Abstract
The genomic era has revolutionized research on secondary metabolites and bioinformatics methods have in recent years revived the antibiotic discovery process after decades with only few new active molecules being identified. New computational tools are driven by genomics and metabolomics analysis, and enables rapid identification of novel secondary metabolites. To translate this increased discovery rate into industrial exploitation, it is necessary to integrate secondary metabolite pathways in the metabolic engineering process. In this review, we will describe the novel advances in discovery of secondary metabolites produced by filamentous fungi, highlight the utilization of genome-scale metabolic models (GEMs) in the design of fungal cell factories for the production of secondary metabolites and review strategies for optimizing secondary metabolite production through the construction of high yielding platform cell factories.
Keywords: Secondary metabolism, Fungi, Biosynthetic gene clusters, Genome mining, Metabolic modeling, Cell factories
1. Introduction
Microbial secondary metabolites are widely exploited for their biological activities to ensure the well-being of humans. Secondary metabolites are used as antibiotics, other medicinals, toxins, pesticides, and animal and plant growth factors [1]. Although the antibiotic effects of certain molds have been reported earlier, it was Flemings' persistence in the usability of the antimicrobial activity of penicillin, which initiated what is known as the golden era of antibiotic discovery [2]. Despite the fungal origin of penicillin, produced by several members of the Penicillium genus [3], most research on secondary metabolites has focused on bacteria, mainly soil isolates of actinomycetes with the majority of compounds originating from the Streptomyces genus [4]. Some of the pioneering work that paved the way for antibiotic discovery was conducted by Nobel laureate Selman Waksman, who's systematic screening of Streptomyces isolates, led to the identification of several antibiotics, including streptomycin and neomycin which have found extensive applications in the treatment of infectious diseases. However, to ensure translation of these findings for commercial production it was necessary with further product optimization and fermentation characterization of microbial physiology, and this resulted in the birth of industrial microbiology as a discipline, with Arnold Demain as one of the founding fathers.
Today we know that although most living organisms can produce secondary metabolites, the ability to produce them is unevenly distributed. Among all known microbial antibiotics and similar bioactive compounds (altogether 22,500), 45% are from actinomycetes, 38% are from fungi and 17% are from unicellular bacteria [4]. Among this wealth of compounds, only about a hundred are in practical use for human therapy, with the majority being derived from actinomycetes [4]. However, it is worth mentioning that in addition to penicillin, several other fungal secondary metabolites have successfully reached the pharmaceutical market, including cholesterol lowering statins [5], the antifungal griseofulvin [6] and the immunosuppressant mycophenolic acid [7].
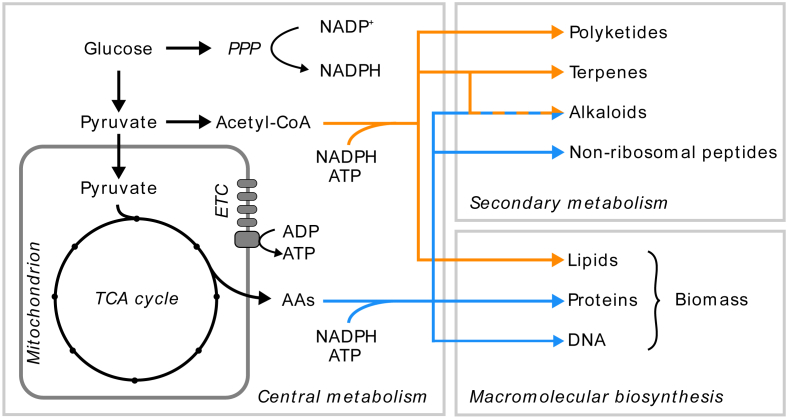
Biosynthesis of secondary metabolites takes place from a limited number of precursor metabolites from the primary metabolism (Fig. 1). In fungi, these precursors are mainly short chain carboxylic acids (e.g. acetyl-CoA) or amino acids, which are linked together by backbone enzymes such as polyketide synthases (PKSs), non-ribosomal peptide synthetases (NRPSs), dimethylallyl tryptophan synthetases (DMATSs) or terpene cyclases (TCs). The resulting oligomers are then subject to chemical modification by tailoring enzymes which are often controlled under common transcriptional regulation as the backbone enzyme [8]. A hallmark trait of the genes involved in a secondary metabolite pathway is that they, in most record cases, physically cluster in the chromosome in biosynthetic gene clusters (BGCs) [9].
Fig. 1.
Biosynthesis of secondary metabolites from precursors of the central carbon metabolism. PPP: Pentose Phosphate Pathway. ETC: Electron Transport Chain. TCA: Tricarboxylic Acid. AAs: Amino Acids.
The characteristic clustering of genes as well as the conserved motifs of backbone genes can be exploited for computational detection of BGCs from sequence data. Tools like SMURF [10], antiSMASH [11], PRISM [12] and SMIPS/CASSIS [13] utilize these features to reliably and with a high accuracy detect BGCs of known compound classes in fungi. Other algorithms detects BGCs without relying on specific motifs or the presence of backbone genes, which enables identification of BGCs beyond PKS, NRPS, DMATS and TCs [14], [15], [16], [17]. Tools and implementations of BGC mining algorithms have been extensively reviewed [18], [19], [20], [21], [22], [23].
A limitation of secondary metabolite production is the low yields that are naturally achieved in most microbes, partly since many secondary metabolites are favored under suboptimal growth conditions [8], [24] and because their biosynthesis compete with essential pathways of metabolism, involved in growth related processes (Fig. 1). Applying metabolic engineering to circumvent these limitations can be greatly assisted by utilization of the mathematical representation of metabolism in genome-scale metabolic models (GEMs), which concepts and applications have been reviewed elswhere [25], [26], [27]. These models, however, often neglect secondary metabolite biosynthesis, hence their potential in studying secondary metabolism has not been fully tapped. Additionally, with the efficient gene editing tool CRISPR-Cas9 being developed for a number of fungal model organisms [28], [29], [30], a great potential exists for implementing the necessary genetic modifications for the development of improved secondary metabolite producers. In this review, we will describe methods for linking BGCs to compounds and show how metabolic modeling can aid in translating the improved secondary metabolite discovery rate into metabolic engineering strategies for the development of fungal platform strains for the production of secondary metabolites.
2. Linking BGCs to compounds
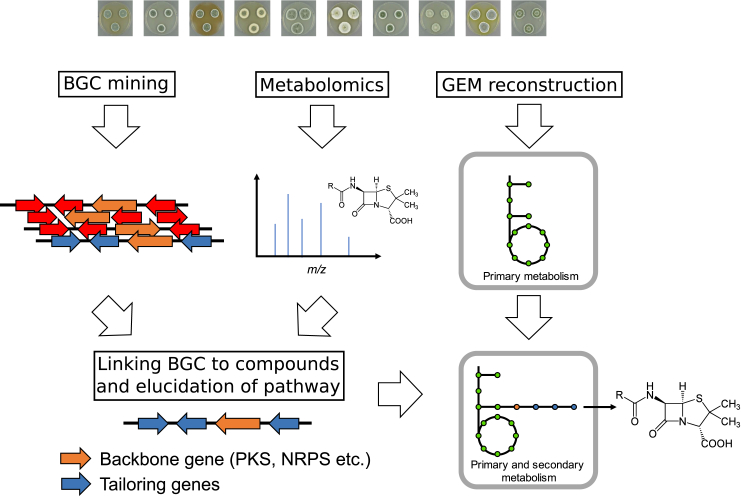
In order to industrially exploit secondary metabolites for production, it is a major advantage to know the genetic basis of the biosynthesis. This allows for employing metabolic engineering strategies for optimizing the production performance of an organism and making the process economically feasible [31]. Among the known secondary metabolites, the vast majority have not had their biosynthetic mechanisms elucidated or linked to a BGC, and are commonly referred to as orphan compounds. Understanding the genetic foundation of secondary metabolite biosynthesis further allows for redesigning the pathways to produce novel compounds [32], as previously shown by widening the product portfolio of β-lactam antibiotics from the penicillin pathway of Penicillium chrysogenum [33]. Genome sequencing combined with genome mining, strongly facilitates the process of connecting BGCs to compounds (Fig. 2) and a number of computational tools have been developed to specifically address this challenge either from a targeted or untargeted approach, or by using metabolomics.
Fig. 2.
Work flow for the integration of secondary metabolite pathways in genome-scale metabolic models (GEMs) based on genomics and metabolomics data. In the top layer, the genome sequence is being mined for the identification of biosynthetic gene clusters (BGCs), metabolomics analysis of culture extract is used for identification of produced secondary metabolites, while GEMs can be reconstructed from an annotated genome. In the second layer, detected BGCs are connected to detected compounds using e.g. by mass spectrometry data. This allows for experimental characterization of the pathways, which then can be implemented in the GEMs and analyzed for improved production performance.
2.1. Targeted approaches
A simple approach, for identifying the BGC of a target compound is to compare the number of similar BGCs between two or more species producing the compound, to narrow down the number of candidate BGCs, which could be responsible for the biosynthesis. Combining this with retro-biosynthetic analysis, which aims at deducing which enzymes and precursors that are likely responsible for the biosynthesis of a given compound, has proved effective in the identification of the genomic loci responsible for production of several secondary metabolites in fungi [34], [35], [36]. Similarly, for an orphan compound, a high similarity to another compound which has been connected to a BGC, can be used for homology search of a similar BGC in the target genome [37].
In some cases, BGCs contain a resistance gene encoding a variant of the enzyme targeted by the pathway product, which is not susceptible to inhibition [38], [39], [40]. This feature was utilized to identify the BGC responsible for mycophenolic acid production in P. brevicompactum, by searching for a resistance gene of the mycophenolic acid target, IMP dehydrogenase [41]. Later the pathway product of the inp BGC in Aspergillus nidulans was predicted to be a proteasome inhibitor, based on the presence of a gene encoding a proteasome subunit in the BGC. The inp BGC was previously shown to be silent [42], but targeted promoter exchange of gene cluster members enabled the expression and isolation of the proteasome inhibitor fellutamide B [43], and these results implied that resistance-gene-guided genome mining can be broadly applied in fungi, as previously demonstrated in bacteria [44].
2.2. Untargeted approaches
Untargeted approaches can be used to assess the entire biosynthetic potential in one or more genomes, by correlating all detected BGCs to databases which links BGCs and compounds. Databases containing fungal BGCs include clustermine360 [45], (297 BGCs), IMG-ABC [46] (2489 BGCs) and MIBiG [47] (1393 BGCs). Recent efforts to increase the number of fungal BGCs in the MIBiG database used text mining to add an additional 197 fungal BGCs to the database [48]. However, reflecting the literature, the number of fungal BGCs in the databases comprises only a fraction of the total number of BGCs, which are mainly of bacterial origin. Assessing the similarity between BGCs and grouping them into gene cluster families e.g. with the scope of mapping newly sequenced BGCs to database entries is not straight forward due to the large size of BGCs, inaccurate definition of boundaries, re-arrangements, and potential presence of non-relevant genes. Some approaches for grouping BGCs have used conserved motifs such as KS and C domain similarity of PKSs and NRPSs [49], number of shared PFAM domains between BGCs [16] or a combination of three different similarity metrics [50]. None of these algorithms, however, were originally developed for comparing BGCs of fungal origin.
Employing mining of BGCs to study the shared and unique features between species, has only been exploited to a limited extent in fungi [15], [51], [52], [53], [54]. These studies however, have mainly concerned few species. In contrast, a number of studies have concerned the comparison of BGCs between hundreds of bacterial species [16], [50], [55], [56], [57], which have led to a characterization of the diversity of BGCs in prokaryotes. Future work should compare secondary metabolism at genus or phylum level in fungi in order to identify global features of secondary metabolism as well as facilitate the discovery of novel compounds.
2.3. Metabolomics approaches
Metabolomics can be utilized to connect mass spectrometry (MS) detected compounds to their corresponding BGCs in a sequenced genome. This approach was first developed using peptidogenomics [58], where tandem MS was used to capture an amino acid sequence tag, from the fragmentation of a given peptide natural product. The sequence tag represents part of a complete peptide, and can be deduced based on the mass shift pattern, and subsequently screened against predicted substrate specificities of NRPSs, obtained from tools such as antiSMASH [59] and NP.searcher [60]. Later Pep2Path [61] was developed to automatize the detection of BGCs responsible for the amino acid sequence tags based on a Bayesian probabilistic scoring algorithm. MS-guided discovery of secondary metabolites has been further extended to glycosylated compounds [62], as well as specific tools for non-ribosomal peptides (NRPs) [63] and ribosomally synthesized and posttranslationally modified peptides (RiPPs) [64].
Recently a pipeline for directly connecting BGCs to a database of known secondary metabolites was published [65]. The pipeline combines three different tools; PRISM [12] for BGC mining and prediction of substrates of PKSs, NRPSs and PKS-NRPSs; GRAPE [65] which automates the process of retro-biosynthesis of polyketides (PKs), NRPs and their hybrids; and GARLIC [65] which compares the substrate predicted by PRISM with the building blocks predicted from the retro-biosynthesis by GRAPE, and hence can assess whether the activity of a backbone enzyme could be responsible for the synthesis of a given compound. The authors tested the pipeline by identifying 16,831 PKS, NRPS and PKS-NRPS BGCs from public data using PRISM, which they compared against a database of 48,222 compounds. Based on known BGC metabolite relationships in the databases, a cut-off was determined which enabled the estimation that 15% of the BGCs had no corresponding product in the compound database. For validation, a BGC from Nocardiopsis potens, without a match in the compound database, was targeted and identified through metabolite profiling. The produced compound was structure elucidated by NMR and indeed proved to be a novel secondary metabolite [65].
3. Genome-scale metabolic modeling of secondary metabolism
With the increasing number of fungal genomes being sequenced [66] and mining strategies for BGC identification being widely accessible [20], the number of characterized biosynthetic pathways and newly discovered antibiotics will likely increase rapidly in the future. To be able to optimize the production of these new compounds, GEMs are useful tools which can aid in the design of metabolic engineering strategies from a global view of metabolism (Fig. 2). The foundation of a GEM is the functional annotation of the genes, and connecting these to the biochemical reactions catalyzed by the corresponding enzymes, provides a comprehensive summary of the metabolic capabilities of an organism [67], [68]. Applications of GEMs are manifold, but commonly include topological network analysis and integration of omics data, or prediction of phenotypic traits through simulations of metabolism e.g. with the goal of designing metabolic engineering strategies [69].
The use of GEMs to predict phenotypic characters of microbes has been successfully demonstrated a number of times [70], [71], [72], [73], and these models serves as a core element of the systems biology toolbox. Despite their widespread usage, only a limited number of studies have applied GEMs for investigating the dynamics of secondary metabolite production in fungi, while more work has focused on prokaryotic secondary metabolite producers. In recent years, secondary metabolism has been studied in GEMs of several actinomycetes [74], including Streptomcyes coelicolor [75], [76], [77], Saccharopolyspora erythraea [78], Streptomyces lividans [79] and Streptomyces tsukubaensis [80], and the first analysis of secondary metabolism in a GEM was conducted with the metabolic network of the antibiotic producer S. coelicolor A3(2) [75]. This S. coelicolor GEM, included two full pathways of secondary metabolites, the PK antibiotic actinorhodin and the NRP, calcium-dependent antibiotic, for which precursor supply was simulated [75]. Later, the network topology of an A. nidulans GEM was utilized to calculate the metabolic fluxes based on 13C labeled glucose upon overexpression of xylulose-5-phosphate phosphoketolases (XPKs) [81]. The analysis suggested that induction of XPKs increase the carbon flux towards acetyl-CoA, the precursor for PK biosynthesis. In a follow-up study, the overexpression of XPKs was combined with the heterologous expression of the PKS 6-methylsalicylic acid (6-MSA) synthase, to investigate the effects on 6-MSA yields. Transcriptome analysis combined with flux and physiological data allowed the proposal of an interaction model describing how the competition between biomass and 6-MSA from the tightly regulated acetyl-CoA node could explain why increased 6-MSA yields were not observed [82]. Exactly the tight regulation and high connectivity of acetyl-CoA in fungal metabolism [83] is likely an important factor why achieving high yields of PKs has proven challenging. Moreover, since secondary metabolite production is highly regulated at multiple different levels, i.e. transcriptional and through epigenetics [8], it is difficult to simulate this part of metabolism using GEMs which does not take these levels of regulations into account.
Production of penicillin by the filamentous fungus P. chrysogenum, is one of the most successful stories of biotechnology, where Classical Strain Improvement (CSI) has been used to increase product titers and productivity by at least three orders of magnitude during 60 years of strain development [84]. Agren et al. (2013) [68] reconstructed a GEM of the CSI developed penicillin over-producing strain, P. chrysogenum Wisconsin54-1255, and used flux balance analysis combined with transcriptome analysis to study metabolic bottlenecks and the influence of co-factor availability on yields of penicillin. Although no experimental validation was performed, the authors suggested increasing NADPH availability as well as modifying different precursor supplying pathways as potential metabolic engineering strategies for increasing the penicillin production [68]. More recently Prauβe et al. (2015) applied elementary flux mode (EFM) analysis on the production of penicillin in a metabolic core model, derived from the same P. chrysogenum GEM. EFM analysis allows for the decomposition of the metabolic network into functional units and represent each a minimal set of reactions that can function at a steady state [85]. A total of 66 EFMs were identified in the network with the glyoxylate shunt being redundant in the highest yielding EFMs, hence it was proposed that disrupting this pathway could result in higher yields of penicillin [86].
An important difference between fungi and bacteria is that bacteria tend to reach higher product yields, which has been speculated to be partly because of the increased complexity, due to the compartmentalization, of metabolism in fungi [87]. In a case study on the production of higher alcohols, Matsuda et al. [88] conducted model simulations of the central metabolism of Escherichia coli and Saccharomyces cerevisiae. The results suggested that a superior production performance of E. coli could be attributed to a higher degree of metabolic flexibility compared with S. cerevisiae, as indicated by the variety of flux distributions taken by the metabolic networks. The production capability in S. cerevisiae was improved in silico, by introducing E. coli reactions in the yeast network [88]. Since secondary metabolite precursors revolve heavily around central metabolism and in particular acetyl-CoA, from which many higher alcohols are derived, a similar engineering strategy might also be used for the improvement of secondary metabolite production.
4. Development of platform cell factories
In many cases, native producers of secondary metabolites are not well suited as industrial cell factories, which depend on features like growth rate, morphology, substrate utilization, by-product formation and product formation. Hence, development of a dedicated plug-and-play platform cell factory for the heterologous production of secondary metabolites is an appealing thought from an industrial point of view. Heterologous expression of fungal secondary metabolite pathways has been successfully achieved in bacteria, yeast and filamentous fungi [18], and each host offer a different set of advantages and disadvantages. Independent of choice, a number of metabolic features influence the production levels of secondary metabolites and the development of a platform strain should consider these, which are described below.
A potential host for expression of fungal secondary metabolites is the yeast S. cerevisiae, which is well-characterized and genetically tractable [89]. In addition, it serves as a minimal fungal host due to its limited native secondary metabolism minimizing interference or competition from other secondary metabolite pathways. Exactly the competition within secondary metabolism has been indicated to be a key determinant on production levels of secondary metabolites. Salo et al. [90] compared secondary metabolite production in the penicillin over-producer P. chrysogenum DS17690, with a derived strain, DS68530, which lost its penicillin gene clusters. They observed that while the derived strain DS68530, had lost the ability to produce penicillin, the production of other NRPs like roquefortines/meleagrin and chrysogines were increased. The explanation was speculated to be caused by a re-direction of nitrogen metabolism toward other NRPs [90]. Similar observations have been reported in bacterial secondary metabolite producing Streptomyces species, where the knock-out of the main secondary metabolite producing BGCs resulted in increased titers of native and heterologous secondary metabolites [91], [92].
Precursor and co-factor availability are important limitations for the production of secondary metabolites. Acetyl-CoA is a key compound in secondary metabolism and serves as the precursor of PKs, often through the carboxylated form malonyl-CoA, as well as terpenes synthesized from isoprene units from the mevalonate pathway. Additionally, acetyl-CoA is a highly connected metabolite in the primary metabolism where it is involved in the biosynthesis of fatty acids and sterols, protein acetylation, energy generation and is compartmentalized in fungi [83], [87]. A number of studies have attempted to increase acetyl-CoA pools for the production of chemicals in yeast including fatty acids [93], butanol [94], sesquiterpenes [95] and PKs [96].
The model PK 6-MSA, is synthesized from one acetyl-CoA and three malonyl-CoA and have been heterologously produced in S. cerevisiae through a 6-MSA synthase from P. patulum and a PPTase [97]. In an attempt to improve 6-MSA production in such a strain by increasing precursor availability, acc1, which corresponding enzyme catalyzes the conversion of acetyl-CoA to malonyl-CoA, was overexpressed from a constitutive promoter and resulted in a 60% increase in 6-MSA titers [96]. Another study aimed at preventing the deactivation of Acc1 by AMP-activated serine/threonine protein kinase (Snf1) upon glucose depletion in a 6-MSA producing S. cerevisiae strain. The authors introduced an amino acid substitution in Acc1, preventing phosphorylation and hence deactivation, which resulted in a 2.8-fold increase in 6-MSA titers compared to the wild type Acc1 strain [98].
A more comprehensive evaluation of metabolic engineering targets to increase acetyl-CoA availability for PK production was conducted by Cardenas and Da Silva [99], in S. cerevisiae producing the plant PK triacetic acid lactone (TAL). Bypassing the native ATP-dependent conversion of pyruvate to acetyl-CoA, with a bacterial NADPH generating pyruvate dehydrogenase (PDHm), resulted in increased TAL titers. The authors further implemented a driving force for NADPH through acetyl-CoA generation, by eliminating NADPH formation via a zwf1 deletion in the pentose phosphate pathway. The resulting strain showed 4.8-fold increased TAL titers. To increase the cytosolic acetyl-CoA pool, a systematic deletion of reactions involved in transport of pyruvate and acetyl-CoA into the mitochondria, was used to identify four gene deletions (Δpor2Δmpc2Δpda1Δyat2) which when combined and introduced in the Δzwf1:PDHm strain, resulted in a 6.4 fold increase in TAL titers, corresponding to 35% of the theoretical yield [99]. Although the above described studies strongly revolve around engineering acetyl-CoA metabolism, the supply of other precursors, including amino acids and co-factors are equally important to consider [100].
Another method to improve secondary metabolite biosynthesis is promoter exchange to construct an inducible pathway. The native promoter acvA in A. nidulans, which express the rate limiting enzyme of the penicillin pathway, was exchanged by an inducible alcohol dehydrogenase 1 promoter and resulted in a 30-fold increase in penicillin yields [101]. More recently Chiang et al. [102] developed a system for the heterologous expression of entire BGCs under control of regulatable promoters, and demonstrated the use of this to express several A. terreus BGCs in A. nidulans [102].
Since many secondary metabolites are toxic to the host, resistance mechanisms are needed to cope with production. Native producers have often evolved specific transporters to secrete [103] or compartmentalize toxic compounds in vesicles [104] or confer self-protection by producing resistant copies of the target enzyme of the pathway product (as described above). In the case of heterologous production, resistance mechanisms need to be considered apart from expression of the biosynthetic genes. This was illustrated in the heterologous expression of a putative efflux pump, mlcE, from the compactin BGC in P. citrinum, which was shown to be a specific transporter, and increased the resistance of S. cerevisiae towards natural and semi-synthetic statins [105].
Combining the above strategies to engineer a secondary metabolite deficient fungal platform strain, exhibiting high precursor supply, for heterologous expression of inducible BGCs, which confers resistance to potential toxic compounds, could serve as a high yielding platform for future production of secondary metabolites. Moreover, such a strain would be useful in the study of lowly expressed or cryptic biosynthetic pathways.
5. Perspectives
Bioinformatics tools enable accurate identification of known and novel BGC classes, and can be utilized in combination with algorithms parsing metabolomics data for connecting BGCs to compounds. Despite the bias in data availability and computational tools towards bacteria, fungi constitute a rich reservoir of pharmaceutically relevant secondary metabolites. Therefore, it is important that future work focus on testing the applicability of developed tools on fungal data, and that the development of novel algorithms, consider the differences that exists between bacteria and fungi.
As a consequence of these bioinformatics tools, and the development of efficient genetic engineering in fungi such as CRISPR-Cas9 [28], it is expected that pathways will be elucidated at a higher pace in the years to come. To maximally profit from this advancement, GEMs will be important assets for better understanding secondary metabolite production and develop metabolic engineering strategies for optimization. Integration of omics data such as transcriptomics to identify which BGCs are being expressed under certain conditions or predict metabolic engineering targets [77], [106], can further aid in understanding how expression of BGCs associated with secondary metabolites of interest is controlled.
A major challenge ahead is, however, that the majority of BGCs are silent under standard laboratory conditions, and efficient procedures to activate these latent pathways is therefore important in order to obtain better description of the secondary metabolome of an organism [107], [108], [109]. This will in turn provide researchers with a greater knowledge base for the selection of computationally identified fungal BGCs which could be of interest for industrial exploitation. Here breakthroughs in synthetic biology, where it is now possible to synthesize whole BGCs in a tailored fashion, e.g. with controllable promoters in front of each of the genes, may address this challenge, as it will hereby be possible to transfer all BGCs identified through genome sequencing to a suitable production host. The benefits of optimizing the metabolism of such a production host, such that it is ensured that metabolism is engineered to efficiently produce all the required precursor metabolites and co-factors, will hereby become even larger and further accelerate advancement of the field. The yeast S. cerevisiae can be an optimal host as it does not produce secondary metabolites endogenously and therefore have few enzymes that may react with pathway intermediates. However, this host may be limited by activities for proper activation of many of the complex enzymes engaged with secondary metabolite production, and establishment of clean hosts where all endogenous BGCs have been removed may therefore be an attractive alternative.
Acknowledgements
This work was supported by the European Commission Marie Curie Initial Training Network Quantfung (FP7-People-2013-ITN, Grant 607332). We also acknowledge funding from the Novo Nordisk Foundation and the Knut and Alice Wallenberg Foundation.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Demain A.L. Small bugs, big business: the economic power of the microbe. Biotechnol Adv. 2000;18:499–514. doi: 10.1016/s0734-9750(00)00049-5. [DOI] [PubMed] [Google Scholar]
- 2.Aminov R.I. A brief history of the antibiotic era: lessons learned and challenges for the future. Front Microbiol. 2010;1:134. doi: 10.3389/fmicb.2010.00134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frisvad J.C., Smedsgaard J., Larsen T.O., Samson R.A. Mycotoxins, drugs and other extrolites produced by species in Penicillium subgenus Penicillium. Stud Mycol. 2004;49:201–241. [Google Scholar]
- 4.Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 5.Barrios-González J., Miranda R.U. Biotechnological production and applications of statins. Appl Microbiol Biotechnol. 2010;85:869–883. doi: 10.1007/s00253-009-2239-6. [DOI] [PubMed] [Google Scholar]
- 6.Finkelstein E., Amichai B., Grunwald M.H. Griseofulvin and its uses. Int J Antimicrob Agents. 1996;6:189–194. doi: 10.1016/0924-8579(95)00037-2. [DOI] [PubMed] [Google Scholar]
- 7.Stassen P.M., Kallenberg C.G.M., Stegeman C.A. Use of mycophenolic acid in non-transplant renal diseases. Nephrol Dial Transpl. 2007;22:1013–1019. doi: 10.1093/ndt/gfl844. [DOI] [PubMed] [Google Scholar]
- 8.Brakhage A.A. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 9.Keller N.P., Turner G., Bennett J.W. Fungal secondary metabolism - from biochemistry to genomics. Nat Rev Microbiol. 2005;3:937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 10.Khaldi N., Seifuddin F.T., Turner G., Haft D., Nierman W.C., Wolfe K.H. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weber T., Blin K., Duddela S., Krug D., Kim H.U., Bruccoleri R. antiSMASH 3.0–a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–W243. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skinnider M.A., Dejong C.A., Rees P.N., Johnston C.W., Li H., Webster A.L.H. Genomes to natural products PRediction informatics for secondary metabolomes (PRISM) Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wolf T., Shelest V., Nath N., Shelest E. CASSIS and SMIPS: promoter-based prediction of secondary metabolite gene clusters in eukaryotic genomes. Bioinformatics. 2016;32:1138–1143. doi: 10.1093/bioinformatics/btv713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Umemura M., Koike H., Nagano N., Ishii T., Kawano J., Yamane N. MIDDAS-M: motif-independent de novo detection of secondary metabolite gene clusters through the integration of genome sequencing and transcriptome data. PLoS One. 2013;8:e84028. doi: 10.1371/journal.pone.0084028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda I., Myco U., Koike H., Asai K., Machida M. Motif-independent prediction of a secondary metabolism gene cluster using comparative genomics: application to sequenced genomes of Aspergillus and ten other filamentous fungal species. DNA Res. 2014;21:447–457. doi: 10.1093/dnares/dsu010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cimermancic P., Medema M.H., Claesen J., Kurita K., Wieland Brown L.C., Mavrommatis K. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vesth T.C., Brandl J., Andersen M.R. FunGeneClusterS: predicting fungal gene clusters from genome and transcriptome data. Synth Syst Biotechnol. 2016;1 doi: 10.1016/j.synbio.2016.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiemann P., Keller N.P. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol. 2014;41:301–313. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 19.Weber T. In silico tools for the analysis of antibiotic biosynthetic pathways. Int J Med Microbiol. 2014;304:230–235. doi: 10.1016/j.ijmm.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Medema M.H., Fischbach M.A. Computational approaches to natural product discovery. Nat Chem Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Lee T.A.J., Medema M.H. Computational strategies for genome-based natural product discovery and engineering in fungi. Fungal Genet Biol. 2016;89:29–36. doi: 10.1016/j.fgb.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Ziemert N., Alanjary M., Weber T. The evolution of genome mining in microbes – a review. Nat Prod Rep. 2016;33:988–1005. doi: 10.1039/c6np00025h. [DOI] [PubMed] [Google Scholar]
- 23.Weber T., Kim H.U. The secondary metabolite bioinformatics portal: computational tools to facilitate synthetic biology of secondary metabolite production. Synth Syst Biotechnol. 2016;1:69–79. doi: 10.1016/j.synbio.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demain A.L. Regulation of secondary metabolism in fungi. Pure Appl Chem. 1986;58:219–226. [Google Scholar]
- 25.Zhang C., Hua Q. Applications of genome-scale metabolic models in biotechnology and systems medicine. Front Physiol. 2016;6:1–8. doi: 10.3389/fphys.2015.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu L., Agren R., Bordel S., Nielsen J. Use of genome-scale metabolic models for understanding microbial physiology. FEBS Lett. 2010;584:2556–2564. doi: 10.1016/j.febslet.2010.04.052. [DOI] [PubMed] [Google Scholar]
- 27.Blazeck J., Alper H. Systems metabolic engineering: genome-scale models and beyond. Biotechnol J. 2010;5:647–659. doi: 10.1002/biot.200900247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nødvig C.S., Nielsen J.B., Kogle M.E., Mortensen U.H. A CRISPR-Cas9 system for genetic engineering of filamentous fungi. PLoS One. 2015;10:e0133085. doi: 10.1371/journal.pone.0133085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsu-ura T., Baek M., Kwon J., Hong C. Efficient gene editing in Neurospora crassa with CRISPR technology. Fungal Biol Biotechnol. 2015:2. doi: 10.1186/s40694-015-0015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pohl C., Kiel J.A.K.W., Driessen A.J.M., Bovenberg R.A.L., Nygård Y. CRISPR/Cas9 based genome editing of Penicillium chrysogenum. ACS Synth Biol. 2016;5:754–764. doi: 10.1021/acssynbio.6b00082. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen J., Keasling J.D. Engineering cellular metabolism. Cell. 2016;164:1185–1197. doi: 10.1016/j.cell.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Medema M.H., van Raaphorst R., Takano E., Breitling R. Computational tools for the synthetic design of biochemical pathways. Nat Rev Microbiol. 2012;10:191–202. doi: 10.1038/nrmicro2717. [DOI] [PubMed] [Google Scholar]
- 33.Crawford L., Stepan A.M., McAda P.C., Rambosek J.A., Confer M.J., Vinci V.A. Production of Cephalosporin intermediates by feeding adipic acid to recombinant Penicillium chrysogenum strains expressing ring expansion activity. Nat Biotechnol. 1995;13:58–62. doi: 10.1038/nbt0195-58. [DOI] [PubMed] [Google Scholar]
- 34.Chooi Y.-H., Cacho R., Tang Y. Identification of the viridicatumtoxin and griseofulvin gene clusters from Penicillium aethiopicum. Chem Biol. 2010;17:483–494. doi: 10.1016/j.chembiol.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao X., Chooi Y.-H., Ames B.D., Wang P., Walsh C.T., Tang Y. Fungal indole alkaloid biosynthesis: genetic and biochemical investigation of the tryptoquialanine pathway in Penicillium aethiopicum. J Am Chem Soc. 2011;133:2729–2741. doi: 10.1021/ja1101085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cacho R.A., Tang Y., Chooi Y.H. Next-generation sequencing approach for connecting secondary metabolites to biosynthetic gene clusters in fungi. Front Microbiol. 2015;6:1–16. doi: 10.3389/fmicb.2014.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grijseels S., Nielsen J.C., Randelovic M., Nielsen J., Nielsen K.F., Workman M. Penicillium arizonense, a new, genome sequenced fungal species, reveals a high chemical diversity in secreted metabolites. Sci Rep. 2016;6:35112. doi: 10.1038/srep35112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowther W.T., McMillen D.A., Orville A.M., Matthews B.W. The anti-angiogenic agent fumagillin covalently modifies a conserved active-site histidine in the Escherichia coli methionine aminopeptidase. Proc Natl Acad Sci U. S. A. 1998;95:12153–12157. doi: 10.1073/pnas.95.21.12153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kennedy J., Auclair K., Kendrew S.G., Park C., Vederas J.C., Richard Hutchinson C. Modulation of polyketide synthase activity by accessory proteins during lovastatin biosynthesis. Science. 1999;284:1368–1372. doi: 10.1126/science.284.5418.1368. [DOI] [PubMed] [Google Scholar]
- 40.Abe Y., Suzuki T., Ono C., Iwamoto K., Hosobuchi M., Yoshikawa H. Molecular cloning and characterization of an ML-236B (compactin) biosynthetic gene cluster in Penicillium citrinum. Mol Genet Genomics. 2002;267:636–646. doi: 10.1007/s00438-002-0697-y. [DOI] [PubMed] [Google Scholar]
- 41.Regueira T.B., Kildegaard K.R., Hansen B.G., Mortensen U.H., Hertweck C., Nielsen J. Molecular basis for mycophenolic acid biosynthesis in Penicillium brevicompactum. Appl Environ Microbiol. 2011;77:3035–3043. doi: 10.1128/AEM.03015-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergmann S., Funk A.N., Scherlach K., Schroeckh V., Shelest E., Horn U. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol. 2010;76:8143–8149. doi: 10.1128/AEM.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh H.-H., Ahuja M., Chiang Y.-M., Oakley C.E., Moore S., Yoon O. Resistance gene-guided genome mining: serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for fellutamide B, a proteasome inhibitor. ACS Chem Biol. 2016;11:2275–2284. doi: 10.1021/acschembio.6b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tang X., Li J., Millán-Aguiñaga N., Zhang J.J., O'Neill E.C., Ugalde J.A. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem Biol. 2015;10:2841–2849. doi: 10.1021/acschembio.5b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Conway K.R., Boddy C.N. ClusterMine360: a database of microbial PKS/NRPS biosynthesis. Nucleic Acids Res. 2013;41:402–407. doi: 10.1093/nar/gks993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hadjithomas M., Chen I.A., Chu K., Ratner A., Palaniappan K., Szeto E. IMG-ABC: a knowledge base to fuel discovery of biosynthetic gene clusters and novel secondary metabolites. MBio. 2015;6 doi: 10.1128/mBio.00932-15. e00932–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medema M.H., Kottmann R., Yilmaz P., Cummings M., Biggins J.B., Blin K. Minimum information about a biosynthetic gene cluster. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Y.F., Tsai K.J.S., Harvey C.J.B., Li J.J., Ary B.E., Berlew E.E. Comprehensive curation and analysis of fungal biosynthetic gene clusters of published natural products. Fungal Genet Biol. 2016;89:18–28. doi: 10.1016/j.fgb.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziemert N., Lechner A., Wietz M., Millán-Aguiñaga N., Chavarria K.L., Jensen P.R. Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci U. S. A. 2014;111:E1130–E1139. doi: 10.1073/pnas.1324161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doroghazi J.R., Albright J.C., Goering A.W., Ju K.-S., Haines R.R., Tchalukov K.A. A roadmap for natural product discovery based on large-scale genomics and metabolomics. Nat Chem Biol. 2014;10:963–968. doi: 10.1038/nchembio.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Condon B.J., Leng Y., Wu D., Bushley K.E., Ohm R.A., Otillar R. Comparative genome structure, secondary metabolite, and effector Coding Capacity across Cochliobolus pathogens. PLoS Genet. 2013;9:e1003233. doi: 10.1371/journal.pgen.1003233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inglis D.O., Binkley J., Skrzypek M.S., Arnaud M.B., Cerqueira G.C., Shah P. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Fewer D.P., Holm L., Rouhiainen L., Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci U. S. A. 2014;111:9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ballester A., Marcet-houben M., Levin E., Sela N., Selma-lázaro C., Carmona L. Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol Plant-Microbe Interact. 2015;28:232–248. doi: 10.1094/MPMI-09-14-0261-FI. [DOI] [PubMed] [Google Scholar]
- 55.Doroghazi J.R., Metcalf W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genomics. 2013;14:611. doi: 10.1186/1471-2164-14-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seipke R.F. Strain-level diversity of secondary metabolism in Streptomyces albus. PLoS One. 2015;10:e0116457. doi: 10.1371/journal.pone.0116457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ju K.S., Gao J., Doroghazi J.R., Wang K.-K.A., Thibodeaux C.J., Li S. Discovery of phosphonic acid natural products by mining the genomes of 10,000 actinomycetes. Proc Natl Acad Sci U. S. A. 2015;112:12175–12180. doi: 10.1073/pnas.1500873112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kersten R.D., Yang Y.-L., Xu Y., Cimermancic P., Nam S.-J., Fenical W. A mass spectrometry-guided genome mining approach for natural product peptidogenomics. Nat Chem Biol. 2011;7:794–802. doi: 10.1038/nchembio.684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Medema M.H., Blin K., Cimermancic P., De Jager V., Zakrzewski P., Fischbach M.A. AntiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39:339–346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li M.H., Ung P.M., Zajkowski J., Garneau-Tsodikova S., Sherman D.H. Automated genome mining for natural products. BMC Bioinforma. 2009:10. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Medema M.H., Paalvast Y., Nguyen D.D., Melnik A., Dorrestein P.C., Takano E. Pep2Path: automated mass spectrometry-guided genome mining of peptidic natural products. PLoS Comput Biol. 2014;10:e1003822. doi: 10.1371/journal.pcbi.1003822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kersten R.D., Ziemert N., Gonzalez D.J., Duggan B.M., Nizet V., Dorrestein P.C. Glycogenomics as a mass spectrometry-guided genome-mining method for microbial glycosylated molecules. Proc Natl Acad Sci U. S. A. 2013;110:E4407–E4416. doi: 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohimani H., Liu W.-T., Kersten R.D., Moore B.S., Dorrestein P.C., Pevzner P.A. NRPquest: Coupling mass spectrometry and genome mining for nonribosomal peptide discovery. J Nat Prod. 2014;77:1902–1909. doi: 10.1021/np500370c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mohimani H., Kersten R.D., Liu W.-T., Wang M., Purvine S.O., Wu S. Automated genome mining of ribosomal peptide natural products. ACS Chem Biol. 2014;9:1545–1551. doi: 10.1021/cb500199h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dejong C.A., Chen G.M., Li H., Johnston C.W., Edwards M.R., Rees P.N. Polyketide and nonribosomal peptide retro-biosynthesis and global gene cluster matching. Nat Chem Biol. 2016;12:1007–1014. doi: 10.1038/nchembio.2188. [DOI] [PubMed] [Google Scholar]
- 66.Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Price N.D., Papin J.A., Schilling C.H., Palsson B.O. Genome-scale microbial in silico models: the constraints-based approach. Trends Biotechnol. 2003;21:162–169. doi: 10.1016/S0167-7799(03)00030-1. [DOI] [PubMed] [Google Scholar]
- 68.Agren R., Liu L., Shoaie S., Vongsangnak W., Nookaew I., Nielsen J. The RAVEN toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput Biol. 2013;9:e1002980. doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brandl J., Andersen M.R. Current state of genome-scale modeling in filamentous fungi. Biotechnol Lett. 2015;37:1131–1139. doi: 10.1007/s10529-015-1782-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Edwards J.S., Ibarra R.U., Palsson B.O. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol. 2001;19:125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- 71.Asadollahi M.A., Maury J., Patil K.R., Schalk M., Clark A., Nielsen J. Enhancing sesquiterpene production in Saccharomyces cerevisiae through in silico driven metabolic engineering. Metab Eng. 2009;11:328–334. doi: 10.1016/j.ymben.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 72.Brochado A.R., Matos C., Møller B.L., Hansen J., Mortensen U.H., Patil K.R. Improved vanillin production in baker's yeast through in silico design. Microb Cell Fact. 2010:9. doi: 10.1186/1475-2859-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xu P., Ranganathan S., Fowler Z.L., Maranas C.D., Koffas M.A.G. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 74.Zakrzewski P., Medema M.H., Gevorgyan A., Kierzek A.M., Breitling R., Takano E. MultiMetEval: comparative and multi-objective analysis of genome-scale metabolic models. PLoS One. 2012;7:e51511. doi: 10.1371/journal.pone.0051511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Borodina I., Krabben P., Nielsen J. Genome-scale analysis of Streptomyces coelicolor A3(2) metabolism. Genome Res. 2005;3:820–829. doi: 10.1101/gr.3364705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim M., Sang Yi J., Kim J., Kim J.N., Kim M.W., Kim B.G. Reconstruction of a high-quality metabolic model enables the identification of gene overexpression targets for enhanced antibiotic production in streptomyces coelicolor A3(2) Biotechnol J. 2014;9:1185–1194. doi: 10.1002/biot.201300539. [DOI] [PubMed] [Google Scholar]
- 77.Kim M., Yi J.S., Lakshmanan M., Lee D.Y., Kim B.G. Transcriptomics-based strain optimization tool for designing secondary metabolite overproducing strains of Streptomyces coelicolor. Biotechnol Bioeng. 2016;113:651–660. doi: 10.1002/bit.25830. [DOI] [PubMed] [Google Scholar]
- 78.Licona-Cassani C., Marcellin E., Quek L.E., Jacob S., Nielsen L.K. Reconstruction of the Saccharopolyspora erythraea genome-scale model and its use for enhancing erythromycin production. Ant Van Leeuwenhoek. 2012;102:493–502. doi: 10.1007/s10482-012-9783-2. [DOI] [PubMed] [Google Scholar]
- 79.D'Huys P.J., Lule I., Vercammen D., Anné J., Van Impe J.F., Bernaerts K. Genome-scale metabolic flux analysis of Streptomyces lividans growing on a complex medium. J Biotechnol. 2012;161:1–13. doi: 10.1016/j.jbiotec.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 80.Huang D., Li S., Xia M., Wen J., Jia X. Genome-scale metabolic network guided engineering of Streptomyces tsukubaensis for FK506 production improvement. Microb Cell Fact. 2013;12:52. doi: 10.1186/1475-2859-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Panagiotou G., Anderson M.R., Grotkjær T., Regueira T.B., Hofmann G., Nielsen J. Systems analysis unfolds the relationship between the phosphoketolase pathway and growth in Aspergillus nidulans. PLoS One. 2008;3:e3847. doi: 10.1371/journal.pone.0003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Panagiotou G., Andersen M.R., Grotkjaer T., Regueira T.B., Nielsen J., Olsson L. Studies of the production of fungal polyketides in Aspergillus nidulans by using systems biology tools. Appl Environ Microbiol. 2009;75:2212–2220. doi: 10.1128/AEM.01461-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Nielsen J. Synthetic biology for engineering acetyl Coenzyme a metabolism in yeast. MBio. 2014;5 doi: 10.1128/mBio.02153-14. e02153–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van den Berg M.A., Albang R., Albermann K., Badger J.H., Daran J.-M., Driessen A.J.M. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol. 2008;26:1161–1168. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- 85.Zanghellini J., Ruckerbauer D.E., Hanscho M., Jungreuthmayer C. Elementary flux modes in a nutshell: properties, calculation and applications. Biotechnol J. 2013;8:1009–1016. doi: 10.1002/biot.201200269. [DOI] [PubMed] [Google Scholar]
- 86.Prauße M.T.E., Schäuble S., Guthke R., Schuster S. Computing the various pathways of penicillin synthesis and their molar yields. Biotechnol Bioeng. 2016;113:173–181. doi: 10.1002/bit.25694. [DOI] [PubMed] [Google Scholar]
- 87.Krivoruchko A., Zhang Y., Siewers V., Chen Y., Nielsen J. Microbial acetyl-CoA metabolism and metabolic engineering. Metab Eng. 2015;28:28–42. doi: 10.1016/j.ymben.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 88.Matsuda F., Furusawa C., Kondo T., Ishii J., Shimizu H., Kondo A. Engineering strategy of yeast metabolism for higher alcohol production. Microb Cell Fact. 2011;10:70. doi: 10.1186/1475-2859-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krivoruchko A., Nielsen J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr Opin Biotechnol. 2015;35:7–15. doi: 10.1016/j.copbio.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 90.Salo O.V., Ries M., Medema M.H., Lankhorst P.P., Vreeken R.J., Bovenberg R.a.L. Genomic mutational analysis of the impact of the classical strain improvement program on β-lactam producing Penicillium chrysogenum. BMC Genomics. 2015;16:937. doi: 10.1186/s12864-015-2154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Komatsu M., Uchiyama T., Omura S., Cane D.E., Ikeda H. Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U. S. A. 2010;107:2646–2651. doi: 10.1073/pnas.0914833107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gomez-Escribano J.P., Bibb M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb Biotechnol. 2011;4:207–215. doi: 10.1111/j.1751-7915.2010.00219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li X., Guo D., Cheng Y., Zhu F., Deng Z., Liu T. Overproduction of fatty acids in engineered Saccharomyces cerevisiae. Biotechnol Bioeng. 2014;111:1841–1852. doi: 10.1002/bit.25239. [DOI] [PubMed] [Google Scholar]
- 94.Krivoruchko A., Serrano-Amatriain C., Chen Y., Siewers V., Nielsen J. Improving biobutanol production in engineered Saccharomyces cerevisiae by manipulation of acetyl-CoA metabolism. J Ind Microbiol Biotechnol. 2013;40:1051–1056. doi: 10.1007/s10295-013-1296-0. [DOI] [PubMed] [Google Scholar]
- 95.Chen Y., Daviet L., Schalk M., Siewers V., Nielsen J. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 96.Wattanachaisaereekul S., Lantz A.E., Nielsen M.L., Nielsen J. Production of the polyketide 6-MSA in yeast engineered for increased malonyl-CoA supply. Metab Eng. 2008;10:246–254. doi: 10.1016/j.ymben.2008.04.005. [DOI] [PubMed] [Google Scholar]
- 97.Kealey J.T., Liu L., Santi D.V., Betlach M.C., Barr P.J. Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc Natl Acad Sci U. S. A. 1998;95:505–509. doi: 10.1073/pnas.95.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi J.W., Da Silva N.A. Improving polyketide and fatty acid synthesis by engineering of the yeast acetyl-CoA carboxylase. J Biotechnol. 2014;187:56–59. doi: 10.1016/j.jbiotec.2014.07.430. [DOI] [PubMed] [Google Scholar]
- 99.Cardenas J., Da Silva N.A. Engineering cofactor and transport mechanisms in Saccharomyces cerevisiae for enhanced acetyl-CoA and polyketide biosynthesis. Metab Eng. 2016;36:80–89. doi: 10.1016/j.ymben.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 100.Wohlleben W., Mast Y., Muth G., Röttgen M., Stegmann E., Weber T. Synthetic Biology of secondary metabolite biosynthesis in actinomycetes: engineering precursor supply as a way to optimize antibiotic production. FEBS Lett. 2012;586:2171–2176. doi: 10.1016/j.febslet.2012.04.025. [DOI] [PubMed] [Google Scholar]
- 101.Kennedy J., Turner G. δ-( L -α-Aminoadipyl)- L -cysteinyl- D -valine synthetase is a rate limiting enzyme for penicillin production in Aspergillus nidulans. Mol Gen Genet MGG. 1996;253:189–197. doi: 10.1007/s004380050312. [DOI] [PubMed] [Google Scholar]
- 102.Chiang Y.-M., Oakley C.E., Ahuja M., Entwistle R., Schultz A., Chang S.-L. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc. 2013;135:7720–7731. doi: 10.1021/ja401945a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Martín J.F., Casqueiro J., Liras P. Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. Curr Opin Microbiol. 2005;8:282–293. doi: 10.1016/j.mib.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 104.Chanda A., Roze L.V., Kang S., Artymovich K.A., Hicks G.R., Raikhel N.V. A key role for vesicles in fungal secondary metabolism. Proc Natl Acad Sci U. S. A. 2009;106:19533–19538. doi: 10.1073/pnas.0907416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ley A., Coumou H.C., Frandsen R.J.N. Heterologous expression of MlcE in Saccharomyces cerevisiae provides resistance to natural and semi-synthetic statins. Metab Eng Commun. 2015;2:117–123. doi: 10.1016/j.meteno.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim M., Sun G., Lee D.-Y., Kim B.-G. BeReTa: a systematic method for identifying target transcriptional regulators to enhance microbial production of chemicals. Bioinformatics. 2016;33:87–94. doi: 10.1093/bioinformatics/btw557. [DOI] [PubMed] [Google Scholar]
- 107.Bergmann S., Schümann J., Scherlach K., Lange C., Brakhage A.A., Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–217. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 108.Schroeckh V., Scherlach K., Nützmann H.-W., Shelest E., Schmidt-Heck W., Schuemann J. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U. S. A. 2009;106:14558–14563. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ochi K., Hosaka T. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol. 2013;97:87–98. doi: 10.1007/s00253-012-4551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]