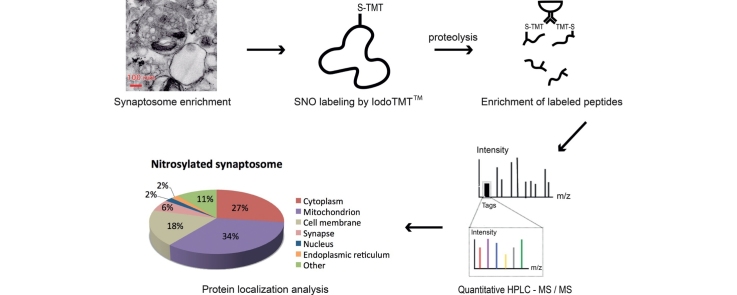
Graphical abstract
Highlights
-
•
Protocol for quantitative proteomics of nitrosylation on synaptosomal proteins.
-
•
Identification of endogenous nitrosylation independent of induction by NO donors.
-
•
Use of iodoTMT sixplex mass tags for stable labeling, enrichment, identification, and multiplex quantitation.
-
•
Applicable on low amounts of sample material of mouse and human brain tissue.
1. Introduction
Nitric oxide (NO) is a free radical that is biosynthesized by nitric oxide synthases. NO contributes to numerous physiologic responses such as vasculature vasodilation, synaptic neurotransmission, respiration, inflammation, and apoptosis (Swomley et al., 2014). One of the important biological modifications induced by NO is S-nitrosylation of cysteine (SNO), which functions in regulating signal transduction. In neurodegenerative disorders, aberrant SNO can propagate protein misfolding, mitochondrial dysfunction, synaptic injury, and neuronal death (Nakamura et al., 2013). For example, increased levels of nitrosylated proteins have been reported in the brain of Alzheimeŕs disease (AD) patients (Heneka et al., 2015). As one of the earliest pathological characteristics of AD is synapse loss, our aim was to establish a robust protocol for the analysis of synaptosomal S-nitrosylation in mouse and human brain samples. Synaptosomes consist of the presynaptic terminal (which contains synaptic vesicles and mitochondria), as well the postsynaptic density and postsynaptic membrane. They are preferably obtained by homogenization of brain tissue and density gradient centrifugation for enrichment of this specific compartment. (Swomley et al., 2014). Numerous proteomics techniques have been developed for detection and quantification of SNO (Qu et al., 2016). However, many of these require high amounts of starting materials in the range of 100–500 μg of protein lysate (Wojdyla and Rogowska-Wrzesinska, 2015). Furthermore, a common procedure applied to analyze SNO modification sites is induction of nitrosylation by use of NO-donors. It is difficult to compare the results from studies using NO donors to the in vivo situation, where cysteine S-nitrosylation depends on the undetermined concentrations of nitrosylating compounds (Zareba-Koziol et al., 2014). Hence, identification of endogenous SNO levels derived from sub-fractionated compartments such as synaptosomes requires high sensitivity and specificity of the detection strategy.; Typical approaches for quantitative SNO proteomics are two-dimensional gel electrophoresis or liquid chromatography (LC) followed by mass spectrometry. Two-dimensional difference in-gel electrophoresis (2D-DIGE) has been described using a modified labeling procedure (NitroDIGE) (Qu et al., 2014b). More frequently used is the biotin switch technique (BST) introduced by Jaffrey and Snyder (2001). It utilizes a replacement strategy to add a stable biotin into a SNO modification, allowing their detection and identification by antibodies or MS (Jaffrey and Snyder, 2001). More recently, an MS-based, isobaric tag labeling strategy was introduced in which peptides are linked with different tags and then fragmented to produce reporter ions of specific masses (Dayon and Sanchez, 2012). Similar to BST, the commercially available labeling reagents (iodoTMT™ 126-131) are used for replacement chemistry of SNO modifications in a 6-plex multiplex (Qu et al., 2014a). Although introduced in 2012, this approach has been barely described in the literature for samples with low amounts of starting material and under basal, endogenous conditions without NO donor treatment. This article provides detailed descriptions of the methodological workflow (Fig. 1) and efficiency of the iodoTMT™ protocol when applied to the analysis of synaptosomal samples, discussing both technical variances of the method as well as the comparability of findings with results previously published on this field.
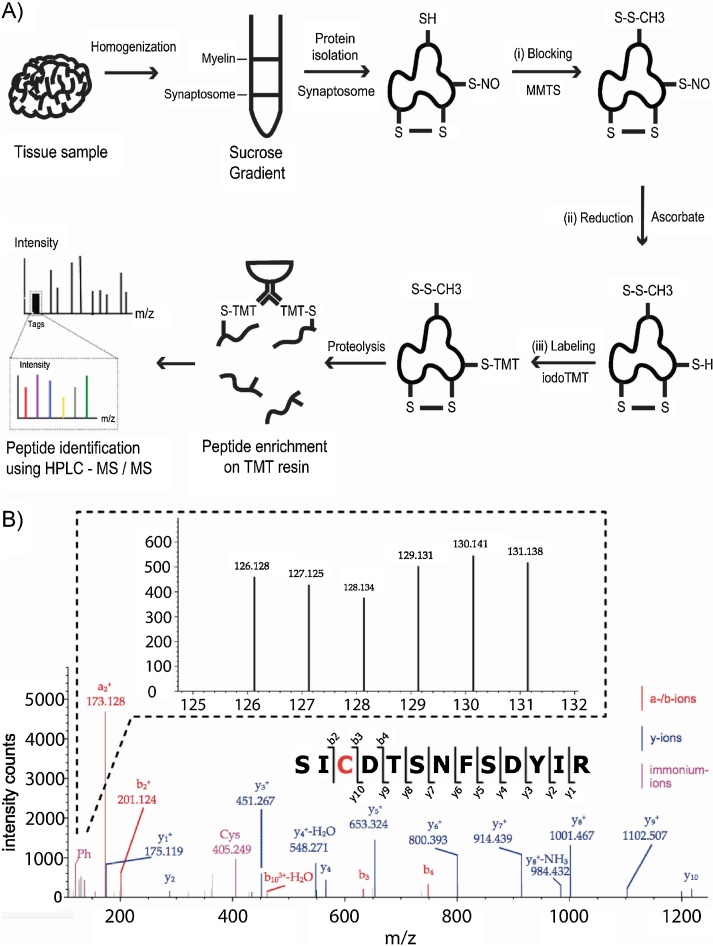
Fig. 1.
The isobaric tag labeling procedure.
(A) Schematic image of the isobaric tag labeling procedure. After synaptosome isolation, SNO proteins are labeled by (i) blocking of free thiols using MMTS; (ii) reduction of cysteine S-nitrosylation with ascorbate, producing new free thiols on previous SNO sites; and (iii) isobaric tag labeling of the new thiols using iodoTMT. iodoTMT-labeled proteins are subjected to in-solution protein digestion. Derived peptides are enriched using affinity-capture on an anti-TMT resin. Eluted peptides are identified by nanoHPLC–MS/MS and quantified using the TMT sixplex. (B) A representative MS/MS spectrum of the iodoTMT-labeled peptide SICDTSNFSDYIR derived from ubiquitin-like modifier-activating enzyme 1. Peaks of the 6 reporter ions are found in the enlarged section of the lower mass range.
2. Materials and methods
2.1. Tissue samples
Three 9-months-old, male wild type mice were euthanized by isoflurance and transcardially perfused with ice-cold normal saline (0.9% w/v NaCl). Mouse brain hemispheres without brainstem and cerebellum were prepared and immediately used for protein extraction. The method was transferred to two human frontal cortex brain samples from non-demented, non-diseased donors. Human brain samples were obtained from unfixed, frozen tissues with a postmortem interval of less than 4 h from the Banner Sun Health Research Institute Brain and Body Donation Program in Sun City, Arizona (Beach et al., 2008). All subjects participating in the Banner Brain and Body Donation Program signed the written informed consent approved by the Banner Health Institutional Review Board.
2.2. Synaptosome isolation
For preparation of synaptosomes, 200 mg tissue where used with equal amounts of white and grey matter. Tissues were homogenized in 9-fold volume of tissue weight in 0.32 M sucrose buffer (50 mM Tris-acetate, 1 mM EDTA, 1 mM EGTA, 5 mM disodium pyrophosphate, 5 mM NaF, 2 mM Na3VO4, 1 mM AEBSF, cOmplete protease inhibitor cocktail (Sigma-Aldrich, Munich, Germany), pH 7.4). Homogenization was performed manually by 15 slow up and down strokes using a Teflon-glass homogenizer (0.1–0.15 mm clearance). Samples were centrifuged to remove nuclei and cellular debris at 800g for 5 min at 4 °C. The collected supernatant was layered on top of a discontinuous sucrose gradient consisting of two layers of sucrose buffer (1.0 M and 1.4 M). Gradient centrifugation was performed using a MLS-50 swinging bucket rotor (Beckman, Krefeld, Germany) at 54000g for 90 min at 4 °C. The synaptosomal fraction was collected from the interface between 1.0 M and 1.4 M sucrose layers. The fraction was diluted with 4-fold volume of HPLC-grade water and sedimented at 54000g for 15 min at 4 °C. The supernatant was discarded and the synaptosomal pellet was stored at −80 °C. Enrichment of pre- and post-synaptic proteins was evaluated using electron microscopy and Western blotting against synaptophysin (1:1000, Abcam, ab52636), post-synaptic density protein (PSD95, 1:2000, Thermo Fisher Scientific, MA1-064), NMDA receptor 2 B (NMDAR2B, 1:1000, Millipore, ab1557p), the nuclear marker Lamin B1 (1:500, Proteintech, 66095-1-Ig) and alpha tubulin (1:2000, Thermo Fisher Scientific, 62204) as a loading control.
2.3. IodoTMT workflow for synaptosomal proteins
2.3.1. Protein labeling with iodoTMT
Three technical replicates were used to evaluate this approach. Synaptosome pellets were resuspended in 1 μg μL−1 HENS lysis buffer (Thermo Fisher Scientific). Each synaptosome sample was prepared for six aliquots with an equal amount of proteins (50 μg), which were then individually labeled with iodoTMT sixplex labeling reagent followed by the optimized protocol. Subsequently, samples were digested, desalted, enriched, and analyzed by MS.
To avoid rearrangements of thiol-modifying groups, the protein mixture was treated with MMTS (20 mM final concentration, stock diluted in dimethyl formamide) at room temperature for 30 min with frequent vortexing (protected from light). Excess MMTS was removed by acetone precipitation at −20 °C for 1 h. The pellets were resuspended in 50 μL of HENS buffer and reduced with 1 μL of 1 M sodium ascorbate in water. Each sample was immediately labeled with 1 μL of iodoTMT labeling reagent (0.2 mg in 10 μL of LC/MS grade methanol) at 37 °C for 1 h, protected from light. Excess iodoTMT reagent was quenched with 0.5 M dithiothreitol (20 mM final concentration) at 37 °C for 15 min. Up to six different samples labeled with iodoTMT 126-131 were combined and alkylated with 10 μL of 0.5 M iodoacetamide in 300 μL of HENS buffer at 37 °C for 1 h.
2.3.2. Tryptic digestion and peptide desalting
Alkylated samples were precipitated with acetone precipitation and dissolved in 50 mM ammonium bicarbonate buffer, pH 8 for overnight digestion using trypsin (protein: trypsin ratio = 1:40 w/w) at 37 °C. The peptides were acidified with 10% trifluoroacetic acid to final concentration of 0.5%, purified using C18 spin columns (Thermo Fisher Scientific) and eluted with 70% (v/v) acetonitrile (ACN).
2.3.3. Enrichment of iodoTMT-labeled peptides
For the enrichment step, the labeled peptides were dried by vacuum centrifugation and resuspended in 100 μL TBS buffer (150 M NaCl, 50 mM Tris, pH 8.0). The peptide samples were incubated with 30 μL anti-TMT resin for 2.5 h at room temperature with end-over-end mixing. The resin was washed three times with 100 μL TBS followed by three washing steps with 100 μL HPLC-grade water. Peptides were eluted twice with 30 μL TMT elution buffer (Thermo Fisher Scientific). Pooled eluates were dried by vacuum centrifugation and redissolved in 8 μL of 0.1% TFA for nanoHPLC MS/MS analysis.
2.3.4. LC–Mass spectrometry analysis
A C18 trap column (20 mm length, 100 μm inner diameter) was coupled to a C18 analytical column (200 mm length, 75 μm inner diameter) packed in-house with 1.9 μm ReproSil-Pur 120 C18-AQ particles (Dr. Maisch, Ammerbuch, Germany). Solvent A consisted of 0.1% formic acid. Solvent B consisted of 90% acetonitrile, 0.1% formic acid. Separation of the peptides used a linear gradient from 4% to 35% solvent B within 240 min at a flow rate of 300 nl/min. The nanoHPLC was coupled online to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Peptide ions between 330 and 1700 m/z were scanned in the Orbitrap detector with a resolution of 30,000 (maximum fill time 400 ms, AGC target 106). The 20 most intense precursor ions (threshold intensity 5000) were subjected to higher energy collision induced dissociation (HCD) and fragments were again analyzed in the Orbitrap. Fragmented peptide ions were excluded from repeated analysis for 15 s.
2.4. Data analysis and protein identification
For data processing and analyses of database searches, Proteome Discoverer software 2.1.0.81 (Thermo Fisher Scientific) was used. Peptide identification was done on an in-house Mascot server, version 2.5.1 (Matrix Science Ltd, London, UK). MS2 data (including a-series ions) were matched against mouse/human sequences from SwissProt (release 2015_04). Precursor Ion m/z tolerance was 10 parts per million (ppm), fragment ion tolerance was 20 millimass units (mmu). Up to two missed cleavages were allowed for tryptic peptides. Low scoring spectrum matches were re-analyzed with semitryptic specificity with up to one missed cleavage. Carbamidomethylation (Cys), Oxidation (Met), acetylation (protein N-terminus), and iodoTMT sixplex (on Cys) were set as dynamic modifications. Mascot results from searches against SwissProt were sent to the percolator algorithm version 2.05 as implemented in Proteome Discoverer. Peptides were identified with a posterior error probability of 1% or better. Proteins were reported with at least one matching peptide because of the experimental design: In cases where only one site in a protein is nitrosylated, only one labeled peptide can be identified after enrichment.
3. Results and discussions
3.1. Method performance
Technical characterization of the method was tested first on wild-type mouse samples (n = 3) and then transferred to human brain samples (n = 2) from donors without CSN disorders. To test reporter ion intensity and determine inter-sample variances, aliquots of the same samples were labeled with IodoTMT™ sixplex reagents, one for each aliquot, and then pooled and analyzed (same-same-experiment). A representative MS/MS spectrum with intense reporter ions and a clear b- and y-series of assigned fragment ions is shown in Fig. 1B. When comparing reporter ion intensities from the same-same-experiment over all identified peptides between all of the 6 labels, there was an average variance of 20% between single reporter ion channels and the mean of all channels, quantified over all of the identified peptides. This indicates that the overall technical variance of the method, including preparative pipetting, sample handling, digestion, IodoTMT™ labeling and enrichment until pooling and analysis of the samples, was within this range.
In theory, all identified peptides should contain a TMT™ label attached to a previously nitrosylated cysteine. Within our dataset, an average of 72% of identified peptides contained cysteine, among which 93%–98% were labeled with IodoTMT™. In conclusion, cysteine-targeted labeling was highly specific, whereas the anti-TMT-antibody based enrichment and purification strategy could not completely remove non-modified peptides. There was barely any unspecific TMT-labeling (<1% of peptides) on the peptide N-terminus (which is the target of the regular TMT™ protocol). Noteworthy, it was not possible to identify nitrosylated peptides form the synaptosome samples without application of the labeling/enrichment strategy.
3.2. Proteome analysis
In murine synaptosomes 1462 unique peptides were identified representing 617 proteins, of which 42% were identified for the first time. In human synaptosomes, 570 unique peptides were identified representing 337 proteins, of which 73% were identified for the first time (by comparing to the SNO database http://dbSNO.mbc.nctu.edu.tw). In comparison, the absolute number of SNO-modified proteins was 2.6-fold higher in mouse proteins compared to human proteins. This matches data found on the SNO database (http://140.138.144.145/∼dbSNO/statistics.php), which at this point lists 1494 murine and 720 human SNO proteins. The lower number of SNO proteins found in human tissue by our approach and described in the database could be attributed to both technical as well as biological variances: Technically, murine brain samples can be prepared directly after euthanizing the animal, which is impossible for human brain samples. Furthermore, due to perfusion with cold saline solution, mouse brain samples are cooled faster. Controlled cold temperature can be used to reduce the activity of nitric oxide synthases, as it has been demonstrated in example for NOS2 (Venturini et al., 1999). For human samples, the post mortem interval was limited to a maximum of 4 h for this study, but this time window might already impact the yield of nitrosylated proteins. Additionally, synaptosomes from mouse brains could be prepared without freeze-thaw cycles, while human brain samples were obtained as frozen material from a biobank, which did not prepare its specimen specifically for this type of analysis. Supposable, the SNO proteome could be further preserved by use of appropriate chemical stabilizers or inhibitors additional to cooling and protease inhibitors, as it is common practice in example for phosphoproteomics. To our knowledge, such substances have not found widespread use yet, although some studies reported use of inhibitors (Koo et al., 2016, Foster et al., 2012). Lastly, there might be substantial species-derived differences between murine and human samples, which cannot be entirely distinguished from method-derived differences to this point.
The composition of the SNO-proteome was analyzed using Gene Ontology enRIchment anaLysis and visuaLiAtion (Gorilla) search engine (Eden et al., 2007) and Uniprot/SwissProt based on the localization classification (UniProt, 2015). The Gorilla search engine result (illustrated in Fig. 2A) indicates enriched GO terms in the Cellular Components of organelle membrane (GO:0031090), oxidoreductase complex (GO:1990204), cytoplasmic part (GO:0044444), organelle inner membrane (GO:0019866), intracellular organelle (GO:0043229), intracellular membrane bounded organelle (GO:0043231) and components of mitochondrial (GO:0044429; GO:0005739; GO:0031966; GO:0044455; GO:0005743). Definitions of the specific components are available from the Gene Ontology Consortium (http://www.geneontology.org/) database. This result revealed the enrichment of synaptosome components such as cytoplasmic part, cell membrane and mitochondria that are enriched in the presynaptic terminal (Swomley et al., 2014).
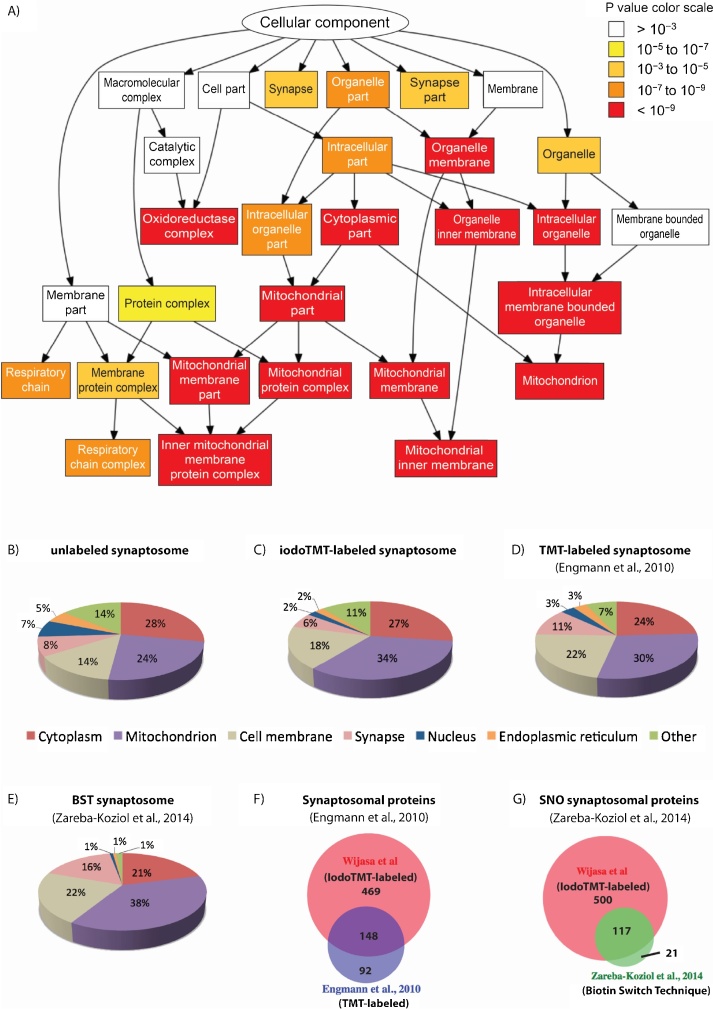
Fig. 2.
Classification of identified synaptosomal proteins based on cellular localization.
(A) Simplified representation of the “cellular component” classes enriched in typical synaptosome components (predominantly from mitochondria) with p −value <10−6 (Eden et al., 2007). The original graph was modified to improve readability. Stronger enrichment is represented by lower p-values; (B) Cellular localization of unlabeled mouse synaptosome; (C) Localization of IodoTMT-labeled mouse nitrosylated synaptosome proteins; (D) Comparison to a TMT-labeled synaptosome whole proteome (independent of modification) (Engmann et al., 2010); (E) Comparison to a biotin switch technique study on synaptosomes (Zareba-Koziol et al., 2014); (F) The overlapping synaptosomal proteins of IodoTMT-labeled and TMT-labeled (Engmann et al., 2010); (G) The overlapping SNO synaptosomal proteins of IodoTMT-labeled and Biotin Switch Technique (Zareba-Koziol et al., 2014).
Classifications based on Uniprot/SwissProt localization entries showed comparable enrichments of typical synaptosome components between the unlabeled synaptosome (2B) and the IodoTMT labeled SNO synaptosome (2C), both obtained from our protocols and results published by others on regular TMT labeling of synaptosome proteomes (2D). The synaptosome SNO-proteome (iodoTMT-labeled) contained a large fraction of mitochondrial proteins (30%), presumably originating from the high amounts of mitochondria located in synapses (Fig. 2C). Additionally, large fractions of cytoplasmic (25%) and cell membrane (20%) associated proteins as well as a small fraction of concrete synapse associated proteins (6–8%) were found, all well in accordance with the expected composition of synaptosomes. The distribution of the identified synaptosomal proteins according to their localization between unlabeled, and labeled synaptosome (iodoTMT or TMT) shows analogous enrichment results with a majority of cytoplasm 24–28%, mitochondria 24–34%, cell membrane 14–22%, and synapse 6–11%. We also found nuclear and endoplasmic reticulum associated proteins resulting from small fragments of these organelles with similar density as synaptosomes.
For the murine samples, these numbers are comparable with, if not higher than, those described in previous reports on synaptosome samples: In example, SNOSID (SNO side identification) analysis using BST identified 138 SNO proteins (Zareba-Koziol et al., 2014) of which 85% were found to overlap with our results (see Fig. 2G); and TMT (Tandem Mass Tag) labeling analysis identified 240 proteins (Fig. 2F), both from murine synaptosome. For human samples, to our knowledge, there are no published data for synaptosome-derived SNO-proteomes. We found that the overall composition of the detected proteome is congruent with previous publications on the synaptosome proteome labeled with TMT independent of their modifications (Fig. 2D) (Engmann et al., 2010) and with BST on SNOSID (Fig. 2E) (Zareba-Koziol et al., 2014).
3.3. Advantages and limitations
The iodoTMT approach showed a large number of identified proteins obtained from a small amount of starting material (50 μg of protein from synaptosome lysate). The number of identified SNO proteins was comparable or higher in comparison to previous studies. Further advantages include high labeling specificity and the use of one stable labeling for enrichment, identification and multiplex quantitation of modified proteins. Also, this protocol was functional for identification of endogenous levels of SNO proteins, without prior induction of nitrosylation by NO donor treatment. Finally, the iodoTMT protocol was easy to adapt and did not require extensive method optimization.
However, this approach also has limitations. The stability of the iodoTMT labeling reagent after reconstitution is limited to 1 week when stored at −20 °C. Therefore, it is recommendable to perform all labeling steps for a study within this time frame.
Another limitation stems from the nature of the labeling and enrichment procedure: As only modified peptides are identified and quantified, the procedure frequently depends on MS/MS-based information implied from a single peptide ion. This limits the number of peptides available for protein identification, which in consequence should be achieved from that single peptide. Therefore, protein identifications are only valid when derived from unique (protein-specific) modified peptides. If identified peptides are not unique, multiple proteins or protein classes have to be considered as potential origin of the peptide.
Although highly efficient, the entire workflow consists of multiple steps (such as blocking, labeling, enrichment, cleaning, digestion) which are time-consuming and can add up to inter-run variances, which were in an average range of 20% in the same experiments. To control this in quantitative studies, we suggest setting up an internal standard that contains pooled and aliquoted proteins of all representative samples. This standard should be added at the beginning of each preparation batch and be analyzed as one of the sixplex samples on each LC-MC-run, hence enabling to control for any technical failures or irregularities that interfere with quantitation. Although this standard cannot reduce inter-run-variances, it could be used to characterize and monitor such variances for quality control purposes.
4. Conclusions
In summary, the protocol possesses many advantages in terms of enrichment, specificity, and quantitation, all based on a single labeling reaction. Additionally, the IodoTMT approach enables sample (6-plex) multiplexing. The protocol was sensitive enough to utilize low amounts of starting material while at the same time providing data comparable to previously published results. Given the increasing scientific interest on protein S-nitrosylation (Qu et al., 2016), we anticipate it to be useful for other researchers analyzing this instable protein modification, especially when dealing with limited amounts of samples such as in analysis of synaptosomes or other cellular compartments.
Conflict of interest
None.
Authors contribution
TSW performed initial method screening, animal and human brain preparations, synaptosome isolation and protein labeling and drafted the manuscript, MS and NBA performed the mass spectrometric measurements and MS data analysis, FB and MTH aimed the study, supervised experimental design and interpretation and drafted the manuscript, VG and MPK approved the study and drafted the manuscript.
Acknowledgements
This work was funded by the German Center for Neurodegenerative Diseases (DZNE e.V.) within the Helmholtz Association, by the German Research Council (DFG, Klinische Forscherguppe 177, TP4), the Institute of Biochemistry and Molecular Biology at the Rheinische Friedrich-Wilhelms University, European Union's Seventh Framework Programme (FP7/2007-2013) under grant agreement no. HEALTH-F2-2011-278850 (INMiND) and the EU-IMI program no. 115568 (AETIONOMY). Michael T. Heneka is a member of the Cluster of Excellence “Immunosensation”.
References
- Beach T.G., Sue L.I., Walker D.G., Roher A.E., Lue L., Vedders L., Connor D.J., Sabbagh M.N., Rogers J. The sun health research institute brain donation program: description and experience, 1987–2007. Cell Tissue Bank. 2008;9:229–245. doi: 10.1007/s10561-008-9067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayon L., Sanchez J.C. Relative protein quantification by MS/MS using the tandem mass tag technology. Methods Mol. Biol. 2012;893:115–127. doi: 10.1007/978-1-61779-885-6_9. [DOI] [PubMed] [Google Scholar]
- Eden E., Lipson D., Yogev S., Yakhini Z. Discovering motifs in ranked lists of DNA sequences. PLoS Comput. Biol. 2007;3:e39. doi: 10.1371/journal.pcbi.0030039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engmann O., Campbell J., Ward M., Giese K.P., Thompson A.J. Comparison of a protein-level and peptide-level labeling strategy for quantitative proteomics of synaptosomes using isobaric tags. J. Proteome Res. 2010;9:2725–2733. doi: 10.1021/pr900627e. [DOI] [PubMed] [Google Scholar]
- Foster M.W., Yang Z., Gooden D.M., Thompson J.W., Ball G.H., Turner M.E., Hou Y., Pi J., Moseley M.A., Que L.G. Proteomic characterization of the cellular response to nitrosative stress mediated by S-nitrosoglutathione reductase inhibition. J. Proteome Res. 2012:2480–2491. doi: 10.1021/pr201180m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heneka M.T., Carson El Khy M.J.J., Landreth G.E., Brosseron F., Feinstein D.L., Jacobs A.H., Wyss-Coray T., Vitorica J., Ransohoff R.M., Herrup K., Frautschy S.A., Finsen B., Brown G.C., Verkhratsky A., Yamanaka K., Koistinaho J., Latz E., Halle A., Petzold G.C., Town T., Morgan D., Shinohara M.L., Perry V.H., Holmes C., Bazan N.G., Brooks D.J., Hunot S., Joseph B., Deigendesch N., Garaschuk O., Boddeke E., Dinarello C.A., Breitner J.C., Cole G.M., Golenbock D.T., Kummer M.P. Neuroinflammation in alzheimer's disease. Lancet Neurol. 2015;14:388–405. doi: 10.1016/S1474-4422(15)70016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffrey S.R., Snyder S.H. The biotin switch method for the detection of S-nitrosylated proteins. Sci. STKE. 2001;2001 doi: 10.1126/stke.2001.86.pl1. PL1. [DOI] [PubMed] [Google Scholar]
- Koo S.J., Spratt H.M., Soman K.V., Stafford S., Gupta S., Petersen J.R., Zago M.P., Kuyumcu-Martinez M.N., Brasier A.R., Wiktorowicz J.E., Garg N.J. S-Nitrosylation proteome profile of peripheral blood mononuclear cells in human heart failure. Int. J. Proteom. 2016:1–19. doi: 10.1155/2016/1384523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Tu S., Akhtar M.W., Sunico C.R., Okamoto S., Lipton S.A. Aberrant protein s-nitrosylation in neurodegenerative diseases. Neuron. 2013;78:596–614. doi: 10.1016/j.neuron.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., Meng F., Bomgarden R.D., Viner R.I., Li J., Rogers J.C., Cheng J., Greenlief C.M., Cui J., Lubahn D.B., Sun G.Y., Gu Z. Proteomic quantification and site-mapping of S-nitrosylated proteins using isobaric iodoTMT reagents. J. Proteome Res. 2014;13:3200–3211. doi: 10.1021/pr401179v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., Meng F., Zhou H., Li J., Wang Q., Wei F., Cheng J., Greenlief C.M., Lubahn D.B., Sun G.Y., Liu S., Gu Z. NitroDIGE analysis reveals inhibition of protein S-nitrosylation by epigallocatechin gallates in lipopolysaccharide-stimulated microglial cells. J. Neuroinflammation. 2014;11:17. doi: 10.1186/1742-2094-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Z., Greenlief C.M., Gu Z. Quantitative proteomic approaches for analysis of protein S-nitrosylation. J. Proteome Res. 2016;15:1–14. doi: 10.1021/acs.jproteome.5b00857. [DOI] [PubMed] [Google Scholar]
- Swomley A.M., Forster S., Keeney J.T., Triplett J., Zhang Z., Sultana R., Butterfield D.A. Abeta, oxidative stress in Alzheimer disease: evidence based on proteomics studies. Biochim. Biophys. Acta. 2014;1842:1248–1257. doi: 10.1016/j.bbadis.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt C. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–12. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini G., Colasanti M., Fioravanti E., Bianchini A., Ascenzi P. Direct effect of temperature on the catalytic activity of nitric oxide synthases types I, II, and III. Nitric Oxide. 1999;3:375–382. doi: 10.1006/niox.1999.0250. [DOI] [PubMed] [Google Scholar]
- Wojdyla K., Rogowska-Wrzesinska A. Differential alkylation-based redox proteomics–Lessons learnt. Redox Biol. 2015;6:240–252. doi: 10.1016/j.redox.2015.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zareba-Koziol M., Szwajda A., Dadlez M., Wyslouch-Cieszynska A., Lalowski M. Global analysis of S-nitrosylation sites in the wild type (APP) transgenic mouse brain-clues for synaptic pathology. Mol. Cell Proteomics. 2014;13:2288–2305. doi: 10.1074/mcp.M113.036079. [DOI] [PMC free article] [PubMed] [Google Scholar]