Abstract
CTCF, Zinc-finger protein, has been identified as a multifunctional transcription factor that regulates gene expression through various mechanisms, including recruitment of other co-activators and binding to promoter regions of target genes. Furthermore, it has been proposed to be an insulator protein that contributes to the establishment of functional three-dimensional chromatin structures. It can disrupt transcription through blocking the connection between an enhancer and a promoter. Previous studies revealed that the onset of various diseases, including breast cancer, could be attributed to the aberrant expression of CTCF itself or one or more of its target genes. In this review, we will describe molecular dysfunction involving CTCF that induces tumorigenesis and summarize the functional roles of CTCF in breast cancer.
Keywords: Breast cancer, Chromatin looping, Co-regulator, CTCF, Insulator
INTRODUCTION
The crucial chromatin organizer, CTCF, is involved in transcriptional regulation through various mechanisms such as enhancer-promoter looping formation and insulation. Genome-wide distributions of CTCF have been analyzed by Chromatin Immunoprecipitation Sequencing (ChIP-Seq), additionally, the dynamic roles of CTCF in gene regulation and genomic interactions have been elucidated by chromosome conformation capture (3C) and by Paired-End Tag Sequencing (ChIA-PET). Global CTCF binding sites are normally conserved within tissues; however, specific CTCF binding sites were identified and regulated by epigenetic factors under specific conditions. CTCF binding is strongly associated with DNA methylation status and aberrant CTCF binding to DNA depends on DNA methylation at the IGF2/H19 locus leads to unexpected transcription that can induce Beckwith-Wiedemann syndrome (BWS) and Silver-Russell syndrome (SRS), respectively. Since genes regulated by CTCF are related to proliferation and apoptosis, the role of this multifunctional protein has been studied in various cancers, including breast cancer. Breast cancer is a heterogeneous disease associated with the activities of hormone receptors for estrogen (ER), progesterone (PR), and HER2 (human epidermal growth factor receptor 2). Hormone receptor status is an important biomarker for breast cancer and genes regulated by hormones are therefore therapeutic targets for breast cancer. Several studies have investigated that genes regulated by ER are modulated by CTCF through binding to ER target genomic regions, constructing higher-order chromatin structures for enhancer and promoter interactions, thereby limiting the influences of ER when CTCF is occupied with other proteins such as cohesin, ER, or the transcription activator, BRG1 (1–4). Here, we will review recent studies of CTCF function to understand mechanisms that regulate gene expression in breast cancer.
DIVERSE ROLES OF CTCF
CTCF, a highly conserved protein which contains 11 zinc fingers in eukaryotes, recognizes and binds to specific sequences in the genome. Genome-wide analysis indicated that half of CTCF-binding sites were observed in intergenic regions, while the remaining sites were present in promoters and intragenic regions (3). This global analysis supports findings of its multifunctional properties as a transcription activator/repressor, insulator, recombination modulator, and architect involved in the establishment of three-dimensional chromatin structures (5, 6).
Several studies have shown that CTCF interacts with other co-factors such as homeodomain transcription factors (HOX-TFs) and glucocorticoid receptor (GR) at promoters or enhancers in order to regulate gene expression in primary mesenchymal limb bud cells and hepatic cells (7, 8). Associations between CTCF and HOXA/D family transcription factors (TFs) have been observed in the chicken genome (7). Of the 9 HOX-TFs, 6 HOX-TFs were grouped with each other based on coincident genomic binding patterns confirmed by ChIP-Seq. These binding patterns were also comparable to CTCF binding sites in the genome. It suggests that CTCF plays a role in transcription regulator with HOX-TFs (7). In addition, GR and CTCF co-localization was confirmed at ANGPL4, a GR target gene. Co-binding of GR and CTCF was involved in chromatin looping structure for communication between enhancer and promoter (8). It was also revealed CTCF functions as an insulator (9–12). CTCF blocks the influence of ER regulation in genome and rearrangement in Tcrd region in germline (10, 12). And CTCF insulator activity involved in B cells developmental stages. Reduced CTCF induced to premature developmental process (11). The function of an insulator, CTCF, is enforced when its target site is also occupied with BRD2, a member of the bromodomain and extra-terminal motif (BET) protein family. Loss of BRD2 coincided with aberrant boundary architecture, even when CTCF occupancy was not altered in the Slc25a37 locus. One explanation is that CTCF and BRD2 could cooperate as insulators to enforce architectural boundaries in the genome in order to block enhancer regulation (9).
The influence of CTCF on V(D)J recombination was manifested via modulation of chromatin loop structures (10). CTCF-depletion studies identified the function of CTCF in association with chromatin structure in mouse embryonic stem cells (2). For maintaining stable genomic complex, CTCF and the cohesin complex, consisting of SMC3, SMC1, RAD21, and STAG1 or STAG2, can co-localize (13). Moreover, global analysis of CTCF, SMC3, and RAD1 shift-banding patterns have demonstrated the proximity of protein-DNA binding motif sequences (13).
In a recent study, multi-functional roles of CTCF were demonstrated in tandem situations. The elimination of CTCF confirmed the multifunctional status of the protein as a prominent factor for transcriptional regulation, distinct looping formation, and maintaining chromatin structure with protein complexes such as cohesin in both inter-chromatin and intra-chromatin looping (2).
DISEASE-RELATED CTCF DYSFUNCTION
It has been discovered that aberrant CTCF induces several diseases or disorders, including mental retardation, Wiedemann syndrome, Silver-Russell syndrome, and various cancers (Table 1) (1, 14–19). Germline CTCF missense and frameshift mutations can result in the syndromic intellectual disability, autosomal dominant mental retardation 21 (MRD21); de novo c.375dupT and c.1186dupA frameshift mutations and Arginine (R) to Tryptophan (W) transitions at amino acid position 567 can lead to weaker binding affinity of CTCF to DNA (14). The distinct phenotypic consequences of these mutations are short stature, microcephaly, mild facial dysmorphisms, and various intellectual disabilities (16). Disruption of the genomic neighborhood through abnormal binding of CTCF to the imprinting control region (ICR) of the IGF2-H19 locus that governs IGF2 and H19 gene expression on chromosome 11p15.5 can result in other diseases such as Beckwith-Wiedemann (BWS) and Silver-Russell syndromes (SRS) (15). This process closely associates with differential DNA methylation of ICR which, in turn, determines the binding affinity of CTCF. Paternal allele normally showed absent CTCF at methylated ICR that leads to activation of IGF2 whereas IGF2 expression was inhibited by CTCF at unmethylated ICR on the maternal allele. Allele-specific modifications of DNA methylation at ICR can result in abnormal binding of CTCF and aberrant transcription of IGF2 and H19. It has been investigated abnormal CTCF bindings depending on DNA methylation status at this locus in testicular germ cell tumors (TGCT), colorectal cancer, bladder cancer, and ovarian cancer. Moreover, SRS by dysfunction of CTCF at the paternal allele has been associated with the development of different cancers, including hepatocellular carcinoma (20), Wilms’ tumor (21), testicular cancer (22) and craniopharyngioma (23).
Table 1.
Diseases caused by CTCF variations
| Disease/Disorder | Cause | Feature | Ref. | |
|---|---|---|---|---|
| Mental retardation, autosomal dominant 21 (MRD21) | Mutation in the CTCF gene | Frameshift mutation; c375dupT, c.1186dupA | (14, 16) | |
| Beckwith-wiedemann syndrome (BWS) | Mutation or deletion of imprinted genes | Igf2 expression, H19 inhibition | (15) | |
| Silver-russell syndrome (SRS) | Igf2 inhibition, H19 expression | |||
| Cancer | Testicular cancer | Methylation in CTCF-binding sites | Igf2 inhibition, H19 inhibition | (20) |
| Colorectal cancer | Hypermethylation of CpG sites in IGF2/H19 | Loss of imprinting of the IGF2 | (21) | |
| Bladder cancer | Hypomethylation of CTCF-binding site | Loss of imprinting of the H19 | (22) | |
| Ovarian cancer | Increase DNA methylation and reduce insulator protein CTCF | H19 inhibition | (23) | |
| Endometrial cancer | CTCF mutation | Missense mutation, R377C | (30) | |
| Prostate cancer | ZF mutation in the CTCF | ZF3 H345R mutation, CAC→CGC | (31) | |
| Wilms’ tumor | ZF3 R339W mutation, CGG→TGG ZF7 R448Q mutation, CGA→CAA |
(32) | ||
| Breast cancer | ZF3 K344E mutation, AAA→GAA | (33, 34) | ||
| Gastrointestinal cancer | CTCF binding site mutations | Mutation at CTCF/cohesion binding site | (19) | |
| Skin cancer | CTCF binding site mutations | Asymmetric mutation style with CTCF | (36) | |
Missense mutations of the zinc-finger domain of the CTCF gene were detected in various cancers, including endometrial cancer, prostate cancer, Wilms’ tumor, and breast cancer (1, 24–28). R377C mutation has been investigated in endometrial cancer (24), and H345R mutation has been identified in prostate cancer (25). Two missense mutations, R339W and R448Q, have also been revealed in Wilm’s tumor (26). Another mutation was also observed in breast cancer which is K344E mutation (AAA→GAA), missense codon mutation (27, 28).
CTCF/cohesin-binding sites (CBSs) mutations were investigated in various cancers including gastrointestinal and skin cancers (29, 30). In gastrointestinal cancer, relatively A·T>C·G and A·T>G·C substitutions were preferentially detected at CBSs (19) and these mutations were related with late replication (29). Mutations arising due to differential nucleotide excision repair (NER) across pyrimidine pairs were also identified at specific CBSs in skin cancer (30).
ABERRANT CTCF FUNCTION IN BREAST CANCER
It has been investigated dysfunction of CTCF caused by mutation and aberrant poly(ADP-Ribosyl)ation (PARlation) in breast cancer cells. Missense codon mutation, K344E, in CTCF zinc finger domain 3, was observed in breast cancer (27, 28). This mutation could affect the ability of CTCF to bind to the promoter/insulator sites of cell-proliferation-related genes such as MYC, PLK, PIM-1, p19ARF, and Igf2/H19 (1). Another mutations were revealed 240G→A transition in the 5′ UTR and S388S mutation (1445C→T) in exon 4 of CTCF gene by the association study between disease and mutations (31). Altered expression levels of CTCF, as a result of mutations in the CTCF coding region, could be a cause of breast cancer development (32). These events are also implicated in altered formations of the protein that may play a role in breast cancer. This study also showed CTCF function as a tumor suppressor gene by mediating significant inhibition of carcinoma cell proliferation (32).
PARlation of CTCF also contributes to apoptosis regulation and cell growth inhibition through translocation from the nucleoplasm to nucleoli in breast cancer cells (33, 34). The presence of two CTCF isoforms, 130 kDa and 180 kDa, in breast cancer compared with one 180 kDa CTCF formed via PARlation in normal tissues, prompted the suggestion that the 130 kDa CTCF could be used as a breast cancer biomarker (35). A 130 kDa CTCF is negatively correlated with tumor size and tumor stage. Increased cell proliferation was observed when the expression of the 130 kDa CTCF isoform increased in cells. NaB treatment of cells yielded the opposite effect that the formation of the 180 kDa CTCF isoform was observed, there was a reduction of MCF-7 cell proliferation, and the induction of apoptosis was verified (35).
Abnormal expression of CTCF which generated breast cancer cells, was regulated by DNA methylation in genome. Here is a study indicated that how DNA methylation and down-regulated CTCF affects to tumorigenesis gene regulation in breast tumors (24). In tumor suppressor genes and the imprinting regulatory regions, aberrant changes of the DNA methylation status had an impact on decreased CTCF expression levels. Down-regulated CTCF, in particular, was positively correlated with dysregulated methylation pattern at CpGs near genes known to be involved in tumorigenesis, such as Trp53, Dnmt4A, RunX1, and Ctcf (24). DNA methylation of CpG regions near CTCF-regulated genes, including oncogenes, results in unavailable CTCF binding. Tumor growth is therefore enhanced in the absence of CTCF modulation of the relevant genes (24). Epigenetic BRCA1 inactivation through methylation of the promoter was associated with absent and novel CTCF expression in nucleus and cytoplasm, respectively, in a separate study (36). However, it was revealed that CTCF gene expression levels were not altered as DNA methylation status of the CTCF promoter region (37). To determine the level of CTCF in breast cancer types, CTCF expression levels were analyzed in different types of breast cancer, invasive lobular carcinoma (ILC), invasive ductal carcinoma (IDC), mucinous carcinoma, metaplastic carcinoma, other invasive carcinomas, and ductal carcinoma in situ (DCIS). There were no differences between the methylation statuses of CTCF promoter sites in breast cancer tissues and adjacent normal breast tissues; however, CTCF gene regulation was greatly diminished in invasive breast cancers. And CTCF expression levels was shown to be negatively correlated with cell proliferation levels, especially in ILC compared to other breast carcinomas (37). In addition to methylation status, CTCF gene expression could also be differentially regulated by female sex hormones. Decreased CTCF expression levels have been reported in a 17β-estradiol(E2)-treated MCF-7 breast cancer cell line. Decreased levels of CTCF were also directly proportional to the action of E2 (38). Taken together, these findings suggest that CTCF regulation could be affected by hormone regulation in breast cancer cell lines and aberrant CTCF regulation could be involved in tumor growth.
DYNAMIC ROLES OF CTCF IN BREAST CANCER
CTCF can affect breast cancer development by regulating target genes, where it can act alone or in combination with other factors to function as a transcription factor, insulator, and/or regulator of chromatin structure (39).
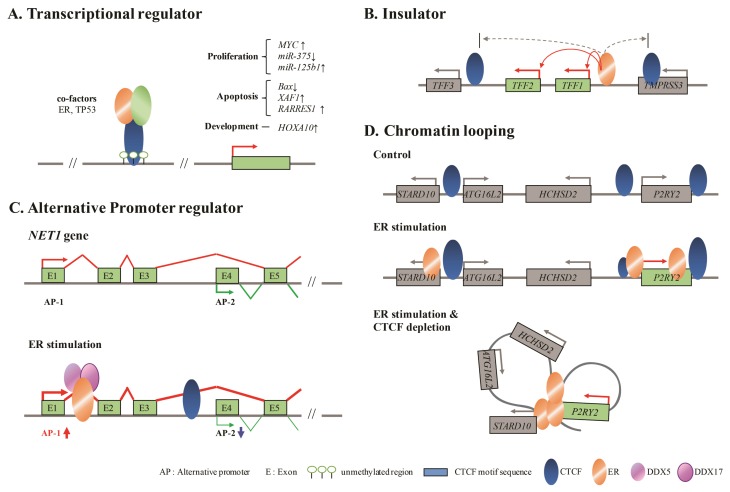
CTCF binding to gene-regulatory regions could be associated with alteration of oncogene expression levels via co-regulatory factors and changes in DNA methylation status (Fig. 1A). A previous study showed alterations of genes could be facilitated by the absence of CTCF binding at target sites in the promoters of cell-proliferation-related genes such as MYC, p19ARF, and Igf2 (1). Consequently, cell growth was differently modulated to result in a malignant phenotype. A correlation was also observed between CTCF depletion and apoptosis in ZR-75-1 cells, but not in HeLa cells (40). The pro-apoptotic related gene, BAX, had an unmethylated promoter in breast cancer cell lines, MCF-7 and ZR-75-1, and non-breast cancer cells, HeLa and 293T. Nevertheless, BAX expression level was increased in breast cancer cells depleted of CTCF through siRNA induced gene knockdown experiments. Double knockdown of BAX and CTCF in ZR-75-1 and MCF-7 breast cancer cell lines caused a decrease in the level of cleavage of PARP-1, hallmark of apoptosis. As the result, apoptosis levels were decreased in double-knockdown cells compared to only CTCF-knockdown cells. These results are consistent with data showing that BAX overexpression leads to apoptosis in CTCF-deficient cells. The BAX gene had two CTCF protein-binding sites in the downstream of the proximal promoter region which were occupied by CTCF in breast cancer cells. It is possible that BAX and CTCF cooperate as negative regulators (40). HOXA10 promoter activity was also modulated by CTCF binding status (41). The HOXA cluster was enriched by the presence of CTCF in MCF-7 breast cancer cells. Of all the genes in the HOXA cluster, HOXA10 expression was altered in a manner that could be inversely correlated to CTCF expression. HOXA10 is sensitively regulated via CTCF binding to CTCF-binding-motif sequences located in promoter regulatory regions. The negative correlation between CTCF enrichment status and HOXA10 expression levels was considered by analyzing two separate histone modifications, H3K27me3 and H3K4me3, repressive marker and active marker. Increased H3K27me3 and decreased H3K4me3 modifications were observed in CTCF-overexpressed MCF-7 cells. CTCF-binding sites were present in promoters in MCF-7 and T-47D, but not in BT-474 cells. It supports that the sensitivity of CTCF binding to the HOXA10 promoter regulatory region depends on the cell type (41).
Fig. 1.
The multiple functions of CTCF in breast cancer cells. (A) CTCF as transcriptional regulator regulates target gene expression through binding with co-factors such as ER and TP53 at unmethylated CpG regions. (B) CTCF as insulator prevents enhancer activity through regulation of ER binding to neighboring genes, TFF3 and TMPRSS3, at the TFF locus. (C) CTCF is involved in inverse expression of genes which have alternative promoters. After ER treatment, long-form transcript of NET1 was highly expressed whereas short-form transcript separated by CTCF was decreased. (D) Decreased CTCF after ER treatment induces chromatin looping to connect distal and proximal ERs at the P2RY2 locus.
It has been identified that DNA methylation status involves in CTCF binding affinity at CpG regions containing CTCF binding motif (42, 43). In addition, their binding affinity was also influenced by histone modifications (44). CTCF occupies to the tumor suppressor gene, X-linked inhibitor of the apoptosis (XIAP)–associated factor 1 (XAF), and controls its expression with a methylation and histone modification sensitive manner. Exposure to demethylation modulators such as 5-aza-2′-deoxycytidine (5-A-DC) could affect the observed DNA methylation status of CpG regions in CTCF-regulated target genes. A negative correlation was observed between XAF1 expression levels and DNA methylation patterns. Enhanced CTCF was associated with a status of demethylation and induced XAF1 translational activity. Histone modifications, H3K27ac, H3K4me3, and H3K4me1, were observed to reflect altered transcriptional activities by CTCF binding and DNA methylation statuses (44). Methylation status could affect CTCF binding and regulation of gene expression. The proximal upstream promoter site of retinoic acid receptor responder 1 (RARRES1), a tumor suppressor gene, was methylated in breast cancer cells such as MCF-7, SK-BR-3 with existing CTCF-binding motifs, but not in other normal breast cell lines such as MCF-10A (45). It was investigated the correlation between the gene expression levels of RARRES1 and methylation status. The gene was down-regulated in breast cancer cells with hypermethylated proximal promoter regions. RARRES1 was reactivated by epigenetic drugs, 5-aza-2′-deoxycytidine (DAC) or Trichostatin A (TSA), or both, in SUM159, but not in SK-BR-3 cells. In SUM159, RARRES1 reactivation led to increased cell death and inhibition of cellular invasive properties. CTCF binding to putative CTCF-binding motif sequences located downstream with H3K4me2 occupancy in the proximal promoter region of RARRES1 gene. It has also been suggested that CTCF prevents DNA methylation and epigenetic silencing. It was confirmed that CTCF-deficient MCF-10A cells had no CTCF present at the RARRES1 promoter region, thereby leading to decreased RARRES1 expression in MCF-10A cells. CTCF binding to RARRES1 promoter may be a key requirement for gene transcription (45). Delta-like ligand-4 (DLL4), a major gene for regulating the Notch-signaling pathway, was also confirmed to be a CTCF target gene (46). CTCF was highly expressed in MCF-7 cells compared to other cancers and CTCF was involved in regulating the DLL4 gene expression levels by binding to the proximal promoter region. Moreover, DLL4 expression was also reactivated in cells deficient for both CTCF and the tumor suppressor gene, p53 (TP53), whereas DLL4 expression was not altered by CTCF alone (46).
Recent studies have also showed the relationship between CTCF binding and miRNAs. The expression levels of miRNAs were also modulated by CTCF binding, DNA methylation, and histone modification (47, 48). In a previous study, it was discovered that increased miR-375 regulation interacted with active ER signaling (47). miR-375 was significantly up-regulated in ER-positive breast cancer cells compared to ER-negative breast cancer cells and normal breast cells. In breast cancer cells, the role of miR-375 was investigated by monitoring decreased cell proliferation as a function of the inhibition of miR-375. Using ER targeted siRNA, decreased signaling of ER was observed to lead to the reduction of miR-375 expression levels by 50%. The authors suggested that CTCF regulated miR-375 gene expression by binding to CpG regions, especially the first CpG islands between two CpG regions. In addition, ER-positive MCF-7 and T-47D cells were confirmed to have one hypermethylated and unmethylated CpG islands, compared to other cell lines. As enriched CTCF binding with H3K9me2 was presented at first CpG region in MCF-12A and MDA-MB-231 ER negative cell lines compared to MCF-7. Through gene expression profiling, Ras dexamethasone-induce 1 (RASD1) was indicated as a miR-375 target gene had negative correlation with ER regulation. Taken together, the loop of ER, miR-375, and RASD1 associates with ER regulation (47). Another miRNA, miR-125b1 was also known to affect to cell proliferation (49). The proximal promoter region of miR-125b1 also has a methylated CpG island in approximately 90% of breast cancers. The expression levels of miR-125b1 was decreased in breast cancer cells (48). It could be illustrated that the DNA methylation statuses of CpG islands are involved in tumorigenesis. The CpG islands has a CTCF-binding site in normal cells, but not in breast cancers. Moreover, H3K9me3 and H3K27me3, repressive histone modification marks, were localized in breast cancers. Epigenetic regulation could be explained by examining methylation status, CTCF binding, and histone modification marks via regulation of miR-125b1 expression. Epigenetic silencing of miR-125b1 in breast cancers could be achieved by a lack of CTCF binding to methylated CpG islands demarcated by histone modifications (48).
CTCF could be involved in regulating the expression levels of CTCF-modulated genes by acting as a transcriptional insulator to induce gene regulation or to block the interaction of enhancers and promoters (12). Another study investigated the binding of CTCF to sites independently regulated by estrogen (50). It was found that estrogen-regulated function of a gene was limited by CTCF-binding sites acting as barriers. Of all the CTCF binding roles, putative insulators were selected at the Trefoil factor (TFF) locus. In this locus, TFF1 and TFF2 were increased by E2, whereas other genes located outside of CTCF binding sites were not significantly induced by E2 (Fig. 1B). And occupied CTCF bindings were not affected by E2 stimulation. It was shown that ERα was located at the TFF1 promoter. This association was hampered when CTCF expression was reduced by siRNA. Even though E2 stimulation could not be involved in CTCF binding alteration, the DNA methylation status at the barrier region could hinder CTCF occupancy. No interactions were observed between the CTCF-binding sites which acted as a barrier and any elements on the outside of these barriers. This boundary or site of CTCF-binding interaction was not affected by ERα or HOA1. These putative insulators may possibly construct initial higher-order chromatin structures in order to restrict ER regulation (50).
One well-known CTCF function is to modulate chromatin structure as a chromatin boundary factor involved in the regulation of transcription. CTCF constructs chromatin architecture with co-factors, such as the cohesion complex, which includes RAD21, SMC1, SMC3, and BRG1 (4, 13, 51–53). Tang and colleagues revealed that the chromatin topological domain could be altered by CTCF binding to specific DNA motif sequences (51). Cell-line specific mutations in CTCF binding motifs resulted in the formation of different chromatin loops. Although BRG1 and CTCF co-localization have not been extensively investigated, it is known that this phenomenon is rare as a function of the entire genome. BRG1 and CTCF interactions could preserve sturdy, topologically-associated domains (TAD) or boundaries (4). BRG1 is a transcriptional co-regulated of the ATP-dependent transcriptional remodeling complex, SWI/SNF. In recent studies, it was shown BRG1 could function to develop cancer cells in a different manner via hormone regulation in breast cancers (54, 55). In previous studies, it was shown BRG1 overexpression had positive correlation with poor prognosis in breast cancer cells (55, 56). Deficient BRG1 was revealed it affected to inhibit cell proliferation through cell cycle arrest, especially G1 phase, caused by decreased cyclin D1 and cyclin E, and increased p27 expression (55). Recently, BRG1 was investigated and the role was confirmed that BRG1 regulation was essential to preserve the topologically-associated domains (TAD) correlated with CTCF binding in the normal MCF-10A and MEF breast cell lines (4). Loss of BRG1 was used to confirm that BRG1 was a causative factor in week formation of a TAD boundary and a reduction in nucleosomes surrounding the CTCF binding site. The strength of the BRG1 and CTCF interaction was enhanced when the event occurred within a range of 1 kb from one another. Dysregulated BRG1 expression may be associated with a disrupted chromatin structure, especially regulatory regions involved in breast cancer cell proliferation. Consequently, it was suggested that related genes could be differentially expressed and that the net result could affect the rate of cell proliferation (4). Co-localization of CTCF with relevant cofactors could stabilize substantial chromatin structures. The direct effect of aberrant chromatin structure and attenuated chromatin looping domains impacted by altered binding of CTCF in breast cancer is the subject of ongoing investigation. It could be expected to affect transcriptional regulation eg, significant alterations in gene expression could lead to tumorigenesis.
Multifunctional CTCF is also associated with the formation of alternative splicing variants. The function of CTCF binding to induce RNA Polymerase II (Pol II) elongation and regulate alternative splicing at exon 5 of CD45 gene in murine splenocytes (6, 57). In a previous study, the relationship between CTCF binding and alternative promoter (AP) activity was also suggested in estrogen-regulated breast cancer cells, MCF-7 (58). AP regulation was observed among estrogen-regulated genes, especially in differentially expressed and regulated genes in MCF-7 cells. Specific ER-regulated regions and CTCF binding sites were observed in most of these genes. For example, NET1 gene had two different AP transcripts. One product was transcribed from exon 1 (AP-1) and the other was transcribed from exon 4 (AP-2), generating NET1 long-form and short-form transcripts (Fig. 1C). AP regulation of NET1 was affected by estrogen stimulation with ER co-regulator factors, DDX5 and DDX17, and CTCF binding. A specific ER-binding site was observed at 8 kb downstream and a CTCF-binding site was observed between exon 3 and exon 4. AP-1 and AP-2 activities could be separated as a function of CTCF binding. AP-1 was highly expressed via ER stimulation and in the absence of the regulation of exon 4 expression, whereas AP-2 expression levels were decreased. Pol II levels and mRNA levels were correlated with the AP regulation. High Pol II levels were observed when AP-1 was increased by ER stimulation, while Pol II activity was decreased in the case of AP-2. Indeed, AP regulation could be possible to produce distinct proteins that are involved in different cellular functions. Cell growth and prognoses of patients were influenced by long-form and short-form expression levels of NET1 transcripts. As a result, it was confirmed that AP activity could lead to aberrant gene regulation and could be regulated by CTCF modulation mediated directly by estrogen or, indirectly, by ER influences on DDX5 and DDX17 (58).
A previous study (59) certified the correlation of the presence of CTCF and ER in breast cancer cell lines. CTCF and ER binding patterns differed in many cell lines, including breast cancers stratified into different subtypes. Cell-line specific colocalization of CTCF and ER binding to target regions were observed in the breast cancer cell line, MCF-7 (59). Most of the ER and CTCF co-localization sites were within 20 kb from estrogen-regulated genes. Therefore, one explanation is that estrogen-mediated gene expression levels could be regulated by ER and CTCF. Fiorito and colleagues studied CTCF-binding enrichment is modulated by ER stimulation to impede the expanded loops of ER regulated enhancers and promoters of genes in MCF-7 (3). Investigators confirmed that the alteration of chromatin looping depended on the timing of an estrogen treatment. CTCF binding sites were increased at intergenic regions with increased duration of the estrogen stimulation time. When estrogen-induced transcripts were highly expressed at specific time points, the CTCF binding density also increased in a proportional manner. ER has an important role in intra-chromosomal interactions to regulate ER target genes. ER-ER chromatin looping was investigated using Chromatin Interaction Analysis by Paired-End Tag Sequencing (ChIA-PET). CTCF binding was found to co-localize with ER binding in ER-ER loops. CTCF occupancy was observed in more than half of the ER looping regions. A 24% increase in ER binding was detected by Chromosome conformation capture (3C)-PCR in CTCF-depleted cells. For example, P2Y purinoceptor 2 (P2RY2) is involved in many functions, including cell proliferation and apoptosis. When compared with controls, this gene is also induced when cells are stimulated by estrogen (Fig. 1D). Estrogen stimulation also appeared to affect three ER binding sites and specific ER-ER looping in P2RY2 was increased under CTCF-depletion conditions. Significantly increased interactions of nuclear-lamina-related proteins and CTCF were observed among the proteins that interacted with CTCF proteins. Interestingly, Lamin B binding was also confirmed at three ER binding sites near P2RY2. This result may be explained as follows: CTCF-repressive ER-ER looping function may be related to nuclear lamina via modulation of the chromatin structure. CTCF involvement in regulating the ER-ER looping structure may modulate ER target gene regulation. The expression levels of cell growth-related genes were significantly changed depending on CTCF and estrogen stimulation. CTCF may modulate functions through ER binding sites located near target genes. The location of CTCF at enhancer regions involved in the transcription of eRNA, could inhibit ER looping formation. As a result, transcription and cell growth could be changed (3).
POTENTIAL THERAPEUTIC TARGET FOR BREAST CANCER
In breast cancer cells, dysregulated gene expression clustered specifically in apoptosis-associated genes, may be a consequence of aberrant CTCF gene regulation. An association was confirmed NaB treatment could affect to apoptosis through CTCF regulation (60). NaB treatment led to increased levels of the 130 kDa CTCF isoform, cell proliferation, reduced apoptosis and CTCF translocation in MCF-7 cells (35). As previously mentioned, CTCF binding was sensitive to DNA methylation status at target binding sites. Indeed, CTCF was not able to occupy methylated regions (40, 44, 45, 47, 48, 50). Therefore, DNA-methylation-related drugs such as 5-A-DC, could be considered as potential targeted therapies in breast cancers. In a previous study, Sulforaphane (SFN) was shown to have an effect on the down-regulation of human telomerase reverse transcriptase (hTERT) gene which is expressed in 90% of cancer cells including breast cancer but not in normal cells. This effect occurs via modulation of the methylation status of the CpG site at its CTCF-binding region (61). SFN inhibited histone deacetylase (HDAC) and induced demethylation of CpGs. SFN-mediated epigenetic regulation affected differentially adjusted transcription factor binding in a sequential manner. As a result, hTERT was down-regulated in MCF-7 and MDA-MB-231 cells, but not significantly changed in MDA-10A cells (61).
However, CTCF is known to be an important factor in the prevention or induction of tumorigenesis, either by oncogenes or tumor suppressor genes. Unlike CTCF targeted therapies, Brother of the Regulator of Imprinted Sites (BORIS) has been studied as a more common target of cancer treatment. BORIS, also called CTCF-like protein (CTCFL), shares a conserved 11 zinc finger domains with CTCF and it is a paralogue with related CTCF-binding motif sequences (62). It has been reported that BORIS and CTCF have a complementary effect (62). Up-regulated CTCF helps protect cells from apoptosis (40, 60). Conversely, induced BORIS expression levels coincided with tumor progression and silenced BORIS expression correlated with increased apoptosis (39). Although BORIS is not observed in most cancers, aberrant expression of BORIS was observed in breast cancer cells (63). It indicates not only CTCF regulation, but also BORIS expression was crucial in modulating cell viability in cancer cells (32). Using zinc-finger-domain targeting, BORIS-targeted siRNA may affect apoptosis and, in fact, reduced BORIS has been implicated in induced cell death (64). This result implies that specific modified CTCF regulation can also be used, possibly as a target to modulate cell apoptosis in breast cancers.
CONCLUSION
CTCF is a dynamic multifunctional protein that regulates transcription via its role as a transcription factor, insulator, and organizer of higher-order chromatin structures or loops. CTCF mutations could alter the protein or its binding sites, resulting in diverse diseases such as different cancers. Missense codon mutations and lowered poly(ADP-Ribosyl)ation of CTCF have been observed to alter CTCF binding to target sites in breast cancer. These events have led to aberrant gene regulation through inhibition of enhancer activity. Although several studies have confirmed a role for CTCF dysfunction in breast cancer, the mechanisms underlying this involvement at specific loci associated with tumorigenesis have yet to be elucidated. The role of CTCF in regulating chromatin loop formation at specific loci could not be confirmed by siRNA disruption of CTCF expression. Continued studies should reveal the potential role of CTCF as therapeutic target for breast cancer, since numerous genes regulated by CTCF are associated with proliferation and apoptosis in breast cancer cells.
ACKNOWLEDGEMENTS
This work was supported by fund from Sookmyung Women’s University (1-1603-2052) and the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2017R1C1B2002806).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Filippova GN, Qi CF, Ulmer JE, et al. Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter tts DNA-binding specificity. Cancer Res. 2002;62:48–52. [PubMed] [Google Scholar]
- 2.Nora EP, Goloborodko A, Valton AL, et al. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944 e922. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiorito E, Sharma Y, Gilfillan S, et al. CTCF modulates Estrogen Receptor function through specific chromatin and nuclear matrix interactions. Nucleic Acids Res. 2016;44:10588–10602. doi: 10.1093/nar/gkw785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barutcu AR, Lajoie BR, Fritz AJ, et al. SMARCA4 regulates gene expression and higher-order chromatin structure in proliferating mammary epithelial cells. Genome Res. 2016;26:1188–1201. doi: 10.1101/gr.201624.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ong CT, Corces VG. CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet. 2014;15:234–246. doi: 10.1038/nrg3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jerkovic I, Ibrahim DM, Andrey G, et al. Genome-Wide Binding of Posterior HOXA/D Transcription Factors Reveals Subgrouping and Association with CTCF. PLoS Genet. 2017;13:e1006567. doi: 10.1371/journal.pgen.1006567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamoto M, Ishihara K, Watanabe T, et al. The Glucocorticoid Receptor Regulates the ANGPTL4 Gene in a CTCF-Mediated Chromatin Context in Human Hepatic Cells. PLoS One. 2017;12:e0169225. doi: 10.1371/journal.pone.0169225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu SC, Gilgenast TG, Bartman CR, et al. The BET Protein BRD2 Cooperates with CTCF to Enforce Transcriptional and Architectural Boundaries. Mol Cell. 2017;66:102–116 e107. doi: 10.1016/j.molcel.2017.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Zhao L, Alt FW, Krangel MS. An Ectopic CTCF Binding Element Inhibits Tcrd Rearrangement by Limiting Contact between Vdelta and Ddelta Gene Segments. J Immunol. 2016;197:3188–3197. doi: 10.4049/jimmunol.1601124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braikia FZ, Oudinet C, Haddad D, et al. Inducible CTCF insulator delays the IgH 3′ regulatory region-mediated activation of germline promoters and alters class switching. Proc Natl Acad Sci U S A. 2017;114:6092–6097. doi: 10.1073/pnas.1701631114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan CS, Song JS. CCCTC-binding factor confines the distal action of estrogen receptor. Cancer Res. 2008;68:9041–9049. doi: 10.1158/0008-5472.CAN-08-2632. [DOI] [PubMed] [Google Scholar]
- 13.Nagy G, Czipa E, Steiner L, et al. Motif oriented high-resolution analysis of ChIP-seq data reveals the topological order of CTCF and cohesin proteins on DNA. BMC Genomics. 2016;17:637. doi: 10.1186/s12864-016-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gregor A, Oti M, Kouwenhoven EN, et al. De novo mutations in the genome organizer CTCF cause intellectual disability. Am J Hum Genet. 2013;93:124–131. doi: 10.1016/j.ajhg.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herold M, Bartkuhn M, Renkawitz R. CTCF: insights into insulator function during development. Development. 2012;139:1045–1057. doi: 10.1242/dev.065268. [DOI] [PubMed] [Google Scholar]
- 16.Bastaki F, Nair P, Mohamed M, et al. Identification of a novel CTCF mutation responsible for syndromic intellectual disability - a case report. BMC Med Genet. 2017;18:68. doi: 10.1186/s12881-017-0429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prawitt D, Enklaar T, Gartner-Rupprecht B, et al. Microdeletion of target sites for insulator protein CTCF in a chromosome 11p15 imprinting center in Beckwith-Wiedemann syndrome and Wilms’ tumor. Proc Natl Acad Sci U S A. 2005;102:4085–4090. doi: 10.1073/pnas.0500037102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aulmann S, Blaker H, Penzel R, Rieker RJ, Otto HF, Sinn HP. CTCF gene mutations in invasive ductal breast cancer. Breast Cancer Res Treat. 2003;80:347–352. doi: 10.1023/A:1024930404629. [DOI] [PubMed] [Google Scholar]
- 19.Katainen R, Dave K, Pitkanen E, et al. CTCF/cohesin-binding sites are frequently mutated in cancer. Nat Genet. 2015;47:818–821. doi: 10.1038/ng.3335. [DOI] [PubMed] [Google Scholar]
- 20.Chitayat D, Friedman JM, Anderson L, Dimmick JE. Hepatocellular carcinoma in a child with familial Russell-Silver syndrome. Am J Med Genet. 1988;31:909–914. doi: 10.1002/ajmg.1320310425. [DOI] [PubMed] [Google Scholar]
- 21.Bruckheimer E, Abrahamov A. Russell-Silver syndrome and Wilms tumor. J Pediatr. 1993;122:165–166. doi: 10.1016/S0022-3476(05)83518-8. [DOI] [PubMed] [Google Scholar]
- 22.Weiss GR, Garnick MB. Testicular cancer in a Russell-Silver dwarf. J Urol. 1981;126:836–837. doi: 10.1016/S0022-5347(17)54773-4. [DOI] [PubMed] [Google Scholar]
- 23.Draznin MB, Stelling MW, Johanson AJ. Silver-Russell syndrome and craniopharyngioma. J Pediatr. 1980;96:887–889. doi: 10.1016/S0022-3476(80)80570-1. [DOI] [PubMed] [Google Scholar]
- 24.Kemp CJ, Moore JM, Moser R, et al. CTCF haploinsufficiency destabilizes DNA methylation and predisposes to cancer. Cell Rep. 2014;7:1020–1029. doi: 10.1016/j.celrep.2014.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suzuki H, Komiya A, Emi M, et al. Three distinct commonly deleted regions of chromosome arm 16q in human primary and metastatic prostate cancers. Genes Chromosomes Cancer. 1996;17:225–233. doi: 10.1002/(SICI)1098-2264(199612)17:4<225::AID-GCC4>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 26.Maw MA, Grundy PE, Millow LJ, et al. A third Wilms’ tumor locus on chromosome 16q. Cancer Res. 1992;52:3094–3098. [PubMed] [Google Scholar]
- 27.Cleton-Jansen AM, Moerland EW, Kuipers-Dijkshoorn NJ, et al. At least two different regions are involved in allelic imbalance on chromosome arm 16q in breast cancer. Genes Chromosomes Cancer. 1994;9:101–107. doi: 10.1002/gcc.2870090205. [DOI] [PubMed] [Google Scholar]
- 28.Lindblom A, Rotstein S, Skoog L, Nordenskjold M, Larsson C. Deletions on chromosome 16 in primary familial breast carcinomas are associated with development of distant metastases. Cancer Res. 1993;53:3707–3711. [PubMed] [Google Scholar]
- 29.Kaiser VB, Taylor MS, Semple CA. Mutational Biases Drive Elevated Rates of Substitution at Regulatory Sites across Cancer Types. PLoS Genet. 2016;12:e1006207. doi: 10.1371/journal.pgen.1006207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poulos RC, Thoms JA, Guan YF, Unnikrishnan A, Pimanda JE, Wong JW. Functional Mutations Form at CTCF-Cohesin Binding Sites in Melanoma Due to Uneven Nucleotide Excision Repair across the Motif. Cell Rep. 2016;17:2865–2872. doi: 10.1016/j.celrep.2016.11.055. [DOI] [PubMed] [Google Scholar]
- 31.Zhou XL, Werelius B, Lindblom A. A screen for germline mutations in the gene encoding CCCTC-binding factor (CTCF) in familial non-BRCA1/BRCA2 breast cancer. Breast Cancer Res. 2004;6:R187–190. doi: 10.1186/bcr774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tiffen JC, Bailey CG, Marshall AD, et al. The cancer-testis antigen BORIS phenocopies the tumor suppressor CTCF in normal and neoplastic cells. Int J Cancer. 2013;133:1603–1613. doi: 10.1002/ijc.28184. [DOI] [PubMed] [Google Scholar]
- 33.Venkatraman B, Klenova E. Role of CTCF poly(ADP-Ribosyl)ation in the regulation of apoptosis in breast cancer cells. Indian J Med Paediatr Oncol. 2015;36:49–54. doi: 10.4103/0971-5851.151784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Torrano V, Navascues J, Docquier F, et al. Targeting of CTCF to the nucleolus inhibits nucleolar transcription through a poly(ADP-ribosyl)ation-dependent mechanism. J Cell Sci. 2006;119:1746–1759. doi: 10.1242/jcs.02890. [DOI] [PubMed] [Google Scholar]
- 35.Docquier F, Kita GX, Farrar D, et al. Decreased poly(ADP-ribosyl)ation of CTCF, a transcription factor, is associated with breast cancer phenotype and cell proliferation. Clin Cancer Res. 2009;15:5762–5771. doi: 10.1158/1078-0432.CCR-09-0329. [DOI] [PubMed] [Google Scholar]
- 36.Butcher DT, Rodenhiser DI. Epigenetic inactivation of BRCA1 is associated with aberrant expression of CTCF and DNA methyltransferase (DNMT3B) in some sporadic breast tumours. Eur J Cancer. 2007;43:210–219. doi: 10.1016/j.ejca.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 37.Wang D, Li C, Zhang X. The promoter methylation status and mRNA expression levels of CTCF and SIRT6 in sporadic breast cancer. DNA Cell Biol. 2014;33:581–590. doi: 10.1089/dna.2013.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Del Campo EP, Marquez JJ, Reyes-Vargas F, et al. CTCF and CTCFL mRNA expression in 17beta-estradiol-treated MCF7 cells. Biomed Rep. 2014;2:101–104. doi: 10.3892/br.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin-Kleiner I. BORIS in human cancers -- a review. Eur J Cancer. 2012;48:929–935. doi: 10.1016/j.ejca.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Mendez-Catala CF, Gretton S, Vostrov A, et al. A novel mechanism for CTCF in the epigenetic regulation of Bax in breast cancer cells. Neoplasia. 2013;15:898–912. doi: 10.1593/neo.121948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mustafa M, Lee JY, Kim MH. CTCF negatively regulates HOXA10 expression in breast cancer cells. Biochem Biophys Res Commun. 2015;467:828–834. doi: 10.1016/j.bbrc.2015.10.058. [DOI] [PubMed] [Google Scholar]
- 42.Teif VB, Beshnova DA, Vainshtein Y, et al. Nucleosome repositioning links DNA (de)methylation and differential CTCF binding during stem cell development. Genome Res. 2014;24:1285–1295. doi: 10.1101/gr.164418.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Maurano MT, Qu H, et al. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012;22:1680–1688. doi: 10.1101/gr.136101.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Victoria-Acosta G, Vazquez-Santillan K, Jimenez-Hernandez L, et al. Epigenetic silencing of the XAF1 gene is mediated by the loss of CTCF binding. Sci Rep. 2015;5:14838. doi: 10.1038/srep14838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peng Z, Shen R, Li YW, Teng KY, Shapiro CL, Lin HJ. Epigenetic repression of RARRES1 is mediated by methylation of a proximal promoter and a loss of CTCF binding. PLoS One. 2012;7:e36891. doi: 10.1371/journal.pone.0036891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao Z, Sherif ZA. The effect of epigenetic silencing and TP53 mutation on the expression of DLL4 in human cancer stem disorder. Oncotarget. 2016;7:62976–62988. doi: 10.18632/oncotarget.11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Souza Rocha Simonini P, Breiling A, Gupta N, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–9184. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 48.Soto-Reyes E, Gonzalez-Barrios R, Cisneros-Soberanis F, et al. Disruption of CTCF at the miR-125b1 locus in gynecological cancers. BMC Cancer. 2012;12:40. doi: 10.1186/1471-2407-12-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang F, Liu T, He Y, et al. MiR-125b promotes proliferation and migration of type II endometrial carcinoma cells through targeting TP53INP1 tumor suppressor in vitro and in vivo. BMC Cancer. 2011;11:425. doi: 10.1186/1471-2407-11-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang Y, Liang J, Li Y, et al. CCCTC-binding factor acts upstream of FOXA1 and demarcates the genomic response to estrogen. J Biol Chem. 2010;285:28604–28613. doi: 10.1074/jbc.M110.149658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tang Z, Luo OJ, Li X, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Seitan VC, Faure AJ, Zhan Y, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome Res. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heidari N, Phanstiel DH, He C, et al. Genome-wide map of regulatory interactions in the human genome. Genome Res. 2014;24:1905–1917. doi: 10.1101/gr.176586.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu Q, Lian JB, Stein JL, Stein GS, Nickerson JA, Imbalzano AN. The BRG1 ATPase of human SWI/SNF chromatin remodeling enzymes as a driver of cancer. Epigenomics. 2017;9:919–931. doi: 10.2217/epi-2017-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bai J, Mei P, Zhang C, et al. BRG1 is a prognostic marker and potential therapeutic target in human breast cancer. PLoS One. 2013;8:e59772. doi: 10.1371/journal.pone.0059772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Q, Sharma S, Cui H, et al. Targeting the chromatin remodeling enzyme BRG1 increases the efficacy of chemotherapy drugs in breast cancer cells. Oncotarget. 2016;7:27158–27175. doi: 10.18632/oncotarget.8384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shukla S, Kavak E, Gregory M, et al. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dutertre M, Gratadou L, Dardenne E, et al. Estrogen regulation and physiopathologic significance of alternative promoters in breast cancer. Cancer Res. 2010;70:3760–3770. doi: 10.1158/0008-5472.CAN-09-3988. [DOI] [PubMed] [Google Scholar]
- 59.Ross-Innes CS, Brown GD, Carroll JS. A co-ordinated interaction between CTCF and ER in breast cancer cells. BMC Genomics. 2011;12:593. doi: 10.1186/1471-2164-12-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Docquier F, Farrar D, D’Arcy V, et al. Heightened expression of CTCF in breast cancer cells is associated with resistance to apoptosis. Cancer Res. 2005;65:5112–5122. doi: 10.1158/0008-5472.CAN-03-3498. [DOI] [PubMed] [Google Scholar]
- 61.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS One. 2010;5:e11457. doi: 10.1371/journal.pone.0011457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klenova EM, Morse HC, 3rd, Ohlsson R, Lobanenkov VV. The novel BORIS + CTCF gene family is uniquely involved in the epigenetics of normal biology and cancer. Semin Cancer Biol. 2002;12:399–414. doi: 10.1016/S1044-579X(02)00060-3. [DOI] [PubMed] [Google Scholar]
- 63.D’Arcy V, Pore N, Docquier F, et al. BORIS, a paralogue of the transcription factor, CTCF, is aberrantly expressed in breast tumours. Br J Cancer. 2008;98:571–579. doi: 10.1038/sj.bjc.6604181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dougherty CJ, Ichim TE, Liu L, et al. Selective apoptosis of breast cancer cells by siRNA targeting of BORIS. Biochem Biophys Res Commun. 2008;370:109–112. doi: 10.1016/j.bbrc.2008.03.040. [DOI] [PubMed] [Google Scholar]