Abstract
Background:
Despite a high prevalence of deleterious missense variants, most studies of RAD51C ovarian cancer susceptibility gene only provide in silico pathogenicity predictions of missense changes. We identified a novel deleterious RAD51C missense variant (p.Arg312Trp) in a high-risk family, and propose a criteria to prioritise RAD51C missense changes qualifying for functional analysis.
Methods:
To evaluate pathogenicity of p.Arg312Trp variant we used sequence homology, loss of heterozygosity (LOH) and segregation analysis, and a comprehensive functional characterisation. To define a functional-analysis prioritisation criteria, we used outputs for the known functionally confirmed deleterious and benign RAD51C missense changes from nine pathogenicity prediction algorithms.
Results:
The p.Arg312Trp variant failed to correct mitomycin and olaparib hypersensitivity and to complement abnormal RAD51C foci formation according to functional assays, which altogether with LOH and segregation data demonstrated deleteriousness. Prioritisation criteria were based on the number of predictors providing a deleterious output, with a minimum of 5 to qualify for testing and a PredictProtein score greater than 33 to assign high-priority indication.
Conclusions:
Our study points to a non-negligible number of RAD51C missense variants likely to impair protein function, provides a guideline to prioritise and encourage their selection for functional analysis and anticipates that reference laboratories should have available resources to conduct such assays.
Keywords: ovarian cancer, whole exome sequencing, RAD51C, missense variant, functional characterisation, pathogenicity prediction
Epithelial ovarian cancer (EOC) is the most deadly of gynaecological malignancies and the fifth most frequent cause of cancer death in women in developed countries (Siegel et al, 2013). Family history of ovarian cancer is one of the strongest risk factors for the disease. Germline deleterious mutations in the BRCA1 and BRCA2 and in the mismatch-repair (MMR) MLH1, MSH2, MSH6, PMS2 genes confer high risk of ovarian cancer while recently described rare mutations in the RAD51C, RAD51D and BRIP1 genes have been associated with moderate risk of ovarian cancer. Eighteen additional common low-penetrance variants have also been identified but altogether, the known genetic risk factors account for less than 50% of the excess familial ovarian cancer risk (Jervis et al, 2014; Kuchenbaecker et al, 2015; Ramus et al, 2015).
RAD51C is one of the novel ovarian cancer moderate-susceptibility genes described in the last few years. In 2010 Meindl et al found six RAD51C deleterious germline mutations (two frameshift, two splice-site and two non-functional missense variants) in 480 German families with occurrence of both breast and ovarian cancer. This accounted for a frequency of 1.3% in such families and for an overall frequency of 0.55% when also breast cancer-only pedigrees were considered. The two missense variants c.374G>T (p.Gly125Val) and c.414G>C (p.Leu138Phe) were demonstrated to be deleterious by performing in vitro functional complementation assays evaluating cell survival and RAD51 foci formation after mitomycin (MMC) treatment. Of eight additional missense variants, four, c.7G>A (p.Gly3Arg), c.376G>A (p.Ala126Thr), c.506T>C (p.Val169Ala), c.791G>T (p.Gly264Val), did not alter RAD51C function and four, c.475G>A (p.Asp159Asn), c.1097G>A (p.Arg366Gln), c.790G>A (p.Gly264Ser), c.859A>G (p.Thr287Ala), showed partial loss of function and their significance could not be unequivocally determined.
Studies conducted in different populations after this initial report generated some controversy. Some of them completely failed to detect obvious protein truncating RAD51C mutations, while others reported lower prevalence of loss-of-function alterations compared to the frequency described by Meindl et al (2010). In 2011 Clague et al identified one likely deleterious mutation, c.458G>A (p.Gly153Asp), as determined through functional yeast two-hybrid analysis of RAD51C interaction with XRCC3 and RAD51B. In a subsequent study carried out by our group that included functional complementation assays, we detected a total of five RAD51C deleterious variants in 785 Spanish breast and/or ovarian cancer families, three of them consisting of amino acid substitutions, c.404G>A (p.Cys135Tyr), c.428A>G (p.Gln143Arg), c.656T>C (p.Leu219Ser) (Osorio et al, 2012). Four out of the five loss-of-function missense changes were found in breast and ovarian cancer families (4 of 300, 1.3%) reproducing the prevalence reported by Meindl et al. In view of these findings we highlighted the predominance of RAD51C pathogenic missense variants in breast and ovarian cancer families, and suggested that lack of functional assessment in other studies might account for the observed discrepancies. We recommended a wider implementation of functional tests to provide a more reliable evaluation of RAD51C significance in breast and ovarian cancer risk (Osorio et al, 2012).
However, despite the observed prevalence of RAD51C pathogenic missense variants, the increasing RAD51C ovarian cancer risk estimations (Pelttari et al, 2011; Loveday et al, 2012; Song et al, 2015) and the foreseen therapeutic impact for mutation carriers in relation to PARP inhibition (Min et al, 2013; Bajrami et al, 2014; Somyajit et al, 2015a), nearly all published studies up to date do not perform functional assays and constrain their scope to provide in silico pathogenicity predictions (Kumar et al, 2009; Adzhubei et al, 2010).
Here, by using different approaches that include a comprehensive functional characterisation, we report a novel deleterious missense RAD51C variant, c.934C>T (p. Arg312Trp), identified by whole exome sequencing (WES) in a Spanish non-BRCA1/2 high-risk breast and ovarian cancer family. Our finding provides yet another evidence of the need to functionally assess all likely pathogenic RAD51C missense variants. In order to outline a possible criteria to select such variants, and to estimate the number of potentially overlooked truly pathogenic changes already found in patients, we have carried out a comprehensive review of the functionally uncharacterised RAD51C missense variants reported in the last years. We have assessed the pathogenicity predictions from eight in silico predictive algorithms as well as the scores derived from the PredictProtein analysis tool (Yachdav et al, 2014). Taking into account the outputs from the so far functionally confirmed RAC51C deleterious and benign changes, we have proposed a prioritisation criteria for directing functional validation of RAD51C missense changes.
Materials and methods
Non-BRCA1/2 family for whole exome sequencing
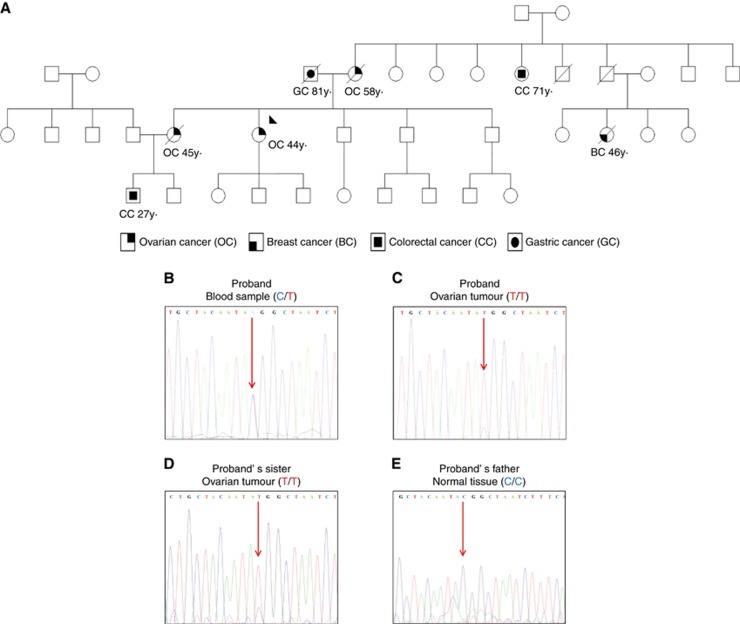
A non-BRCA1/2 breast and ovarian cancer family assessed at the CNIO Familial Cancer Clinical Unit (FCCU) was selected along with other families to search for novel ovarian cancer risk variants through WES. Whole exome sequencing study was approved by the Ethics Committee of the Instituto de Salud Carlos III (CEI PI 21-2013). In this specific family the proband, her mother and sister were affected at ages 44 years, 58 years and 45 years by high-grade endometrioid, endometrioid, and undifferentiated ovarian cancer, respectively. A proband’s cousin was affected by breast cancer at age 46 years. Other cancers in the family included a gastric carcinoma in the proband’s father (at age 81 years) and two colorectal carcinomas in a maternal aunt (at age 71 years) and in a nephew (at age 27 years.) who had been previously diagnosed as a carrier of a de novo APC mutation (mutation was discarded in his father and mother). All affected members with ovarian cancer were deceased except for the proband. Formalin-fixed paraffin embedded (FFPE) tumour samples were available for the proband, her sister and father (Figure 1). Tumours were provided by CNIO Biobank and Biobanco i+12 in the Hospital 12 de Octubre integrated in the Spanish Hospital Biobanks network (RetBioH; www.redbiobancos.es) following standard procedures with appropriate approval of the Ethical and Scientific Committees.
Figure 1.
Spanish non-BRCA1/2 high-risk breast and ovarian cancer family selected for Whole Exome Sequencing (WES) study. (A) Proband is highlighted by a black arrow. Individuals with tumours and ages of diagnosis are shown. (B) Electropherograms of RAD51C sequence spanning the c.934C>T variant show the presence of the c.934C>T variant in the proband’s blood sample. Sequence traces confirms loss of heterozygosity (LOH) in the proband’s (C) and her sister’s (D) ovarian tumours and RAD51C wild-type sequence in normal tissue from the proband’s father (E). A full colour version of this figure is available at the British Journal of Cancer journal online.
Case-control study: Kaspar genotyping
The RAD51C p.Arg312Trp variant was genotyped in cases (n=1490 probands from non-BRCA1/2 families from CNIO (n=613) and Hospital Clínico San Carlos (HCSC) (n=877)) and controls (n=646 healthy women from CNIO (n=327) and HCSC (n=319)) to find additional carriers and determine its frequency in Spanish general population. Genotyping was performed by the KBiosciences (Herts, UK) fluorescence-based competitive allele-specific PCR assay (KASPar). Details of the KASPar methodology can be found at http://www.kbioscience.co.uk/.
Whole exome sequencing
Genomic DNA from the proband of the selected non-BRCA1/2 family (Figure 1) was isolated from peripheral blood lymphocytes using FlexiGene DNA Kit (Qiagen, Hilden, Germany) and DNA concentration was determined using PicoGreen dsDNA quantification reagent (Invitrogen, Carlsbad, CA, USA). Exome from the sample was fully captured, enriched and sequenced using the SureSelect Human All Exon Kit for 51 Mb (Agilent Technologies, Santa Clara, CA, USA). Whole exome sequencing was performed in Sistemas Genómicos (Valencia, Spain) using the SOLiD 5500XL sequencing platform. ‘Paired end’ reads of 101 nt long were generated. Reads were aligned against the human reference genome version GRCh37/hg19. Read alignment was performed using BWA and ‘in-house’ scripts. In all, 96.27% of target regions were observed with coverage >20 ×. Variant calling was performed using a combination of two different algorithms: VarScan (Koboldt et al, 2009) and GATK (McKenna et al, 2010). Custom scripts were developed to combine and filter variants. Identified variants were annotated using the Ensembl (release 64) database (Flicek et al, 2012). We focused on single nucleotide substitutions and small insertions and deletions (indels) found in heterozygosity. Only rare variants (MAF<0.01) or novel variants (not described in dbSNP130, HapMap controls and 1000 Genomes data retrieved through Ensembl) were considered. Then, single nucleotide substitutions were filtered by discarding (I) variants in intergenic or intronic regions, (II) variants not mapping to the canonical isoforms, (III) variants predicted to produce synonymous amino acid changes (we only selected high/medium impact variants: frameshift indels and non-sense, missense and canonical splice site variants). We next selected (IV) variants with potential damaging effect based on in silico predictors (predicted to be pathogenic by at least SIFT and Polyphen-2) (V) in genes with a potential cancer-related function (with particular focus on DNA-repair pathways) that presented sufficient depth and quality.
Loss of heterozygosity (LOH) and segregation analysis
DNA was extracted from tumoural FFPE sections (with >80% tumour cellularity) and from normal FFPE tissue from the proband’s father using the DNeasy Blood & Tissue Kit (Qiagen). A sequence segment spanning the nucleotide change that originates the RAD51C p.Arg312Trp variant was amplified by PCR using the oligonucleotide pair RAD51C-F (5′-GATCAGAGGCGTTCTGAGAAAT-3′) and RAD51C-R (5′-ACATTTCTATGTTTGCTCTAGGTGA-3′). For loss of heterozygosity (LOH) assessment germline (from peripheral blood or normal tissue) and tumoural DNAs were amplified and sequenced with ABI sequencer 3730XL simultaneously. Loss of heterozygosity in tumours was scored by a significant reduction of at least 30% of the wild allele peak-height relative to the sequence trace from normal tissue.
Complementation of CL-V4B RAD51C-deficient cells
RAD51C-wild-type V79B and RAD51C-deficient CL-V4B Chinese hamster cells (Zdzienicka, 1987; Godthelp et al, 2002) were kindly provided by Drs Albert Pastink and Haico van Attikum (Leiden University Medical Center, Leiden, the Netherlands). Both cell lines were cultured in DMEM and HAM-F10 medium respectively, at 37 °C in a humidified 5% CO2 atmosphere. A DNA sequence consisting of the full-length human RAD51C cDNA (Ensembl Transcript ID ENST00000337432, RAD51C-001) flanked by XbaI and BamHI restriction sites was designed, synthesised and purchased from IDT Technologies, and directionally cloned into the pUltra lentiviral vector that contains GFP as selection marker (Addgene, Cambridge, MA, USA). The c.934C>T nucleotide change originating the p.Arg312Trp variant was then introduced in the RAD51C-wild-type lentiviral vector (RAD51C.WT) by using the Quick Change-Site directed Mutagenesis kit (Agilent Technologies) with the forward 5′-GGGACATGCTGCTACAATATGGCTAATCTTTCATTGGGAC-3′ and reverse 5′-GTCCCAATGAAAGATTAGCCATATTGTAGCAGCATGTCCC-3′oligonucleotides. This modified vector was designated as RAD51C.p.Arg312Trp. The RAD51C.WT and RAD51C.p.Arg312Trp lentiviral vectors were co-transfected with the VSV-G envelope and PAX packaging plasmids (Addgene) into 293FT cells (Invitrogen) in the presence of Fugene 6 (Promega, Madison, WI, USA) and lentivirus-containing supernatants were collected after 36 h of transfection and filtered. Then the RAD51C-deficient CL-V4B cells were infected with RAD51C.WT and RAD51C.p.Arg312Trp-expressing viruses as well as with viruses containing the empty p-Ultra vector as control. RAD51C-wild-type V79B cells were transduced with the empty vector. GFP-positive infected cells were sorted by FACS and stably maintained in culture. We confirmed the presence of the human RAD51C transgenes (wild-type and modified with the variant) in the transduced CL-V4B cells by using RT–PCR and specific primers against human RAD51C. Amplified products were sequenced and the presence of the RAD51C wild-type cDNA or the c.934C>T nucleotide change was verified. Primers used in the RT–PCR were as follows: Transgenic human RAD51C cDNA, forward 5′-ACCGAAAAGCTTTGGAGGAT-3′ and reverse 5′-TTGCAATGAACATGCAGAAG-3′ oligonucleotides; Actin cDNA, forward 5′-CGAGCGCGGCTACAGCTT-3′ and reverse 5′-TCCTTAATGTCACGCACGATTT-3′.
Immunoblotting
The expression of endogenous (V79B cells) and transgenic (CL-V4B cells) RAD51C protein was also confirmed by western blot. Briefly, cell lysates were prepared in RIPA buffer (Sigma-Aldrich, St Louis, MO, USA) and protease inhibitors cocktail (Roche, Basel, Switzerland). Protein content was determined by Lowry analysis (Bio-Rad, Hercules, CA, USA). Eighty micrograms of proteins were analysed by SDS–PAGE on polyacrylamide gels and transferred to Immobilon-P membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 5% nonfat milk for 1 h at RT. Blots were probed with primary antibodies anti-RAD51C (Novus Biologicals, Littleton, CO, USA, NB100-177) that detects human and hamster RAD51C protein, at 1/250 dilution or anti-Actin (Sigma-Aldrich, A2228) at 1/5000 dilution in TBS-T (50 mM Tris-HCl, 150 mM NaCl, pH 7.5 plus 0.1% Tween 20) containing 5% nonfat milk. Anti-mouse IgG-HRP secondary antibody was visualised using the ECL detection kit (Supersignal WestFEMTO Maximum Sensitivity Substrate, Thermo Fisher Scientific, Boston, MA, USA).
Cell survival assays
Cells seeded onto six-well plates at a density of 100 000 per well were treated with Mitomycin (MMC, Sigma-Aldrich) and Olaparib (Axon Medchem, Groningen, Netherlands) at different doses (MMC, up to 500 nM and Olaparib, up to 20 μM), during 5 days and then tripsinised, washed in PBS and counted using a Countess automated cell counter (Invitrogen). For visual examination, colonies from replicate plates described above were stained with crystal violet 0.05% (Sigma-Aldrich). Relative IC50 values were determined by nonlinear regression of variable slope model by GraphPad Prism 7.00 Software for Windows.
Cell cycle
For cell cycle analysis, 10 000 cells from plates treated with vehicle (DMSO) and MMC (50 nM) during 5 days were collected by centrifugation, fixed with pre-cooled 70% ethanol overnight at 4 °C, washed once with PBS and incubated with propidium bromide 1 μg ml−1 (BD Biosciences, San Jose, CA, USA) and 1 mg ml−1 RNase (Sigma-Aldrich). Fluorescence was detected on a FACSCanto II flow cytometer (BD Biosciences) and analysed with FlowJo v10 software. All experiments were performed in quadruplicate.
Formation of pH2AX and RAD51 foci
In order to examine pH2AX and RAD51 foci formation we seeded V79B RAD51C-proficient cells transduced with empty vector (EV) and RAD51C-deficient CL-V4B cells complemented with RAD51C.WT, RAD51C.pArg312Trp or EV onto 96-well Black/Clear Tissue Culture Treated Imaging Plate (BD Falcon, San Jose, CA, USA). After 24 h cells were irradiated with gamma-rays at 10 Gy and left 6 h of recovery period. Then, non-irradiate (control) and irradiated plates were fixed with 4% paraformaldehyde (Electron Microscopy Science, Hatfield, PA, USA) for 15 min at room temperature (r.t.) and permeabilised with 0.5% Triton X-100 for 20 min. After 30 min in blocking buffer (20% fetal bovine serum in PBS-Tween 0.1%) cells were incubated at 4 °C with mouse anti-pH2AX (Millipore) at 1 : 5000 dilution or rabbit anti-RAD51 (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 1 : 100 dilution for 1 h at r.t. Cells were washed in PBS-Tween and then incubated with secondary antibodies Alexa 568 and Alexa 488 at 1 : 250 (Invitrogen), respectively during 1 h at r.t. After this period, cells were washed with PBS-Tween and were incubated with 4,6-diamidino-2-phenylindole (Sigma-Aldrich) 10 min at r.t. Cell were washed once in PBS-Tween and left in PBS prior to microscope analysis. Images were automatically acquired from each well by an Opera High-Content Screening System (Perkin Elmer, Waltham, MA, USA). A × 40 magnification lens was used and pictures were taken at non-saturating conditions. We determined the percentage of foci-positive cells (cells with at least one foci per nucleus) using Acapella Analysis Software (Perkin Elmer). For each experiment, ∼10 000 nucleus were analysed. pH2AX and RAD51 foci were evaluated from three independent experiments.
Chromosome instability test
Cell were seeded onto 100 mm plates and 24 h after culture set-up, cells were treated with diethyl epoxy butane (DEB) at a final concentration of 0.06 μg ml−1 (Sigma-Aldrich), and vehicle for spontaneous chromosome fragility evaluation. Forty-eight hours after DEB treatment, colcemid was added at a final concentration of 0.1 μg ml−1. Cultures were collected 2 h later when metaphase spreads were obtained according to standard cytogenetic methods and finally, stained with 5% Giemsa in pH 6.8 buffer for 10 min. For chromosome fragility evaluation, 100–200 metaphases with 46 centromeres were analysed per duplicate for each culture. The main criteria for the determination of chromosome fragility were as follows: only chromatid aberrations were considered (gaps, breaks), and interchanges (radial figures) were converted to the minimum number of breaks necessary to form each figure. Cell images were captured at × 100 using an Axioplan 2 imaging Microscope (Zeiss) and Axiocam MRm camera (Zeiss).
Pathogenicity prediction tools
In order to propose a criterion that helps to prioritise RAD51C missense for functional validation we reviewed all reported missense changes classified according to functional assays. We also selected all other described RAD51C missense variants classified as likely pathogenic by at least two in silico predictors in the relevant original manuscript. We only considered studies that analysed a minimum number of 100 cases. Functional effect of selected RAD51C missense variants was then assessed by using eight in silico predictors: the individual predictors SIFT (Ng and Henikoff, 2001) (using ‘SIFT sequence’ method), FATHMM (Shihab et al, 2013) (using ‘FATHMM inherited disease unweighted’ method), SNP&GO (Calabrese et al, 2009), MutationAssessor (Reva et al, 2011), MutPred (Li et al, 2009), MutationTaster (Schwarz et al, 2010), Polyphen-2 (Adzhubei et al, 2010) and the metapredictor Condel (González-Pérez and López-Bigas, 2011). For simplicity different outcomes displayed by each predictor were unified as follows: SIFT ‘affected protein function’, FATHMM ‘damaging’, SNP&GO ‘disease’, Mutation Assessor functional impact ‘high’ and ‘medium’, MutPred scores with g>0.5 in ‘Probably of deleterious mutation’ and P<0.05 in ‘Molecular mechanism disrupted’ outputs, MutationTaster ‘disease causing’ and ‘disease causing automatic’, Polyphen-2 ‘probably damaging’ and ‘possibly damaging’, and Condel ‘deleterious’ were all designated as ‘deleterious’. On the other hand, SIFT ‘tolerated’, FATHMM ‘tolerated’, SNP&GO ‘neutral’, Mutation Assessor functional impact ‘low’, MutPred scores with g<0.5 in ‘Probably of deleterious mutation’ and P>0.05 in ‘Molecular mechanism disrupted’ outputs, MutationTaster ‘polymorphism’ and ‘polymorphism automatic’, Polyphen-2 ‘benign’, and Condel ‘neutral’ were all designated as ‘tolerated’. These predictors were selected according to previous studies that evaluated prediction congruency and accuracy in different data sets with known pathogenic and neutral variants (Thusberg et al, 2011; Thompson et al, 2013; Martelotto et al, 2014). The total number of predictors out of the eight tested that yielded a ‘deleterious’ output was assigned to each variant and denominated ‘combined in silico prediction score’ (CPS). In order to get a more complete and consistent pathogenicity prediction for RAD51C we also used the PredictProtein software (Yachdav et al, 2014), a comprehensive meta-service for sequence analysis that predicts structural and functional features of proteins, including effects of point mutations. PredictProtein uses the trained classifier SNAP2 and assigns a score that ranges from −100 for strong ‘no-effect’ prediction to +100 for a strong ‘effect’, the prediction score being to some extent correlated with the severity of the effect (score>50, strong signal for effect; score<−50, strong signal for neutral/no effect) (Martelotto et al, 2014). The sensitivity (SEN), specificity (SPC), Positive Predictive Value (PPV) and Negative Predictive Value (NPV) of the CPS and the ‘PredictProtein score’ (PPS) were calculated based on the functionally characterised variants and a prioritisation criteria for all other variants was proposed. All selected tools, including PredictProtein, are publicly available with a user-friendly web-based performance and fast generating outcomes, all of them important features for clinical diagnostic laboratories.
Statistical analysis
The data are shown as the mean±standard error of the mean (s.e.m.). Statistical significance was estimated with the Student’s t-test. All statistical tests were two-sided and nominal p-values less than 0.05 were considered statistically significant.
Results
Whole exome sequencing (WES)
A non-BRCA1/2 Spanish family with three ovarian and one breast cancer cases across generations was selected to define novel ovarian cancer susceptibility genetic variants by using WES (Figure 1A). DNA from peripheral blood of the only alive ovarian cancer-affected member was isolated and sequenced. After stringent filtering, 206 heterozygous variants fulfilled our prioritisation criteria I to III (see Methods). Of these, 5 out of 92 variants predicted to be likely pathogenic were located in genes with a potential cancer-related function (filtering criteria IV–V) and were validated by Sanger sequencing: RAD51C (c.934C>T, p.Arg312Trp), PML (c.1961C>G, p.Ser654Cys), EXO1 (c.2212-1G>C), DCHS2 (c.6024dupG, p.Tyr2009ValfsTer15) and MZF1 (c.1223_1233delAGGAGCGGCCG, p.Glu408ValfsTer33). The fact that RAD51C is a known moderate-high ovarian cancer susceptibility gene prompted us to give priority to the RAD51C variant for further validation analysis. The c.934C>T variant (rs730881932) creates an amino acid substitution in the RAD51C protein (p.Arg312Trp) not previously reported in the literature or in the 1000 Genome Project, ESP or ExAC browsers. It had been reported as a single submission to the ClinVar database (SCV000211628) (Landrum et al, 2014, 2016) by ‘GenDx’ having been found in an individual as a germline change and in the COSMIC database (COSM292595) (Forbes et al, 2008, 2017) as a somatic mutation in a large intestine carcinoma. The change (p.Arg312Trp) was classified as deleterious according to the three predictors integrated in our standard WES analysis pipeline (SIFT, Polyphen-2 and Condel) (Kumar et al, 2009; Adzhubei et al, 2010; González-Pérez and López-Bigas, 2011). Sequence alignment revealed that the reference amino acid is highly conserved among RAD51C paralogues and orthologues (Supplementary Figure 1).
Loss-of-heterozygosity and segregation analysis
In order to gather further evidence about the pathogenicity of the p.Arg312Trp variant, we analysed if the proband’s tumour exhibited loss of the RAD51C wild-type allele. In the sequence trace corresponding to the tumour (Figure 1C) we observed a major reduction of the wild-type allele peak intensity compared to that of the trace from the proband’s blood sample (Figure 1B) indicating the existence of LOH. Owing to the impossibility to obtain blood samples of the proband’s sister and parents, we could only perform segregation analysis by using the ovarian tumour from the proband’s sister and the gastric tumour and normal tissue from the proband’s father. We could not retrieve the ovarian tumour or other tissue sample from the proband’s affected mother. We detected the p.Arg312Trp variant (c.934 C<T) in the tumour of the proband’s sister (which also exhibited LOH) (Figure 1D), while we did not observe the presence of the variant either in the normal tissue (Figure 1E) or the gastric tumour from the father (not shown). These results would imply that the proband’s mother is an obligate carrier and would confirm the segregation of the p.Arg312Trp variant with the disease (ovarian cancer).
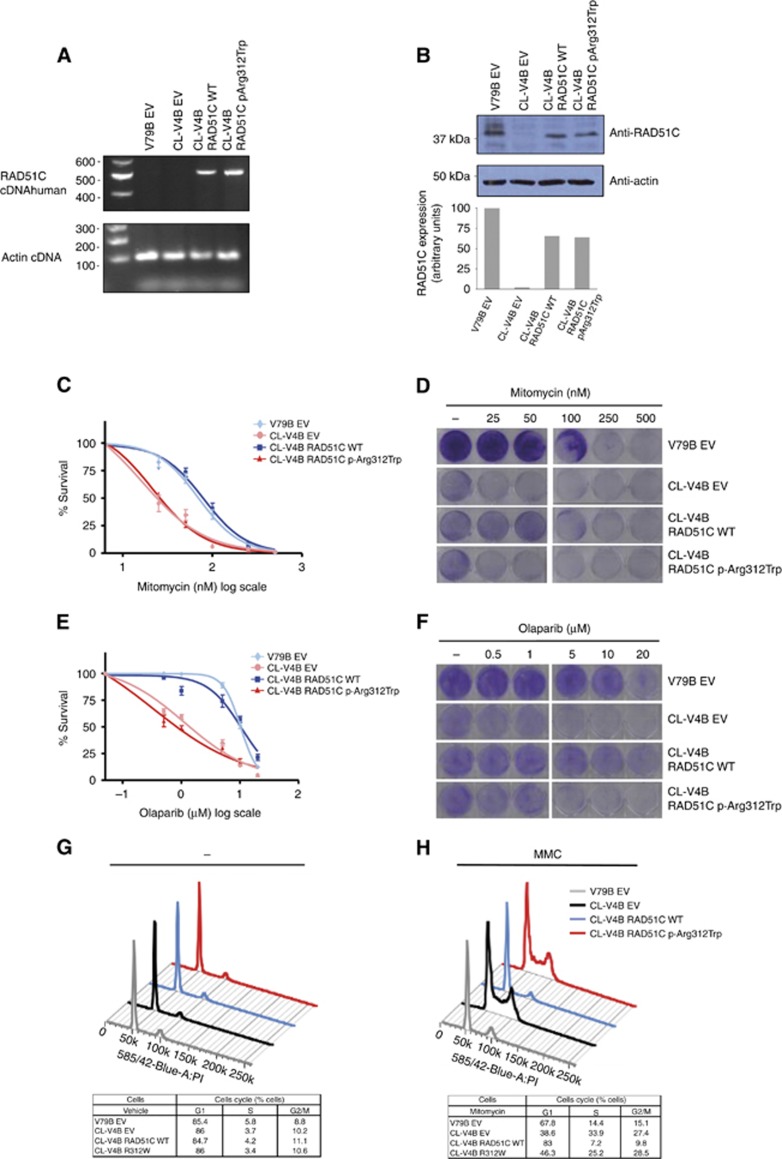
Characterisation of transduced cells
Prior to performing functional studies in the RAD51C-wild-type V79B and the RAD51C-deficient CL-V4B hamster cell lines, we confirmed the expression of transgenic RAD51C in our transduced cell models. By RT–PCR using specific primers against human RAD51C we verified the presence of amplification product in the human RAD51C-transduced CL-V4B cells and the expected absence of a PCR band in the V79B EV control cells since our primers against human RAD51C should not anneal against hamster RAD51C sequence (Figure 2A). Amplified products were sequenced and the presence of the c.934 C<T nucleotide change was confirmed (not shown). Subsequently, expression of endogenous and transgenic RAD51C protein was assessed by immunoblotting in V79B and CL-V4B cells, respectively, using an antibody that detects both human and hamster RAD51C. Levels of exogenous RAD51C in RAD51C.WT and RAD51C.p.Arg312Trp -transduced CL-V4B cells were quantified, representing in both cell line models about 65% of endogenous RAD51C expressed in V79B cells (Figure 2B).
Figure 2.
Functional characterisation of RAD51C p.Arg312Trp missense variant. (A) RT–PCR expression analysis of RAD51C transgenic cDNA in RAD51C-deficient CL-V4B cells transduced with wild-type (CL-V4B RAD51C WT) or p.Arg312Trp–containing (CL-V4B RAD51C.p.Arg312Trp) human RAD51C. CL-V4B (CL-V4B EV) and RAD51C wild-type expressing V79B cells (V79B EV) transduced with empty vector were included as controls. Primers were designed to specifically amplify human RAD51C. (B) Verification and quantification of RAD51C protein expression levels in transgenic cells by using immunoblotting analysis with an anti-RAD51C antibody that recognises human and hamster RAD51C. CL-V4B RAD51C WT, CL-V4B RAD51C.p.Arg312Trp and control CL-V4B EV and V79B EV cells were treated with increasing MMC (C) and Olaparib (E) doses and survival was examined after 5 days of treatment. Values represent means±standard error of the mean (s.e.m.) of four independent experiments. Representative visual examination of cell survival assays by using crystal violet staining of MMC-treated (D), Olaparib-treated (F) and vehicle-treated cells. Plating efficiency differences between CL-V4B and V79B were accounted for by normalising each treated V79B and CL-V4B cell counting with its corresponding untreated (control) point. Cell cycle analysis of CL-V4B RAD51C WT, CL-V4B RAD51C.p.Arg312Trp and control cells treated with MMC (50 nM) (H) or vehicle (–) (G). Low panels show percentage of cells in G1, S and G2/M phases in each cell line. Data are means of four independent experiments. A full colour version of this figure is available at the British Journal of Cancer journal online.
Cell survival analysis
RAD51C deficiency, as well as defects in other Fanconi Anemia genes, triggers extreme sensitivity to DNA interstrand cross-link agents such as MMC. To determine whether the p.Arg312Trp missense variant impairs RAD51C function, we treated RAD51C.WT and RAD51C.p.Arg312Trp-transduced RAD51C-deficient CL-V4B cells with MMC and conducted cell survival assays. CL-V4B and V79B cells transduced with empty vector (CL-V4B EV and V79B EV) were used as controls. Expression of the p.Arg312Trp variant failed to correct the MMC hypersensitivity of CL-V4B EV cells (Figure 2C and D).The half maximal inhibitory concentration (IC50) value for each cell line and MMC treatment was 74.2 nM for V79B EV, 21.7 nM for CL-V4B EV, 89.5 nM for CL-V4B RAD51C.WT, and 23.1 nM for CL-V4B RAD51C.p.Arg312Trp cells, respectively. In order to test clinically relevant ovarian cancer treatments such as PARP inhibition, we also decided to explore Olaparib sensitivity in RAD51C.WT and RAD51C.p.Arg312Trp-transduced RAD51C-deficient CL-V4B cells. As shown in Figure 2E and F, RAD51C.p.Arg312Trp-transduced cells were sensitive to Olaparib compared with RAD51C.WT transduced cell line. The IC50 values for each cell line and Olaparib exposure were 10.2 μM for V79B EV, 1.1 μM for CL-V4B EV, 10.8 μM for CL-V4B RAD51C.WT and 0.4 μM for RAD51C.p. Arg312Trp-transduced RAD51C-deficient CL-V4B cell lines, respectively. In summary, we were able to show that RAD51C.p.Arg312Trp-transduced CL-V4B cells exhibited survival levels and colony formation similar to those observed in CL-V4B EV cells, and presented a significant reduction in IC50 upon MMC and Olaparib treatment (P<0.001) compared with V79B EV and wild-type RAD51C-expressing CL-V4B cells.
Cell cycle analysis
RAD51C-defective cells are characterised by G2/M cell cycle arrest and genomic instability (French et al, 2002; Kuznetsov et al, 2007; Smeenk et al, 2010). V79B cells and CL-V4B cells transduced with wild-type RAD51C, p.Arg312Trp variant and empty vector were treated with 50 nM MMC, and together with the corresponding controls without treatment were subjected to cell cycle analysis (Figure 2G and H). Empty vector-transduced CL-V4B cells showed an increase of G2/M phase cells (27.4±1.1) after exposure to MMC. This phenotype was reverted when CL-V4B cells were transduced with wild-type RAD51C under the same conditions (9.8±1, P<0.001). However, expression of the p.Arg312Trp variant did not rescue the G2/M arrest exhibited by CL-V4B EV cells (28.5 ±0.9) (Figure 2H). The population differences observed in G2/M phase were statistically significant (P<0.001) in RAD51C.p.Arg.312Trp expressing CL-V4B cells compared with V79B EV and RAD51C.WT-transduced RAD51C-deficient CL-V4B cell lines. Additionally, we also observed a statistically significant (P<0.01) increase of S population in CL-V4B EV and in CL-V4B cell expressing the p.Arg312Trp variant (33.9±2.8 and 25.2±1.9 respectively) compared with V79B EV and CL-V4B RAD51C WT complemented cells (14.4±1 and 7.2±0.9, respectively).
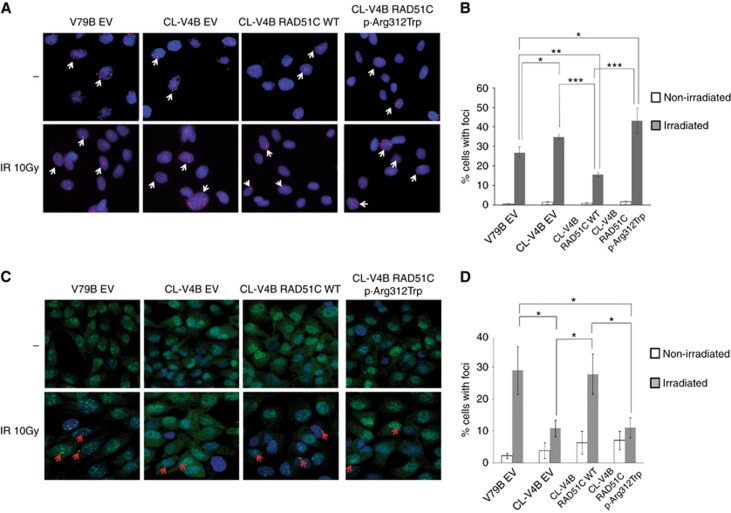
pH2AX and RAD51 foci formation
DNA double strand breaks (DSB) induction increases levels of the DNA damage marker pH2AX in DSB sites, and this accumulation might be more evident in cells with a functional impairment of proteins involved in HR repair of DBS such as BRCA1/2, PALB2 and RAD51C (Vaclová et al, 2015; Somyajit et al, 2015b; Ahlskog et al, 2016; Zamborszky et al, 2017). In addition, the inability to form RAD51 foci after DNA damage constitutes a readout of HR defects and therefore, functional impairment of RAD51C protein causes a reduction of RAD51 foci formation at sites of DSB (Godthelp et al, 2002; Lio et al, 2004; Liu et al, 2007; Min et al, 2013). Accordingly, when we induced DNA damage by ionising irradiation, we observed a significant increase (P<0.05) of pH2AX foci in RAD51C-deficient CL-V4B EV compared to V79B EV RAD51C-proficient cells (34.8±1.3 vs 26.9±2.8). Subsequently, RAD51C.pArg312Trp-transduced RAD51C-deficient CL-V4B cells showed greater accumulation of pH2AX foci compared with CL-V4B RAD51C.WT complemented cells (43.1±6.5 vs 15.5±1.1; P<0.001, respectively) (Figure 3A and B). When we assessed RAD51 foci, as shown in Figure 3C and D, CL-V4B EV cells displayed a significant reduction in the number of RAD51 foci-positive cells compared to V79B EV RAD51C-proficient cells (10.8±2.6 vs 28.9±7.5, respectively; P<0.05). While complementation of CL-V4B cells with wild-type RAD51C cDNA rescued the observed phenotype, expression of p.Arg312Trp mutant protein did not rescue the reduced RAD51 foci formation capacity (27.8±6.2 vs 10.9±3.1; P<0.05, respectively), confirming their impairment in HR.
Figure 3.
Analysis of pH2AX and RAD51 foci formation. (A) Inmunofluorescence analysis of pH2AX induction after gamma-ray irradiation (10 Gy) was assessed in CL-V4B RAD51C WT, CL-V4B RAD51C.p.Arg312Trp, CL-V4B EV and V79B EV cells. (B) Percentage of cells with pH2AX foci is shown. (C) RAD51 foci formation was evaluated in V79B EV, CL-V4B EV, CL-V4B RAD51C WT and CL-V4B RAD51C.p.Arg312Trp cells after gamma-ray irradiation (10 Gy). (D) Percentage of cells with at least one RAD51 foci is presented. Non-irradiated cells (–) were included as controls. Representative images (40 ×) of each experiment are exposed. * P<0.05; ** P<0.01; *** P<0.001. Values correspond to the mean±s.e.m. of three independent experiments. A full colour version of this figure is available at the British Journal of Cancer journal online.
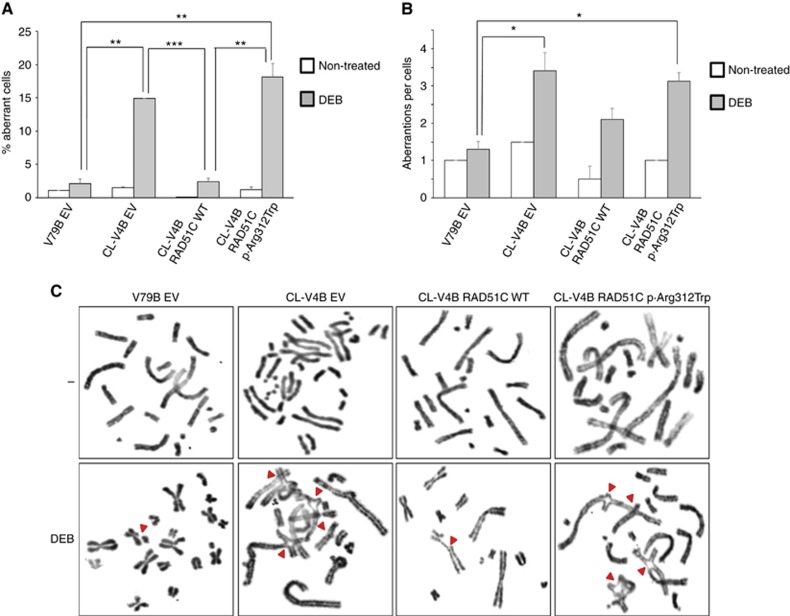
Chromosomal instability test
Bialleic mutations in the RAD51C gene cause Fanconi anaemia, characterised by chromosomal instability and predisposition to leukaemia and other cancers (Wang, 2007; Meindl et al, 2010; Vaz et al, 2010). Here, we assessed the sensitivity of RAD51C wild-type expressing V79B cells (V79B EV) and RAD51C-deficient CL-V4B cells complemented with RAD51C.WT, RAD51C.pArg312Trp or empty vector (EV) to the diethyl epoxy butane (DEB) chromosome fragility-inducing reagent. DEB-treatment induced a significant increase in the percentage of aberrant CL-V4B EV RAD51C-deficient cells compared with RAD51C wild-type expressing V79B cells (V79B EV) (14.9±0 vs 2.1±0.7; P<0.01), and presented a higher number of chromatide aberrations per aberrant cell (3.4±0.5 vs 1.3±0.2; P<0.05) (Figure 4A–C). Expression of RAD51C.pArg312Trp protein did not rescue the chromosomal instability observed in CL-V4B RAD51C-deficient cells, in contrast with CL-V4B cells expressing RAD51C WT, that were able to complement DEB toxicity. As we show in Figure 4, RAD51C.pArg312Trp-transduced RAD51C-deficient CL-V4B cells exhibited a percentage of aberrant cells significantly higher than that observed in V79B cells (18.2±2 vs 2.1±0.7, P<0.01) and in RAD51C WT-transduced RAD51C-deficient CL-V4B cells (18.2±2 vs 2.4±0.5, P<0.01). The number of aberrations per cell was also higher than that shown in V79B control cells.
Figure 4.
DEB-induction chromosomal fragility test. Percentage of aberrant cells (A) and number of aberrations per aberrant cell (B) were evaluated in CL-V4B RAD51C WT, CL-V4B RAD51C.p.Arg312Trp and control CL-V4B EV and V79B EV cells, after 48 h of treatment with DEB (0.06 μg ml−1) or with vehicle (non-treated). * P<0.05; ** P<0.01; *** P<0.001. Results are shown as mean±s.e.m. of two replicates. (C) Representative images (× 100) of each condition are shown. A full colour version of this figure is available at the British Journal of Cancer journal online.
Case-control study: Kaspar genotyping
A case-control study was designed by using a kaspar genoyping assay to determine frequency of the p.Arg312Trp variant in Spanish population. None of the analysed 646 healthy Spanish women and 1490 probands from Spanish non-BRCA1/2 high-risk breast and/or ovarian cancer families presented the variant.
Prioritisation criteria for functional analysis of RAD51C missense variants
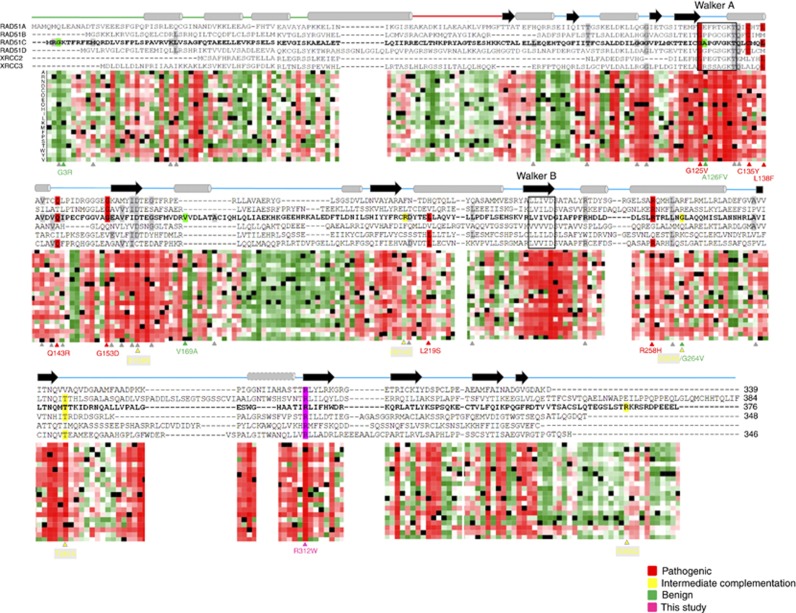
After revision of literature we found 17 functionally characterised RAD51C missense variants (7 classified as pathogenic, 4 as benign and 5 as undetermined/hypomorphic) and 23 missense variants in silico classified as potentially deleterious by at least two predictors (usually SIFT and Polyphen-2) in the relevant original report (Table 1). All seven functionally assessed pathogenic variants and the p.Arg312Trp reported here were classified by at least six of the eight selected predictors as deleterious, with five variants obtaining this classification by all eight classifiers. On the other hand, PPS was positive (signal for effect) for the eight pathogenic missense variants with a value higher than 50 (strong signal for effect) in six of them and with a minimum value of 33. In particular, the p.Arg312Trp variant rendered a score of 95, close to 100, the maximum pathogenicity value provided by PredictProtein software. In contrast, five of the nine functionally characterised as benign or undetermined variants exhibited negative PPS (signal for ‘no effect’) and although four exhibited positive values, only a maximum of four predictors classified them as deleterious. Exception was the undetermined p.Asp159Asn missense variant, with a deleterious prediction from all eight predictors and a PPS value of 80. According to these results, by using a ‘combined in silico prediction score (CPS)’ of ⩾5, sensitivity and specificity of the classification was 100% and 89%, respectively, with a Positive Predictive Value (PPV) of 89% (11% false positive rate) and a Negative Predictive Value (NPV) of 100% (no false negatives). By using a PPS of ⩾33, sensitivity was 100% and specificity 55% with a PPV of 66% (33% false positive rate) and a NPV of 100% (no false negatives). In order to give primacy to not leaving any true positive variant without being tested and to minimise the number of false positives qualifying for a functional test, we propose (i) to perform functional assays in variants with a CPS⩾5; (ii) prioritise them from the highest to the lowest CPS (i.e. 8 predictors=priority A; 5 predictors=priority D); (iii) for variants with same CPS sub-rank them according to their PPS (PPS⩾33, priority 1; 0<PPS<33, priority 2; PPS⩽0, priority 3). According to this criterion nearly half of the 23 selected unclassified variants would qualify for functional tests with the highest priority (A1) (Table 1). Among the highest-priority variants are the p.Thr132Pro and the p. Gln133Lys, the former located within the Walker A domain of the protein and both situated in the vicinity of three functionally characterised pathogenic variants in a highly conserved region. Similarly, the p.Val156G, p.Asp159Tyr and Gly162Glu variants are in or near a β-strand and close to the pathogenic change p. Gly153Asp (Figure 5).
Table 1. RAD51C missense variants: review, in silico predictions and prioritisation criteria for directing functional validation.
| Nucleotide change | Protein change | Protein change | rs number | Study | Functionally characterised | Final categorisation in study | SIFT | Polyphen-2 | FATHMM | SNP&GO | Mutation assessor | MUTPRED | MUTTASTER | Condel | Combined in silico prediction score (CPS) (1–8) | Predict Protein Score (PPS) (−100–+100) | In-silico prioritisation score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| c.374G>T | G125V | p.Gly125Val | rs267606998 | Meindl et al | Yes | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 83 | A1 |
| c.404G>A | C135Y | p.Cys135Tyr | rs767796996 | Osorio et al | Yes | Deleterious | Tolerated | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Tolerated | 6 | 75 | C1 |
| c.414G>C | L138F | p.Leu138Phe | rs267606999 | Meindl et al | Yes | Deleterious | Deleterious | Deleterious | Deleterious | Tolerated | Deleterious | Deleterious | Deleterious | Deleterious | 7 | 33 | B1 |
| c.428A>G | Q143R | p.Gln143Arg | rs587780255 | Osorio et ala | Yes | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 49 | A1 |
| c.458G>A | G153D | p.Gly153Asp | rs765730332 | Clague et al | Yes | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 75 | A1 |
| c.656T>C | L219S | p.Leu219Ser | rs201529791 | Osorio et al | Yes | Deleterious | Deleterious | Deleterious | Deleterious | Tolerated | Deleterious | Deleterious | Deleterious | Deleterious | 7 | 56 | B1 |
| c.773G>A | R258H | p.Arg258His | rs267606997 | Jonson et al | Yes | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 68 | A1 |
| c.934C>T | R312W | p.Arg312Trp | rs730881932 | This study | Yes | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 95 | A1 |
| c.7G>A | G3R | p.Gly3Arg | rs376403182 | Meindl et al | Yes | Tolerated | Deleterious | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | 1 | −75 | NP |
| c.376G>A | A126T | p.Ala126Thr | rs61758784 | Meindl et al | Yes | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Deleterious | Tolerated | Deleterious | Tolerated | 2 | 90 | NP |
| c.506T>C | V169A | p.Val169Ala | rs587780256 | Meindl et al | Yes | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Deleterious | Deleterious | Tolerated | 2 | −67 | NP |
| c.791G>T | G264V | p.Gly264Val | - | Meindl et al | Yes | Tolerated | Deleterious | Tolerated | Tolerated | Tolerated | Deleterious | Tolerated | Deleterious | Tolerated | 3 | 37 | NP |
| c.475G>A | D159N | p.Asp159Asn | - | Meindl et al | Yes | Undetermined | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 80 | A1 |
| c.640C>T | R214C | p.Arg214Cys | rs140804406 | Clague et al | Yes | Undetermined | Tolerated | Tolerated | Deleterious | Tolerated | Tolerated | Deleterious | Deleterious | Tolerated | 3 | 0 | NP |
| c.790G>A | G264S | p.Gly264Ser | rs147241704 | Meindl et al | Yes | Undetermined | Deleterious | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Deleterious | Tolerated | 2 | −3 | NP |
| c.859A>G | T287A | p.Thr287Ala | rs28363317 | Meindl et al | Yes | Undetermined | Deleterious | Deleterious | Tolerated | Tolerated | Deleterious | Tolerated | Deleterious | Tolerated | 4 | 89 | NP |
| c.1097G>A | R366Q | p.Arg366Gln | rs577852020 | Meindl et al | Yes | Undetermined | Tolerated | Deleterious | Tolerated | Tolerated | Deleterious | Tolerated | Deleterious | Tolerated | 3 | −7 | NP |
| c.4C>G | R2G | p.Arg2Gly | - | Pang et al | No | Likely pathogenic | Deleterious | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | Tolerated | 1 | −14 | NP |
| c.29T>G | M10R | p.Met10Arg | rs730881936 | Vuorela et al | No | Undetermined | Deleterious | Tolerated | Tolerated | Deleterious | Deleterious | Deleterious | Deleterious | Tolerated | 5 | 68 | D1 |
| 77A>T | K26M | p.Lys26Met | rs746026526 | Song et al | No | Potentially deleterious | Deleterious | Tolerated | Deleterious | Tolerated | Deleterious | Deleterious | Deleterious | Tolerated | 5 | 43 | D1 |
| 80T>C | L27P | p.Leu27Pro | rs587781309 | Song et al | No | Potentially deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 73 | A1 |
| 271C>T | L91F | p.Leu91Phe | - | Song et al | No | Potentially deleterious | Deleterious | Deleterious | Tolerated | Tolerated | Deleterious | Tolerated | Deleterious | Deleterious | 5 | 11 | D2 |
| c.305C>T | T102I | p.Thr102Ile | - | Loveday et al | No | - | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 73 | A1 |
| 335G>T | G112V | p.Gly112Val | - | Song et al | No | Potentially deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 67 | A1 |
| c.341G>T | G114V | p.Gly114Val | - | Thompson et al | No | Undetermined | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 81 | A1 |
| 394A>C | T132P | p.Thr132Pro | - | Cunningham et al | No | Potentially deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 82 | A1 |
| 397C>A | Q133K | p.Gln133Lys | - | Cunningham et al | No | Potentially deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 61 | A1 |
| 419T>G | V140G | p.Val140Gly | - | Song et al | No | Potentially deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 66 | A1 |
| c.431T>C | I144T | p.Ile144Thr | rs28363307 | Loveday et al | No | Putative pathogenic | Tolerated | Deleterious | Tolerated | Deleterious | Deleterious | Tolerated | Deleterious | Tolerated | 4 | −53 | NP |
| c.461A>G | E154G | p.Glu154Gly | rs758847241 | Cunningham et al | No | Undetermined | Deleterious | Deleterious | Tolerated | Tolerated | Tolerated | Deleterious | Deleterious | Tolerated | 4 | −29 | NP |
| c.467T>G | V156G | p.Val156Gly | - | Cunningham et al | No | Undetermined | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 69 | A1 |
| c.472A>T | I158F | p.Ile158Phe | rs771888635 | Cunningham et al | No | Undetermined | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 15 | A2 |
| c.475G>A | D159Y | p.Asp159Tyr | rs775213492 | Cunningham et al | No | Undetermined | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 81 | A1 |
| c.485G>A | G162E | p.Gly162Glu | rs35151472 | Thompson et al | No | Undetermined | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 62 | A1 |
| c.523G>A | A175T | p.Ala175Thr | rs587780838 | Thompson et al | No | Undetermined | Deleterious | Deleterious | Tolerated | Tolerated | Deleterious | Tolerated | Deleterious | Deleterious | 5 | 1 | D2 |
| c.644A>G | D215G | p.Asp215Gly | - | Pang et al | No | Likely pathogenic | Deleterious | Deleterious | Deleterious | Tolerated | Deleterious | Deleterious | Deleterious | Deleterious | 7 | 57 | B1 |
| 677T>C | L226P | p.Leu226Pro | - | Song et al | No | Potentially deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | 8 | 22 | A2 |
| c.745C>T | R249C | p.Arg249Cys | rs28363311 | Thompson et al | No | Putative pathogenic | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Tolerated | Deleterious | Deleterious | 7 | 52 | B1 |
| c.784T>G | L262V | p.Leu262Val | rs149331537 | Thompson et al | No | Putative pathogenic | Tolerated | Deleterious | Deleterious | Tolerated | Deleterious | Deleterious | Deleterious | Deleterious | 6 | −21 | C3 |
| c.835G>C | A279P | p.Ala279Pro | rs587782507 | Loveday et al | No | Potentially deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Deleterious | Tolerated | 7 | 13 | B2 |
Abbreviation: NP=No priority.
The variant p.Gln143Arg had been previously reported by Romero et al (2011) but its pathogenicity was not characterised.
Figure 5.
In silico studies. Sequence alignment of human RAD51C and its paralogues RAD51A, RAD51B, RAD51D, XRCC2 and XRCC3. Secondary structure obtained from P.furiosus (Pf) RAD51 and described by Miller et al (2004) is shown schematically above the sequences with cylinders for α-helices and arrows for β-strands. The helix showed with a dashed line is predicted in the Rad51 paralogues using PSI-PRED, but is absent in the P. furiosus Rad51 crystal structure. The Walker A and Walker B ATP binding motifs are indicated by rectangles. Horizontal line linking α-helixes and β-strands correspond to the N-terminal domain (green), linker region (red) and C-terminal domain (blue). A heat map that shows the tolerance to independent amino acid (aa) substitutions (y-axis) for each position of the RAD51C protein (x-axis) generated using PredictProtein tool (Yachdav et al, 2014) is shown below the sequence alignment. Red indicates strong signal for a deleterious effect (score>50); white indicates a small effect; green indicates a strong signal for neutral effect/no effect (score<−50); and black represents the corresponding wild-type residue. RAD51C missense substitutions functionally characterised described in the literature and listed in Table 1 are specified below the heatmap and shown in different colours: Residues highlighted in red, green and yellow correspond to pathogenic, benign and undetermined/hypomorphic missense variants, respectively. Variants highlighted in grey correspond to the functionally uncharacterised changes detailed in Table 1. The novel RAD51C p.Arg312Trp pathogenic variant described in this study is highlighted in purple. A full colour version of this figure is available at the British Journal of Cancer journal online.
Discussion
RAD51C is one of the novel ovarian cancer moderate-susceptibility genes described in the last years. Discrepancies across different studies have been observed regarding the prevalence of RAD51C pathogenic mutations in high-risk breast and ovarian cancer families (Léveillé et al, 2004; Honrado et al, 2006; Beauchamp et al, 2007; Forget and Kowalczykowski, 2010; Meindl et al, 2010; Zheng et al, 2010; Romero et al, 2011; Vuorela et al, 2011). In a prior work (Osorio et al, 2012), we emphasised the relatively high frequency of unique RAD51C missense mutations found in patients and highlighted that the fact that most conducted studies do not assess pathogenicity, which might account for the inconsistency of mutation prevalence data. We recommended that the so far partially disregarded rare RAD51C missense changes should be functionally evaluated in order to have a complete and precise picture of the significance of RAD51C mutations.
In line with our observations, a recent study that analysed the coding sequence and splice site boundaries of RAD51C in 3429 OC patients and 2772 controls described that the burden of missense variants predicted to be deleterious was significantly higher in cases than controls, indicating that at least a proportion of them might be pathogenic (Song et al, 2015). The same study reported an odds ratio of 5.2 (95% CI 1.1–24) associated with RAD51C mutations (Song et al, 2015). Such risk, calculated only with clearly pathogenic variants in unselected OC cases, is similar to prior estimations by Pelttari et al (6.31, 95% CI 1.15–34.6; unselected OC) (Pelttari et al, 2011) and Loveday et al (5.88, 95% CI 2.91–11.88) (Loveday et al, 2012). These estimates need to be confirmed with large international studies but already suggest a level of risk of EOC that warrants the inclusion of RAD51C in routine genetic tests. Importantly, clinical utility of the identification of RAD51C mutation carriers would be beyond prediction of ovarian cancer susceptibility as, similar to BRCA1/2 mutation carriers, patients carrying germline RAD51C mutations could also benefit from targeted PARP inhibition-based treatments (Min et al, 2013; Bajrami et al, 2014; Somyajit et al, 2015a). Despite these major implications, virtually all published studies still limit the analysis of missense variants to a categorisation based on the probability of being deleterious according to prediction programs (Kumar et al, 2009; Adzhubei et al, 2010).
Here we report yet another example of a rare RAD51C missense change (p.Arg312Trp) that impairs protein function. The variant was identified in a Spanish high-risk breast and ovarian cancer family and sequence alignment tools indicated high-conservation of the affected nucleotide across paralogues and orthologues and prediction programs classified the variant as deleterious. We demonstrated segregation with the disease and, established by in vitro functional assays, inability to complement the phenotype of RAD51C-deficient CL-V4B cells, both regarding extreme sensitivity to MMC and sensitivity to the PARP inhibitor Olaparib. In addition, cells expressing p.Arg312Trp variant, presented increased levels of DNA damage, abnormal RAD51 foci formation upon irradiation and higher chromosomal instability. These findings add further evidence to previous observations that cells harbouring mutations in HR proteins, in addition to DNA-repair impairment also show greater accumulation of DNA damage (Vaclová et al, 2015; Ahlskog et al, 2016; Zamborszky et al, 2017). The accumulation of DNA damage, particularly DBS, and the incapability to restart replication stalled forks, might be implicated in the primary steps of breast and ovarian cancer development (Vaclová et al, 2015; Somyajit et al, 2015b). In agreement with a prior study that found loss of the wild allele in tumours from carriers of clearly deleterious mutations (protein truncating variants) (Pelttari et al, 2011), we observed LOH in tumours from the p.Arg312Trp carriers, which would support its deleterious effect. Taken all together, these results indicate that the missense p.Arg312Trp substitution causes a loss of function of the RAD51C protein and most probably is the main cause of three ovarian cancer cases detected in the family.
This new pathogenic RAD51C missense variant adds to other seven missense changes earlier functionally classified as deleterious (Meindl et al, 2010; Vaz et al, 2010; Clague et al, 2011; Osorio et al, 2012) and reinforces the necessity to unequivocally determine the pathogenic nature of any potentially damaging RAD51C missense variant found in breast and ovarian cancer high-risk families. However, since functional complementation analysis are expensive and require time and expertise is fundamental to prioritise which variants qualify for this type of screening.
For this reason, we performed an extensive review of RAD51C unclassified missense variants described in the literature, and by assessing the performance of nine in silico prediction tools in a set of functionally characterised variants we have outlined a criteria to direct RAD51C functional assays. We selected previously well-rated prediction methods (Thusberg et al, 2011; Thompson et al, 2013; Martelotto et al, 2014; Dong et al, 2015) and among them we favoured user-friendly free-online tools vs complex computational algorithms as this would be key for a practical implementation in non-bioinformatics-oriented laboratories and in the clinical diagnosis routine. We have proposed a prioritisation criteria based first on the number of selected predictors providing a deleterious output, with a minimum of 5 in order to qualify for testing, subsequently refined by PredictProtein score, this having to be greater than 33 in order to assign high-priority label. With this criteria, all known pathogenic mutations would have been selected for high-priority testing while only one of the undetermined/hypomorphic variants (p.Asp159Asn) would have qualified for further analysis. Interestingly, the p.Asp159Asn was classified as pathogenic by all eight predictors and exhibited a PredictProtein score close to the maximum signal for effect in the protein function (80 of a maximum of 100). Taking into account that according to the original report this variant segregated with the disease in a breast cancer family (Meindl et al, 2010) it might be worth a functional re-evaluation of its partial activity.
It is beyond doubt that the set of RAD51C functionally characterised variants in which our prioritisation criteria is based is far too small to build a robust classifier, but it delineates a starting point to guide and encourage selection of already described and of novel missense variants for complementation assays. Our review points to a non-negligible number of reported variants likely to impair protein function, but the reliability of our criteria and the extent to which RAD51C pathogenic missense changes have been overlooked will only be determined by the systematic incorporation of functional testing over time. If the prevalence of damaging RAD51C amino acid substitutions is confirmed, a comprehensive list of RAD51C missense mutations already classified as benign or pathogenic based on functional analysis may facilitate the diagnosis of new families referred for RAD51C screening. However, given the unique nature of most reported pathogenic variants, we foresee that reference laboratories performing RAD51C screening should be prepared to have readily available resources and knowledge to conduct and interpret RAD51C functional tests.
Acknowledgments
We particularly want to acknowledge the patients enrolled in this study for their participation and Drs Albert Pastink and Haico van Attikum (Leiden University Medical Center, Leiden, Netherlands) for providing RAD51C-wild type V79B and RAD51C-deficient CL-V4B Chinese hamster cells. We also want to thank the Spanish National Genotyping Center (CEGEN) for excellent technical support, Ma Carmen Gómez Neira, CNIO Confocal Microscopy Unit, Diego Megías and CNIO Flow cytometry Unit, also for great help and technical support, CNIO Cytogenetic Unit, and CNIO Biobank, particularly to Ma Jesús Artiga, and the Biobanco i+12 in the Hospital 12 de Octubre integrated in the Spanish Hospital Biobanks network (RetBioH; www.redbiobancos.es) for facilitating access to samples. Biobank is supported by Instituto de Salud Carlos III.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Meth 7(4): 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlskog JK, Larsen BD, Achanta K, Sørensen CS (2016) ATM/ATR-mediated phosphorylation of PALB2 promotes RAD51 function. EMBO Rep 17(5): 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajrami I, Frankum JR, Konde A, Miller RE, Rehman FL, Brough R, Campbell J, Sims D, Rafiq R, Hooper S, Chen L, Kozarewa I, Assiotis I, Fenwick K, Natrajan R, Lord CJ, Ashworth A (2014) Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res 74(1): 287–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauchamp NJ, Taybert J, Champion MP, Layet V, Heinz-Erian P, Dalton A, Tanner MS, Pronicka E, Sharrard MJ (2007) High frequency of missense mutations in glycogen storage disease type VI. J Inherit Metab Dis 30(5): 722–734. [DOI] [PubMed] [Google Scholar]
- Calabrese R, Capriotti E, Fariselli P, Martelli PL, Casadio R (2009) Functional annotations improve the predictive score of human disease-related mutations in proteins. Human Mutat 30(8): 1237–1244. [DOI] [PubMed] [Google Scholar]
- Clague J, Wilhoite G, Adamson A, Bailis A, Weitzel JN, Neuhausen SL (2011) RAD51C germline mutations in breast and ovarian cancer cases from high-risk families. PLoS ONE 6(9): e25632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C, Wei P, Jian X, Gibbs R, Boerwinkle E, Wang K, Liu X (2015) Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet 24(8): 2125–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Gordon L, Hendrix M, Hourlier T, Johnson N, Kähäri AK, Keefe D, Keenan S, Kinsella R, Komorowska M, Koscielny G, Kulesha E, Larsson P, Longden I, McLaren W, Muffato M, Overduin B, Pignatelli M, Pritchard B, Riat HS, Ritchie GRS, Ruffier M, Schuster M, Sobral D, Tang YA, Taylor K, Trevanion S, Vandrovcova J, White S, Wilson M, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Durbin R, Fernández-Suarez XM, Harrow J, Herrero J, Hubbard TJP, Parker A, Proctor G, Spudich G, Vogel J, Yates A, Zadissa A, Searle SMJ (2012) Ensembl 2012. Nucleic Acids Res 40(D1): D84–D90.22086963 [Google Scholar]
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45(D1): D777–D783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR (2008) The catalogue of somatic mutations in cancer (COSMIC). Curr Protoc Hum Genet; Chapter 10: Unit 10.11. [DOI] [PMC free article] [PubMed]
- Forget AL, Kowalczykowski SC (2010) Single-molecule imaging brings Rad51 nucleoprotein filaments into focus. Trends in Cell Biol 20(5): 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CA, Masson J-Y, Griffin CS, O'Regan P, West SC, Thacker J (2002) Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J Biol Chem 277(22): 19322–19330. [DOI] [PubMed] [Google Scholar]
- Godthelp BC, Wiegant WW, van Duijn-Goedhart A, Schärer OD, van Buul PPW, Kanaar R, Zdzienicka MgZ (2002) Mammalian Rad51C contributes to DNA cross-link resistance, sister chromatid cohesion and genomic stability. Nucleic Acids Res 30(10): 2172–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Pérez A, López-Bigas N (2011) Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet 88(4): 440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honrado E, Osorio A, Palacios J, Benitez J (2006) Pathology and gene expression of hereditary breast tumors associated with BRCA1, BRCA2 and CHEK2 gene mutations. Oncogene 25(43): 5837–5845. [DOI] [PubMed] [Google Scholar]
- Jervis S, Song H, Lee A, Dicks E, Tyrer J, Harrington P, Easton DF, Jacobs IJ, Pharoah PPD, Antoniou AC (2014) Ovarian cancer familial relative risks by tumour subtypes and by known ovarian cancer genetic susceptibility variants. J Med Genet 51(2): 108–113. [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L (2009) VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25(17): 2283–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchenbaecker KB, Ramus SJ, Tyrer J, Lee A, Shen HC, Beesley J, Lawrenson K, McGuffog L, Healey S, Lee JM, Spindler TJ, Lin YG, Pejovic T, Bean Y, Li Q, Coetzee S, Hazelett D, Miron A, Southey M, Terry MB, Goldgar DE, Buys SS, Janavicius R, Dorfling CM, van Rensburg EJ, Neuhausen SL, Ding YC, Hansen TVO, Jonson L, Gerdes A-M, Ejlertsen B, Barrowdale D, Dennis J, Benitez J, Osorio A, Garcia MJ, Komenaka I, Weitzel JN, Ganschow P, Peterlongo P, Bernard L, Viel A, Bonanni B, Peissel B, Manoukian S, Radice P, Papi L, Ottini L, Fostira F, Konstantopoulou I, Garber J, Frost D, Perkins J, Platte R, Ellis S, Embrace Godwin AK, Schmutzler RK, Meindl A, Engel C, Sutter C, Sinilnikova OM, Collaborators GS, Damiola F, Mazoyer S, Stoppa-Lyonnet D, Claes K, De Leeneer K, Kirk J, Rodriguez GC, Piedmonte M, O’Malley DM, de la Hoya M, Caldes T, Aittomaki K, Nevanlinna H, Collee JM, Rookus MA, Oosterwijk JC, Breast Cancer Family R, Tihomirova L, Tung N, Hamann U, Isaccs C, Tischkowitz M, Imyanitov EN, Caligo MA, Campbell IG, Hogervorst FBL, Hebon, Olah E, Diez O, Blanco I, Brunet J, Lazaro C, Pujana MA, Jakubowska A, Gronwald J, Lubinski J, Sukiennicki G, Barkardottir RB, Plante M, Simard J, Soucy P, Montagna M, Tognazzo S, Teixeira MR, Investigators KC, Pankratz VS, Wang X, Lindor N, Szabo CI, Kauff N, Vijai J, Aghajanian CA, Pfeiler G, Berger A, Singer CF, Tea M-K, Phelan CM, Greene MH, Mai PL, Rennert G, Mulligan AM, Tchatchou S, Andrulis IL, Glendon G, Toland AE, Jensen UB, Kruse TA, Thomassen M, Bojesen A, Zidan J, Friedman E, Laitman Y, Soller M, Liljegren A, Arver B, Einbeigi Z, Stenmark-Askmalm M, Olopade OI, Nussbaum RL, Rebbeck TR, Nathanson KL, Domchek SM, Lu KH, Karlan BY, Walsh C, Lester J, Australian Cancer S, Australian Ovarian Cancer Study G, Hein A, Ekici AB, Beckmann MW, Fasching PA, Lambrechts D, Van Nieuwenhuysen E, Vergote I, Lambrechts S, Dicks E, Doherty JA, Wicklund KG, Rossing MA, Rudolph A, Chang-Claude J, Wang-Gohrke S, Eilber U, Moysich KB, Odunsi K, Sucheston L, Lele S, Wilkens LR, Goodman MT, Thompson PJ, Shvetsov YB, Runnebaum IB, Durst M, Hillemanns P, Dork T, Antonenkova N, Bogdanova N, Leminen A, Pelttari LM, Butzow R, Modugno F, Kelley JL, Edwards RP, Ness RB, du Bois A, Heitz F, Schwaab I, Harter P, Matsuo K, Hosono S, Orsulic S, Jensen A, Kjaer SK, Hogdall E, Hasmad HN, Azmi MAN, Teo S-H, Woo Y-L, Fridley BL, Goode EL, Cunningham JM, Vierkant RA, Bruinsma F, Giles GG, Liang D, Hildebrandt MAT, Wu X, Levine DA, Bisogna M, Berchuck A, Iversen ES, Schildkraut JM, Concannon P, Weber RP, Cramer DW, Terry KL, Poole EM, Tworoger SS, Bandera EV, Orlow I, Olson SH, Krakstad C, Salvesen HB, Tangen IL, Bjorge L, van Altena AM, Aben KKH, Kiemeney LA, Massuger LFAG, Kellar M, Brooks-Wilson A, Kelemen LE, Cook LS, Le ND, Cybulski C, Yang H, Lissowska J, Brinton LA, Wentzensen N, Hogdall C, Lundvall L, Nedergaard L, Baker H, Song H, Eccles D, McNeish I, Paul J, Carty K, Siddiqui N, Glasspool R, Whittemore AS, Rothstein JH, McGuire V, Sieh W, Ji B-T, Zheng W, Shu X-O, Gao Y-T, Rosen B, Risch HA, McLaughlin JR, Narod SA, Monteiro AN, Chen A, Lin H-Y, Permuth-Wey J, Sellers TA, Tsai Y-Y, Chen Z, Ziogas A, Anton-Culver H, Gentry-Maharaj A, Menon U, Harrington P, Lee AW, Wu AH, Pearce CL, Coetzee G, Pike MC, Dansonka-Mieszkowska A, Timorek A, Rzepecka IK, Kupryjanczyk J, Freedman M, Noushmehr H, Easton DF, Offit K, Couch FJ, Gayther S, Pharoah PP, Antoniou AC, Chenevix-Trench G The Consortium of Investigators of Modifiers of B, Brca (2015) Identification of six new susceptibility loci for invasive epithelial ovarian cancer. Nat Genet 47(2): 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4(8): 1073–1081. [DOI] [PubMed] [Google Scholar]
- Kuznetsov S, Pellegrini M, Shuda K, Fernandez-Capetillo O, Liu Y, Martin BK, Burkett S, Southon E, Pati D, Tessarollo L, West SC, Donovan PJ, Nussenzweig A, Sharan SK (2007) RAD51C deficiency in mice results in early prophase I arrest in males and sister chromatid separation at metaphase II in females. J Cell Biol 176(5): 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Gu B, Hart J, Hoffman D, Hoover J, Jang W, Katz K, Ovetsky M, Riley G, Sethi A, Tully R, Villamarin-Salomon R, Rubinstein W, Maglott DR (2016) ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res 44(Database issue): D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR (2014) ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res 42(Database issue): D980–D985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léveillé F, Blom E, Medhurst AL, Bier P, Laghmani EH, Johnson M, Rooimans MA, Sobeck A, Waisfisz Q, Arwert F, Patel KJ, Hoatlin ME, Joenje H, de Winter JP (2004) The Fanconi anemia gene product FANCF is a flexible adaptor protein. J Biol Chem 279(38): 39421–39430. [DOI] [PubMed] [Google Scholar]
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P (2009) Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 25(21): 2744–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lio Y-C, Schild D, Brenneman MA, Redpath JL, Chen DJ (2004) Human Rad51C deficiency destabilizes XRCC3, impairs recombination, and radiosensitizes S/G2-phasecells. J Biol Chem 279(40): 42313–42320. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tarsounas M, O'Regan P, West SC (2007) Role of RAD51C and XRCC3 in genetic recombination and DNA repair. J Biol Chem 282(3): 1973–1979. [DOI] [PubMed] [Google Scholar]
- Loveday C, Turnbull C, Ruark E, Xicola RMM, Ramsay E, Hughes D, Warren-Perry M, Snape K, Eccles D, Evans DG, Gore M, Renwick A, Seal S, Antoniou AC, Rahman N (2012) Germline RAD51C mutations confer susceptibility to ovarian cancer. Nat Genet 44(5): 475–476. [DOI] [PubMed] [Google Scholar]
- Martelotto LG, Ng CK, De Filippo MR, Zhang Y, Piscuoglio S, Lim RS, Shen R, Norton L, Reis-Filho JS, Weigelt B (2014) Benchmarking mutation effect prediction algorithms using functionally validated cancer-related missense mutations. Genome Bio 15(10): 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20(9): 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, Freund M, Lichtner P, Hartmann L, Schaal H, Ramser J, Honisch E, Kubisch C, Wichmann HE, Kast K, Deiszler H, Engel C, Muller-Myhsok B, Neveling K, Kiechle M, Mathew CG, Schindler D, Schmutzler RK, Hanenberg H (2010) Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet 42(5): 410–414. [DOI] [PubMed] [Google Scholar]
- Miller KA, Sawicka D, Barsky D, Albala JS (2004) Domain mapping of the Rad51 paralog protein complexes. Nucleic Acids Res 32(1): 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min A, Im S-A, Yoon Y-K, Song S-H, Nam H-J, Hur H-S, Kim H-P, Lee K-H, Han S-W, Oh D-Y, Kim T-Y, O'Connor MJ, Kim W-H, Bang Y-J (2013) RAD51C-deficient cancer cells are highly sensitive to the PARP inhibitor olaparib. Mol Cancer Ther 12(6): 865–877. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11(5): 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio A, Endt D, Fernández F, Eirich K, de la Hoya M, Schmutzler R, Caldés T, Meindl A, Schindler D, Benitez J (2012) Predominance of pathogenic missense variants in the RAD51C gene occurring in breast and ovarian cancer families. Hum Mol Genet 21(13): 2889–2898. [DOI] [PubMed] [Google Scholar]
- Pelttari LM, Heikkinen T, Thompson D, Kallioniemi A, Schleutker J, Holli K, Blomqvist C, Aittomaki K, Butzow R, Nevanlinna H (2011) RAD51C is a susceptibility gene for ovarian cancer. Hum Mol Genet 20(16): 3278–3288. [DOI] [PubMed] [Google Scholar]
- Ramus SJ, Song H, Dicks E, Tyrer JP, Rosenthal AN, Intermaggio MP, Fraser L, Gentry-Maharaj A, Hayward J, Philpott S, Anderson C, Edlund CK, Conti D, Harrington P, Barrowdale D, Bowtell DD, Alsop K, Mitchell G, Group AS, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Sieh W, McGuire V, Lester J, Bogdanova N, Dürst M, Hillemanns P, Consortium OCA, Odunsi K, Whittemore AS, Karlan BY, Dörk T, Goode EL, Menon U, Jacobs IJ, Antoniou AC, Pharoah PDP, Gayther SA (2015) Germline mutations in the BRIP1, BARD1, PALB2, and NBN genes in women with ovarian cancer. J Natl Cancer Inst 107(11): djv214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C (2011) Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res 39(17): e118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero A, Pérez-Segura P, Tosar A, García-Saenz JÁ, Díaz-Rubio E, Caldés T, de la Hoya M (2011) A HRM-based screening method detects RAD51C germ-line deleterious mutations in Spanish breast and ovarian cancer families. Breast Cancer Res Treat 129(3): 939–946. [DOI] [PubMed] [Google Scholar]
- Schwarz JM, Rodelsperger C, Schuelke M, Seelow D (2010) MutationTaster evaluates disease-causing potential of sequence alterations. Nat Meth 7(8): 575–576. [DOI] [PubMed] [Google Scholar]
- Shihab HA, Gough J, Cooper DN, Stenson PD, Barker GLA, Edwards KJ, Day INM, Gaunt TR (2013) Predicting the functional, molecular, and phenotypic consequences of amino acid substitutions using hidden Markov models. Human Mutat 34(1): 57–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63: 11–30. [DOI] [PubMed] [Google Scholar]
- Smeenk G, de Groot AJL, Romeijn RJ, van Buul PPW, Zdzienicka MZ, Mullenders LHF, Pastink A, Godthelp BC (2010) Rad51C is essential for embryonic development and haploinsufficiency causes increased DNA damage sensitivity and genomic instability. Mutation Res 689(1–2): 50–58. [DOI] [PubMed] [Google Scholar]
- Somyajit K, Mishra A, Jameei A, Nagaraju G (2015. a) Enhanced non-homologous end joining contributes toward synthetic lethality of pathological RAD51C mutants with poly (ADP-ribose) polymerase. Carcinogenesis 36(1): 13–24. [DOI] [PubMed] [Google Scholar]
- Somyajit K, Saxena S, Babu S, Mishra A, Nagaraju G (2015. b) Mammalian RAD51 paralogs protect nascent DNA at stalled forks and mediate replication restart. Nucleic Acids Res 43(20): 9835–9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Dicks E, Ramus SJ, Tyrer JP, Intermaggio MP, Hayward J, Edlund CK, Conti D, Harrington P, Fraser L, Philpott S, Anderson C, Rosenthal A, Gentry-Maharaj A, Bowtell DD, Alsop K, Cicek MS, Cunningham JM, Fridley BL, Alsop J, Jimenez-Linan M, Høgdall E, Høgdall CK, Jensen A, Kjaer SK, Lubiński J, Huzarski T, Jakubowska A, Gronwald J, Poblete S, Lele S, Sucheston-Campbell L, Moysich KB, Odunsi K, Goode EL, Menon U, Jacobs IJ, Gayther SA, Pharoah PDP (2015) Contribution of germline mutations in the RAD51B, RAD51C, and RAD51D genes to ovarian cancer in the population. J Clin Oncol 33(26): 2901–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson BA, Greenblatt MS, Vallee MP, Herkert JC, Tessereau C, Young EL, Adzhubey IA, Li B, Bell R, Feng B, Mooney SD, Radivojac P, Sunyaev SR, Frebourg T, Hofstra RMW, Sijmons RH, Boucher K, Thomas A, Goldgar DE, Spurdle AB, Tavtigian SV (2013) Calibration of multiple in silico tools for predicting pathogenicity of mismatch repair gene missense substitutions. Hum Mutat 34(1): 255–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thusberg J, Olatubosun A, Vihinen M (2011) Performance of mutation pathogenicity prediction methods on missense variants. Hum Mutat 32(4): 358–368. [DOI] [PubMed] [Google Scholar]
- Vaclová T, Gómez-López G, Setién F, Bueno JMG, Macías JA, Barroso A, Urioste M, Esteller M, Benítez J, Osorio A (2015) DNA repair capacity is impaired in healthy BRCA1 heterozygous mutation carriers. Breast Cancer Res Treat 152(2): 271–282. [DOI] [PubMed] [Google Scholar]
- Vaz F, Hanenberg H, Schuster B, Barker K, Wiek C, Erven V, Neveling K, Endt D, Kesterton I, Autore F, Fraternali F, Freund M, Hartmann L, Grimwade D, Roberts RG, Schaal H, Mohammed S, Rahman N, Schindler D, Mathew CG (2010) Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat Genet 42(5): 406–409. [DOI] [PubMed] [Google Scholar]
- Vuorela M, Pylkäs K, Hartikainen JM, Sundfeldt K, Lindblom A, von Wachenfeldt Wäppling A, Haanpää M, Puistola U, Rosengren A, Anttila M, Kosma V-M, Mannermaa A, Winqvist R (2011) Further evidence for the contribution of the RAD51C gene in hereditary breast and ovarian cancer susceptibility. Breast Cancer Res Treat 130(3): 1003–1010. [DOI] [PubMed] [Google Scholar]
- Wang W (2007) Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet 8(10): 735–748. [DOI] [PubMed] [Google Scholar]
- Yachdav G, Kloppmann E, Kajan L, Hecht M, Goldberg T, Hamp T, Hönigschmid P, Schafferhans A, Roos M, Bernhofer M, Richter L, Ashkenazy H, Punta M, Schlessinger A, Bromberg Y, Schneider R, Vriend G, Sander C, Ben-Tal N, Rost B (2014) PredictProtein – an open resource for online prediction of protein structural and functional features. Nucleic Acids Res 42(Web Server issue): W337–W343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamborszky J, Szikriszt B, Gervai JZ, Pipek O, Poti A, Krzystanek M, Ribli D, Szalai-Gindl JM, Csabai I, Szallasi Z, Swanton C, Richardson AL, Szuts D (2017) Loss of BRCA1 or BRCA2 markedly increases the rate of base substitution mutagenesis and has distinct effects on genomic deletions. Oncogene 36(6): 746–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zdzienicka MgZ (1987) Mutagen-sensitive cell lines are obtained with high frequency in V79 Chinese cells. Mutat Res 178(2): 235–244. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Zhang J, Hope K, Niu Q, Huo D, Olopade OI (2010) Screening RAD51C nucleotide alterations in patients with a family history of breast and ovarian cancer. Breast Cancer Res Treat 124(3): 857–861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.