Abstract
The rapid development of synthetic biology enables the design, construction and optimization of synthetic microbial consortia to achieve specific functions. In China, the “973” project-“Design and Construction of Microbial Consortia” was funded by the National Basic Research Program of China in January 2014. It was proposed to address the fundamental challenges in engineering natural microbial consortia and reconstructing microbial consortia to meet industrial demands. In this review, we will introduce this “973” project, including the significance of microbial consortia, the fundamental scientific issues, the recent research progresses, and some case studies about synthetic microbial consortia in the past two and a half years.
Keywords: Microbial consortia, Synthetic biology, “973” project of China
1. Introduction
In natural environments, 99% microorganisms exist in the form of microbial consortia. However, some defects of naturally occurring microbial consortia, such as difficulty in culturing, long operation cycle, low conversion efficiency, and poor stability and controllability, limited their practical applications in biotechnology industries. Synthetic microbial consortia constructed via synthetic biology approaches would be an alternative for programming novel complex behaviors and optimal features for practical biotechnology applications. Arnold [1] and Weiss [2], [3] pointed out that synthetic microbial consortia could perform even more complicated tasks and endure more changeable environments than that of monocultures, thus providing an important new frontier for synthetic biology. A better knowledge of the multicellular systems that drive cell-cell interactions in the consortia was highly needed [4], [5]. Engineering novel cell-cell interaction capabilities became crucial in the nascent field of synthetic biology [2].
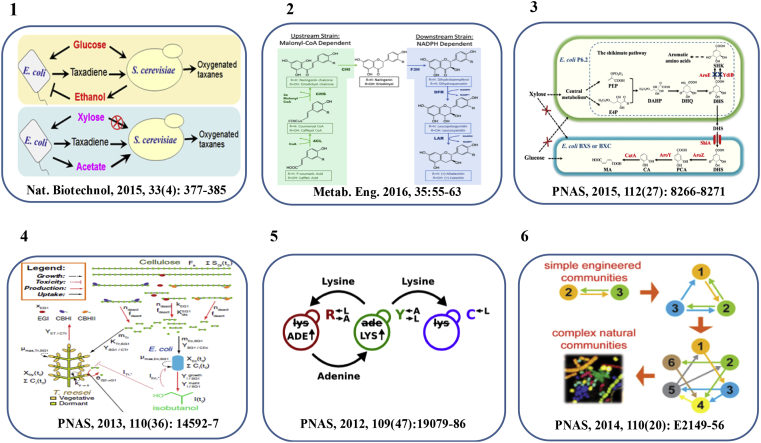
Recently, scientists have made great progresses about analysis, design and construction of microbial consortia (Fig. 1). Stephanopoulos et al. constructed an Escherichia coli- Saccharomyces cerevisiae consortium to successfully produce oxygenated taxanes [6], and an E. coli- E. coli consortium to produce muconic acid [7], [8] and 3-amino-benzoic acid [9]. Jones and his colleges [10] constructed an E. coli- E. coli co-culture for the efficient production of flavonoids. Lin et al. [11] designed and constructed a fungal-bacterial consortium to efficiently produce isobutanol from cellulose. Shou et al. [12], [13], [14] have been focused on engineering and analyzing the underlying mechanisms of cell-cell communication for many years. A series of synthetic syntrophic communities were constructed to probe the metabolic cross feeding principles underlying the complex microbial consortia [15], [16], [17], [18], [19].
Fig. 1.
Applications of synthetic microbial consortia. Part 1 is adapted by permission from Nature Biotechnology [6]©. Part 2 is adapted by permission from Metabolic Engineering [10]©. Part 3 is adapted by permission from Proc Natl Acad Sci USA [7]©. Part 4 is adapted by permission from Proc Natl Acad Sci USA [11]©. Part 5 is adapted by permission from Proc Natl Acad Sci USA [12]©. Part 6 is adapted by permission from Proc Natl Acad Sci USA [15]©.
In the USA, Defense Advanced Research Projects Agency (DARPA) announced a funding entitled “Biological Robustness in Complex Settings (BRICS)” in August 2014. The BRICS program aimed to design synthetic communities consisting of multiple organisms and to elucidate the design principles of engineering robust microbial consortia. The end-program objective is to engineer robust, stable, and safe bio-systems. In China, the “973” project-“Design and Construction of Microbial Consortia”, funded by the National Basic Research Program of China in January 2014 was proposed to address fundamental challenges in engineering natural microbial consortia and reconstructing artificial microbial consortia to meet industrial demands. Great progresses were made in China about the analysis, design, construction of microbial consortia related to microbial fuel cells (MFCs) [20], vitamin C fermentation [21], [22], polyhydroxyalkanoate (PHA) production [23], methane production [24], wastewater treatment [25], biodegradation [26], etc. For further design and construction of synthetic microbial consortia, a synthetic biology module library SynbioML@TJU (http://www.synbioml.org/) that contains more than 5000 artificial synthetic genes and functional modules for diverse products was established by Tianjin University, China. SynbioML covers synthetic genes and functional modules for biosynthesis of natural products (terpenes, flavonoid, polyketones, alkaloids, etc), chemical products (steroids, aminoglycosides, polypeptides, etc), nutrition and health care products (fatty acids, vitamins, etc), biofuels (bioethanol, aliphatic alcohols, butanediol, etc), environmental biological sensors, microbial fuel cells, etc. All of the physical modules, preserved at Tianjin University, can be freely obtained through SynbioML website. Users can search for modules in the website by gene name, protein name, E.C. number, metabolic pathway, enzyme reaction etc.
2. “973” project about microbial consortia in China
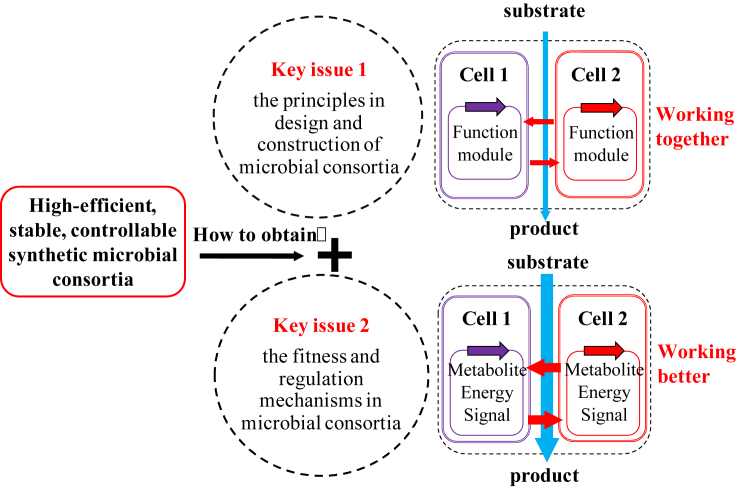
To obtain high-efficient, stable, and controllable synthetic microbial consortia, two predominant, fundamental scientific issues were needed to be addressed in this “973” project (Fig. 2): (1) the principles in the design and construction of microbial consortia to make microbes work together; (2) the fitness and regulation mechanisms in synthetic microbial consortia to make microbes work better.
Fig. 2.
Key issues for achieving high-efficient, stable, and controllable synthetic microbial consortia.
The design principles for synthesizing microbial consortia mainly based on the interaction modes among microbes, including cell-cell communications, and exchange of metabolites and energy, etc. In the natural microbial consortia, there are many ways of interactions according to the modes of metabolic exchange, including commensalism, synergism, mutualism, competition, neutralism, parasitism, and predation, etc. The stable interaction generally relied on the intercellular communication among cells by means of co-utilization of different substrates in the environment, sequential conversion of substrates and reutilization, complement of metabolite, and other ways to meet normal growth of individual cells in multicellular systems. Understanding the cooperative mechanisms in naturally occurring communities would be helpful for designing synthetic microbial consortia. Systems biology could offer insights into the rational design and construction of microbial consortia, and provide detailed molecular understanding of the synthesized microbial ecosystems by rational design strategies [27].
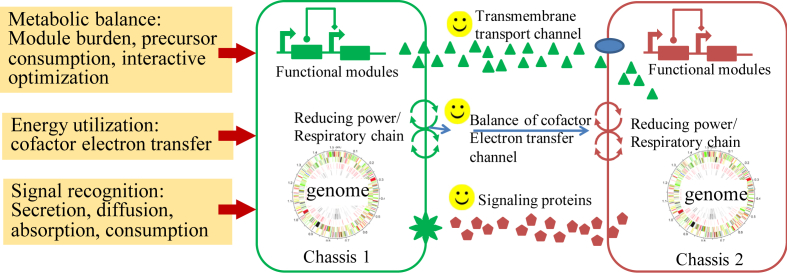
In the synthetic microbial consortia, the partition of different functional modules is benefit for the achievement of orthogonality among modules. However, the partition that achieved by cell membrane would affect the recognition of signal molecule, and the transformation of metabolite, the restriction on transformation of energy and signal molecules. Thus, optimization of synthetic microbial consortia on metabolic balance, energy utilization and signal recognition would be beneficial for the synergistic effect between cells (Fig. 3). Interactions between microbial species are generally mediated by the exchange of small molecules, secreted by one species and consumed by another. In addition, the balance of the electron transfer system and the energy barrier, and signal molecules interaction are also important for the construction of microbial ecosystems. Focusing on the key signal molecules and the production, secretion, diffusion, absorption, consumption and response of metabolites, systematic analysis is essential for design, construction and improvement of the microbial consortia.
Fig. 3.
Fitness and regulation of synthetic microbial consortia.
3. Research progress in “973” project about microbial consortia
3.1. Case study 1: one-step fermentation of vitamin C
To achieve rational design and construction of a target microbial consortium, a detailed understanding of the multicellular interaction is a prerequisite. Systematic understanding of the interaction can provide comprehensive and in-depth clues into the design and construction of the microbial consortia [28], [29], [30], [31], [32]. On the other hand, systems-level knowledge of synthetic microbial consortia will provide in-depth understanding of the consortia and thus redesigning the biological functions of artificial consortia via synthetic biology [33], [34].
3.1.1. Genomics analysis of the bacteria in microbial consortium
The conventional two-step fermentation, consisting of Gluconobacter oxydans, Bacillus spp. and Ketogulonicigenium vulgare, was widely adopted in the industrial production of 2-keto-l-gulonic acid (2-KGA), the precursor of vitamin C. During the second step, K. vulgare is responsible for the biosynthesis of 2-KGA from l-sorbose, and Bacillus spp., as a companion, promotes the growth and production efficiency of K. vulgare. The in-depth analysis of the interaction between Bacillus spp. and K. vulgare was conducted. Firstly, the genome sequence of B. megaterium [35] and K. vulgare [36], [37] was analyzed and annotated, which provided a better-defined genetic background for studies in gene expression and regulation, especially the genome-scale metabolic network construction. The metabolomic and proteomic analyses were also carried out on the B. megaterium-K. vulgare consortium [29], [30], [31], [32]. The systematic analyses on vitamin C-producing strains provided deeply insights into the relationship between the two microorganisms, which benefited our reconstruction of the microbial consortium.
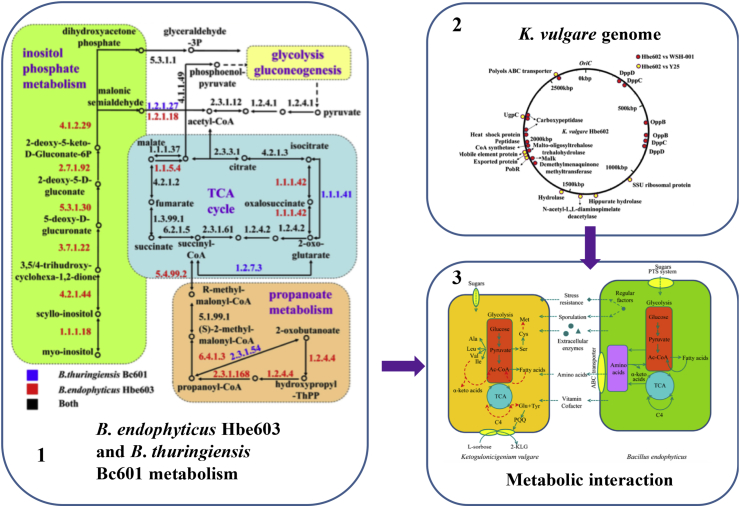
The complete genome of K. vulgare Hbe602 was deciphered to provide insights into the symbiosis mechanism and the versatile metabolism [36]. Comparative genomic analyses of K. vulgare Hbe602, WSH-001 and Y25 were carried out, and the differences with WSH-001 and Y25 were labeled in the red and yellow dots, respectively (Fig. 4). Through the whole genome comparison, the different genes with the nucleotide identities lower than 90% were obtained, mostly focusing on proteolytic enzymes and transporters. The complete genome sequencing of B. thuringiensis Bc60135 and B. endophyticus Hbe60337 were reported, and the comparative genomics analysis was carried out, which enabled a deeper understanding of the cooperative mechanism with K. vulgare, and facilitated the optimization of bacterial consortium. In all, B. endophyticus provided essential functions that K. vulgare lacked to reach its maximum growth rate and acted as an alternative source of environmental nutrients in the consortium.
Fig. 4.
Genomic and metabolomic analysis of bacteria and the cell-cell interaction in vitamin C fermentation. Part 1 is adapted by permission from Scientific Reports [35]©. Part 2 is adapted by permission from Scientific Reports [36]©. Part 3 is adapted by permission from PLoS One [37]©.
Furthermore, metabolic network of B. thuringiensis Bc601 and B. endophyticus Hbe603s was obtained, including the central carbon metabolism, amino acid metabolism and cofactor metabolism (Fig. 4). In the central carbon metabolism, the complete glycolysis, citrate cycle (TCA cycle) and pentose phosphate pathway in the two microbial species were identified. In the amino acid metabolism, B. endophyticus Hbe603 has more lysine degradation related genes than B. thuringiensis Bc601. In the cofactor and vitamin metabolism, both species have the complete biosynthesis pathway of folate, protoheme, pantothenate and CoA, while defect in the lipoic acid and biotin biosynthesis pathway.
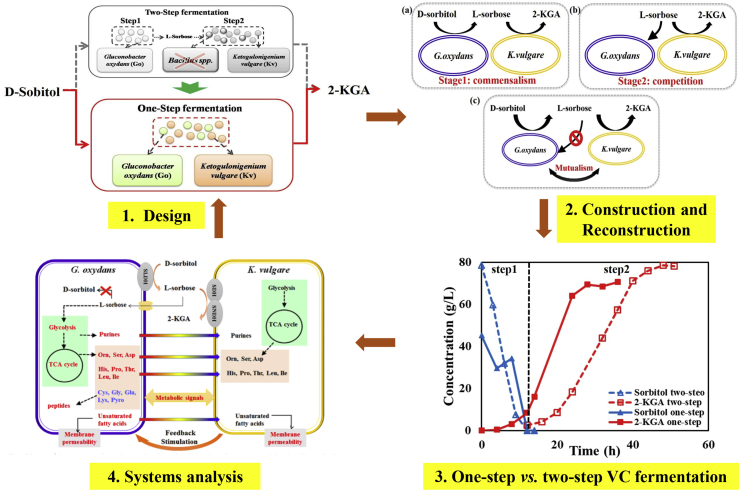
3.1.2. Reconstruction of one-step vitamin C fermentation
During the conventional two-step fermentation of vitamin C, the long incubation period and twice sterilization processes inhibited the further optimization significantly. Therefore, production of 2-KGA by mono-cultured G. oxydans [38] and a synthetic G. oxydans-K. vulgare consortium [22] were constructed and then enhanced by genetic modification. In the one-step fermentation of 2-KGA by G. oxydans-K. vulgare, the final titer of 2-KGA is 76.6 g/L, and the yield of 2-KGA against D-sorbitol reached 89.7% within 36 h, which was comparable to that of the conventional two-step fermentation (about 90% within 48 h)21. Meanwhile, comparing with the traditional method, one-step fermentation shortens the fermentation time by about 25% and eliminates the need for a second sterilization process. In this way, the rate of equipment utilization could be significantly improved and the production cost could be notably saved. Moreover, the symbiotic interaction between the two microorganisms was optimized to perform better (Fig. 5). The relationship between the two microbes changed from commensalism & competition to mutualism. The metabolic interaction between the strains was further investigated by metabolomics, which verified the enhancement of the mutualism between the microbes and provided us potential strategies for further improving the synthetic consortium.
Fig. 5.
Reconstruction of vitamin C one-step fermentation and optimization of the relationship between bacteria. Parts 1, 2, 3, 4 are adapted by permission from Microbial Cell Factories [22]©.
3.2. Synthesizing microbial consortia for power generation in microbial fuel cells
Microbial fuel cells (MFCs) were capable of converting chemical energy stored in chemical compounds to electrical energy by the metabolism of microorganisms. MFCs have functional and operational advantages over the technologies currently used for generating energy from a variety of organic substrates, including sugars, cellulose, organic acids and wastewater pollution. First, the direct conversion of varies of substrate energy to electricity enables high conversion efficiency much better than wind and solar energy. Second, MFCs can operate efficiently at ambient, and even at low, temperatures distinguishing them from all current bio-energy processes. Third, MFCs have potential for widespread application in locations lacking electrical infrastructures and also to expand the diversity of fuels we use to satisfy our energy requirements. Shewanella oneidensis MR-1 was one of the well-established model exoelectrogen, which was widely used in MFCs for wastewater treatment and power generation. Single-species MFCs faced many practical barriers in industrial applications, especially its low extracellular electron transfer (EET) rate and narrow range of substrates such as lactate, pyruvate and formate [39], [40]. In addition, engineering single strain to perform multi-tasks would induce metabolic burden and low viability for power production in MFCs.
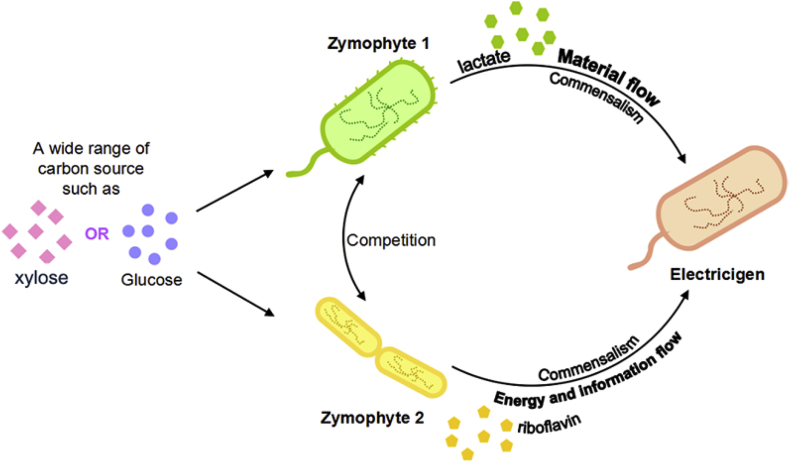
To solve these issues, extending engineering capabilities from single cells to multicellular microbial consortia brought new inspiration and strategies for improving performance in MFCs. Yang et al. [20] recently designed an E. coli-S. oneidensis co-culture system, in which the genetically engineered E. coli played as a fermenter to consume xylose for the synthesis of formate and flavins to feed the exoelectrogen (S. oneidensis) as the carbon source and electron donor, respectively. Thus, a high-performance xylose-fed MFC system for bioelectricity production in MFCs was developed. However, the fermenter E. coli would form biofilms to occupy active anode surfaces, which would reduce the power generation by exoelectrogen. Therefore, we used S. cerevisiae incapable of forming biofilm at anaerobic MFC conditions to replace E. coli to develop a fungus (S. cerevisiae)-bacterium (S. oneidensis) co-culture, which avoided the fermenter cells to occupy anode surfaces. By engineering S. cerevisiae to metabolize glucose to produce lactate but not ethanol, the engineered S. cerevisiae-S. oneidensis consortium enabled a glucose-fed high performance MFCs.
To take advantage of the capability of B. subtilis for riboflavin (the electron shuttle of S. oneidensis) biosynthesis, the TJU iGEM Team designed and constructed a three-species co-culture system with E. coli, B. subtilis and S. oneidensis by the principle of “division of labor” (Fig. 6). To enable the three-species microbial consortium with high-performance MFC was established by optimizing the interaction of material, information and energy interactions between the three microbial species. This project won the best energy project in the 2015 iGEM competition (http://2015.igem.org/Team:TJU/Overview).
Fig. 6.
A synthetic 3-species microbial consortium for high-performance microbial fuel cell (MFC) system. The three microbial species were E. coli, B. subtilis, and S. oneidensis.
4. Perspective
Natural ecosystems have complex microbial community compositions and functions. Environmental variation may exert significant influence upon the metabolic exchange, energy flux and nutrient cycling within even the simplest microbial consortia. So far, most studies about synthetic microbial consortia mainly focus on “build a consortium to understand it”. For synthetic microbial consortia, more design principles and models are required to reveal the interactions and dynamic changes of high-performance synthetic systems. Based on the understanding of the interaction among microbes, the improvement of robustness, stability and reproducibility should be further explored. Further development on ecological principles, engineered strategies and modeling tools will help us to achieve significant progress in designing diverse synthetic ecology with the reproducibility, robustness, controllable and even self-regulating characterizations.
Acknowledgements
This work was funded by the Ministry of Science and Technology of China (“973” Program: 2014CB745100).
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Brenner K., You L., Arnold F.H. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Gerchman Y., Weiss R. Teaching bacteria a new language. Proc Natl Acad Sci U. S. A. 2004;101:2221–2222. doi: 10.1073/pnas.0400473101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purnick P.E., Weiss R. The second wave of synthetic biology: from modules to systems. Nat Rev Mol Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. [DOI] [PubMed] [Google Scholar]
- 4.Mee M.T., Wang H.H. Engineering ecosystems and synthetic ecologies. Mol Biosyst. 2012;8:2470–2483. doi: 10.1039/c2mb25133g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Widder S., Allen R.J., Pfeiffer T., Curtis T.P., Wiuf C., Sloan W.T. Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 2016 doi: 10.1038/ismej.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou K., Qiao K., Edgar S., Stephanopoulos G. Distributing a metabolic pathway among a microbial consortium enhances production of natural products. Nat Biotechnol. 2015;33(4):377–383. doi: 10.1038/nbt.3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang H., Pereira B., Li Z., Stephanopoulos G. Engineering Escherichia coli coculture systems for the production of biochemical products. Proc Natl Acad Sci U. S. A. 2015;112(27):8266–8271. doi: 10.1073/pnas.1506781112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang H., Li Z., Pereira B., Stephanopoulos G., Engineering E. coli-E. coli cocultures for production of muconic acid from glycerol. Microb Cell Fact. 2015;14:134. doi: 10.1186/s12934-015-0319-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang H., Stephanopoulos G. Co-culture engineering for microbial biosynthesis of 3-amino-benzoic acid in Escherichia coli. Biotechnol J. 2016;11(7):981–987. doi: 10.1002/biot.201600013. [DOI] [PubMed] [Google Scholar]
- 10.Jones J.A., Vernacchio V.R., Sinkoe A.L., Collins S.M., Ibrahim M.H., Lachance D.M. Experimental and computational optimization of an Escherichia coli co-culture for the efficient production of flavonoids. Metab Eng. 2016;35:55–63. doi: 10.1016/j.ymben.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Minty J.J., Singer M.E., Scholz S.A., Bae C.H., Ahn J.H., Foster C.E. Design and characterization of synthetic fungal-bacterial consortia for direct production of isobutanol from cellulosic biomass. Proc Natl Acad Sci U. S. A. 2013;110(36):14592–14597. doi: 10.1073/pnas.1218447110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Waite A.J., Shou W. Adaptation to a new environment allows cooperators to purge cheaters stochastically. Proc Natl Acad Sci U. S. A. 2012;109(47):19079–19086. doi: 10.1073/pnas.1210190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shou W., Ram S., Vilar J.M. Synthetic cooperation in engineered yeast populations. Proc Natl Acad Sci U. S. A. 2007;104(6):1877–1882. doi: 10.1073/pnas.0610575104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Momeni B., Waite A.J., Shou W. Spatial self-organization favors heterotypic cooperation over cheating. Elife. 2013;2:e00960. doi: 10.7554/eLife.00960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mee M.T., Collins J.J., Church G.M., Wang H.H. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U. S. A. 2014;111(20):E2149–E2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pande S., Shitut S., Freund L., Westermann M., Bertels F., Colesie C. Metabolic cross-feeding via intercellular nanotubes among bacteria. Nat Commun. 2015;6:6238. doi: 10.1038/ncomms7238. [DOI] [PubMed] [Google Scholar]
- 17.Benomar S., Ranava D., Cárdenas M.L., Trably E., Rafrafi Y., Ducret A. Nutritional stress induces exchange of cell material and energetic coupling between bacterial species. Nat Commun. 2015;6:6283. doi: 10.1038/ncomms7283. [DOI] [PubMed] [Google Scholar]
- 18.Zelezniak A., Andrejev S., Ponomarova O., Mende D.R., Bork P., Patil K.R. Metabolic dependencies drive species co-occurrence in diverse microbial communities. Proc Natl Acad Sci U. S. A. 2015;112:6449–6454. doi: 10.1073/pnas.1421834112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarosz D.F., Brown J.C., Walker G.A., Datta M.S., Ung W.L., Lancaster A.K. Cross-kingdom chemical communication drives a heritable, mutually beneficial prion-based transformation of metabolism. Cell. 2014;158:1083–1093. doi: 10.1016/j.cell.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y., Wu Y.C., Hu Y.D., Cao Y.X., Loo P.C., Bin C. Synthetic bacterial consortium for high-performance microbial fuel cell using xylose as carbon source. ACS Catal. 2015;5(11):6937–6945. [Google Scholar]
- 21.Wang E.X., Ding M.Z., Ma Q., Dong X.T., Yuan Y.J. Reorganization of a synthetic microbial consortium for one-step vitamin C fermentation. Microb Cell Fact. 2016;15:21. doi: 10.1186/s12934-016-0418-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye C., Zou W., Xu N., Liu L. Metabolic model reconstruction and analysis of an artificial microbial ecosystem for vitamin C production. J Biotechnol. 2014;182–183:61–67. doi: 10.1016/j.jbiotec.2014.04.027. [DOI] [PubMed] [Google Scholar]
- 23.Huang L., Liu C., Liu Y., Jia X. The composition analysis and preliminary cultivation optimization of a PHA-producing microbial consortium with xylose as a sole carbon source. Waste Manag. 2016;52:77–85. doi: 10.1016/j.wasman.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 24.Yang Z., Guo R., Xu X., Wang L., Dai M. Enhanced methane production via repeated batch bioaugmentation pattern of enriched microbial consortia. Bioresour Technol. 2016;216:471–477. doi: 10.1016/j.biortech.2016.05.062. [DOI] [PubMed] [Google Scholar]
- 25.Pan X., Qi H., Mu L., Wen J., Jia X. Comparative metabolomic-based metabolic mechanism hypothesis for microbial mixed cultures utilizing cane molasses wastewater for higher 2-phenylethanol production. J Agric Food Chem. 2014;62(40):9927–9935. doi: 10.1021/jf502239d. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y., Li C., Zhou Z., Wen J., You X., Mao Y. Enhanced biodegradation of alkane hydrocarbons and crude oil by mixed strains and bacterial community analysis. Appl Biochem Biotechnol. 2014;172(7):3433–3447. doi: 10.1007/s12010-014-0777-6. [DOI] [PubMed] [Google Scholar]
- 27.Song H., Ding M.Z., Jia X.Q., Ma Q., Yuan Y.J. Synthetic microbial consortia from systematic analysis to construction and applications. Chem Soc Rev. 2014;43:6954–6981. doi: 10.1039/c4cs00114a. [DOI] [PubMed] [Google Scholar]
- 28.Kleiner M., Wentrup C., Lott C., Teeling H., Wetzel S., Young J. Metaproteomics of a gutless marine worm and its symbiotic microbial community reveal unusual pathways for carbon and energy use. Proc Natl Acad Sci U. S. A. 2012;109:E1173–E1182. doi: 10.1073/pnas.1121198109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding M.Z., Zou Y., Song H., Yuan Y.J. Metabolomic analysis of cooperative adaptation between co-cultured Bacillus cereus and Ketogulonicigenium vulgare. PloS One. 2014;9(4):e94889. doi: 10.1371/journal.pone.0094889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma Q., Zhou J., Zhang W., Meng X., Sun J., Yuan Y.J. Integrated proteomic and metabolomic analysis of an artificial microbial community for two-step production of vitamin C. PLoS One. 2011;6(10):e26108. doi: 10.1371/journal.pone.0026108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J., Ma Q., Yi H., Wang L., Song H., Yuan Y.J. Metabolome profiling reveals metabolic cooperation between Bacillus megaterium and Ketogulonicigenium vulgare during induced swarm motility. Appl Environ Microbiol. 2011;77(19):7023–7030. doi: 10.1128/AEM.05123-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du J., Zhou J., Xue J., Song H., Yuan Y.J. Metabolomic profiling elucidates community dynamics of the Ketogulonicigenium vulgare–Bacillus megaterium consortium. Metabolomics. 2012;8(5):960–973. [Google Scholar]
- 33.Lindemann S.R., Bernstein H.C., Song H.S., Fredrickson J.K., Fields M.W., Shou W. Engineering microbial consortia for controllable outputs. ISME J. 2016 doi: 10.1038/ismej.2016.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu P., Bhan N., Koffas M.A. Engineering plant metabolism into microbes: from systems biology to synthetic biology. Curr Opin Biotechnol. 2013;24(2):291–299. doi: 10.1016/j.copbio.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 35.Jia N., Ding M.Z., Gao F., Yuan Y.J. Comparative genomics analysis of the companion mechanisms of Bacillus thuringiensis Bc601 and Bacillus endophyticus Hbe603 in bacterial consortium. Sci Rep. 2016;6:28794. doi: 10.1038/srep28794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia N., Ding M.Z., Du J., Pan C.H., Tian G., Lang J.D. Insights into mutualism mechanism and versatile metabolism of Ketogulonicigenium vulgare Hbe602 based on comparative genomics and metabolomics studies. Sci Rep. 2016;6:23068. doi: 10.1038/srep23068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jia N., Du J., Ding M.Z., Gao F., Yuan Y.J. Genome sequence of Bacillus endophyticus and analysis of its companion mechanism in the Ketogulonigenium vulgare-Bacillus strain consortium. PLoS One. 2015;10(8):e0135104. doi: 10.1371/journal.pone.0135104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao L., Hu Y., Liu J., Du G., Zhou J., Chen J. Stepwise metabolic engineering of Gluconobacter oxydans WSH-003 for the direct production of 2-keto-L-gulonic acid from D-sorbitol. Metab Eng. 2014;24:30–37. doi: 10.1016/j.ymben.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 39.Yang Y., Ding Y., Hu Y., Cao B., Rice S.A., Kjelleberg S. Enhancing bidirectional electron transfer of Shewanella oneidensis by a synthetic flavin pathway. ACS Synth Biol. 2015;4(7):815–823. doi: 10.1021/sb500331x. [DOI] [PubMed] [Google Scholar]
- 40.Liu T., Yu Y.Y., Deng X.P., Ng C.K., Cao B., Wang J.Y. Enhanced Shewanella biofilm promotes bioelectricity generation. Biotechnol Bioeng. 2015;112(10):2051–2059. doi: 10.1002/bit.25624. [DOI] [PubMed] [Google Scholar]