Abstract
Background:
Tyrosine kinase inhibitors (TKIs) have demonstrated clinical benefits in the treatment of several tumour types. However, the emergence of TKI resistance restricts the therapeutic effect. This study uses non-small cell lung cancer (NSCLC) to explore the mechanisms contributing to TKI resistance in tumours.
Methods:
Biological phenotypes and RNA microarray expression data were analysed in NSCLC cells with and without TKI pretreatment. Specific inhibitors and siRNAs were used to validate the direct involvement of an AKT/FOXM1/STMN1 pathway in TKI resistance. Patients’ tissues were analysed to explore the clinical importance of FOXM1 and STMN1.
Results:
In vitro and in vivo studies showed that TKIs induced the enrichment of cancer stem cells (CSC), promoted epithelial to mesenchymal transition (EMT), and conferred multidrug resistance on NSCLC cells in a cell type- and TKI class-dependent manner. Mechanistically, TKIs activated an AKT/FOXM1/STMN1 pathway. The crucial role of this pathway in TKI-induced enrichment of CSC and drug resistance was verified by silencing FOXM1 and STMN1 or blocking the AKT pathway. Additionally, overexpression of STMN1 was associated with upregulation of FOXM1 in advanced NSCLC patients, and STMN1/FOXM1 upregulation predicted a poor outcome.
Conclusions:
Our findings elucidate an additional common mechanism for TKI resistance and provide a promising therapeutic target for reversing TKI resistance in NSCLC.
Keywords: tyrosine kinase inhibitors, non-small cell lung cancer, cancer stem cell, resistance, AKT pathway, FOXM1, STMN1
Tyrosine kinase inhibitors (TKIs) are the first group of compounds to be used successfully for target-based cancer therapy (Ma and Adjei, 2009). A series of TKIs, including imatinib, gefitinib, erlotinib, sorafenib, regorafenib, and so on, has been used in the clinic to treat various kinds of tumour (Sivendran et al, 2012; Jackson and Chester, 2015; Kircher et al, 2016). However, resistance to TKI therapy, either intrinsic or acquired, is a major clinical problem (Kuwano et al, 2016).
Several molecular mechanisms have been demonstrated to contribute to TKI resistance: (1) secondary mutations in tyrosine kinase receptors, such as EGFR T790M, which confers resistance to gefitinib and erlotinib in lung cancer (Morgillo et al, 2016; Piotrowska and Sequist, 2016); (2) oncogene amplification, such as MET gene amplification in gefitinib-resistant lung cancer (Tan et al, 2015); (3) activation of alternative pathways and signalling molecules, such as the PI3K pathway, which confers resistance to sorafenib in hepatic cancer (Chen et al, 2011), and IGF1R, which confers gefitinib resistance in head and neck squamous carcinoma (Jameson et al, 2011); and (4) aberrant or impaired downstream pathways, for example, loss of PTEN, which confers resistance to erlotinib in lung cancer (Bidkhori et al, 2012). However, there is still a need for an improved understanding of the complex mechanisms that are involved in TKI resistance, especially the mechanisms that may be shared by different TKIs. There is also a requirement for the development of validated predictive markers to allow better treatment decisions based on the probability of a positive response.
In this study, we used non-small cell lung cancer (NSCLC) as a model to explore common mechanisms that contribute to TKI resistance in tumours. We investigated gefitinib, which specifically inhibits EGFR (epidermal growth factor receptor), and sorafenib and regorafenib, which are multikinase inhibitors. All three TKIs are approved or under investigation for the treatment of NSCLC (Hellmann et al, 2015; Paz-Ares et al, 2015; Piotrowska and Sequist, 2016). Our results show that TKI pretreatment significantly enriched cancer stem cells (CSC), promoted epithelial to mesenchymal transitions (EMT), and induced multidrug resistance in NSCLC cells. Importantly, a novel AKT/FOXM1/STMN1 biological axis was identified to mediate this phenotypic transformation. Moreover, the present data suggest that high expression of both STMN1 and FOXM1 may be a valuable predictive biomarker for determining the prognosis of NSCLC patients.
Materials and methods
Cell lines, cell culture, and treatment
Human lung cancer cell lines NCI-H460, A549, and NCI-H1299 were obtained from the American Type Culture Collection (Manassas, VA, USA). These cancer cells were routinely cultured in RPMI-1640 or DMEM medium supplemented with 10% foetal bovine serum and maintained at 37 °C in a humidified incubator with 5% CO2. The cells were treated with TKIs at pharmacological concentrations (5 μM sorafenib, 5 μM regorafenib, and 4 μM gefitinib) for 48 h, and then allowed to recover for 48 h. After two–four of these treatment cycles, the cells were prepared for biological studies.
Patients and therapy
A total of 72 patients with advanced NSCLC (stage IIIB and stage IV) were enroled between January 2004 and June 2012 from Wuhan General Hospital (Wuhan, China). Detailed information is shown in Supplementary Table 1. Ethical oversight and approval was obtained from the Institutional Review Board of Wuhan General Hospital.
Immunohistochemistry
A tissue microarray was constructed (in collaboration with the Shanghai Biochip Company Ltd.) as described previously (Wang et al, 2016). Paraffin sections were dewaxed with xylene and rehydrated in descending concentrations of ethanol. The endogenous peroxidase was inhibited, and the slides were incubated with antibody against FOXM1 (1 : 100; Abcam, Inc. Cambridge, MA, USA) and STMN1 (1 : 100; Cell Signaling Technology, Beverly, MA, USA). The expression level of FOXM1 and STMN1 was graded as described previously (Wang et al, 2007).
Compounds and reagents
Gefitinib was purchased from J&K Chemical (Beijing, China). Sorafenib, regorafenib, MK2206 (AKT inhibitor), and PD98059 (ERK inhibitor) were obtained from MedChem Express (Princeton, NJ, USA). The primary antibodies against Sox2, Oct4, Nanog, STMN1, E-cadherin, N-cadherin, vimentin, ERK, phosphor–ERK, Akt, phosphor–Akt, JNK, phosphor–JNK, MMP-9, Histone3, and β-actin were obtained from Cell Signaling Technology. The primary antibodies against FOXM1, E2F1, and MMP2 were purchased from Abcam. Silencer Select Validated siRNAs against STMN1 and FOXM1 were obtained from Life Technologies, Carlsbad, CA, USA.
Clonogenicity assay
The cells were treated with TKIs (5 μM sorafenib, 5 μM regorafenib, and 4 μM gefitinib) for 48 h, and then recovered for 48 h. After two–four treatment cycles (two cycles for sorafenib, four cycles for regorafenib and gefitinib), the cells were resuspended and reseeded in 30 mm dishes (500 cells per dish). Next, the cells were incubated for an additional 7 days. The colonies obtained were fixed in paraformaldehyde, stained with crystal violet, and counted. The number of colonies was compared with the number obtained from untreated cells.
Tumoursphere formation assay
Single cells prepared from mechanical and enzymatic dissociation were seeded in six-well ultra-low attachment plates (Corning, NY) at 3000 cells per well for about 2 weeks. The cells were cultured in serum-free DMEM/F-12 medium containing B27 supplement (1 × Invitrogen, Carlsbad, CA, USA), 20 ng ml−1 human recombinant bFGF (PeproTech, Rocky Hill, NJ, USA), and 20 ng ml−1 EGF (PeproTech).
Western blot analysis
About 1–5 × 106 cells were gathered after pretreatment for the indicated time periods as described above. Western blotting was performed as previously described (Wang et al, 2015). In brief, equal amounts of total protein extracts from cultured cells or tissues were fractionated by 10–15% SDS–PAGE and electrically transferred onto polyvinylidene difluoride (PVDF) membranes. Mouse or rabbit primary antibodies and horseradish peroxidase-conjugated appropriate secondary antibodies were used to detect the designated proteins. The antibody concentrations are listed in Supplementary Table 2. The bound secondary antibodies on the PVDF membrane were reacted with ECL detection reagents (Pierce, Rockford, IL, USA) and exposed in a dark room. Results were normalised to the internal control (β-actin or Histone3).
Flow cytometry analysis
Aldefluor assays were done according to the manufacturer’s guidelines (Stem Cell Technologies, Cambridge, MA, USA). Single cells obtained from cell cultures were incubated in an Aldefluor assay buffer containing an ALDH substrate, bodipy-aminoacetaldehyde (1 μmol l−1 per 1 000 000 cells), for 30–60 min at 37 °C. As a negative control, a fraction of the cells from each sample was incubated under identical conditions in the presence of the ALDH inhibitor diethylaminobenzaldehyde. Flow cytometry was used to measure the ALDH-positive cell population. Desired cell populations were isolated using a FACSAria III flow cytometer (BD Biosciences, San Jose, CA, USA).
Real-time cell analysis (RTCA)
RTCA assays were used to detect cell proliferation or cell migration. The xCELLigence system (Real Time Cell Analyzer Single Plate, RTCA SP) allows real-time monitoring of cell proliferation or migration based on impedance measurement. The technology uses specific cell culture E-Plates with the bottoms covered with microelectrodes that act as an electrical impedance-based cell sensor. The analysis is based on the measurement of electrical impedance created by attached cells across the high-density electrode array coating the bottom of the wells. The impedance value is automatically converted to a dimensionless parameter, cell index (CI), which is defined as the relative change in electrical impedance created by the attached cells. As a quantitative measure of cellular status, CI value represents the extent of the cell-covered area and is directly related to the number of the proliferative or migratory cells.
Cell viability assay
The in vitro cell viability was determined by MTT assay. Cells (8 × 104 cells ml−1) were seeded into 96-well culture plates. After overnight incubation, the cells were treated with various concentrations of agents for 48 h. Then 10 μl MTT solution (2.5 mg ml−1 in PBS) was added to each well, and the plates were incubated for an additional 4 h at 37 °C. After centrifugation (2500 rpm, 10 min), the medium containing MTT was aspirated, and 100 μl DMSO was added. The optical density of each well was measured at 570 nm with a Biotek SynergyTM HT Reader.
Microarray gene expression analysis
Total RNA was isolated from TKI-pretreated cells using an RNeasy Mini Kit (Qiagen, Düsseldorf, Germany) as described in the product insert. Array hybridisation was performed according to the Affymetrix FS450_0002 Hybridisation Protocol for gene expression. The Affymetrix GeneChip PrimeView Human Gene Expression Arrays were scanned with an Affymetrix Genechip Scanner 7G.
Chromatin immunoprecipitation assay (ChIP)
NCI-H460 cells were prepared for the ChIP assay according to the manufacturer’s instructions (ChIP Assay Kit, Beyontime, China). A FOXM1 antibody was used for immunoprecipitation. STMN1 promoter primers were used to carry out PCR analysis of DNA isolated from the ChIP experiment. The sequences of PCR primers are as follows: Forward: 5′-ATACGTGGATTGAGGACCACT-3′, Reverse: 5′-TCCAATGTCAAGTAGCGGTTG-3′. PCR products were analysed by 1.5% agarose/ethidium bromide gel electrophoresis. Data are presented as %input, which was calculated from the integral optical density value of the PCR bands as: IP/input × 100.
Mouse xenograft tumour model
To assess the characteristics of chemotherapy-resistant tumours, viable NCI-H460 cells (5 × 106 per 100 μl PBS per mouse), as confirmed by trypan blue staining, were subcutaneously injected into the right flank of 7- to 8-week-old male BALB/c nude mice. When the average tumour volume reached 50 mm3, the mice were randomly divided into three treatment groups: control (saline only, n=6); sorafenib (100 mg kg−1 per day, i.g.; n=6); and gefitinib (32 mg kg−1 per days, i.g.; n=6). The dose of sorafenib and gefitinib was calculated according to the clinically equivalent amount. After 3 weeks, the mice were killed and the tumours were excised and stored at −80 °C until used for western blotting. These studies were performed in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Committee on the Ethics of Animal Experiments of Shenyang Pharmaceutical University.
Statistical analysis
Differences between experimental groups were evaluated by one-way ANOVA or Turkey’s post hoc test using the SPSS11.5 software package for Windows (SPSS, Chicago, IL, USA). Survival curves were constructed using the Kaplan–Meier method. Statistical significance was based on a P-value of 0.05 (P<0.05, two-tailed test).
Results
Pretreatment with TKIs induces a CSC phenotype in NSCLC cells
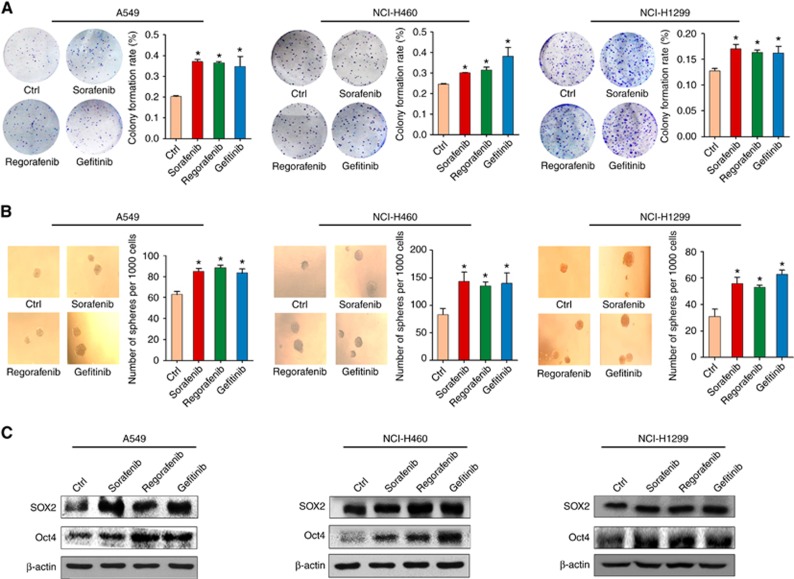
Enrichment of CSC is considered to be a characteristic of chemotherapy-resistant cells (Zhao, 2016), so we first measured whether three TKIs, sorafenib, regorafenib, and gefitinib, affected the CSC phenotypes of NSCLC cells. Our results showed that, at pharmacological concentrations, pretreatment with TKIs significantly increased the number of colonies formed by A549 cells (p53 wild type) as compared to control cells treated with DMSO (Figure 1A). A similar pattern was also observed in NCI-H460 (p53 wild type) and NCI-H1299 (p53 deletion) cells. Next, we examined the effects of TKIs on tumoursphere formation, which mimics the self-renewal capability of CSC. As shown in Figure 1B, pretreatment with all three TKIs promoted the formation of tumourspheres in the three NSCLC cell lines (Figure 1B).
Figure 1.
The effects of TKI agents on CSCs’ characteristics in NSCLC cells. (A) The formation of colonies by A549, NCI-H460, and NCI-H1299 cells after pretreatment with sorafenib (5 μM), regorafenib (5 μM), and gefitinib (4 μM). (B) The formation of tumourspheres by the three cell lines after pretreatment with sorafenib (5 μM), regorafenib (5 μM), and gefitinib (4 μM). (C) The effects of pretreatment with TKI agents on the expression of Sox2 and Oct4 in A549, NCI-H460, and NCI-H1299 cell lines. β-actin was used as a loading control. All error bars are s.e.m. *P<0.05, compared with the control (Ctrl).
To further confirm the phenotypic change in TKI-pretreated cells, the levels of the pluripotency transcription factors, Oct4 and Sox2, were also assessed. As shown in Figure 1C, the expression levels of Sox2 and Oct4 in NSCLC cells were upregulated by the three TKI agents, which is consistent with our phenotypic data. The CSC traits were also confirmed by ALDH assessment. Flow cytometry data indicated that pretreatment with sorafenib and gefitinib, but not regorafenib, enhanced the activity of ALDH in A549 cells (Supplementary Figure S1A). Further, our results revealed that sorafenib and gefitinib pretreatment led to the upregulation of certain ALDH subtypes in a cell type-dependent manner (Supplementary Figure S1B). Next, we further verified the above data using an in vivo model. NCI-H460 xenograft mice were administered with sorafenib and gefitinib for 3 weeks, and the levels of Sox2 and Oct4 in tumours were assessed by western blot. Both proteins were upregulated in the NCI-H460 xenograft tumour tissues (Supplementary Figure S2). Taken together, these results illustrate that continuous treatment with TKIs can induce the enrichment of CSC in NSCLC cell lines, which might be associated with TKI resistance.
TKI pretreatment triggers EMT and confers multidrug resistance in NSCLC
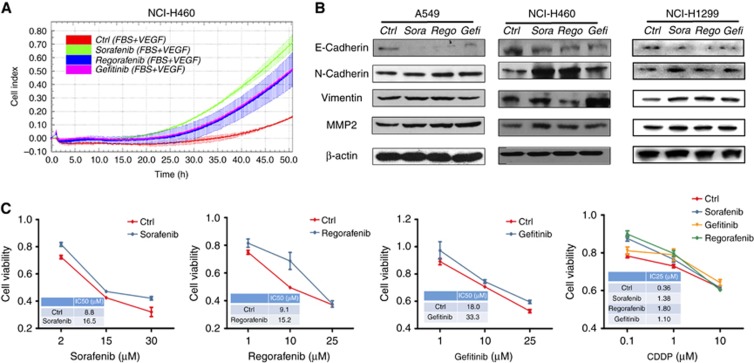
Epithelial to mesenchymal transition (EMT) is considered to be an important characteristic of TKI resistance (Morgillo et al, 2011a; Jakobsen et al, 2016). We therefore detected the migration status of cells after treatment with TKIs. RTCA data revealed that TKI pretreatment of NCI-H460 cells resulted in an enhancement of cell migration as compared with DMSO-treated controls (Figure 2A). The effect was strongest for sorafenib treatment. Consistent with the cell migration data, sorafenib pretreatment of NCI-H460 cells also led to a marked decrease of E-cadherin and an increase of N-cadherin, vimentin and MMP2 (Figure 2B), which are recognised as EMT-related proteins (Tania et al, 2014). A similar pattern was also found in NSCLC cells pretreated with regorafenib and gefitinib (Figure 2B). Moreover, an in vivo study in NCI-H460 xenograft mice indicated that the sorafenib administration decreased the level of E-cadherin and increased the level of vimentin in the xenograft tumour tissue, while gefitinib decreased the level of E-cadherin and increased the level of both vimentin and MMP-9 (Supplementary Figure S2). The above analyses of phenotypes and biomarkers demonstrate that TKIs trigger EMT in NSCLC cells.
Figure 2.
The effects of TKI agents on cell migration, EMT markers, and drug resistance in NSCLC cells. (A) The migration of NCI-H460 cells after pretreatment with sorafenib (5 μM), regorafenib (5 μM), and gefitinib (4 μM). The CI represents the capacity for cell migration, and the slope of the curve is related to the migration velocity of the tumour cells. (B) The effects of pretreatment with TKI agents on the expression of E-cadherin, N-cadherin, vimentin, and MMP2 in A549, NCI-H460, and NCI-H1299 cells. β-actin was used as a loading control. (C) The viability of TKI-pretreated or control NCI-H460 cells after treatment with different concentrations of TKIs or CDDP for 48 h.
The EMT phenotypic transformation might affect the drug sensitivity of the cells. Therefore, we next measured whether TKI-treated and untreated NSCLC cells differ in drug sensitivity. NCI-H460 cells were pretreated with TKIs, then exposed to TKIs or cisplatin (CDDP), a first-line chemotherapeutic drug for NSCLC (Fennell et al, 2016). Cell viability assays showed that TKI pretreatment of NCI-H460 cells resulted in resistance to TKIs and CDDP (Figure 2C). Similarly, TKI pretreatment partially reduced the sensitivity of A549 and NCI-H1299 cells to TKIs (Supplementary Figure S3). These results indicate that TKI pretreatment of NSCLC cells can promote resistance to multiple drugs.
TKI pretreatment of NSCLC cells promotes the upregulation of STMN1 mediated by FOXM1
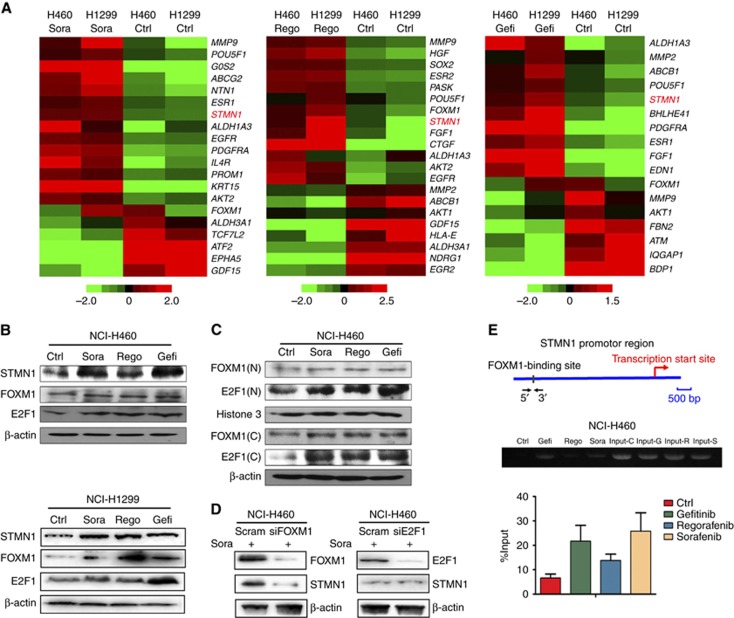
To explore the mechanisms underlying the TKI-induced phenotypic changes in NSCLC cells, we performed microarray gene expression analysis using Affymetrix PrimeView chips. As shown in the heat plots in Figure 3A, some oncogenesis-related genes (red colour) were upregulated in TKI-pretreated NCI-H460 and NCI-H1299 cells. The drug efflux genes ABCG2 and ABCB1 were upregulated in sorafenib- and gefitinib-treated cells; the CSC biomarker ALDH1A3 was upregulated in cells treated with all three TKIs; and the EMT-related gene MMP-9 and the cell proliferation-related gene EGFR were upregulated in sorafenib- and regorafenib-treated cells. STMN1, a phosphoprotein associated with cancer metastasis (Kuang et al, 2016), was upregulated in cells treated with all three TKIs (Figure 3A). For validation, we analysed the expression level of STMN1 in TKI-treated and untreated NCI-H460, NCI-H1299, and A549 cell lines, and found that the STMN1 protein was upregulated in all three NSCLC cell lines following pretreatment with TKIs (Figure 3B and Supplementary Figure S4). This suggests that STMN1 may play a crucial role in TKI-induced phenotypic change.
Figure 3.
Microarray gene expression analysis and STMN1 regulation in TKI-pretreated NSCLC cells. (A) Heat maps depicting the differential expression of tumour-related signature genes in TKI-pretreated and control NCI-H460 and NCI-H1299 cells. Red and green indicate high and low mRNA expression levels, respectively. (B) The expression levels of STMN1, FOXM1, and E2F1 were detected by western blot in TKI-pretreated and control NCI-H460 and NCI-H1299 cells. β-actin was used as a loading control. (C) The subcellular distribution of FOXM1 and E2F1 in TKI-pretreated NCI-H460 cells. Histone3 was detected as a nuclear control, and β-actin was used as a cytoplasmic control. N, nuclear; C, cytoplasmic. (D) The expression of STMN1 and FOXM1, or STMN1 and E2F1, was assessed in sorafenib-pretreated NCI-H460 cells after knockdown of FOXM1 or E2F1, respectively, by siRNA (50 nM). (E) ChIP assays confirm the binding of FOXM1 to the promoter region upstream of the STMN1 gene. All error bars are s.e.m.
The transcription factors, FOXM1 and E2F1, were reported to positively regulate the expression level of STMN1 (Petrovic et al, 2008; Chen et al, 2013). To address whether the overexpression of STMN1 is related to FOXM1 and E2F1, we measured their expression levels in TKI-treated and -untreated NSCLC cell lines. Our results revealed that pretreatment with TKIs also induced, to different extents, the expression of FOXM1 and E2F1 in the three NSCLC cell lines (Figure 3B and Supplementary Figure S2). Additionally, subcellular distribution results indicated that pretreatment with the three TKIs led to the accumulation of FOXM1 and E2F1 in the nucleus as well as the cytoplasm in NCI-H460 cells (Figure 3C). To further confirm the role of the transcription factors in STMN1 regulation, the expression of STMN1 was evaluated in sorafenib-pretreated NCI-H460 and A549 cells after siRNA knockdown of FOXM1 or E2F1. As shown in Figure 3D and Supplementary Figure S4B, knockdown of FOXM1 by siRNA reduced the expression of STMN1 compared with the scramble control. In contrast, knockdown of E2F1 did not result in the downregulation of STMN1. The above data indicate that FOXM1, but not E2F1, plays an important role in the sorafenib-induced upregulation of STMN1. The direct regulatory relationship between FOXM1 and STMN1 was further verified by ChIP assay. Our data demonstrated that pretreatment with the three TKIs contributed to a substantial enrichment of FOXM1 on the STMN1 promoter in NCI-H460 cells (Figure 3E). This suggests a potential role for FOXM1 in transcriptional regulation of STMN1. Taken together, the results demonstrate that overexpression of STMN1 in TKI-pretreated NSCLC cells is dependent on FOXM1 regulation.
STMN1 and FOXM1 are involved in TKI-induced CSC enrichment and drug resistance in NSCLC cells
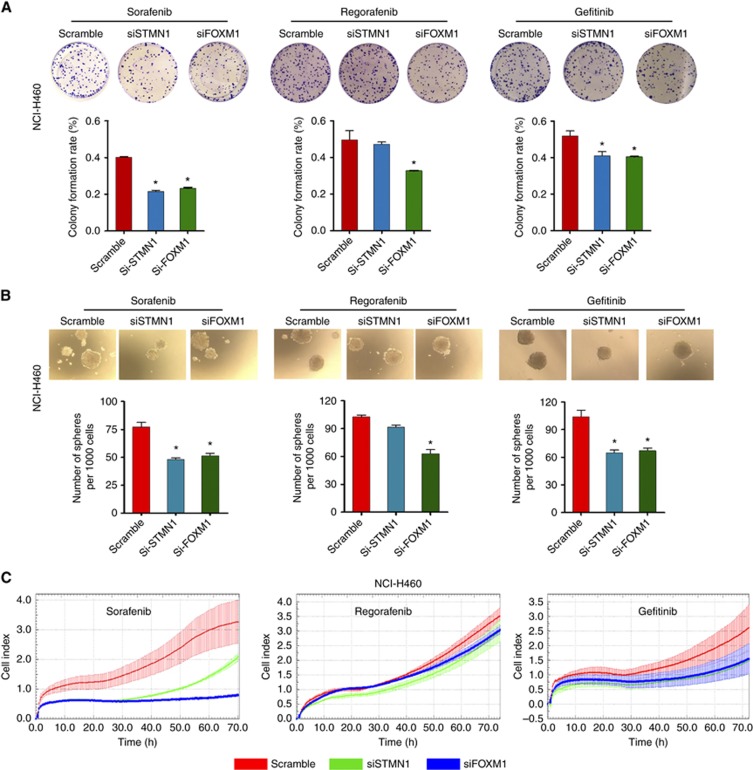
To understand the role of STMN1 and FOXM1 in TKI-induced enrichment of CSCs and drug resistance, we next knocked down STMN1 and FOXM1 by siRNA. Colony formation assay data showed that knockdown of FOXM1 contributed to a marked reduction in the number of colonies, compared to the scramble control, in NCI-H460 cells pretreated with the three TKIs (Figure 4A). Knockdown of STMN1 also resulted in a decrease in the colony number in sorafenib- and gefitinib-pretreated NCI-H460 cells (Figure 4A), and the efficacy of STMN1 knockdown was similar to that of FOXM1 knockdown, suggesting the possibility of a dependent relationship between FOXM1 and STMN1. In contrast to FOXM1 knockdown, STMN1 knockdown had no significant effect on colony formation in regorafenib-pretreated NCI-H460 cells. Knockdown of FOXM1 also led to an obvious reduction in TKI-induced sphere formation in NCI-H460 cells (Figure 4B). Consistent with the colony formation data, knockdown of STMN1 resulted in a decrease in the number of tumourspheres in sorafenib- and gefitinib-pretreated NCI-H460 cells, but had no significant effect on regorafenib-pretreated cells. We also investigated the effect of STMN1 and FOXM1 knockdown on the sensitivity of TKI-pretreated cells to TKIs. The results revealed that both STMN1 siRNA and FOXM1 siRNA enhanced, to different extents, the sensitivity of TKI-pretreated NCI-H460 cells to TKIs (Figure 4C). In particular, knockdown of STMN1 and FOXM1 in sorafenib-pretreated cells resulted in an obvious improvement in sensitivity to sorafenib. Gefitinib- and regorafenib-pretreated cells showed a moderately and weakly increased sensitivity to TKIs, respectively. Taken together, our results indicate the possibility that STMN1 and FOXM1 mediate the TKI-induced CSC enrichment and drug resistance in NSCLC cells.
Figure 4.
The effects of knockdown of STMN1 and FOXM1 on TKI-induced CSC enrichment and drug resistance in NSCLC cells. Colony formation (A) and sphere formation (B) were assessed in TKI-pretreated NCI-H460 cells after knockdown of STMN1 or FOXM1 by siRNA (50 nM). (C) The sensitivity to TKIs was assessed by RTCA proliferation assays in TKI-pretreated NCI-H460 cells after single knockdown of STMN1 and FOXM1 by siRNA (50 nM). All error bars are s.e.m. *P<0.05, compared with the control.
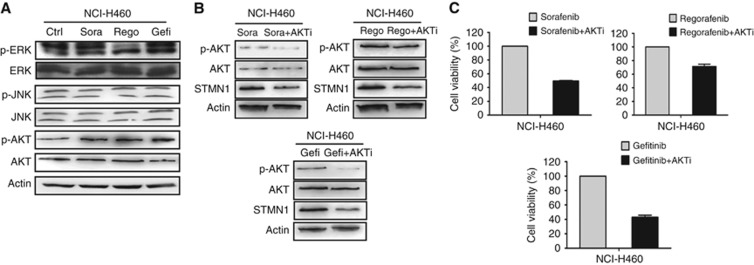
The PI3K/AKT pathway, but not the MAPK pathway, mediates TKI-induced STMN1 upregulation and drug resistance
TKI resistance was reported to be related to feedback activation of the PI3K/AKT and MAPK pathways (Morgillo et al, 2011b), which are considered to be upstream of FOXM1 (Miyashita et al, 2015). Therefore, we next asked whether the FOXM1-dependent upregulation of STMN1 is associated with the PI3K/AKT and MAPK pathways. Western blot data showed that the three TKIs can induce the upregulation of phosphorylated AKT, but have no obvious effect on phosphor–JNK expression (Figure 5A). The three TKIs had different effects on activation of the MAPK/ERK pathway. Sorafenib and gefitinib, but not regorafenib, induced an upregulation of phosphor–ERK in NCI-H460 cells (Figure 5A). To further explore the role of PI3K/AKT and MAPK/ERK in STMN1 regulation and drug resistance, we pretreated NCI-H460 cells with TKIs and measured the expression of STMN1 and cell viability after treatment with inhibitors of AKT or ERK. As shown in Figure 5B, treatment with MK2206, an AKT inhibitor, resulted in decreased levels of STMN1 and phosphor–AKT in NCI-H460 cells pretreated with the three TKIs. Consistent with the STMN1 downregulation, treatment with the AKT pathway inhibitor also led to growth inhibition of TKI-pretreated NCI-H460 cells (Figure 5C). In contrast, the ERK pathway inhibitor PD98059 did not affect the expression of STMN1 or the viability of TKI-pretreated cells (Supplementary Figure S5A and B). Taken together, the above data suggest that the PI3K/AKT pathway, but not the MAPK pathway, mediates TKI-induced STMN1 upregulation and drug resistance in NSCLC cells.
Figure 5.
The effects of TKIs on the AKT and MAPK pathways, and the effect of blocking the AKT pathway on STMN1 expression and cell viability in NSCLC cells. (A) The expression levels of phosphor–ERK, ERK, phosphor–JNK, JNK, phosphor–AKT, and AKT were detected by western blot in TKI-pretreated and control NCI-H460 cells. β-actin was used as a loading control. (B) The effect of the AKT inhibitor MK2206 (10 μM) on the expression levels of phosphor–AKT, AKT, and STMN1 in TKI-pretreated NCI-H460 cells. β-actin was used as a loading control. (C) The effect of MK2206 (10 μM) on the viability of TKI-pretreated NCI-H460 cells.
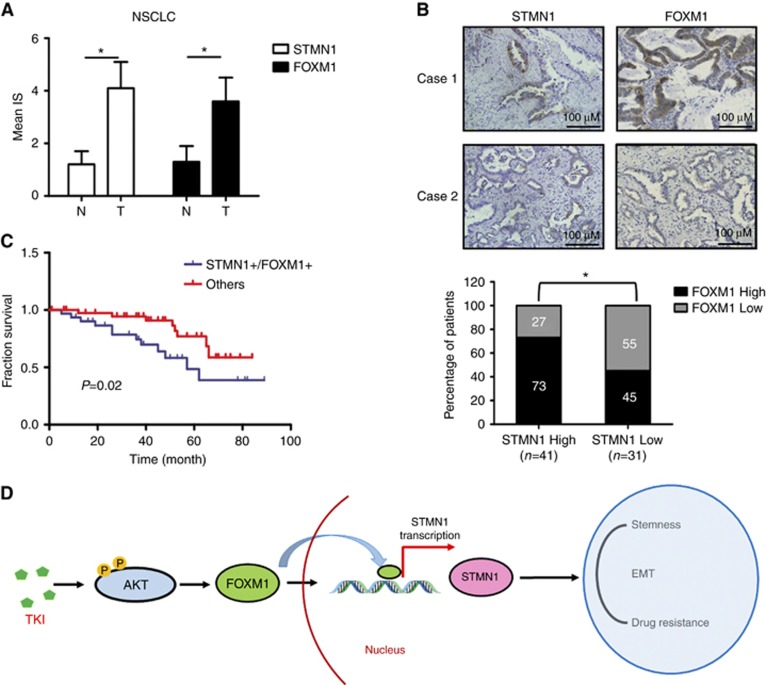
Overexpression of STMN1 and FOXM1 is associated with unfavourable prognosis in NSCLC patients
To address whether the expression of STMN1 and FOXM1 is related to the development and progression of disease in NSCLC patients, immunohistochemistry was performed on 72 lung tumours from NSCLC patients and 10 paired normal tissue samples. Both STMN1 and FOXM1 proteins were elevated in the NSCLC samples compared with the adjacent normal tissues (Figure 6A), suggesting a pro-oncogenic role of STMN1 and FOXM1 in NSCLC. In addition, our data showed that 41 tumours (56.9%) had a higher level of STMN1. Out of 41 tumours with higher levels of STMN1, 30 (73%) also expressed FOXM1 at a relatively high level (Figure 6B). Statistical analysis of the data indicated that there was a positive correlation between STMN1 and FOXM1 in NSCLC tumour tissues (P<0.05, Figure 6B). Notably, the group of lung cancer patients with higher expression of both STMN1 and FOXM1 had poorer overall survival than other groups with lung cancer (Figure 6C). Therefore, STMN1 together with FOXM1 may serve as a predictive marker of outcome in NSCLC patients.
Figure 6.
The expression of STMN1 and FOXM1 in normal human lung tissues and NSCLCs. (A) The expression levels of STMN1 and FOXM1 in NSCLC tissues and adjacent normal tissues. Abbreviations: N, normal tissue; T, tumour tissue. *P<0.05, compared with normal(N) tissues. (B) The expression of STMN1 and FOXM1 in representative NSCLC tissues (top). Images are magnified × 200. The correlation between STMN1 and FOXM1 expression in NSCLC tumour tissues (bottom). *P<0.05, compared between two groups. (C) Overall survival of patients according to expression of STMN1 together with FOXM1 in NSCLC tumour tissues. (D) Schematic figure illustrating how TKIs enrich CSCs, induce EMT, and trigger drug resistance in NSCLC cells. Phosphorylation is indicated by P in yellow circles.
Discussion
Here, our in vitro and in vivo studies showed that TKI pretreatment led to enrichment of CSC, induction of EMT, and triggering of multidrug resistance in NSCLC cells. Further studies elucidated that TKI pretreatment resulted in the activation of an AKT/FOXM1/STMN1 pathway. The crucial role of FOXM1 and STMN1 in TKI-induced enrichment of CSC and drug resistance was demonstrated by knockdown of STMN1 and FOXM1 in NSCLC cells. Interestingly, blocking the PI3K/AKT pathway with a specific inhibitor led to the downregulation of STMN1 and reversed the drug resistance. Importantly, we found that overexpression of STMN1 was correlated with upregulation of FOXM1 and was associated with poor prognosis in advanced NSCLC patients. Our findings illustrate a novel common mechanism for TKI resistance and provide a promising biomarker and therapeutic target for NSCLC.
Drug resistance is considered as one of the most important challenges to cancer therapy (Tan et al, 2015). Multiple mechanisms are involved in resistance to various drugs, and only a limited number of common mechanisms have been reported. Recently, we and others have reported that cytotoxic drugs can enrich CSC and subsequently result in drug resistance (Zhao, 2016; Wang et al, 2017). Here we verified that, similar to some cytotoxic drugs, TKI pretreatment at pharmacological concentrations also resulted in CSC enrichment and induction of EMT in NSCLC cells. Consistent with our findings, several clinical studies have shown that phenotypic and histological transformations frequently occurred during TKI treatment, especially in resistant patients. Sequist et al (2011) revealed that 14% of studied TKI-resistant cases showed histological transformation from NSCLC to small cell lung cancer (SCLC), and 6% of cases displayed the EMT phenotypic transformation. Furthermore, Ren et al (2014) reported that, compared with the epithelial phenotype, the EMT phenotype predicted a poor response to TKI and a poor outcome. Due to the diverse differentiation potential of CSC, it is possible that the enrichment of CSC after TKI treatment leads to histological or phenotypic transformation, and consequently contributes to TKI resistance. In fact, the results showing that TKI pretreatment induces CSC phenotypes in NSCLC cells can be explained by two different mechanisms, which are not mutually exclusive: (1) TKIs actively induce CSC phenotypes; (2) CSC are more resistant to TKIs, and therefore CSC are enriched in the cell population after treatment.
STMN1, a 19 KD cytosolic protein, was first identified as a microtubule-destabilising phosphoprotein, which is involved in the construction and function of the mitotic spindle. Overexpression of STMN1 was found in various tumours, including neuroblastoma, ovarian cancer, breast cancer, and lung cancer (Cassimeris, 2002; Rana et al, 2008). Recently, several studies revealed that STMN1 plays an important role in the tumour cell cycle (Rubin and Atweh, 2004), metastasis (Wu et al, 2014), and paclitaxel resistance (Lin et al, 2016), which further verifies the oncogenic role of STMN1. In the present study, our microarray mRNA expression data demonstrated that STMN1 was upregulated in three TKI-pretreated NSCLC cell lines. Further study showed that siRNA-mediated STMN1 knockdown resulted in the reversal of the TKI-induced CSC phenotypes and resistance to TKI agents. Our data demonstrated that upregulation of STMN1 plays a common role in regulating the TKI-induced CSC enrichment and drug resistance. The potential value of analysing STMN1 in NSCLC was also confirmed by examination of tumour tissues. The expression of STMN1 was found to be upregulated in tumour tissues, as compared with normal tissues, and higher expression of both STMN1 and FOXM1 was associated with unfavourable prognosis. These findings suggest the prognostic and therapeutic value of analysing STMN1 and FOXM1 in NSCLC.
Clarifying the mechanism of STMN1 upregulation is very important for understanding TKI resistance and developing effective therapeutic approaches. Previous studies showed that FOXM1 and E2F1, as transcription factors, can regulate the expression of STMN1 (Petrovic et al, 2008; Chen et al, 2013). In our study, gene manipulation and ChIP assay data indicated that FOXM1, but not E2F1, plays an important role in the upregulation of STMN1 induced by sorafenib in NCI-H460 cells. Furthermore, the immunohistochemistry data further confirmed that overexpression of STMN1 was associated with the upregulation of FOXM1 in tumour tissues from NSCLC patients. To further extend the regulatory network containing FOXM and STMN1, we also investigated the upstream pathways of FOXM1, including PI3K/AKT and MAPK (Miyashita et al, 2015). Our data revealed that the PI3K/AKT pathway, but not the MAPK pathway, mediated TKI-induced STMN1 upregulation and drug resistance in NSCLC cells. This suggests that, besides manipulation of STMN1 or FOXM1, regulation of the PI3K/AKT pathway by specific inhibitors can also eliminate TKI-induced CSC enrichment and drug resistance in NSCLC.
In conclusion, the present study shows that TKIs can enrich CSCs, induce EMT, and subsequently result in drug resistance, and these properties are associated with aberrant activation of the AKT/FOXM1/STMN1 axis (Figure 6E). Genetic manipulation of FOXM1 and STMN1, or blocking the AKT pathway by specific inhibitors, may eliminate CSC and reverse drug resistance. Our results not only elucidate an additional common mechanism for TKI resistance, but also provide a promising therapeutic target to reverse TKI resistance in NSCLC.
Acknowledgments
The authors gratefully acknowledge financial support from National Natural Science Foundation of China (No. 81572947), Generic Technology Innovative Platform of Liaoning Province (No. 2015010101), and Natural Science Foundation of Hubei Province, China (No. 2015CFB280).
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
The authors declare no conflict of interest.
Supplementary Material
References
- Bidkhori G, Moeini A, Masoudi-Nejad A (2012) Modeling of tumor progression in NSCLC and intrinsic resistance to TKI in loss of PTEN expression. PLoS One 7: e48004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L (2002) The oncoprotein 18/stathmin family of microtubule destabilizers. Curr Opin Cell Biol 14: 18–24. [DOI] [PubMed] [Google Scholar]
- Chen KF, Chen HL, Tai WT, Feng WC, Hsu CH, Chen PJ, Cheng AL (2011) Activation of phosphatidylinositol 3-kinase/Akt signaling pathway mediates acquired resistance to sorafenib in hepatocellular carcinoma cells. J Pharmacol Exp Ther 337: 155–161. [DOI] [PubMed] [Google Scholar]
- Chen YL, Uen YH, Li CF, Horng KC, Chen LR, Wu WR, Tseng HY, Huang HY, Wu LC, Shiue YL (2013) The E2F transcription factor 1 transactives stathmin 1 in hepatocellular carcinoma. Ann Surg Oncol 20: 4041–4054. [DOI] [PubMed] [Google Scholar]
- Fennell DA, Summers Y, Cadranel J, Benepal T, Christoph DC, Lal R, Das M, Maxwell F, Visseren-Grul C, Ferry D (2016) Cisplatin in the modern era: the backbone of first-line chemotherapy for non-small cell lung cancer. Cancer Treat Rev 44: 42–50. [DOI] [PubMed] [Google Scholar]
- Hellmann MD, Sturm I, Trnkova ZJ, Lettieri J, Diefenbach K, Rizvi NA, Gettinger SN (2015) Preliminary safety, pharmacokinetics, and efficacy of regorafenib, cisplatin, and pemetrexed in patients with advanced nonsquamous non-small-cell lung cancers. Clin Lung Cancer 16: 514–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SE, Chester JD (2015) Personalised cancer medicine. Int J Cancer 137: 262–276. [DOI] [PubMed] [Google Scholar]
- Jakobsen KR, Demuth C, Sorensen BS, Nielsen AL (2016) The role of epithelial to mesenchymal transition in resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Transl Lung Cancer Res 5: 172–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson MJ, Beckler AD, Taniguchi LE, Allak A, Vanwagner LB, Lee NG, Thomsen WC, Hubbard MA, Thomas CY (2011) Activation of the insulin-like growth factor-1 receptor induces resistance to epidermal growth factor receptor antagonism in head and neck squamous carcinoma cells. Mol Cancer Ther 10: 2124–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher SM, Nimeiri HS, Benson AB (2016) Targeting angiogenesis in colorectal cancer: tyrosine kinase inhibitors. Cancer J 22: 182–189. [DOI] [PubMed] [Google Scholar]
- Kuwano M, Sonoda K, Murakami Y, Watari K, Ono M (2016) Overcoming drug resistance to receptor tyrosine kinase inhibitors: learning from lung cancer. Pharmacol Ther 161: 97–110. [DOI] [PubMed] [Google Scholar]
- Kuang XY, Jiang HS, Li K, Zheng YZ, Liu YR, Qiao F, Li S, Hu X, Shao ZM (2016) The phosphorylation-specific association of STMN1 with GRP78 promotes breast cancer metastasis. Cancer Lett 377: 87–96. [DOI] [PubMed] [Google Scholar]
- Lin X, Liao Y, Chen X, Long D, Yu T, Shen F (2016) Regulation of oncoprotein 18/stathmin signaling by ERK concerns the resistance to taxol in nonsmall cell lung cancer cells. Cancer Biother Radiopharm 31: 37–43. [DOI] [PubMed] [Google Scholar]
- Ma WW, Adjei AA (2009) Novel agents on the horizon for cancer therapy. CA Cancer J Clin 59: 111–137. [DOI] [PubMed] [Google Scholar]
- Morgillo F, Della Corte CM, Fasano M, Ciardiello F (2016) Mechanisms of resistance to EGFR-targeted drugs: lung cancer. ESMO Open 1: e000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgillo F, Cascone T, D'Aiuto E, Martinelli E, Troiani T, Saintigny P, De Palma R, Heymach JV, Berrino L, Tuccillo C, Ciardiello F (2011. a) Antitumour efficacy of MEK inhibitors in human lung cancer cells and their derivatives with acquired resistance to different tyrosine kinase inhibitors. Br J Cancer 105: 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgillo F, Martinelli E, Troiani T, Orditura M, De Vita F, Ciardiello F (2011. b) Antitumor activity of sorafenib in human cancer cell lines with acquired resistance to EGFR and VEGFR tyrosine kinase inhibitors. PLoS One 6: e28841. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Miyashita A, Fukushima S, Nakahara S, Yamashita J, Tokuzumi A, Aoi J, Ichihara A, Kanemaru H, Jinnin M, Ihn H (2015) Investigation of FOXM1 as a potential new target for melanoma. PLoS ONE 10: e0144241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska Z, Sequist LV (2016) Treatment of EGFR-mutant lung cancers after progression in patients receiving first-line EGFR tyrosine kinase inhibitors: a review. JAMA Oncol 2: 948–954. [DOI] [PubMed] [Google Scholar]
- Paz-Ares L, Hirsh V, Zhang L, de Marinis F, Yang JC, Wakelee HA, Seto T, Wu YL, Novello S, Juhász E, Arén O, Sun Y, Schmelter T, Ong TJ, Peña C, Smit EF, Mok TS (2015) Monotherapy administration of sorafenib in patients with non-small cell lung cancer (MISSION) trial: a phase III, multicenter, placebo-controlled trial of sorafenib in patients with relapsed or refractory predominantly nonsquamous non-small-cell lung cancer after 2 or 3 previous treatment regimens. J Thorac Oncol 10: 1745–1753. [DOI] [PubMed] [Google Scholar]
- Petrovic V, Costa RH, Lau LF, Raychaudhuri P, Tyner AL (2008) FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin (KIS) to promote cell cycle progression. J Biol Chem 283: 453–460. [DOI] [PubMed] [Google Scholar]
- Ren S, Su C, Wang Z, Li J, Fan L, Li B, Li X, Zhao C, Wu C, Hou L, He Y, Gao G, Chen X, Ren J, Li A, Xu G, Zhou X, Zhou C, Schmid-Bindert G (2014) Epithelial phenotype as a predictive marker for response to EGFR-TKIs in non-small cell lung cancer patients with wild-type EGFR. Int J Cancer 135: 2962–2971. [DOI] [PubMed] [Google Scholar]
- Rana S, Maples PB, Senzer N, Nemunaitis J (2008) Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev Anticancer Ther 8: 1461–1470. [DOI] [PubMed] [Google Scholar]
- Rubin CI, Atweh GF (2004) The role of stathmin in the regulation of the cell cycle. J Cell Biochem 93: 242–250. [DOI] [PubMed] [Google Scholar]
- Sivendran S, Liu Z, Portas LJ Jr, Yu M, Hahn N, Sonpavde G, Oh WK, Galsky MD (2012) Treatment-related mortality with vascular endothelial growth factor receptor tyrosine kinase inhibitor therapy in patients with advanced solid tumors: a meta-analysis. Cancer Treat Rev 38: 919–925. [DOI] [PubMed] [Google Scholar]
- Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, Bergethon K, Shaw AT, Gettinger S, Cosper AK, Akhavanfard S, Heist RS, Temel J, Christensen JG, Wain JC, Lynch TJ, Vernovsky K, Mark EJ, Lanuti M, Iafrate AJ, Mino-Kenudson M, Engelman JA (2011) Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med 3: 75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tania M, Khan MA, Fu J (2014) Epithelial to mesenchymal transition inducing transcription factors and metastatic cancer. Tumour Biol 35: 7335–7342. [DOI] [PubMed] [Google Scholar]
- Tan CS, Gilligan D, Pacey S (2015) Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol 16: 447–459. [DOI] [PubMed] [Google Scholar]
- Wang L, Li H, Ren Y, Zou S, Fang W, Jiang X, Jia L, Li M, Liu X, Yuan X, Chen G, Yang J, Wu C (2016) Targeting HDAC with a novel inhibitor effectively reverses paclitaxel resistance in non-small cell lung cancer via multiple mechanisms. Cell Death Dis 7: e2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Kim SH, Lee JH, Choi YL, Kim YC, Park TS, Hong YC, Wu CF, Shin YK (2007) Inactivation of SMAD4 tumor suppressor gene during gastric carcinoma progression. Clin Cancer Res 13: 102–110. [DOI] [PubMed] [Google Scholar]
- Wang L, Chen G, Chen K, Ren Y, Li H, Jiang X, Jia L, Fu S, Li Y, Liu X, Wang S, Yang J, Wu C (2015) Dual targeting of retinoid X receptor and histone deacetylase with DW22 as a novel antitumor approach. Oncotarget 6: 9740–9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Liu X, Ren Y, Zhang JY, Chen JL, Zhou WL, Guo W, Wang X, Chen H, Li M, Yuan X, Zhang X, Yang JY, Wu CF (2017) Cisplatin enriching cancer stem cells confer multidrug-resistance in non-small cell lung cancer via enhancing TRIB1/HDAC activity. Cell Death Dis 8: e2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Tan XF, Tan HT, Lim TK, Chung MC (2014) Unbiased proteomic and transcript analyses reveal that stathmin-1 silencing inhibits colorectal cancer metastasis and sensitizes to 5-fluorouracil treatment. Mol Cancer Res 12: 1717–1728. [DOI] [PubMed] [Google Scholar]
- Zhao J (2016) Cancer stem cells and chemoresistance: The smartest survives the raid. Pharmacol Ther 160: 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.