectious varicella zoster virus in astronauts. J Med Virol 80:1116–1122. doi: 10.1002/jmv.21173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Henderson HH, Timberlake KB, Austin ZA, Badani H, Sanford B, Tremblay K, Baird NL, Jones K, Rovnak J, Frietze S, Gilden D, Cohrs RJ. 2015. Occupancy of RNA polymerase II phosphorylated on serine 5 (RNAP S5P) and RNAP S2P on varicella-zoster virus genes 9, 51, and 66 is independent of transcript abundance and polymerase location within the gene. J Virol 90:1231–1243. doi: 10.1128/JVI.02617-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gilden DH, Shtram Y, Friedmann A, Wellish M, Devlin M, Cohen A, Fraser N, Becker Y. 1982. Extraction of cell-associated varicella-zoster virus DNA with triton X-100-NaCl. J Virol Methods 4:263–275. doi: 10.1016/0166-0934(82)90073-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.