Abstract
The transcription repressor Bach2 has been proposed as a regulator of T cell quiescence, but the underlying mechanism is not fully understood. Given the importance of interleukin-2 in T cell activation, we investigated whether Bach2 is a component of the network of factors that regulates interleukin-2 expression. In primary and transformed CD4+ T cells, Bach2 overexpression counteracted T cell receptor/CD28- or PMA/ionomycin-driven induction of interleukin-2 expression, and silencing of Bach2 had the opposite effect. Luciferase and chromatin immunoprecipitation assays revealed that Bach2 binds to multiple Maf-recognition element-like sites on the interleukin-2 proximal promoter in a manner competitive with AP-1, and thereby represses AP-1-driven induction of interleukin-2 transcription. Thus, this study demonstrates that Bach2 is a direct repressor of the interleukin-2 gene in CD4+ T cells during the immediate early phase of AP-driven activation, thereby playing an important role in the maintenance of immune quiescence in the steady state.
Keywords: Bach2, CD4+ T cells, IL-2, Repressor
INTRODUCTION
Interleukin-2 (IL-2) is produced by CD4+ T cells immediately after activation through the T cell receptor (TCR)/CD28, and it plays diverse roles in the immune response. In addition to its well-known activity as a T cell growth factor, it has broad crucial roles in not only stimulating but also limiting immune reactions (1–3), so that production of IL-2 needs to be tightly regulated. Studies have indicated that IL-2 production is both positively and negatively regulated at the transcriptional level. The regulatory network for IL-2 gene transcription is known to include positive regulators, such as NFAT, AP-1, NF-κB, and Oct-1, and negative regulators, such as TCF-8, Satb1, Blimp-1, and Aiolos (4–7).
Bach2 is a transcriptional repressor containing a basic-region leucine zipper domain (8). It forms heterodimers with small Maf proteins, and these dimers bind to Maf-recognition elements (MAREs) on target genes. Bach2 was initially characterized as a B cell-specific factor critical to somatic hypermutation and class-switch recombination of Ig-encoding genes during plasma cell differentiation (9, 10). However, Bach2 has recently been shown to participate in T cell-mediated immune responses in a cell-intrinsic manner (11–13). Bach2 represses the genes associated with the differentiation of effector cells and effector memory cells, including Blimp-1 and Gata3 (11–13), enabling T cells to maintain a quiescent state. Although these findings establish Bach2 as a key regulator of T cell-mediated immune homeostasis, it remains to be determined whether Bach2 is a component of the network of factors that regulates IL-2 expression in conventional T cells.
In the current study, we investigated whether Bach2 regulates expression of the IL-2 gene. Our results reveal that Bach2 binds to MARE-like sites in the IL-2 proximal promoter in competition with AP-1, and thereby represses the AP-1-driven induction of IL-2 gene transcription in CD4+ T cells. Thus, we identify a novel component for the regulatory network of IL-2 expression, and describe a mechanism by which naive CD4+ T cells maintain quiescence in the steady state.
RESULTS AND DISCUSSION
IL-2 varies inversely with Bach2 in transformed CD4+ T cells
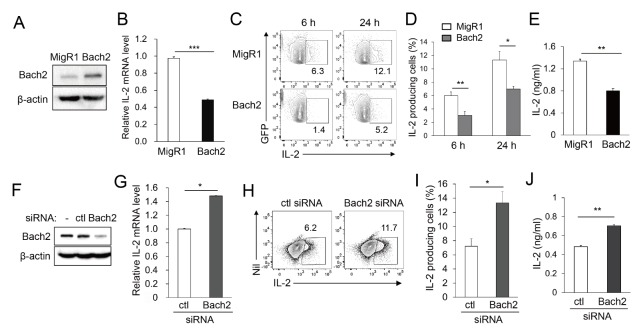
Given that the T cell receptor (TCR)-driven downregulation of Bach2 coincides with enhanced induction of IL-2 expression in mouse CD4+ T cells (data not shown), we investigated whether Bach2 participates in the regulation of IL-2 expression using a human transformed Jurkat T cell line. The cells were transfected with Bach2-expressing vector (MigR1–Bach2) and were assayed to confirm enhanced expression of Bach2 (Fig. 1A). The cells transfected with MigR1–Bach2 and stimulated with PMA/ionomycin for 16 hours produced significantly reduced amounts of IL-2 mRNA than those transfected with empty vector (Fig. 1B). Consistent with this, in intracellular FACS analysis gated on only GFP+ transfected cells, fewer Bach2-transfected cells became IL-2-producing cells at 6 and 24 hours after stimulation than empty vector-transfected (control) cells (Fig. 1C and D). The media collected from the culture of Bach2-transfected cells contained a reduced amount of IL-2 than media collected from vehicle-transfected cells (Fig. 1E). These data demonstrate that Bach2 counteracts the PMA/ionomycin-mediated induction of IL-2 gene expression at the transcriptional and/or post-transcriptional levels.
Fig. 1.
IL-2 varies inversely with Bach2. Jurkat T cells were transfected with MigR1, MigR1–Bach2, or Bach2-specific or control (ctl) siRNA and assayed by immunoblotting (A and F). The transfectants were stimulated with PMA/ionomycin for 16 hours and assayed by qRT-PCR (B and G), for 6 or 24 hours and assayed by FACS (C, D, H and I). The culture supernatants were collected and assayed by ELISA (E and J). Representative FACS profiles with the percentage of cells in the indicated area (C and H) and percentages of IL-2-producing cells (D and I) are shown. All data are representative of three independent experiments. Nil, no staining. *P < 0.05, **P < 0.01 and ***P < 0.001 by Student’s t-test.
Bach2 involvement in the regulation of IL-2 expression was further investigated in Jurkat T cells by silencing endogenous Bach2 expression. Jurkat T cells were transfected with Bach2-specific or control siRNA and then stimulated to induce IL-2 expression. The Jurkat T cells expressed a substantial amount of endogenous Bach2 protein, and Bach2-specific siRNA reduced the expression of Bach2 protein (Fig. 1F). Silencing of Bach2 resulted in increased IL-2 production, as judged by measuring IL-2 mRNA and secreted proteins and percentages of IL-2-producing cells (Fig. 1G–J). This result supports our hypothesis that Bach2 regulates IL-2 expression at the transcriptional and/or post-transcriptional level.
Bach2 represses transcription of IL-2 in Jurkat T cells by binding to the IL-2 promoter
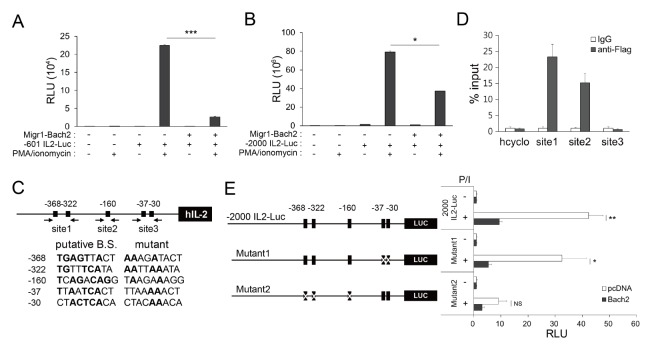
To test the hypothesis that Bach2 binds to the IL-2 promoter and represses transcription rather than regulating IL-2 at the post-transcriptional level (e.g., by affecting mRNA stability), we first conducted transient transfection and reporter assays using the reporter vector −601 IL2–Luc constructs, comprising 601 bp of the murine IL-2 promoter upstream of the translation start site. Jurkat T cells were transfected with the reporter vector together with MigR1–Bach2 or empty vector, and cultured in the presence or absence of PMA/ionomycin. In the absence of stimulation, luciferase expression driven by the IL-2 proximal promoter was very low and there was no significant difference between cells transfected with MigR1–Bach2 or the vehicle (Fig. 2A). Activation with PMA/ionomycin, however, strongly induced the expression of luciferase, which was dramatically reduced by Bach2 overexpression, indicating that Bach2 inhibits the activation of the IL-2 proximal promoter. We obtained similar results when we replaced the −601 IL2–Luc construct with the −2000 IL2–Luc construct, comprising 2000 bp of the human IL-2 promoter upstream of the translation start site (Fig. 2B). This suggests that murine Bach2 can also exert effects on the human IL-2 promoter, and that Bach2 binding sites are present within 601 bp upstream of the IL-2 translation start site.
Fig. 2.
Bach2 is a repressor of IL-2. (A, B and right panel of E) Jurkat T cells were transfected with MigR1 (−) or MigR1–Bach2 (+) plasmids, together with reporter constructs, cultured with or without PMA/ionomycin (P/I), and analyzed using luciferase assays. (C) The human IL-2 promoter region including WT and mutated sequences of putative Bach2-binding sites (B.S.) are shown. Conserved sequences of the canonical MARE are indicated by bold. The numbers indicate bp upstream of the translation start site. Arrows indicate the annealing sites of primers for ChIP-qPCR. (D) Jurkat T cells transfected with pcDNA3-2×FLAG–Bach2 were stimulated with PMA/ionomycin and assayed by ChIP-qPCR. (The left panel of E) Schematic representation of mutated reporter constructs. All data are representative of three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 by Student’s t-test. NS, not significant.
With the help of previous investigations (14–16) and in silico analysis, we identified five MARE-like sequences as putative Bach2-binding sites, at −368, −322, −160, −37 and −30 bp upstream of the human IL-2 translation start site (Fig. 2C). These sequences have approximately 55–67% nucleotide sequence homology with the canonical sequences of MARE (TGA(C/G)TCAGC). To investigate whether Bach2 binds to the IL-2 proximal promoter in the cell, and exactly which MARE-like sites are involved in this association, we transfected Jurkat T cells with Flag-tagged Bach2-expressing vector or empty vector, cultured the cells with or without PMA/ionomycin, and then conducted chromatin immunoprecipitation (ChIP) assays. Immunoprecipitation of the cross-linked chromatin with anti-Flag antibody (Ab), but not with IgG, significantly enriched the genome regions located between −408 and −303 (referred to as Site1) and between −250 and −149 (referred to as Site2) upstream from the IL-2 translation start site in stimulated cells (Fig. 2D). Interestingly, the genome region containing −37 and −30 MARE-like sequences (referred to as Site3) was not enriched. This result suggests that Bach2 binds to −368/−322 and −160 MARE-like sites, but not −37/−30 MARE-like sites.
To confirm the binding of Bach2 to −368/−322 and −160 MARE-like sites, but not −37/−30 MARE-like sites, we performed reporter assays with multiple mutated IL-2 promoter–luciferase constructs as depicted in Fig. 2C and E. We found that both luciferase induction by PMA/ionomycin and suppression by Bach2 were intact when using the mutant1 construct (mutations in −37 and −30 MARE-like sites), but highly impaired when using the mutant2 construct (mutations in −368, −322, and −160 MARE-like sites) (Fig. 2E). This result suggests that promoter sites, including the MARE-like sequences at −368, −322, and/or −160, but not at −37/−30, are crucial not only for PMA/ionomycin-mediated induction but also for Bach2-mediated repression of IL-2 transcription, which is consistent with our results from the ChIP assay. This result also implies that the repressor Bach2 and activator(s) induced by PMA/ionomycin share the same binding sites.
Bach2 competes with AP-1 for binding to MARE-like sites of the IL-2 promoter
AP-1 is a critical transcription factor that regulates the IL-2 promoter. Given that the canonical AP-1 recognition motif (TGA(C/G)TCA) is embedded in MARE (14, 15), we wanted to know whether there is any functional interference between AP-1 and Bach2 on the IL-2 proximal promoter. To investigate this, we transfected Jurkat T cells with c-Jun- and c-Fosexpressing vectors and conducted luciferase assays with the −601 IL2–Luc construct. Luciferase expression was induced without any exogenous stimulation when transfected with c-Jun- and c-Fos-expressing vectors (Fig. 3A). Importantly, this induction was significantly downregulated by expression of Bach2. Conversely, the repressive effect of Bach2 on PMA/ionomycin-induced IL-2 transcription was totally counteracted by AP-1 overexpression (Fig. 3B). These results demonstrate mutual functional interference between Bach2 and AP-1 on the IL-2 proximal promoter.
Fig. 3.
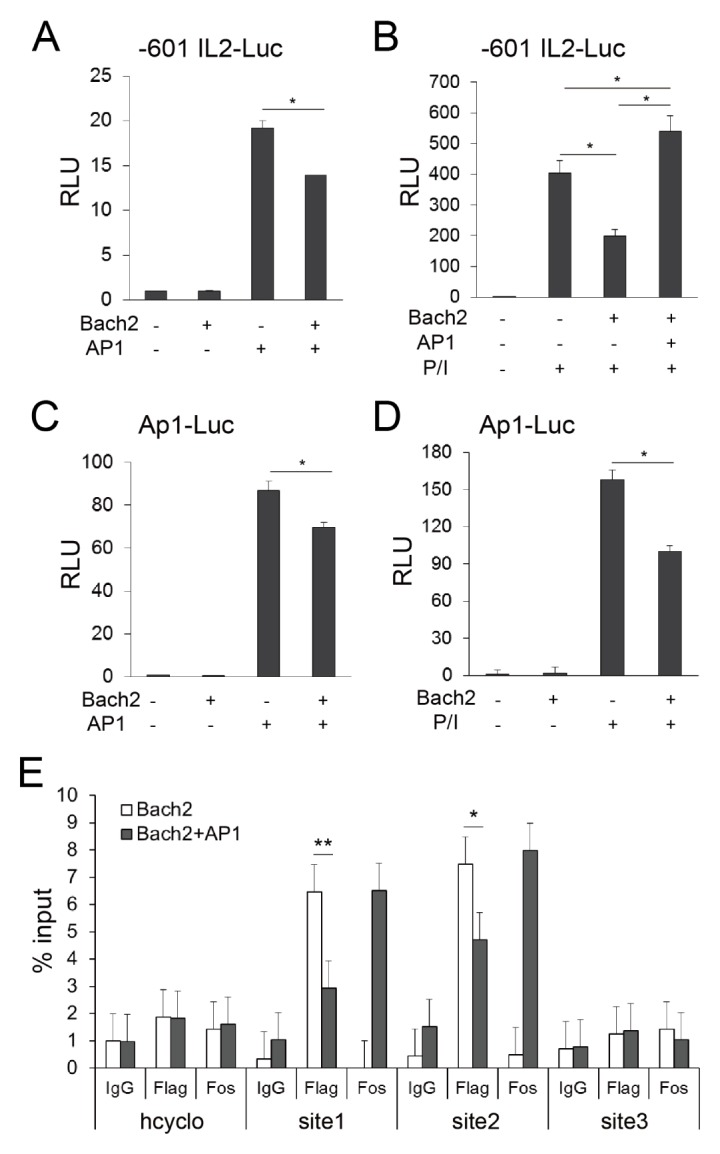
Bach2 competes with AP-1 for binding to the IL-2 proximal promoter. (A to D) Jurkat T cells were transfected with Bach2-and/or AP-1-expressing vectors, along with reporter constructs, as indicated. The cells were cultured in the presence (B and D) or absence (A and C) of PMA/ionomycin (P/I) for 24 hours and analyzed using luciferase assays. (E) Jurkat T cells were transfected with pcDNA3-2×FLAG-Bach2 alone or together with AP-1-expressing vectors and assayed by ChIP-qPCR methods. The data are representative of three independent experiments. *P < 0.05 and **P < 0.01 by Student’s t-test.
Next, we tested whether Bach2 can elicit its activity via the canonical AP-1 recognition motif. AP-1 overexpression alone was sufficient to induce luciferase expression from the Ap1–Luc construct, and this effect was partially but significantly reduced by Bach2 overexpression (Fig. 3C). We also obtained similar results when the cells were stimulated with PMA/ionomycin (Fig. 3D). These results suggest that the functional interference between Bach2 and AP-1 is caused because they share the same binding sites on the IL-2 proximal promoter.
To evaluate this hypothesis, we conducted ChIP assays using Jurkat T cells which were transfected with Bach2-expressing vector alone or together with AP-1-expressing vectors (Fig. 3E). The enrichment of Bach2 to the Site1 and Site2 of the IL-2 promoter was reduced by AP-1 overexpression, suggesting that Bach2 suppress IL-2 production via competition with AP-1 for binding to the −368/−322 and −160 MARE-likes sites of IL-2 promoter.
Bach2 regulates IL-2 expression through direct and indirect (Blimp-1-mediated) pathways in primary CD4+ T cells
The aforementioned results obtained using Jurkat T cells suggested that Bach2 is a repressor of the IL-2 gene in CD4+ T cells. To evaluate whether this is also true in primary CD4+ T cells, CD4+ T cells sorted from normal mice were transduced with a retrovirus carrying the Bach2-overexpressing vector (MigR1–Bach2) and IL-2 production in response to stimulation with anti-CD3/CD28 monoclonal Abs (mAbs) was assessed. Bach2-transduced cells gave rise to a lower percentage of IL-2-producing cells than cells transduced with vehicle alone (Fig. 4A).
Fig. 4.
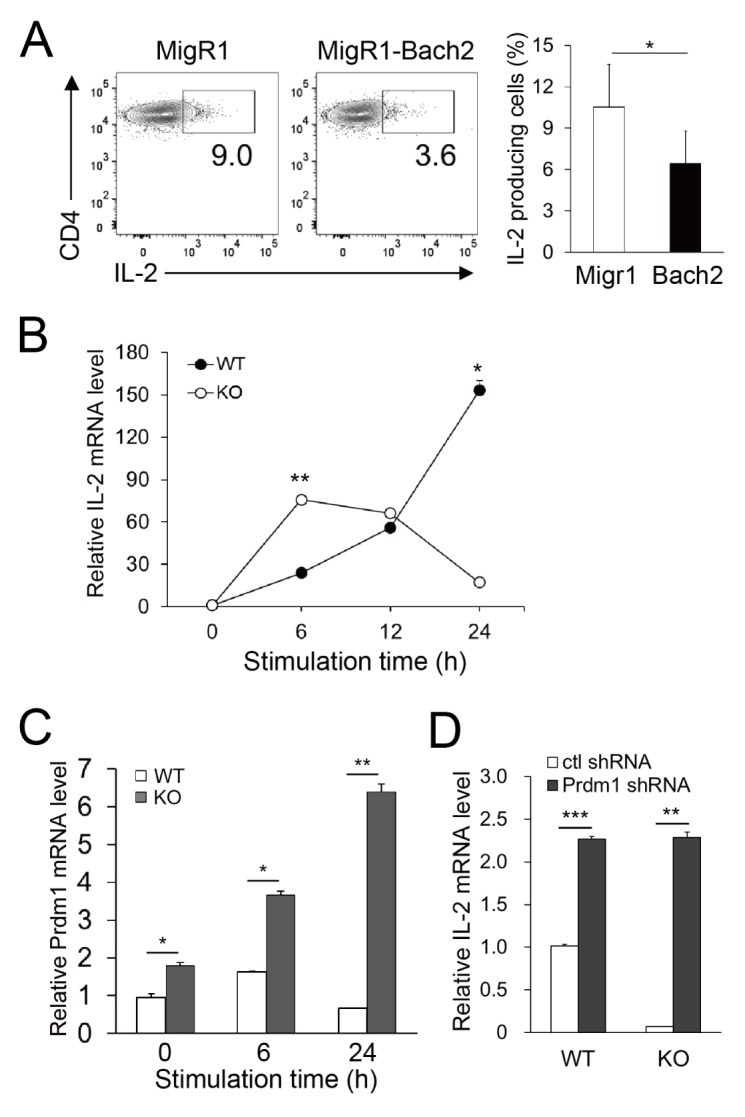
Bach2 regulates IL-2 expression in mouse primary CD4+ T cells. (A) CD4+ T cells were transduced with retroviruses carrying either empty vector (MigR1) or MigR1–Bach2, stimulated with anti-CD3 and anti-CD28 mAbs for 24 hours, and assayed by FACS. Representative FACS profiles gated on GFP+ cells and the percentages of IL-2-producing cells are shown. (B and C) Naive CD4+ T cells sorted from WT or Bach2 KO mice were stimulated for the time indicated and assayed by qRT-PCR to measure the mRNA levels of IL-2 (B) and Prdm1 (C). (D) CD4+ T cells sorted from WT or Bach2 KO mice were infected with Prdm1-specific or control shRNA-containing retrovirus and stimulated for 24 hours. GFP+ cells were sorted and assayed by qRT-PCR. All data represent three independent experiments. *P < 0.05, **P < 0.01 and ***P < 0.001 by Student’s t-test.
We further evaluated the action of Bach2 on the repression of IL-2 using Bach2-deficient cells. We found that CD4+ T cells from Bach2 knockout (KO) mice produced approximately 3-fold more IL-2 mRNA 6 hours after stimulation than those from wild-type (WT) littermates. However, IL-2 expression decreased in KO cells between 6 and 24 hours, whereas it was increased gradually in WT cells (Fig. 4B). This might be due to other regulators that are induced when Bach2 is deficient. Because Bach2 has been proven to repress Prdm1 (which encodes Blimp-1, a known repressor of IL-2) (7), we suggest that Blimp-1 might be one such regulator. In agreement with this hypothesis, the level of Prdm1 mRNA in naive CD4+ T cells from Bach2 KO mice was higher than that from WT mice and gradually increased with time when stimulated with anti-CD3/CD28 mAbs (Fig. 4C). To evaluate our hypothesis, we transduced WT and KO CD4+ T cells with a retrovirus delivering Prdm1-specific shRNA and assessed IL-2 expression. Blimp-1-silencing in Bach2 KO cells resulted in enhanced expression of IL-2 mRNA at the level equivalent to that of WT cells (Fig. 4D), indicating that Blimp-1 is overexpressed in Bach2 KO cells during the later phase of activation, and participates in the regulation of IL-2 expression. Taken together, these results suggest that Bach2 expressed in primary naive CD4+ T cells directly represses IL-2 gene transcription during the early phase of T cell activation. Although Bach2 is also suggested to have the potential to drive IL-2 gene transcription by repressing Blimp-1 during later phases of activation, this is unlikely because of the activation-driven downregulation of Bach2. We did not detect such an effect of Blimp-1 in Jurkat T cells, as shown in Fig. 1G to J, presumably due to the undetectable level of Blimp-1 in Jurkat T cells (data not shown).
We found that Bach2 and AP-1 competed to bind to the same sites in the IL-2 proximal promoter. This appears to be due to sequence similarity between their binding sites. If that is the case, repression by Bach2 ought to be observed for other genes with AP-1 binding sites in their promoter regions. Indeed, many genes associated with the terminal differentiation of CD8+ T cells have AP-1 binding sites and have been found to be repressed by Bach2 (14).
Lesniewski et al.’s results obtained from umbilical cord blood CD4+ T cells (16) are in accordance with our description of the association of Bach2 on the IL-2 promoter, but, in contrast to the current results, Bach2 was found to promote directly IL-2 transcription. This discrepancy may stem from differences in the types of cells studied, as fetal CD4+ T cells have many features distinct from adult CD4+ cells (16). In addition, the authors of the study focused on the role of Bach2 in the spontaneous production of IL-2 without any stimulus during a resting state, which is different from the current study, which investigated TCR/CD28-driven induction of IL-2 gene expression.
In conclusion, we demonstrate here that Bach2 acts as a direct repressor of IL-2 that prevents the AP-1-driven induction of IL-2 transcription. Therefore, the current study is important, as it is the first to identify the competition between Bach2 and AP-1 at the binding site of the IL-2 gene and it also describes a novel mechanism by which Bach2 contributes to CD4+ T cell quiescence. In addition, given the association of Bach2 polymorphism with many immune disorders (17–20), the current study may help our understanding of the pathophysiology of these disorders.
MATERIALS AND METHODS
Mice
Bach2−/− mice and their littermate controls (9) were bred an animal facility at Hanyang University under specific pathogenfree conditions. The study protocol was approved by the Institutional Animal Care and Use Committee (IACUC) of Hanyang University. All animal experiments were carried out in strict accordance with the guidelines and regulations.
Plasmid constructs
Mouse Bach2 cDNA was reverse-transcribed from RNA isolated from mouse T cells, amplified by PCR and inserted into pcDNA3-2 × FLAG and MigR1 (carrying IRES–GFP cDNA) vectors. Plasmids carrying c-Jun and c-Fos cDNA were purchased from Korea Human Gene Bank. The reporter constructs −601 IL2–Luc and Ap1–Luc (comprising copies of AP-1 recognition motifs) (21) were provided by Dr. Young Dae Yun (Ewha Womans University, Seoul, Korea). The construct −2000 IL2–Luc was generated by PCR and cloned in a pGL4 vector (Promega). Mutated IL-2 promoter–luciferase constructs (referred to as Mutant1 and Mutant2) were generated by introducing substitution mutations into the construct −2000 IL2–Luc, as depicted in Fig. 2C and E. pLMPd plasmids carrying DNA sequences encoding Prdm1- or Cd19-specific shRNA (22) were a kind gift from Dr. Youn Soo Choi (Seoul National University, Seoul, Korea).
Primary mouse cell culture
Spleen cells from mice were prepared as described previously (23). Mouse CD4+ T cells were purified by negative selection using MACS columns (eBioscience or Stemcell). To purify naive CD62L+CD44loCD4+ T cells, MACS-sorted CD4+ T cells were further sorted using FACSAria III (BD Biosciences). Total or naive CD4+ T cells were cultured in the presence of immobilized 5 μg/ml anti-CD3 mAb (145-2C11; eBioscience) and soluble 1 μg/ml anti-CD28 mAb (37.51; eBioscience) unless indicated otherwise, followed by further assays.
Jurkat T cell culture and transfection
One million Jurkat T cells (ATCC) were transfected with 10 μg plasmid DNA or 100 nM siRNA by electroporation using NEPA21 Super Electroporator (Nepa Gene). Human Bach2 siRNA and negative control siRNA were purchased from Qiagen. Twenty-four hours after transfection, whole cells or FACS-sorted GFP+ cells were used in further experiments. The level of Bach2 protein in transfected cells was assessed by standard immunoblotting using anti-Bach2 Ab (Cell Signaling).
Retroviral transduction
PLAT-E retroviral packaging cells were transfected with MigR1–Bach2 and pCL–eco, and the supernatants containing retroviruses were collected, as described previously (24). Pre-activated CD4+ T cells were spin-infected with retrovirus supernatants as described (24).
FACS
To induce IL-2 expression, Jurkat T cells were stimulated with 40 ng/ml PMA plus 1 μM ionomycin (both from Sigma-Aldrich) for either 6 or 24 hours. Mouse CD4+ T cells were stimulated as described above. Brefeldin-A (BD Biosciences) was added for the last 4 hours of simulation. Cells were stained with anti-mouse CD4 mAb (RM4-5; eBioscience) and anti-mouse IL-2 mAb (MQ1-17H12; eBioscience) or anti-human IL-2 mAb (JES6-5H4; BD Biosciences), and assayed using a FACSCanto II (BD Biosciences), as described previously (25).
ELISA
Jurkat T cells transfected with MigR1 or MigR1–Bach2 were stimulated with PMA and ionomycin for 24 hours. The concentration of human IL-2 in the culture supernatants was determined by ELISA using IL-2 ELISA Ready-SET-Go kit (eBioscience), according to the manufacturer’s instructions.
Quantitative RT-PCR
Quantitative RT-PCR was conducted as described previously (23). The primer sequences used were described in Supplementary Table 1.
Luciferase assay
Jurkat T cells were transiently cotransfected with plasmids expressing cDNA and reporter constructs. Twenty-four hours after transfection, live cells were cultured for 24 hours with or without PMA/ionomycin, and assayed with a Luciferase Assay System (Promega) according to the manufacturer’s protocol. Firefly luciferase activity was normalized to either β-galactosidase activity or Renilla luciferase activity and recorded as relative luciferase units (RLUs) (26).
Chromatin immunoprecipitation–quantitative PCR (ChIP-qPCR)
Jurkat cells were assayed by the standard ChIP-qPCR methods, as described (27). Anti-Flag (F1804; Sigma-Aldrich), anti-c-Fos (Sc-8047; Santa Cruz) and control IgG Abs (Sc-2027; Santa Cruz) were used. The primer sequences used were described in Supplementary Table 1.
Supplementary Information
ACKNOWLEDGEMENTS
We thank Drs Young Dae Yun, Mi-La Cho, and Youn Soo Choi for providing reagents and Yun-Seung Jeong for technical assistance. We thank the Analytical Instrumental Center (Seoul) at Hanyang University for technical support. This work was supported by a NRF grant (NRF-2014R1A2A1A11052070).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Siegel JP, Sharon M, Smith PL, Leonard WJ. The IL-2 receptor beta chain (p70): role in mediating signals for LAK, NK, and proliferative activities. Science. 1987;238:75–78. doi: 10.1126/science.3116668. [DOI] [PubMed] [Google Scholar]
- 2.Mingari MC, Gerosa F, Carra G, et al. Human interleukin-2 promotes proliferation of activated B cells via surface receptors similar to those of activated T cells. Nature. 1984;312:641–643. doi: 10.1038/312641a0. [DOI] [PubMed] [Google Scholar]
- 3.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams TM, Moolten D, Burlein J, et al. Identification of a zinc finger protein that inhibits IL-2 gene expression. Science. 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 6.Pavan Kumar P, Purbey PK, Sinha CK, et al. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22:231–243. doi: 10.1016/j.molcel.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 7.Martins GA, Cimmino L, Liao J, Magnusdottir E, Calame K. Blimp-1 directly represses Il2 and the Il2 activator Fos, attenuating T cell proliferation and survival. J Exp Med. 2008;205:1959–1965. doi: 10.1084/jem.20080526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oyake T, Itoh K, Motohashi H, et al. Bach proteins belong to a novel family of BTB-basic leucine zipper transcription factors that interact with MafK and regulate transcription through the NF-E2 site. Mol Cell Biol. 1996;16:6083–6095. doi: 10.1128/MCB.16.11.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muto A, Tashiro S, Nakajima O, et al. The transcriptional programme of antibody class switching involves the repressor Bach2. Nature. 2004;429:566–571. doi: 10.1038/nature02596. [DOI] [PubMed] [Google Scholar]
- 10.Muto A, Ochiai K, Kimura Y, et al. Bach2 represses plasma cell gene regulatory network in B cells to promote antibody class switch. EMBO J. 2010;29:4048–4061. doi: 10.1038/emboj.2010.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roychoudhuri R, Hirahara K, Mousavi K, et al. BACH2 represses effector programs to stabilize T(reg)-mediated immune homeostasis. Nature. 2013;498:506–510. doi: 10.1038/nature12199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsukumo S, Unno M, Muto A, et al. Bach2 maintains T cells in a naive state by suppressing effector memory-related genes. Proc Natl Acad Sci U S A. 2013;110:10735–10740. doi: 10.1073/pnas.1306691110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim EH, Gasper DJ, Lee SH, Plisch EH, Svaren J, Suresh M. Bach2 regulates homeostasis of Foxp3+ regulatory T cells and protects against fatal lung disease in mice. J Immunol. 2014;192:985–995. doi: 10.4049/jimmunol.1302378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roychoudhuri R, Clever D, Li P, et al. BACH2 regulates CD8+ T cell differentiation by controlling access of AP-1 factors to enhancers. Nat immunol. 2016;17:851–860. doi: 10.1038/ni.3441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muto A, Tashiro S, Tsuchiya H, et al. Activation of Maf/AP-1 repressor Bach2 by oxidative stress promotes apoptosis and its interaction with promyelocytic leukemia nuclear bodies. J Biol Chem. 2002;277:20724–20733. doi: 10.1074/jbc.M112003200. [DOI] [PubMed] [Google Scholar]
- 16.Lesniewski ML, Haviernik P, Weitzel R, et al. Regulation of IL-2 expression by transcription factor BACH2 in umbilical cord blood CD4+ T cells. Leukemia. 2008;22:2201–2207. doi: 10.1038/leu.2008.234. [DOI] [PubMed] [Google Scholar]
- 17.International Multiple Sclerosis Genetics C, Wellcome Trust Case Control C. Sawcer S, et al. Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature. 2011;476:214–219. doi: 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferreira MA, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13. 5 as risk loci for asthma. Lancet. 2011;378:1006–1014. doi: 10.1016/S0140-6736(11)60874-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke A, McGovern DP, Barrett JC, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper JD, Smyth DJ, Smiles AM, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi YB, Kim CK, Yun Y. Lad, an adapter protein interacting with the SH2 domain of p56lck, is required for T cell activation. J Immunol. 1999;163:5242–5249. [PubMed] [Google Scholar]
- 22.Chen R, Belanger S, Frederick MA, et al. In vivo RNA interference screens identify regulators of antiviral CD4+ and CD8+ T cell differentiation. Immunity. 2014;41:325–338. doi: 10.1016/j.immuni.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oh Y-K, Jang E, Paik D-J, Youn J. Early growth response-1 plays a non-redundant role in the differentiation of B cells into plasma cells. Immune Netw. 2015;15:161–166. doi: 10.4110/in.2015.15.3.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi YS, Gullicksrud JA, Xing S, et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho WS, Jang E, Kim H-Y, Youn J. Interleukin 17-expressing innate synovial cells drive K/BxN serum-induced arthritis. Immune Netw. 2016;16:366–372. doi: 10.4110/in.2016.16.6.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Na HH, Noh HJ, Cheong HM, Kang Y, Kim KC. SETDB1 mediated FosB expression increases the cell proliferation rate during anticancer drug therapy. BMB Rep. 2016;49:238–243. doi: 10.5483/BMBRep.2016.49.4.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeong Y-S, Kim D, Lee YS, et al. Integrated expression profiling and genome-wide analysis of ChREBP targets reveals the dual role for ChREBP in glucose-regulated gene expression. PloS One. 2011;6:e22544. doi: 10.1371/journal.pone.0022544. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.