Abstract
Budding yeast has dozens of prions, which are mutually dependent on each other for the de novo prion formation. In addition to the interactions among prions, transmissions of prions are strictly dependent on two chaperone systems: the Hsp104 and the Hsp70/Hsp40 (J-protein) systems, both of which cooperatively remodel the prion aggregates to ensure the multiplication of prion entities. Since it has been postulated that prions and the remodeling factors constitute complex networks in cells, a quantitative approach to describe the interactions in live cells would be required. Here, the researchers applied dual-color fluorescence cross-correlation spectroscopy to investigate the molecular network of interaction in single live cells. The findings demonstrate that yeast prions and remodeling factors constitute a network through heterogeneous protein-protein interactions.
Keywords: Fluorescence cross-correlation spectroscopy, Live cell, Protein interaction, Remodeling factors, Yeast prions
INTRODUCTION
Prions are protein-based infectious factors (1). In the prion, the altered conformation of a protein converts its normal structure to an altered form, resulting in the formation of ordered aggregates called amyloids (1). Although this concept was originally developed for mammalian neurodegenerative diseases, such as scrapie, it has been applied to non-Mendelian genetic elements in budding yeast cells, [PSI+] and [URE3] (2). In [PSI+] and [URE3] cells, amyloid structures of Sup35 and Ure2 are the prion determinants and are transmitted to daughter cells through a dynamic remodeling of amyloid aggregates (3–8). Extensive studies using genetically tractable budding yeast as a model system have revealed that more than several dozen yeast proteins, including Rnq1, New1, and Swi1, behave as prions (9–14).
Although amyloids are kinds of homo-oligomers formed via cross-beta interactions, yeast prions are related each other in de novo formation and transmission. It has been demonstrated that prions affect the appearance of other prions (10, 15). As a well-known example, it was demonstrated that the prion form of Rnq1 is required for the de novo appearance of [PSI+], and vice versa (10, 15). Therefore, it has been suggested that there are interactions among prions either in a direct or indirect manner. In addition to the mutual dependence among prions, the transmission of yeast prions is strictly dependent on two chaperone systems: the Hsp104 and the Hsp70/Hsp40 (J-protein) systems. Hsp104, which is a homohexameric ATPase involved in the thermotolerance of yeast, is an essential factor to maintain yeast prions (16, 17). Unlike Hsp104, which is unique in yeast, the Hsp70/Hsp40 system is diverse (18). An essential J-protein Sis1 (Hsp40), cochaperone of Ssa Hsp70, is a critical factor for the transmission of several yeast prions, including [PSI+] (19, 20). Hsp104, a molecular disaggregase, remodels the amyloid aggregates to ensure the multiplication of prion entities in cooperation with the Hsp70/Hsp40 system (18, 21–23).
As described above, it seems that prions are in a complex network with the assistance of remodeling factors to propagate and transmit into the cells. To better understand the protein network, it would be important to quantitatively detect the physical interaction among prions and/or between prions, and the remodeling factors. In fact, previous studies have reported physical interactions among prions as well as between prions, and the remodeling factors in lysates and in cells (24, 25). Affinity purification of Sup35 aggregates from [PSI+] lysates to identify associated proteins revealed that the Sup35 aggregates contained Rnq1. It was also revealed that the major components of [PSI+] aggregates were Sup35 and Ssa1/2 (Hsp70) (25, 26).
Fluorescence microscopic imaging of GFP- or RFP-tagged prions and remodeling factors were also used for detecting colocalzation of the proteins on a large and immobile aggregate formed in live prion cells (27–30). However, there is no quantitative analysis of such interactions between mobile proteins in other regions of live cells surrounding the immobile aggregate.
In this study, the researchers applied fluorescence correlation spectroscopy (FCS) and fluorescence cross correlation spectroscopy (FCCS) to detect the interaction network of related proteins in single live cells. FCS is a technique used to determine the diffusional mobility of fluorescence molecules, providing information about the size of the molecules. Therefore, this technique is useful to investigate dynamic property of prions by discriminating prion aggregates from the monomeric state (31–34). Additionally, FCCS is an advanced FCS method that uses two different colored proteins to directly detect an interaction between proteins, in addition to the acquisition of each of the FCS parameters of the two proteins (35). Since FCS and FCCS are usually combined with confocal laser scanning microscopy (CLSM), the researchers can define the detection volume at any position of interest inside cells in a noninvasive manner. Interestingly, FCS and FCCS successfully detected the physical interaction among prions (Sup35, Ure2, Rnq1, New1) and remodeling factors (Hsp104 and Sis1) in the freely mobile and oligomer states, and showed that they specifically interacted with each other in the prion state. These results suggest that there is a dynamic and heterogeneous network of prions, and remodeling factors composed of various physical interactions.
RESULTS
FCS analysis of the hydrodynamic properties of yeast prions and the remodeling factors
Previous studies based on the CLSM observation combined with FCS have already demonstrated that FCS is a powerful method to discriminate prion oligomers (i.e. small and mobile aggregates) from the monomeric form, by quantitative analysis of diffusional behavior of the GFP-tagged N and M prion domains of Sup35 (Sup35NM-GFP) as well as the intact Sup35 containing GFP (Sup35NGMC) (31–33). In this study, the researchers further characterized diffusional behaviors of Sup35 variants and other yeast prions such as Rnq1, Ure2, and New1, and the representative remodeling factors, Hsp104 and Sis1 (Table S1).
The diffusion coefficients (Ds) of each protein in the supernatant solution obtained from cell lysates and the cytosol of live cells were summarized in Table 1. Monomer GFP (mGFP) in the lysis solution prepared from non-prion [psi−] and prion [PSI+] cells show a single D value expected from their molecular weights in spherical shape, indicating that mGFP molecules diffuse in a monomeric state without aggregates (Fig. 1A black, Table S1). Only one diffusional component was detected in the lysis solution of the yeast prion proteins from non-prion [psi−] ([gpsi−]) cell lysate (Fig. 1A blue and red, Table S1), even though the diffusion of Sup35NM-GFP (Sup35NGMC) was slower than expected, as calculated from its theoretical molecular weight. In addition to a previous study (31), this result suggests that monomers diffuse in a nonspherical shape.
Table 1.
Summary of D values for GFP, prion proteins and remodeling factors
| Type of sample | Probe molecule | Dfast (μm2s−1)a (%)b | Dslow (μm2s−1)a (%)b |
|---|---|---|---|
| [psi−] | GFP (control)c | 21.0 ± 3.8 (90) | 0.26 ± 0.3 (10) |
| [psi−] | Sup35NM-GFP | 9.1 ± 1.5 (87) | 0.26 ± 0.2 (13) |
| [gpsi−] | NGMC | 5.3 ± 2.1 (85) | 0.18 ± 0.2 (15) |
| [rnq−] | Rnq1-GFP | 4.3 ±1.2 (90) | 0.23 ± 0.2 (10) |
| [PSI+] | GFP (control)c | 18.2 ± 4.3 (89) | 0.2 ± 0.3 (11) |
| [PSI+] | Sup35NM-GFP | 9.3 ± 2.8 (26) | 0.12 ± 0.1 (74) |
| [GPSI+] | NGMC | 4.8 ± 1.9 (47) | 0.15 ± 0.1 (53) |
| [RNQ+] | Rnq1-GFP | 4.8 ± 2.2 (23) | 0.16 ± 0.1 (77) |
| Lysis solution | GFP ([psi−], [PSI+])c | 77.0 ± 3.0 (100) | n.d. |
| Sup35NM-GFP ([psi−]) | 51.3 ± 3.7 (100) | n.d. | |
| Sup35NM-GFP ([PSI+]) | 43.5 ± 12 (30) | 3.3 ± 1.1 (70) | |
| NGMC ([gpsi−]) | 31 ± 2.1 (100) | n.d. | |
| NGMC ([GPSI+]) | 33 ± 3.1 (9) | 6.2 ± 2.0 (91) | |
| Rnq1-GFP ([rnq−]) | 56.3 ± 10.6 (100) | n.d. | |
| Rnq1-GFP ([RNQ+]) | 55.7 ± 6.4 (15) | 2.9 ± 2.0 (85) | |
| Non-prion cell | Hsp104-GFP | 3.3 ± 1.6 (42) | 0.34 ± 0.2 (58) |
| Sis1-mCherry | 11.0 ± 8.6 (65) | 0.6 ± 0.3 (35) | |
| Lysis solution | Hsp104-GFP | 10.5 ± 1.0 (100) | n.d. |
| Sis1-mCherry | 18.0 ± 4.0 (100) | n.d. |
Diffusion coefficients were calculated from the FAF fitting result (means ± s.d.; cell number n=5).
Fractional ratios corresponding to the diffusion coefficients are represented by percentage.
Diffusion of GFP monomer in live yeast cells is shown as a reference.
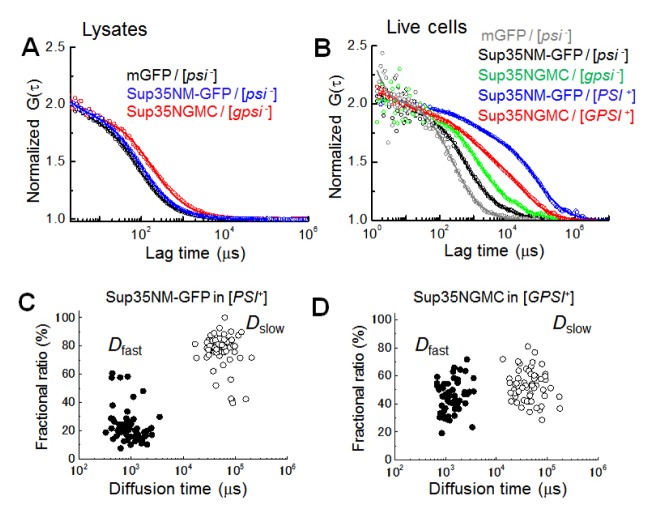
Fig. 1.
Unique and similar diffusional properties of prion oligomers in live cells. (A) Representative normalized fluorescence auto-correlation functions (FAFs) of mGFP (black), Sup35NM-GFP (blue), Sup35-GFP (red) in yeast lysates prepared from non-prion cells are shown (circle symbol). (B) Representative normalized FAFs of mGFP (gray) detected in the [psi−] cell, Sup35NM-GFP in the [psi−] (black) and the [PSI+] (blue) cells, and Sup35-GFP in the [gpsi−] (green) and the [GPSI+] (red) cells are respectively shown. For comparison of mobility, all the curves were normalized to the same amplitude, G (0) = 2. Solid lines indicate fitting of two-component models to the results. (C) and (D) Plot of diffusion times versus the corresponding fractional ratios of Sup35NM-GFP and Sup35-GFP (Sup35NMGC) in the [PSI+] and [GPSI+] prion cells are respectively shown. Two types of diffusion times (τi, i = 2) were evaluated from two-component model fitting (see also Methods). Fast diffusion time (solid circle) and slow diffusion time (open circle) are inversely proportional to diffusional coefficients of Dfast and Dslow, respectively (Table 1).
In contrast, two diffusional components were detected in the lysis solution of the yeast prion proteins from the prion cells (Table 1). Fast diffusion of the proteins from yeast prion cells, represented by Dfast, corresponded to the diffusion of the proteins in the lysis solution from non-prion cells and constituted a small proportion, ranging from 9 to 30%. Slow diffusion of the proteins, represented by Dslow, corresponded with large spherical complexes with hydrodynamic sizes, ranging from 60 to 500 MD and covered a large proportion of lysis samples, ranging from 70 to 91%. It is notable that the molecular weight of the large complexes can be much smaller than the estimated values, if the molecular shape of the complexes is not spherical, but rod-like (32).
For cellular analysis, FCS measurements for yeast prion proteins were carried out on other positions of cells surrounding a dot-like immobile aggregate (fluorescent foci) (31, 33). Fast diffusion occupied a large proportion of the cellular diffusion of mGFP in [psi−] cells and was well characterized by their hydrodynamic size as demonstrated in a previous study (Fig. 1B gray) (31). Diffusions of Sup35NM-GFP in [psi−] and Sup35NGMC in [gpsi−] cells also consisted of a large proportion of fast diffusion ranging from 85 to 90%. Although the Dfast values of the two proteins were slightly smaller than those expected from the D values in the lysis solution and cellular viscosity (Fig. 1B black and green, Table 1), it is suggested that the fast diffusions present monomeric states of each protein, respectively. Slow diffusion of mGFP and yeast prion proteins equally occupied a small proportion ranging from 10 to 15%, and the Dslow values were not different from each other. Slow diffusion detected in non-prion cells could have originated from a non-specific cellular interaction or from a very slow photobleaching of GFP (35).
The diffusion of Sup35NM-GFP and Sup35NGMC in [PSI+] and [GPSI+] cells were much slower than those of the Sup35 molecules in the non-prion cells, indicating that large oligomers were detected and the diffusion was mainly represented by a large proportion of the mobile oligomers (Fig. 1B blue and red). The diffusion profiles (i.e. FAFs) of Sup35NM-GFP were different from those of Sup35NGMC. Distribution of Dfast and Dslow values obtained from the [PSI+] cells were not much different from those of the [GPSI+] cells (Fig. 1C, D). Instead, the distribution of the fractional ratio of Dfast and Dslow was largely different between the [PSI+] and the [GPSI+] cells. The large difference could be explained by the abundance of Sup35 proteins, since Sup35NM-GFP was overexpressed, whereas Sup35NGMC was endogenously expressed. In practice, it was found that the concentration of overexpressed Sup35NM-GFP was 2.4-fold higher than that of the endogenous Sup35NGMC for non-prion cells (Table S2). Therefore, the number of Sup35NM-GFP aggregates in a cell would be relatively higher than that of the Sup35NGMC aggregates. Alternatively, the difference of expression could reflect the tendency to form foci: Sup35NMGFP might be more prone to form amyloids than Sup35NGMC, resulting in an accumulation of Sup35NM-GFP oligomers and formation of foci with the fibrillar structure (31–33). The result demonstrates that the hydrodynamic property (i.e. size) of the Sup35 aggregates (the mobile oligomers) in the prion cells are almost the same regardless of the expression level of Sup35-GFP. Rnq1 tagged with mGFP (Rnq1-GFP) was also characterized using FCS (Fig. S1, Table 1) and the diffusion profile were comparable to those of Sup35NM-GFP or Sup35NGMC (Table 1). The results suggest that the biochemical balance between the elongation and fragmentation of amyloids might be common among yeast prions.
The researchers also expressed Hsp104-mCherry (Hsp104-GFP) or Sis1-mCherry, which are known as prion-remodeling factors in non-prion cells. FCS analyses showed that around half of the Hsp104-GFP and the Sis1-mCherry molecules diffused very slowly, even in non-prion cells (Fig. S2, Table 1). The Dslow values for Hsp104-GFP and Sis1-mCherry were 0.34 and 0.6 μm2/s, respectively. The values were 30-fold slower than the diffusion coefficient of those proteins in the lysates (Table 1). The slow diffusions of Hsp104 and Sis1 in the non-prion cells may result from a strong interaction with an immobile cellular compartment, such as the cytoskeleton. Consequently, the FCS analyses in this study demonstrated that a large proportion of mobile and small aggregates or oligomers in yeast prion cells exists in the other region of cells surrounding an immobile large aggregate. Moreover, the hydrodynamic property of the oligomers in live cells can be fully differentiated from monomer molecules.
FCCS analysis of the interactions among prions in live cells
Yeast genetic experiments have suggested that there are interactions among prions as well as between prions, and remodeling factors such as Hsp104. In fact, fluorescence microscopic studies based on confocal imaging using GFP and RFP confirmed colocalizations of prion proteins and remodeling factors, such as Sup35-Rnq1 or Sup35-Hsp104. It was suggested that transmission or propagation of the prion state is accomplished by the dynamic property of prion oligomers, such as diffusion and rapid transmission of small oligomers from the mother cell to the daughter cell (31). Nevertheless, the interactions were only observed in highly bright and very large foci. The details of the physical interactions between them in living cells are still unclear. Interactions between highly mobile monomeric and oligomeric proteins in live cells are hardly traceable by conventional imaging methods such as confocal microscopy. Therefore, the researchers applied the CLSM-based FCCS technique to investigate physical interactions among highly mobile prions and remodeling factors.
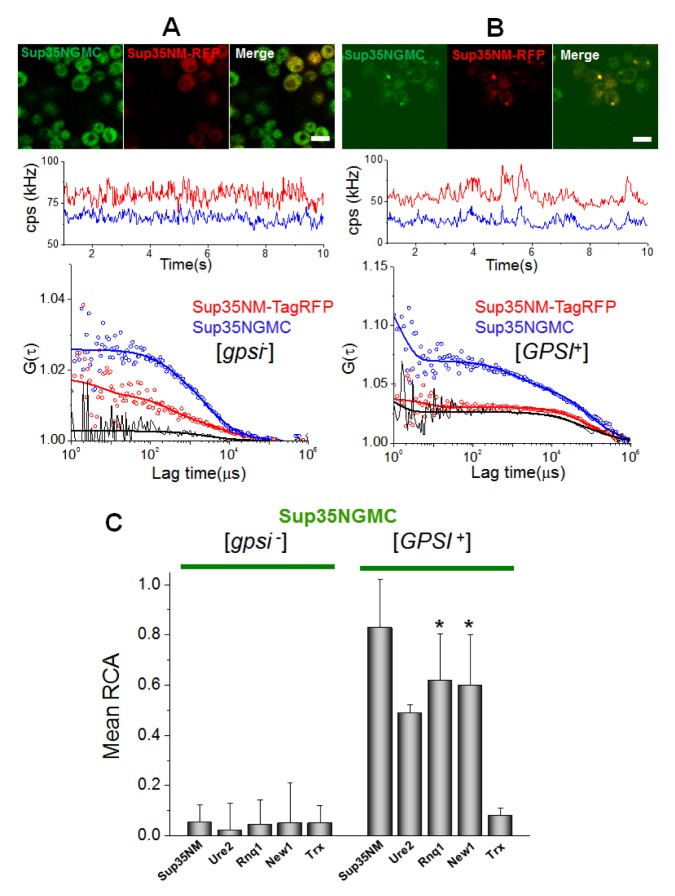
Firstly, the researchers examined interactions among prions, using Sup35NGMC with overexpressed RFP (mCherry or TagRFP), TagRFP tagged Sup35NM, Ure2N, Rnq1C, and New1N prion domains in the [gpsi−] or the [GPSI+] cells (Fig. 2 and Fig. S3). Fig. 2A and 2B show representative confocal images of cells co-expressing TagRFP- and GFP-tagged proteins, time traces of average fluorescence intensities, and corresponding correlation functions. The correlation function, G(τ), contains two auto-correlation functions (blue and red), which represent the behaviors of individual proteins, and one cross-correlation function (black), which represents the codiffusion of two fluorescent proteins (i.e. physical interaction, see also Methods section). An FCCS analysis between Sup35NGMC and TagRFP-tagged Sup35NM in the [gpsi−] cells showed only background levels of relative cross-correlation amplitude (Fig. 2A). In contrast, a high positive RCA value was detected in the analysis of Sup35NGMC and Sup35NM -TagRFP in the [GPSI+] cells (Fig. 2B), indicating that Sup35 molecules strongly interact with each other and form stable oligomers in prion cells. In addition, cross-correlation signals between Sup35-GFP and other prion proteins tagged with TagRFP in the [gpsi−] cells showed RCA values similar with background levels, whereas those in the [GPSI+] cells showed all high RCA values (Fig. S3A–D). This result indicates that Sup35 and other prion proteins strongly interact with each other (Fig. 2C).
Fig. 2.
Stable interaction among prion proteins in live cells. (A) Representative confocal image of [gpsi−] cells co-expressing Sup35NM-TagRFP and Sup35NGMC and (B) image of [GPSI+] cells co-expressing Sup35NM-TagRFP and Sup35NGMC are shown (upper). Scale bar, 5 μm. Representative FCCS measurement carried on cells are respectively shown (central and bottom). (Central) Time trace of average fluorescence intensity (counts per second; cps in kHz) of the two proteins. (Bottom) Two corresponding fluorescence auto-correlation functions (FAFs) of the TagRFP signal (red) and GFP signal (blue), and one fluorescence cross-correlation function (FCF) are shown. Fit curves (solid line) from two-component analysis are also shown. (C) The mean values of relative cross-correlation amplitudes (RCA), representing interactions between Sup35NGMC with other prion proteins of Sup35NM-TagRFP, Ure2-TagRFP, Rnq-TagRFP, and New1-TagRFP, Trx-TagRFP in [gpsi−] and [GPSI+] cells, are shown. Thioredoxin-fused TagRFP (Trx-TagRFP), a non-prion related protein, expressed in [gpsi−] and [GPSI+] was used as negative control. Error bars represent the s.d. *P < 0.05 compared with that of Sup35NM-TagRFP in [GPSI+] cells.
FCCS analysis of the interactions between prions and remodeling factors in live cells
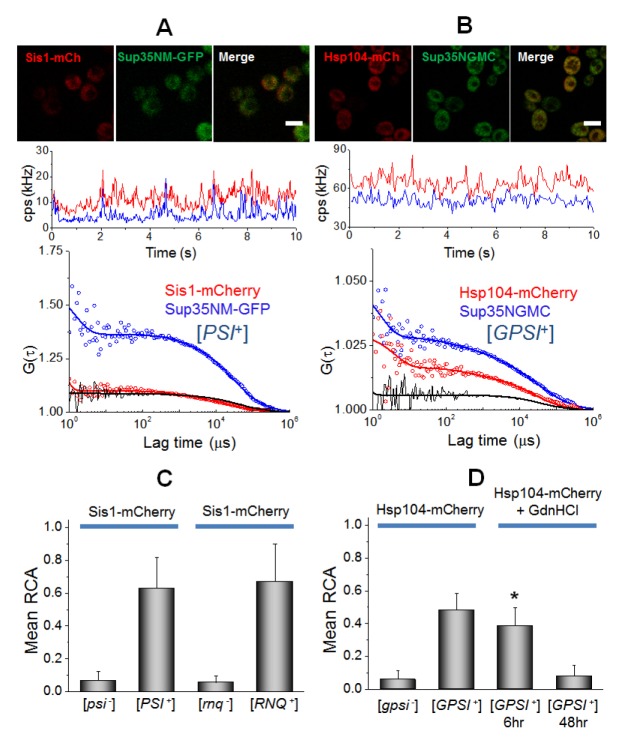
Secondly, the interactions of Sup35 with two remodeling factors, Hsp104 and Sis1, were examined (Fig. 3). The FCCS analysis revealed a strong interaction between Sup35NM-GFP and Sis1-mCherry as well as Hsp104-mCherry in prion cells (black curves in Fig. 3A, B). In the [psi−] ([gpsi−]) non-prion cells, small values of the mean RCA were observed (Fig. 3C, D and Fig. S3E, F). Moreover, FCCS showed a strong interaction between Rnq1-GFP and Sis1-mCherry in the [RNQ+] cells, while no significant RCA value was detected in the [rnq−] cells (Fig. 3C). This result demonstrates that the interaction of Sup35 with remodeling factors depends on the prion state in cells. Histograms of RCA in Sis1-Sup35 or Sis1-Rnq1 analyses showed that the overall RCA values in Sis1-Rnq1 were higher than those in Sis1-Sup35 (Fig. S4), even though the mean values of RCA were like each other. This suggests that the remodeling factor, Sis1, binds to Rnq1 in a more stable manner than Sup35. The FCCS also revealed a significant interaction between Sup35 and Hsp104 (Fig. 3D). The mean values of RCA for the interaction between Sis1 and Sup35 (or Rnq1) were larger than those between Hsp104 and Sup35 (or Rnq1) (Fig. 3C, 3D). This finding is consistent with previous studies, in which Sis1 are preferentially bound to either Sup35 or Rnq1 aggregates (20, 25). Since GdnHCl is known to cure [PSI+] due to the perturbation of Hsp104 as shown by FCS analyses of single cells (33), the researchers measured FCCS for the interaction between Sup35-Hsp104 in the GdnHCl-treated [GPSI+] cells. Strikingly, the mean values of RCA were gradually reduced with incubation time after GdnHCl treatment (Fig. 3D). The fractional ratio of Dslow was also reduced from 53% into 15% after 48 hr. The result indicates that the reduction of RCA values is correlated with the decrement of fractional ratio of the Dslow component (33). The result supports that GdnHCl treatment could inactivate Hsp104 through the inhibition of the interaction between Hsp104 and Sup35 oligomers, eventually leading to the curing of [PSI+] (22, 33).
Fig. 3.
Quantification of interaction between Sup35 oligomers and remodeling factors, Sis1 and Hsp104 in live cells. (A) Confocal image of [PSI+] cells co-expressing Sis1-mCherry and Sup35NM-GFP and (B) image of [GPSI+] cells co-expressing Hsp104-mCherry and Sup35NGMC are shown (upper). Scale bar, 5 μm. Representative FCCS measurement carried on the cells are shown (central and bottom). (C) The mean values of relative cross-correlation amplitudes (RCA) between Sis1-mCherry and Sup35NM-GFP in [psi−] and [PSI+] cells and between Sis1-mCherry and Rnq1-GFP in [rnq−] and [RNQ+] cells are shown. (D) The mean RCA values between Hsp104-mCherry and Sup35-GFP in [gpsi−] and [GPSI+], and GdnHCl treated [GPSI+] cells during 6 hr and 48 hr are shown. Error bars represent the s.d. *P < 0.05 compared with that in non-treated [GPSI+] cells.
DISCUSSION
Previous genetic experiments showed that there are interactions between prions (10). However, studies based on genetics could not directly answer whether those interactions are due to the physical prion-prion interactions or an indirect consequence via trans-acting factors like remodeling ones. Although colocalization analyses of fluorescent foci of prion proteins in cells has already suggested a direct association of a prion with other types (27, 28), such confocal imaging-based analysis cannot examine highly mobile and small prion oligomers. It is emphasized that the FCCS analysis clearly demonstrated that strong physical interactions among prions existed in cells under the condition where those prions were freely diffuse as oligomers in the cytoplasm.
The researchers note that prion-prion interactions were only detected in prion cells, but not in non-prion cells. The result demonstrates that prion interactions are dependent on the amyloid states. It also suggests that prion-induced de novo formation of other prion is due to a cross-seeding mechanism (8), by which pre-existing amyloids were used as seeds of other prion proteins. Moreover, a comparison of the RCA values in the FCCS analysis showed that Sis1-prion interactions are stronger than Hsp104-prion interactions. This result is consistent with a previous biochemical observation that the major components of cellular Sup35 aggregates were Sup35 and Sis1 (25). Consequently, this study demonstrats that the formation of the heterocomplex of prion oligomers may be a common event in prion cells, even though only a part of the known prion proteins were examined in this study.
MATERIALS AND METHODS
Detailed information is provided in the Supplementary Material.
Supplementary Information
ACKNOWLEDGEMENTS
We would like to thank Roger Tsien (UCSD) for the mCherry gene. This work was supported by National Institutes of Health Grant GM53655 and the USDA Cooperative State Research, Education and Extension Service (CSREES) project WISO4769 (to E.A.C.); Human Frontier Science Program Long-Term Fellowship (T.H.).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 3.Tuite MF, Cox BS. Propagation of yeast prions. Nat Rev Mol Cell Biol. 2003;4:878–890. doi: 10.1038/nrm1247. [DOI] [PubMed] [Google Scholar]
- 4.Tuite MF, Serio TR. The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat Rev Mol Cell Biol. 2010;11:823–833. doi: 10.1038/nrm3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wickner RB, Edskes HK, Shewmaker F, et al. Yeast prions: evolution of the prion concept. Prion. 2007;1:94–100. doi: 10.4161/pri.1.2.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inoue Y. Life cycle of yeast prions: propagation mediated by amyloid fibrils. Protein Pept Lett. 2009;16:271–276. doi: 10.2174/092986609787601796. [DOI] [PubMed] [Google Scholar]
- 7.Taguchi H, Kawai-Noma S. Amyloid oligomers: diffuse oligomer-based transmission of yeast prions. FEBS J. 2010;277:1359–1368. doi: 10.1111/j.1742-4658.2010.07569.x. [DOI] [PubMed] [Google Scholar]
- 8.Liebman SW, Chernoff YO. Prions in yeast. Genetics. 2012;191:1041–1072. doi: 10.1534/genetics.111.137760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sondheimer N, Lindquist S. Rnq1: an epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/S1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 10.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: the story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/S0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 11.Osherovich LZ, Weissman JS. Multiple Gln/Asn-rich prion domains confer susceptibility to induction of the yeast [PSI(+)] prion. Cell. 2001;106:183–194. doi: 10.1016/S0092-8674(01)00440-8. [DOI] [PubMed] [Google Scholar]
- 12.Du Z, Park KW, Yu H, Fan Q, Li L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. doi: 10.1038/ng.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suzuki G, Shimazu N, Tanaka M. A yeast prion, Mod5, promotes acquired drug resistance and cell survival under environmental stress. Science. 2012;336:355–359. doi: 10.1126/science.1219491. [DOI] [PubMed] [Google Scholar]
- 15.Bradley ME, Edskes HK, Hong JY, Wickner RB, Liebman SW. Interactions among prions and prion “strains” in yeast. Proc Natl Acad Sci U S A. 2002;99(Suppl 4):16392–16399. doi: 10.1073/pnas.152330699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bosl B, Grimminger V, Walter S. The molecular chaperone Hsp104--a molecular machine for protein disaggregation. J Struct Biol. 2006;156:139–148. doi: 10.1016/j.jsb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 17.Doyle SM, Wickner S. Hsp104 and ClpB: protein disaggregating machines. Trends Biochem Sci. 2009;34:40–48. doi: 10.1016/j.tibs.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 18.Masison DC, Kirkland PA, Sharma D. Influence of Hsp70s and their regulators on yeast prion propagation. Prion. 2009;3:65–73. doi: 10.4161/pri.3.2.9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aron R, Higurashi T, Sahi C, Craig EA. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007;26:3794–3803. doi: 10.1038/sj.emboj.7601811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higurashi T, Hines JK, Sahi C, Aron R, Craig EA. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc Natl Acad Sci U S A. 2008;105:16596–16601. doi: 10.1073/pnas.0808934105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. EMBO J. 2008;27:2712–2724. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romanova NV, Chernoff YO. Hsp104 and prion propagation. Protein Pept Lett. 2009;16:598–605. doi: 10.2174/092986609788490078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler J, Tyedmers J, Bukau B, Mogk A. Chaperone networks in protein disaggregation and prion propagation. J Struct Biol. 2012;179:152–160. doi: 10.1016/j.jsb.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 24.Allen KD, Wegrzyn RD, Chernova TA, et al. Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+] Genetics. 2005;169:1227–1242. doi: 10.1534/genetics.104.037168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagriantsev SN, Gracheva EO, Richmond JE, Liebman SW. Variant-specific [PSI+] infection is transmitted by Sup35 polymers within [PSI+] aggregates with heterogeneous protein composition. Mol Biol Cell. 2008;19:2433–2443. doi: 10.1091/mbc.E08-01-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salnikova AB, Kryndushkin DS, Smirnov VN, Kushnirov VV, Ter-Avanesyan MD. Nonsense suppression in yeast cells overproducing Sup35 (eRF3) is caused by its non-heritable amyloids. J Biol Chem. 2005;280:8808–8812. doi: 10.1074/jbc.M410150200. [DOI] [PubMed] [Google Scholar]
- 27.Derkatch IL, Uptain SM, Outeiro TF, Krishnan R, Lindquist SL, Liebman SW. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A. 2004;101:12934–12939. doi: 10.1073/pnas.0404968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454:1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Winkler J, Tyedmers J, Bukau B, Mogk A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J Cell Biol. 2012;198:387–404. doi: 10.1083/jcb.201201074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saibil HR, Seybert A, Habermann A, et al. Heritable yeast prions have a highly organized three-dimensional architecture with interfiber structures. Proc Natl Acad Sci U S A. 2012;109:14906–14911. doi: 10.1073/pnas.1211976109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawai-Noma S, Ayano S, Pack CG, et al. Dynamics of yeast prion aggregates in single living cells. Genes Cells. 2006;11:1085–1096. doi: 10.1111/j.1365-2443.2006.01004.x. [DOI] [PubMed] [Google Scholar]
- 32.Kawai-Noma S, Pack CG, Kojidani T, et al. In vivo evidence for the fibrillar structures of Sup35 prions in yeast cells. J Cell Biol. 2010;190:223–231. doi: 10.1083/jcb.201002149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kawai-Noma S, Pack CG, Tsuji T, Kinjo M, Taguchi H. Single mother-daughter pair analysis to clarify the diffusion properties of yeast prion Sup35 in guanidine-HCl-treated [PSI] cells. Genes Cells. 2009;14:1045–1054. doi: 10.1111/j.1365-2443.2009.01333.x. [DOI] [PubMed] [Google Scholar]
- 34.Greene LE, Park YN, Masison DC, Eisenberg E. Application of GFP-labeling to study prions in yeast. Protein Pept Lett. 2009;16:635–641. doi: 10.2174/092986609788490221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pack CG, Yukii H, Toh-e A, et al. Quantitative live-cell imaging reveals spatio-temporal dynamics and cytoplasmic assembly of the 26S proteasome. Nat Commun. 2014;5:3396. doi: 10.1038/ncomms4396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.