Abstract
Pachytene piRNAs are MIWI-/MILI-bound small RNAs abundantly expressed in pachytene spermatocytes and round spermatids in adult mouse testes. Miwi knockout (KO) male mice are sterile due to spermiogenic arrest. In Caenorhabditis elegans, sperm-borne piRNAs appear to have an epigenetic role during fertilization and development because progeny of individuals with piRNA-deficient gametes display a progressive loss of fertility after several generations. In mice, it remains unknown whether pachytene piRNA-deficient round spermatids can produce offspring, and whether the progeny of Miwi mutants also exhibits transgenerational, progressive fertility loss. Here, we report that Miwi KO round spermatids could fertilize both wild-type (WT) and Miwi KO oocytes through round spermatid injection, and could produce healthy and fertile offspring despite the global downregulation of both MIWI-/MILI-bound pachytene piRNAs. Progeny of ROSI-derived heterozygotes, both male and female, displayed normal fertility for at least three generations when bred with either WT or Miwi KO females. Our data indicate that aberrant MIWI-/MILI-bound pachytene piRNA profiles in spermatids do not affect fertilization, early embryonic development, or fertility of the offspring, suggesting that pachytene piRNAs might not be required for paternal transgenerational epigenetic inheritance in mice.
Keywords: PIWI, MIWI, pachytene piRNA, fertilization, embryonic development, fertility, epigenetic inheritance
Introduction
The Argonaute family proteins consists of two subclasses, the Argonaute (AGO) and the PIWI [1], both of which are highly conserved in vertebrates [2]. AGO proteins are ubiquitously expressed and functionally associated with both microRNAs (miRNAs) and small interfering RNAs (siRNAs) in the RNA-induced silencing complex during posttranscriptional regulation of gene expression [3]. PIWI proteins, on the other hand, are exclusively expressed in the germline and form specific protein complexes with PIWI-interacting RNAs (piRNAs). piRNAs appear to have several major functions, including repression of retrotransposons in mammalian testes [4, 5], control of germline stem cell self-renewal in Drosophila [6], and epigenetic memory in the Caenorhabditiselegans germline [7, 8]. In mice, three PIWI family proteins: MILI (PIWIL2), MIWI2 (PIWIL4) and MIWI (PIWIL1) have been identified to express in male germ cells during different stages of spermatogenesis [9]. MILI is expressed from pro-spermatogonia to round spermatids, whereas MIWI2 expression is restricted to gonocytes/prospermatogonia in fetal testes and MIWI is exclusively expressed from the late pachytene spermatocyte to round spermatid stages in postnatal testes [10–13]. These differential expression patterns suggest that the three PIWI family proteins may have different functions during male germ cell development. Indeed, while MILI tends to associate with piRNAs that are ∼26 nucleotides (nt) long, MIW2 and MIWI bind ∼28nt and ∼30nt pachytene piRNAs, respectively [5, 14–17]. Genetic disruptions of each of the three PIWI family proteins lead to different phenotypes. Mili or Miwi2 knockout (KO) male mice exhibit meiotic prophase I defects that are attributed to genetic damages caused by de-suppression of retrotransposon activity in the absence of a piRNA silencing mechanism [12, 18–20], whereas deletion of Miwi in mice causes a spermiogenic arrest of the developing germ cells at the step 4 round spermatid stage, which is associated with aberrant mRNA transcriptome and up-regulation of postmeiotic LINE1 activities [13, 15, 21].
In Drosophila, piRNAs appear to have an epigenetic role in maternal inheritance by silencing paternal transposons, and this phenomenon has been termed hybrid dysgenesis [22, 23]. In C. elegans, piRNAs can trigger a transgenerational inheritance of epigenetic memory in the germline through RNA-induced epigenetic gene silencing (RNAe) and RNA-induced epigenetic gene activation (RNAa) [7, 8, 24]. Genetically, functional loss of two of Argonaute proteins in C .elegans, ALG-3/4 and CSR-1, leads to a male spermatogenic arrest phenotype, which is similar to that of Miwi mutant mice [25, 26]. Interestingly, in the absence of paternal CSR-1 activity in C. elegans, the males displayed normal fertility initially but progressively became sterile over a period of five to six generations [27]. This phenomenon has been termed ‘germline mortal’, which is believed to result from disruptions of the Argonaute silencing pathways leading to a gradual loss of the ‘adapted’ epigenetic state reinforced by small-RNAs pathways [8, 28]. It remains unknown, however, whether this phenomenon exists in mammals. Given that Miwi mutant mice have a similar phenotype to that of CSR-1 mutant C. elegans, we sought to explore whether paternal MIWI and MIWI-associated pachytene piRNAs play a similar role in mice. In Miwi KO males, round spermatid production in the seminiferous tubules proceed up to step 4, which provides us with an opportunity to use steps 1–4 round spermatids to fertilize eggs through round spermatid injection (ROSI). In this study, we aimed to answer the following questions: (i) is the pachytene piRNA profile altered in Miwi-deficient round spermatids? (ii) Are the Miwi-deficient round spermatids competent for fertilization? (iii) If so, are the progeny derived from ROSI offspring using Miwi-deficient round spermatids fertile? (iv) Does paternal MIWI inactivation in mice phenocopy CSR-1 inactivation in C. elegans and display the ‘gremlin mortal’ phenomenon? Here, we report that MIWI deficiency indeed leads to altered pachytene piRNAs profiles and Miwi-deficient round spermatids can fertilize the eggs and produce normal offspring. Unlike C. elegans, the absence of paternal MIWI did not induce epigenetic ‘germline mortal’ phenotype in mice.
Results
The Pachytene piRNA Profile Is Altered in Miwi KO Round Spermatids
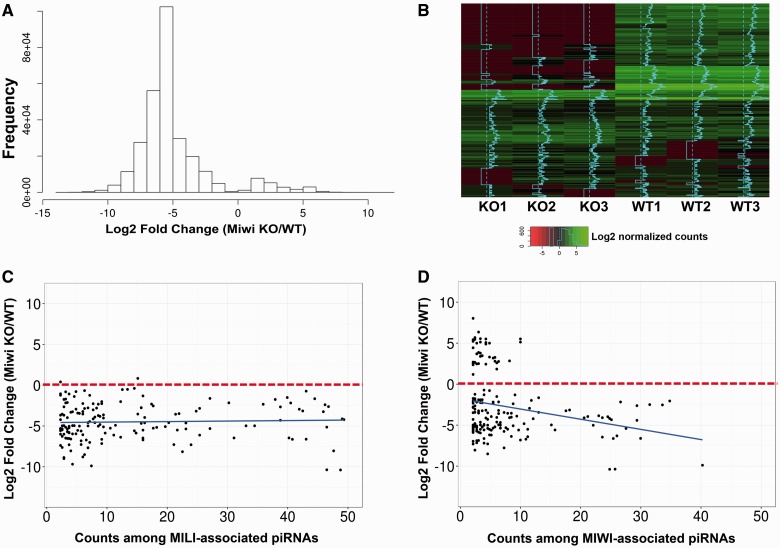
Previous reports have shown that global inactivation of Miwi leads to spermiogenic arrest at step 4 round spermatids, which is associated with dysregulation of the mRNA transcriptome and LINE1 transposon silencing [13, 21, 29]. To further investigate the function of Miwi in pachytene piRNA biogenesis, we purified round spermatids from Miwi KO and WT testes using the STA-PUT method [30] and performed small noncoding RNA deep sequencing (sncRNA-Seq) analyses. Annotation for piRNAs in the sncRNA-Seq reads revealed that levels of ∼90% of piRNAs in the Miwi KO round spermatids were drastically downregulated compared with those in WT round spermatids (Fig. 1A and B, Supplementary Fig. S1). We further analysed the dysregulated piRNAs by mapping these piRNA sequences to all known piRNA clusters collected in the piRNA Cluster Database [31] (Fig. 1C and D). Almost all of the MILI-associated piRNAs identified in Miwi KO round spermatids were downregulated (Fig. 1C, Supplementary Table S1), so were the majority of the MIWI-bound piRNAs identified in Miwi KO round spermatids (Fig. 1D, Supplementary Table S1). Similarly, we found that the number of dysregulated piRNAs in Miwi KO round spermatids that could mapped to piRNAs expressed in postnatal day 20 (P20) testes (enriched in pachytene piRNAs) was much greater that that matched to piRNAs expressed in P2 testes (abundant in pre-pachytene piRNAs), suggesting that the dysregulated piRNAs in Miwi KO round spermatids are mostly pachytene piRNAs (Supplementary Figure. S2). We also annotated other sncRNAs, including endo-siRNAs, miRNAs, mitochondrial small RNAs, rRNAs-derived small RNAs, small nucleolar RNAs (snoRNA), small nuclear RNAs (snRNA) and tRNA-derived small RNAs, in the sncRNA-Seq reads (Supplementary Fig. S3). Interestingly, we found a large number of miRNAs and endo-siRNAs were upregulated in Miwi KO round spermatids (Supplementary Fig. S3 and Table S1).
Figure 1:
Dysregulated pachytene piRNAs in Miwi KO round spermatids. (A) Fold change distribution of dysregulated piRNAs in Miwi KO round spermatids. Note that ∼90% of the piRNAs were downregulated in Miwi KO round spermatids compared with WT controls. (B) Heatmap showing expression levels of 1000 randomly chosen piRNAs in Miwi KO and WT round spermatids. The average log2 values of normalized piRNA counts in sncRNA-Seq reads represent piRNA expression levels. SncRNA–Seq was performed in biological triplicate [i.e. 3 Miwi KO round spermatid samples (KO1-3) with each purified from 4 Miwi KO mice, and 3 WT round spermatid samples (WT1-3) with each purified from 3 WT mice]. (C) Scatter blot showing that almost all of the MILI-associated piRNAs identified in Miwi KO round spermatids were downregulated. (D) Scatter blot showing that the majority of MIWI-associated piRNAs in Miwi KO round spermatids were downregulated. Counts (>2) were plotted against log2 fold changes of piRNAs in Miwi KO round spermatids. All data points represent average values of samples in triplicate (n = 3).
Miwi KO Round Spermatids Can Fertilize Both WT and Miwi KO Oocytes and Support Embryonic Development to Term
Miwi KO females are fertile, whereas Miwi KO males are infertile due to defective spermatogenesis [13]. The most advanced spermatogenic cells in the testes of Miwi KO males are step 4 round spermatids (Fig. 2A–D). To assess effects of paternal piRNAs, we next performed ROSI and evaluated the fertilization efficiency and developmental potential by counting the number of fertilized eggs and pre-implantation embryos that reached 2-cell and blastocyst stages. We found that Miwi KO round spermatids could fertilize both WT and Miwi KO oocytes, and no differences in pre-implantation embryonic development (from 2-cell to blastocyst stages) were observed between the WT and Miwi KO ROSI groups (Tables 1 and 2). To evaluate the post-implantation development of the embryos derived from ROSI using Miwi KO round spermatids, we transferred the 2-cell embryos into the oviduct of surrogate mothers and allowed them to develop to term. When Miwi KO round spermatids were injected into WT oocytes, ∼7.6% of transferred 2-cell embryos developed into live offspring (Fig. 2E–F), showing no significant difference compared with the rate of WT ROSI s (∼9.6% of transferred two-cells developed into live offspring) (Table 1). Similarly, when Miwi KO round spermatids were injected into Miwi KO oocytes, ∼9.6% of transferred 2-cell embryos led to live-born pups, and this rate was also comparable to that of injection of WT round spermatids into Miwi KO oocytes (∼8.3% birth rate) (Table 2). Overall, our ROSI results suggest that Miwi KO round spermatids, despite significantly downregulated pachytene piRNAs, could fertilize the egg and support embryonic development to term, with a similar efficiency compared with WT round spermatids.
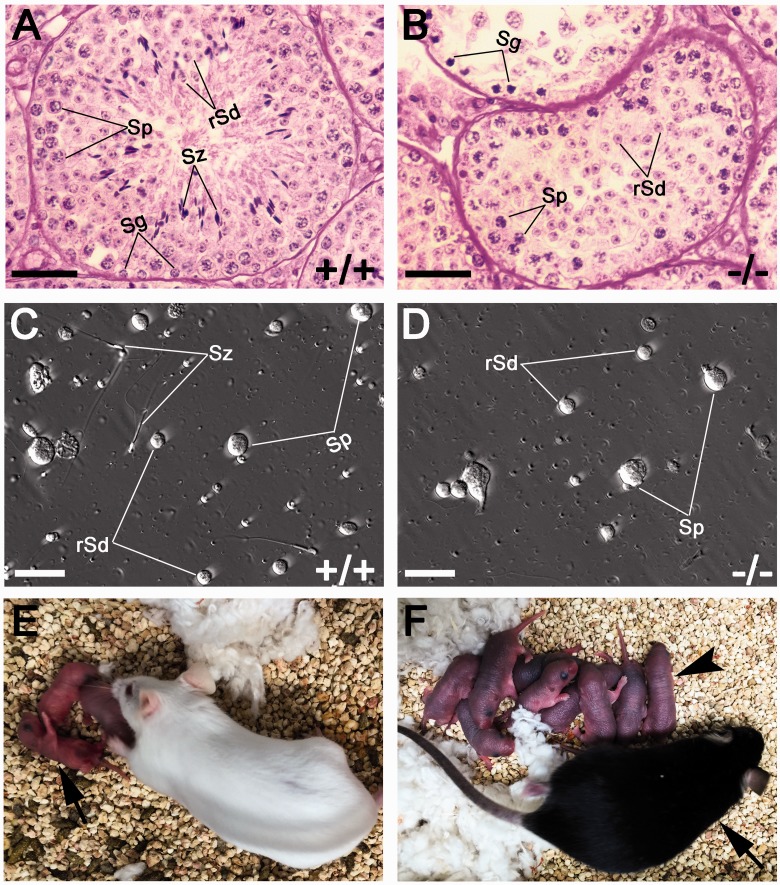
Figure 2:
Spermiogenic arrest in Miwi KO testes. (A) PAS staining of a WT stage IV seminiferous tubule showing normal spermatogenesis. (B) PAS staining of seminiferous tubules of Miwi KO testes showing spermiogenic arrest at step 4. (C) Phase-contrast image showing all stages of germ cells are present in WT testis suspension. (D) Phase-contrast image of Miwi KO testis suspension showing absence of elongated spermatids. (E) Live offspring produced through ROSI using Miwi KO round spermatids and WT oocytes. Arrow points to F0 offspring. (F) A ROSI-derived F1 adult mouse (arrow) and his viable F2 progeny (arrowhead). Sg, Spermatogonia; Sp, spermatocytes; rSd, round spermatids; Sz, spermatozoa. Scale bar: 50 µm in (A) and (B), 25 µm in (C) and (D).
Table 1:
Fertilization and development of WT oocytes after ROSI using WT and Miwi KO round spermatids
| Genotype of round spermatids injected | Total no. oocytes injected (No. exp.) | Experimental series 1 |
Experimental series 2 |
|||||
|---|---|---|---|---|---|---|---|---|
| Number of preimplantation embryos (%) |
No. 2-cell embryos transferred (No. exp.) | No. recipients | No. live born pups (%) | Offspring genotype | ||||
| Fertilized eggs (%) | 2-cell embryos (%) | Blastocysts (%) | ||||||
| WT | 140 (5) | 119 (85.0)a | 106 (89.1)a | 27 (25.5)a | 104 (4) | 4 | 10 (9.6)a | Miwi +/+ |
| Miwi KO | 169 (5) | 146 (86.4)a | 129 (88.4)a | 35 (27.1)a | 158 (4) | 7 | 12 (7.6)a | Miwi +/- |
Note: Fertilized eggs refer to 2PN + PN eggs after ROSI. Statistical analyses were conducted using χ2 test between WT and Miwi KO group. Values with the same superscripts have no statistic difference (P > 0.05).
Table 2:
Fertilization and development of Miwi KO oocytes after ROSI using WT and Miwi KO round spermatids
| Genotype of round spermatids injected | Total no. oocytes injected (no. exp.) | Experimental series 1 |
Experimental series 2 |
|||||
|---|---|---|---|---|---|---|---|---|
| No. (%) of |
No. of 2-cell embryos transferred (no. exp.) | No. of recipients | No. (%) of live offspring | Offspring genotype | ||||
| Fertilized eggs (% of) | 2-cell (% of) | Blastocyst (% of) | ||||||
| WT | 65 (3) | 54 (83.1)a | 46 (85.2)a | 13 (24.1)a | 48 (3) | 3 | 4 (8.3)a | Miwi +/− |
| Miwi KO | 70 (3) | 58 (82.9)a | 51 (87.9)a | 15 (29.4)a | 73 (3) | 4 | 7 (9.6)a | Miwi−/− |
Note: Fertilized egg is defined as 2PN + PN eggs after ROSI. Five experiments were carried out for WT and miwi KO ROSI. Statistical analyses were conducted using χ2 test between WT and miwi KO group. Values with the same superscripts are no any significantly differences (P > 0.05).
ROSI Progeny Display Normal Development and Fertility
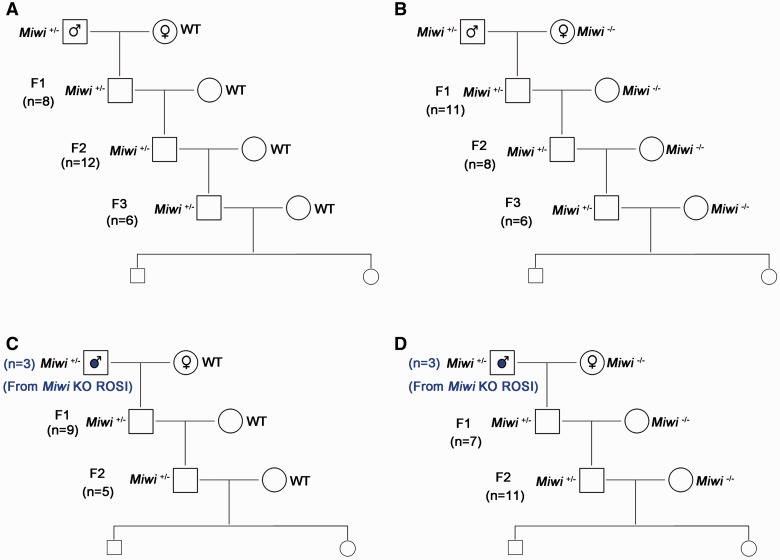
To determine whether the ‘germline mortal’ phenomenon also exists in the progeny of Miwi mutant mice, we set up four types of breeding schemes, i.e. Miwi+/− males derived from natural breeding mated with WT females (Fig. 3A) or Miwi−/− females (Fig. 3B), and Miwi+/− males derived from Miwi KO ROSI mated with WT females (Fig. 3C) or Miwi−/− females (Fig. 3D). Interestingly, unlike C. elegans Csr-1 mutants, the Miwi+/− male offspring do not display fertility decrease in all four breeding schemes tested (Fig. 3), and the F1, F2 and F3 generations of Miwi+/− males derived from natural mating all exhibited normal litter size and litter interval (Supplementary Table S2). Moreover, the Miwi+/− males derived from Miwi KO round spermatids injection were all fertile when they were bred with either WT or Miwi−/− females. Similarly, all progeny derived from ROSI using the Miwi KO round spermatid displayed normal fertility. Collectively, these breeding data suggest that the progeny of Miwi KO round spermatids all have normal fertility, and aberrant paternal pachytene piRNA profiles do not appear to cause the ‘germline mortal’ phenomenon in mice.
Figure 3:
Normal fertility of Miwi heterozygous progeny derived from both natural and ROSI founders in various breeding schemes. (A) The male Miwi heterozygous F1, F2, and F3 progeny derived from Miwi +/− males mated with WT females all displayed normal fertility. (B) The male Miwi heterozygous F1, F2, and F3 progeny derived from Miwi +/− males mated with Miwi−/− females displayed normal fertility. (C) The male Miwi heterozygous F1, F2 and F3 progeny derived from Miwi KO ROSI mated with WT females all exhibited normal fertility. (D) The male Miwi heterozygous F1, F2, and F3 progeny derived from Miwi KO ROSI males mated with Miwi−/− females all exhibited normal fertility. n” denotes the total number of offspring studied in each of mating schemes (A–D).
Discussion
Unlike pre-pachytene piRNAs that are mainly derived from repetitive sequences, pachytene piRNAs are mostly encoded by several huge piRNA gene clusters in the genome in mice [13, 21, 29]. In addition to MIWI and MILI, several other proteins have been found to be involved in pachytene piRNA biogenesis as well, including tudor-domain proteins (TDRD 1–5), MOV10L1, GASZ, Maelstrom (MAEL) [20, 32–39]. Interestingly, inactivation of Mov10l1 completely abolishes pachytene piRNA biogenesis [38, 39], whereas Miwi KO round spermatids appear to be able to still produce pachytene piRNAs, but at drastically reduced levels, based on the present study. Therefore, Miwi KO round spermatids are not totally lacking pachytene piRNAs, but rather containing an aberrant pachytene piRNA profile. The fact that spermiogenesis is arrested at step 4 in Miwi KO males suggests that a normal piRNA profile is essential for the progress of spermiogenesis beyond step 4. Two previous studies have already shown that the Miwi KO testes display an accumulation of transcripts expressed abundantly in haploid spermatids, which suggest that pachytene piRNAs may be responsible for eliminating those haploid transcripts by targeted degradation through binding to the 3’UTRs, a manner like miRNAs-induced mRNA degradation [40, 41]. However, it remains puzzling that both those pachytene piRNAs and the spermiogenic transcripts are expressed as early as the pachytene spermatocyte stage, but no degradation occurs. Nevertheless, the pachytene piRNA profile is altered in Miwi KO round spermatids.
Pachytene piRNAs have been shown to be encoded by several large piRNA gene clusters based on RIP-Seq analyses using MIWI and MILI antibodies [15, 29]. The fact that both MIWI- and MILI-bound piRNAs identified in Miwi KO round spermatids are mostly downregulated suggests that Miwi inactivation mainly implicates pachytene piRNAs. However, many of the piRNAs identified in Miwi KO round spermatids are not included in the MIWI- or MILI-bound piRNA databases. The finding that dysregulated piRNAs in Miwi KO round spermatids are mostly mapped to piRNAs expressed in P20 (enriched in pachytene spermatocytes and first appearance of round spermatids), rather than P2 (spermatogonia as the only male germ cell type) testes, further supports that it is the pachytene piRNAs that are dysregulated in the absence of MIWI in round spermatids. Another interesting finding is that Miwi inactivation not only affects pachytene piRNA production, but also causes upregulation of many miRNAs and endo-siRNAs. It is likely that this effect is secondary to the overly downregulated pachytene piRNA production given that MIWI is well documented to bind and regulate pachytene piRNA biogenesis [13, 15, 29]. But this result indeed suggests a connection between pachytene piRNAs and miRNAs/endo-siRNAs. Given that miRNAs/endo-siRNAs are mostly involved in post-transcriptional regulation, our data further support notions from previous studies suggesting that pachytene piRNAs also function as a post-transcriptional regulator [15, 21, 29]. It remains an interesting topic to dissect the relationship between pachytene piRNAs and miRNAs/endo-siRNAs in the control of gene expression during spermatogenesis.
Despite the aberrant piRNA profile in Miwi KO round spermatids, ROSI revealed similar fertilization rates and embryonic development potential between WT and Miwi KO round spermatids. Interestingly, Mael-null round spermatids are also capable of fertilizing eggs and supporting embryonic development [33]. Given the similar spermiogenic arrest phenotype in both Miwi and Mael KO males, these data indicate that normal pachytene piRNA profiles are not essential for fertilization and embryonic development.
Pachytene piRNAs appear to have a role in epigenetic inheritance in C. elegans [42, 43]. Miwi KO males display a spermatogenic arrest at the round spermatid stage, which is similar to that of Argonaute proteins ALG-3/4 and CSR-1 mutant C. elegans [13, 27]. Interestingly, the heterozygous offspring derived from Alg-3/4 or Csr-1 homozygous males exhibit reduced fertility and repeatedly backcrossing heterozygous to homozygous leads to the ‘germline-mortal’ phenomenon, i.e. a progressive loss of fertility after several generations [27]. These findings suggest that ALG-3/4 and CSR-1 and their associated piRNAs are required for the transgenerational inheritance of paternal epigenetic information. Much like ALG-3/4-associated 26G-RNAs, mouse MIWI-associated pachytene piRNAs are expressed exclusively in spermatocytes and spermatids [5]. Surprisingly, we did not observe the ‘germline-mortal’ phenomenon in heterozygous progeny derived from ROSI offspring using Miwi KO round spermatids in this study. There are two possible explanations for the lack of paternal piRNA-mediated epigenetic inheritance in mice. First, the epigenetic regulation by the Argonaute/small RNA pathway may be different, as C. elegans employs AGO proteins to protect paternal gene expression from silencing by interactions between an RNAa allele and an RNAe allele. In contrast, PIWI proteins are used for silencing transposons and mRNA transcripts in mice [14, 40, 41]. Second, mice may have much more complex genetic and epigenetic regulations compared with C. elegans. It will be interesting in the future to determine whether other PIWI family/Argonaute members have a role in paternal piRNA-mediated epigenetic inheritance in mice.
Previous reports have shown that in Miwi KO round spermatids, the retrotransposon (LINE1) is upregulated, which is accompanied by increased DNA double strand breaks [13, 21, 29]. Normal fertilization and development of embryos derived from Miwi KO ROSI suggest that LINE1 activation in Miwi KO round spermatids might have been suppressed by the global genomic reprogramming during fertilization and pre-implantation embryonic development. In Drosophila, maternally deposited piRNAs are important for mounting an effective silencing response and can epigenetically mediate transposon suppression in the offspring, and a lack of maternal piRNA inheritance appears to lead to hybrid dysgenesis [22]. Interestingly, in mice, maternal piRNAs seems to have nothing to do with transposable element silencing or epigenetic regulation because female mice deficient in either of the PIWI family proteins (i.e. MIWI, MILI and MIWI2) all display normal developmental competence and fertility [12, 13, 44]. Our ROSI results, together with earlier data [33], demonstrate that neither paternal nor maternal normal pachytene piRNAs are required for fertilization and embryonic development. Furthermore, the progeny of offspring derived from ROSI using Miwi KO round spermatids all displayed normal development and fertility for at least three generations, suggesting a lack of transgenerational effects when paternal piRNAs are dysregulated.
In summary, our data provides physiological evidence showing that the MIWI-/MILI-bound pachytene piRNAs may not be essential for normal fertilization and embryonic development in mice. Unlike Argonaute proteins CSR-1 or ALG-3/4 in C. elegans, a loss of paternal MIWI activity dose not lead to the ‘germline-mortal’ phenotype in mice.
Materials and Methods
Animals
All animal work was performed following the protocol approved by the Institutional Animal Care and Use Committee of the University of Nevada, Reno. Mice were housed and maintained under specific pathogen-free conditions with a temperature- and humidity-controlled animal facility in the University of Nevada, Reno. Global Miwi KO mouse line was recovered using cryopreserved sperm, purchased from the Mutant Mouse Resource & Research Center (Item no. 029995-MU-SPERM) in the Genetic Engineering Center of the University of Nevada, Reno. All mice used in this study were on the C57BL/6J background.
Chemicals and Media
All chemicals in this study were purchased from Sigma (St Louis, MO) unless otherwise stated. For collecting oocytes and round spermatids, a modified CZB-HEPES medium containing 20mM HEPES-Na, 5mM NaHCO3, and 0.1 mg/ml polyvinyl alcohol (cold water soluble) was used. For culturing oocytes before ROSI, a CZB medium supplemented with 5.56mM D-glucose and 4mg/ml BSA (Fraction V, Calbiochem, Temecula, CA) was used as described [45, 46]. For culturing fertilized embryos after ROSI, the KSOM medium supplemented with amino acids (KSOM + AA, Cat no. MR-121-D, Millipore, Temecula, CA) was used.
Histological Analysis
Testes were dissected from WT or Miwi KO mice. After fixation in the Bouin’s solution overnight at 4 °C, the samples were embedded into paraffin. Sections (5 µm) were cut and then stained with the Periodic acid Schiff and Hematoxylin (Sigma) for histological analyses.
Preparation of Oocytes
Six to eight-week-old WT and Miwi KO female mice were each injected with 5 IU of pregnant mare’s serum gonadotropin, followed by injection of 5 IU of human chorionic gonadotropin (hCG) 48 h apart. Mature oocytes were collected from oviducts 14–16 h after hCG injection and cumulus cells were removed by treatment with 0.1% bovine testicular hyaluronidase in HEPES-CZB at 37 °C for 2–3 min. Cumulus-free oocytes were rinsed and kept in CZB at 37 °C under 5% CO2 in air before ROSI.
Round Spermatid Injection
ROSI was performed as described [47, 48] with slight modifications. Briefly, testicular cell suspension was diluted in HEPES-CZB containing 1% (w/v) polyvinyl pyrrolidone on the injection dish. Round spermatids were identified by size and morphology in WT or Miwi KO testicular cell suspension. Individual round spermatids were injected into the oocytes. The total duration of ROSI was no longer than 2 h. The oocytes were activated shortly after injection by incubation in Ca2+-free CZB medium supplemented with 10 mM SrCl2 at 37°C, 5% CO2 for 4 h, after which they were transferred into KSOM + AA medium for subsequent culture. Fertilization was confirmed after 6–8 h of injection, and embryonic development following ROSI was assessed every 24 h up to 5 days.
Embryo Transfer to Surrogate Mothers
To examine the developmental potential of ROSI embryos, two-cell embryos (15–20) were transferred into the oviducts of a pseudopregnant CD1 female as described [47]. Surrogate mothers were subjected to caesarian section on day 20 of pregnancy and the pups were transferred to foster mothers. All offspring from Miwi KO round spermatids were genotyped and some raised by foster mothers until weaning, were allowed to mature and breed.
Small RNA Sequencing
Round spermatids were purified from WT and Miwi KO adult testes using a mini-STA-PUT method [30] and the purity was >95% based microscopic examination, as described previously [49]. Small RNA was isolated from round spermatids using the mirVana RNA isolation kit (Ambion) according to the manufacturer’s instructions. RNA quality and quantity were assessed using the Agilent 2100 Bioanalyzer. Small RNA libraries were prepared using the Ion Total RNA-Seq Kit v2 (Invitrogen) followed by deep sequencing using an Ion Proton sequencer (Life Technologies), as described previously [49–52]. SncRNA–Seq was performed in biological triplicate, i.e., 3 Miwi KO round spermatid samples with each purified from 4 Miwi KO mice, and 3 wild-type (WT) round spermatid samples with each purified from 3 WT mice.
Bioinformatic Analysis
The Miwi- and Mili-bound piRNA cluster datasets were downloaded from the piRNA Cluster Database (http://www.smallrnagroup.uni-mainz.de/piRNAclusterDB.html) [31], and used as the reference piRNA cluster datasets. The sncRNA-Seq reads were mapped to the reference datasets using our in-house small RNA alignment software, Sequery v.2 [53, 54]. The mapped counts were normalized based on the sequencing depth (counts per million reads) using DESeq2. Both normalization and differential expression analyses were performed using DESeq2 (significant changes were defined as P < 0.05) [55]. Log2 values of fold change of dysregulated piRNAs in Miwi KO round spermatids were plotted against their counts in MIWI- and MILI-bound piRNA clusters using the R script of the ggplot2 package. The Miwi KO and WT round spermatid sncRNA-Seq data have been deposited into the GEO database with an accession number of GSE86319.
Statistic Analysis
All data were presented as mean ± SEM, and statistical differences between datasets were assessed by one-way ANOVA or t-test using the SPSS16.0 software. P value ≤ 0.05 was considered as significant differences, and P value ≤ 0.01 was considered as highly significant differences between groups compared. ROSI data were analysed using χ2 tests, compared with the WT group, and P value ≤ 0.05 was regarded as significant differences.
Supplementary Material
Acknowledgements
This study was supported, in part, by grants from the NIH (HD060858, HD071736 and HD085506 to W. Y.) and the Templeton Foundation (PID: 50183 to WY).
Supplementary data
Supplementary data is available at EnvEpig online.
Conflict of interest statement. None declared.
References
- 1. Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell 2007;26:611–23. [DOI] [PubMed] [Google Scholar]
- 2. Hutvagner G, Simard Mj. Argonaute proteins: key players in RNA silencing. Nat Rev Mol Cell Biol 2008;9:22–32. [DOI] [PubMed] [Google Scholar]
- 3. Hock J, Meister G. The Argonaute protein family. Genome Biol 2008;9:210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aravin Aa, Hannon Gj, Brennecke J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007;318:761–4. [DOI] [PubMed] [Google Scholar]
- 5. Girard A, Sachidanandam R, Hannon Gj, Carmell Ma. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 2006;442:199–202. [DOI] [PubMed] [Google Scholar]
- 6. Cox Dn, Chao A, Baker J, Chang L, Qiao D, Lin H. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 1998;12:3715–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shirayama M, Seth M, Lee Hc, Gu W, Ishidate T, Conte D, Jr, Mello Cc. piRNAs initiate an epigenetic memory of nonself RNA in the C. elegans germline. Cell 2012;150:65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Seth M, Shirayama M, Gu W, Ishidate T, Conte D, Jr, Mello Cc. The C. elegans CSR-1 argonaute pathway counteracts epigenetic silencing to promote germline gene expression. Dev Cell 2013;27:656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bao J, Yan W. Male germline control of transposable elements. Biol Reprod 2012;86:162, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kuramochi-Miyagawa S, Kimura T, Yomogida K, Kuroiwa A, Tadokoro Y, Fujita Y, Sato M, Matsuda Y, Nakano T. Two mouse piwi-related genes: miwi and mili. Mech Dev 2001;108:121–33. [DOI] [PubMed] [Google Scholar]
- 11. Bao J, Zhang Y, Schuster As, Ortogero N, Nilsson Ee, Skinner Mk, Yan W. Conditional inactivation of Miwi2 reveals that MIWI2 is only essential for prospermatogonial development in mice. Cell Death Differ 2014;21:783–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carmell Ma, Girard A, van de Kant Hj, Bourc'his D, Bestor Th, de Rooij Dg, Hannon Gj. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 2007;12:503–14. [DOI] [PubMed] [Google Scholar]
- 13. Deng W, Lin H. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2002;2:819–30. [DOI] [PubMed] [Google Scholar]
- 14. Siomi Mc, Sato K, Pezic D, Aravin Aa. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol 2011;12:246–58. [DOI] [PubMed] [Google Scholar]
- 15. Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory Bd, Mourelatos Z. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 2012;19:773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fu Q, Wang Pj. Mammalian piRNAs: Biogenesis, function, and mysteries. Spermatogenesis 2014;4:e27–89.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein Mj, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006;442:203–07. [DOI] [PubMed] [Google Scholar]
- 18. Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai Rs. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 2009;16:639–46. [DOI] [PubMed] [Google Scholar]
- 19. Wang J, Saxe Jp, Tanaka T, Chuma S, Lin H. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 2009;19:640–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 2009;17:775–87. [DOI] [PubMed] [Google Scholar]
- 21. Grivna St, Pyhtila B, Lin H. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci U S A 2006;103: 13415–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brennecke J, Malone Cd, Aravin Aa, Sachidanandam R, Stark A, Hannon Gj. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 2008;322:1387–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grentzinger T, Armenise C, Brun C, Mugat B, Serrano V, Pelisson A, Chambeyron S. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Res 2012;22:1877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ashe A, Sapetschnig A, Weick Em, Mitchell J, Bagijn Mp, Cording Ac, Doebley Al, Goldstein Ld, Lehrbach Nj, Le Pen J, et al. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell 2012;150:88–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Claycomb Jm, Batista Pj, Pang Km, Gu W, Vasale Jj, van Wolfswinkel Jc, Chaves DA, Shirayama M, Mitani S, Ketting Rf, et al. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 2009;139:123–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Conine Cc, Batista Pj, Gu W, Claycomb Jm, Chaves DA, Shirayama M, Mello Cc. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci U S A 2010;107:3588–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conine Cc, Moresco Jj, Gu W, Shirayama M, Conte D, Jr., Yates Jr, 3rd, Mello Cc. Argonautes promote male fertility and provide a paternal memory of germline gene expression in C. elegans. Cell 2013;155:1532–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buckley Ba, Burkhart Kb, Gu Sg, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature 2012;489:447–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Reuter M, Berninger P, Chuma S, Shah H, Hosokawa M, Funaya C, Antony C, Sachidanandam R, Pillai Rs. Miwi catalysis is required for piRNA amplification-independent LINE1 transposon silencing. Nature 2011;480:264–67. [DOI] [PubMed] [Google Scholar]
- 30. Bellve Ar. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 1993;225:84–13. [DOI] [PubMed] [Google Scholar]
- 31. Rosenkranz D. piRNA cluster database: a web resource for piRNA producing loci. Nucleic Acids Res 2016;44:D223–D30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ma L, Buchold Gm, Greenbaum Mp, Roy A, Burns Kh, Zhu H, Han Dy, Harris Ra, Coarfa C, Gunaratne Ph, et al. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet 2009;5:e1000635.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Castaneda J, Genzor P, van der Heijden Gw, Sarkeshik A, Yates Jr, 3rd, Ingolia Nt, Bortvin A. Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. Embo J 2014;33:1999–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zheng K, Wang Pj. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 2012;8:e10030–38.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pek Jw, Anand A, Kai T. Tudor domain proteins in development. Development 2012;139:2255–66. [DOI] [PubMed] [Google Scholar]
- 36. Saxe Jp, Chen M, Zhao H, Lin H. Tdrkh is essential for spermatogenesis and participates in primary piRNA biogenesis in the germline. Embo J 2013;32:1869–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 2011;20:364–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Frost Rj, Hamra Fk, Richardson Ja, Qi X, Bassel-Duby R, Olson En. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci U S A 2010;107:11847–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin Kj, Stark A, Sachidanandam R, Pillai Rs, Wang Pj. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci U S A 2010;107:11841–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang P, Kang Jy, Gou Lt, Wang J, Xue Y, Skogerboe G, Dai P, Huang Dw, Chen R, Fu Xd, et al. MIWI and piRNA-mediated cleavage of messenger RNAs in mouse testes. Cell Res 2015;25:193–07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Watanabe T, Cheng Ec, Zhong M, Lin H. Retrotransposons and pseudogenes regulate mRNAs and lncRNAs via the piRNA pathway in the germline. Genome Res 2015;25:368–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ku Hy, Lin H. PIWI proteins and their interactors in piRNA biogenesis, germline development and gene expression. Natl Sci Rev 2014;1:205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Peng Jc, Lin H. Beyond transposons: the epigenetic and somatic functions of the Piwi-piRNA mechanism. Curr Opin Cell Biol 2013;25:190–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kuramochi-Miyagawa S, Kimura T, Ijiri Tw, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 2004;131:839–49. [DOI] [PubMed] [Google Scholar]
- 45. Chatot Cl, Lewis Jl, Torres I, Ziomek Ca. Development of 1-cell embryos from different strains of mice in CZB medium. Biol Reprod 1990;42:432–40. [DOI] [PubMed] [Google Scholar]
- 46. Yanagimachi R, Wakayama T, Kishikawa H, Fimia Gm, Monaco L, Sassone-Corsi P. Production of fertile offspring from genetically infertile male mice. Proc Natl Acad Sci U S A 2004;101:1691–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biol Reprod 1995;52:709–20. [DOI] [PubMed] [Google Scholar]
- 48. Yuan S, Tang C, Zhang Y, Wu J, Bao J, Zheng H, Xu C, Yan W. mir-34b/c and mir-449a/b/c are required for spermatogenesis, but not for the first cleavage division in mice. Biol Open 2015;4:212–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu Q, Song R, Ortogero N, Zheng H, Evanoff R, Small Cl, Griswold Md, Namekawa Sh, Royo H, Turner Jm, Yan W. The RNase III enzyme DROSHA is essential for microRNA production and spermatogenesis. J Biol Chem 2012;287: 25173–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schuster A, Skinner Mk, Yan W. Ancestral vinclozolin exposure alters the epigenetic transgenerational inheritance of sperm small noncoding RNAs. Environ Epigenet 2016;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yuan S, Schuster A, Tang C, Yu T, Ortogero N, Bao J, Zheng H, Yan W. Sperm-borne miRNAs and endo-siRNAs are important for fertilization and preimplantation embryonic development. Development 2016;143:635–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ortogero N, Schuster As, Oliver Dk, Riordan Cr, Hong As, Hennig Gw, Luong D, Bao J, Bhetwal Bp, Ro S, et al. A novel class of somatic small RNAs similar to germ cell pachytene PIWI-interacting small RNAs. J Biol Chem 2014;289: 32824–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ortogero N, Hennig Gw, Luong D, Yan W. Computer-assisted annotation of small RNA transcriptomes. Methods Mol Biol 2015;1218:353–64. [DOI] [PubMed] [Google Scholar]
- 54. Ortogero N, Hennig Gw, Langille C, Ro S, McCarrey Jr, Yan W. Computer-assisted annotation of murine Sertoli cell small RNA transcriptome. Biol Reprod 2013;88:3.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550 http:// download. springer. com/ static/ pdf/ 68/ art% 253A10. 1186% 252Fs13059- 014- 0550- 8. pdf? originUrl= http% 3A% 2F% 2Fgenomebiology. biomedcentral. com% 2Farticle% 2F 10.11186% 2Fs13059- 014- 0550- 8& token2= exp= 1476470239~acl= %2Fstatic% 2Fpdf% 2F68% 2Fart% 25253 A10. 1186% 25252Fs13059- 014- 0550- 8. pdf*~ hmac= 23dc02970c 4518ec697 821ba7f31b74067 0e31a27839c33871dd3f 2585a2 3cb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.