Significance
T-lymphocyte activation during immune surveillance begins with recognition by its T-cell receptor (TCR) of a peptide ligand bound to an MHC molecule (pMHC) on infected or otherwise altered cells. Using optical traps, we show that activation is fostered at the single-molecule level through synergy between external load on the TCR–pMHC bond and internal, sustained stepping via motor-dependent transport. Chemical thresholds in the absence of load require much higher pMHC density than observed physiologically. With load, however, T lymphocytes can be reliably activated with 10 pN per TCR molecule, mimicking native shear motions involving a mere two pMHCs at the interaction surface. Initial TCR triggering sensitivity results from synergistic mechanosensing rather than previously postulated serial engagement.
Keywords: mechanosensor, T-cell receptor, T-cell activation, optical tweezers, cellular force relaxation
Abstract
T lymphocytes use surface T-cell receptors (TCRs) to recognize peptides bound to MHC molecules (pMHCs) on antigen-presenting cells (APCs). How the exquisite specificity of high-avidity T cells is achieved is unknown but essential, given the paucity of foreign pMHC ligands relative to the ubiquitous self-pMHC array on an APC. Using optical traps, we determine physicochemical triggering thresholds based on load and force direction. Strikingly, chemical thresholds in the absence of external load require orders of magnitude higher pMHC numbers than observed physiologically. In contrast, force applied in the shear direction (10 pN per TCR molecule) triggers T-cell Ca2+ flux with as few as two pMHC molecules at the interacting surface interface with rapid positional relaxation associated with similarly directed motor-dependent transport via 8-nm steps, behaviors inconsistent with serial engagement during initial TCR triggering. These synergistic directional forces generated during cell motility are essential for adaptive T-cell immunity against infectious pathogens and cancers.
The T-cell receptor (TCR) expressed on T lymphocytes of the adaptive immune system is a stout and squat (12-nm wide 8-nm tall) multisubunit surface complex with a ligand binding moiety that is an disulfide-linked heterodimer buttressed by the associated invariant CD3 subunits (1–3). The chains are each encoded by V and J gene segments and in the case of the , a D segment as well (4). The clone-specific TCR unique to each T lymphocyte endows mammals with the capacity to detect perturbations in host cellular function resulting from myriad infectious pathogens, physical damage (thermal, irradiation, etc.), or premalignant or malignant cellular transformations while averting strong self-reactivities that could induce autoimmunity (5).
Signaling is initiated through ligation of the clonotype by its cognate antigenic peptide bound to an MHC molecule (pMHC) and displayed on the surface of antigen-presenting cells (APCs) (6, 7). Ligation impacts disposition and function of the associated CD3 dimers (CD3, CD3, and CD3) as well as the transmembrane domains of the TCR heterodimer and CD3 subunits that interdigitate in the membrane to signal into the cytoplasm (8, 9). A cascade of intracellular events involving phosphorylation of immunoreceptor tyrosine-based activation motifs (ITAMs) on the CD3 cytoplasmic domains with subsequent ZAP70 activation and downstream signaling follows (10, 11). Membrane-associated CD8 and CD4 coreceptors, marking cytotoxic T lymphocytes (CTLs) and helper T cells, respectively, function to bring the membrane-anchored Src family tyrosine kinase Lck to the TCR–pMHC complex for the initiation of ITAM phosphorylation. Signaling, in turn, leads to a transient rise of cytosolic Ca2+ and other biochemical events resulting in transcriptional activation, ultimately resulting in developmental decisions or effector functions (11–13).
A CTL must search for and destroy a target expressing a handful of “foreign” (i.e., aberrant) peptides of the relevant specificity displayed among a sea of self-pMHC molecules (100,000) on the surface of an altered host cell. Confounding immune surveillance is an apparent weak affinity of TCRs for pMHC, often with micromolar to undetectable affinity in solution, behaviors not reconciled with functional outcomes when examined by 2D affinity measurements (14). T lymphocytes do not use somatic hypermutation to strengthen TCR affinity as do B cells for generation of high-affinity antibodies (15). Nevertheless, T cells confronting “weakly” interacting ligands are able to sense and discriminate antigens with exquisite specificity, even detecting single-amino acid differences between two peptides bound to the same MHC allele product (16). A serial engagement process where a single (or low copy number) antigenic pMHC binds and unbinds multiple receptors (hundreds) on a T cell to amplify and sustain its activation has been proposed to generate signaling through fleeting encounters, including those important for antitumor immunity (17, 18). The original concept of serial engagement (or serial triggering) was based on reutilization of pMHC over a period of hours that has been recently redefined to encompass shorter cycles of TCR stimulation over a period of seconds (19).
Of note, however, T cells exert physical force on the TCR–pMHC complex at the T cell–APC interface through both internal and external processes that impact the above calculus. During immune surveillance, T cells scan their environment, crawling over tissue components in various organs and adopting elongated shapes typical of polarized cells (20). Surveillance-based motility generates tensile, shear, and compressive stresses over a wide range of forces (piconewton to nanonewton) (21–23). Internally, cytoskeletal machinery drives the relative movements of cell membranes, including rearrangement, transport, and organization of TCR complex constituents and other cell surface molecules (24–27). As a consequence, the immunological synapse (IS) is formed with TCRs surrounded by segregated clusters of other proteins (28). The transportation of TCRs to the IS center is caused by the centripetal or retrograde actin flow (29, 30).
Experiments directly testing the role of mechanical force in triggering TCRs were first provided by our own studies using an optically trapped bead to present pMHC to a T cell (31). Such measurements used an oscillating shear force with a 50-pN amplitude to trigger T-cell Ca2+ flux, with as few as 10 pMHCs per bead. Experiments with the same beads but force application normal to the cell surface did not lead to triggering. How T cells might use mechanical force and direction for triggering was conceptually proposed to involve nonlinear bonding kinetic mechanisms, including conformational change allostery and bond strengthening (32). This paradoxical extension of TCR–pMHC-bond lifetime under force, so-called catch bond behavior, was then observed experimentally (19, 33). The importance of force on TCR triggering has been confirmed in other work (34–40). However, during the initial T-cell surface contact, external (scanning) and internal (retrograde flow) force appear to operate in opposing directions (41). How these directional forces cooperate with each other and orchestrate the TCR triggering is still obscure.
Here, we actively present pMHC-bound beads to T cells, controlling the pMHC surface concentration, force magnitude, and direction using an optical trap. A chemical threshold is identified in the absence of trapping force that is well above expected physiological numbers required for triggering by foreign ligands. In contrast, under proper loading profiles, as few as two pMHC molecules are sufficient to trigger a T cell. Applying force in the shear direction more readily triggers than does application along the normal direction, highlighting the anisotropic feature of this mechanosensor. Rather than serial engagement, during receptor–pMHC ligation, active transport of beads with discrete 8-nm steps through an actomyosin-based mechanism is directly visualized. The regular stepping of this motility process is in direct conflict with features of abrupt unbinding, irregular snapback, and multiple binding signatures expected for serial engagement-based mechanisms, none of which are observed. Our measurements support a model where force-stabilized pMHC–TCR ligation of as few as two pMHC molecules under a load of 10–20 pN per complex is sufficient for a T-cell activation process associated in seconds with processive actomyosin-based displacement. These findings are relevant to establishing signatures of protective T cells arising from natural infection as well as vaccination efforts for eliciting protective CTL-based immunity and tumor immunotherapy.
Results
Antigen Presentation Assay to Mimic Cell Triggering Through Optically Trapped Beads.
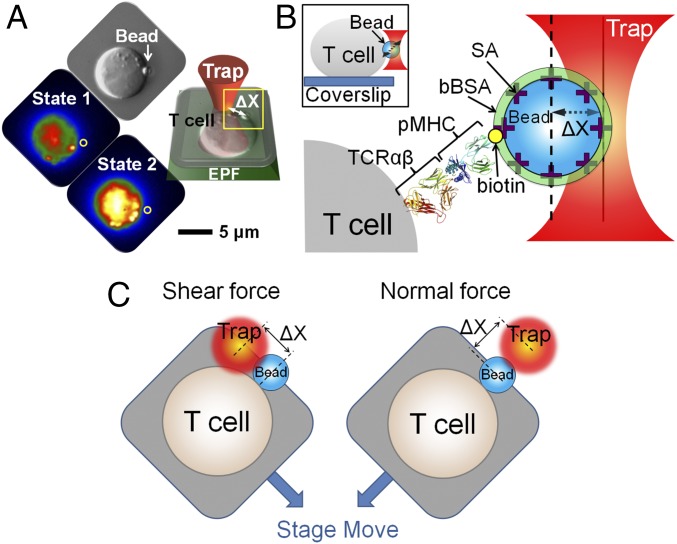
TCR triggering was investigated using optical tweezers combined with fluorescence visualization that permits piconewton step force application while simultaneously monitoring Ca2+ flux linked to T-cell triggering (31, 42, 43). Our study used individual T cells immobilized on the coverslip surface (Fig. 1A and SI Appendix, Fig. S1). Before immobilization, cells were preloaded with Quest-Rhod 4 as a Ca2+ indicator. Thus, Ca2+ flux, an early T-cell activation process, could be visualized by fluorescence enhancement. Under bright-field illumination, functionalized beads were held by the fixed trap, and the surface-immobilized T cell was gently repositioned using a piezo stage to facilitate bead–cell attachment. Binding was visualized directly and through displacement of the bead out of the trap indicated by changes in signal of our position sensing subsystem, which records the bead position at 3 kHz. After binding, the instrument was transitioned to low-light imaging to monitor fluorescence excited by a 532-nm laser with a low light-sensitive Andor Ixon camera. Images were recorded at 0.25 Hz for 10 min. Under optimal conditions, triggered cells exhibited fluorescence levels that steadily increased, whereas nontriggered cells showed a steady drop in fluorescence caused by photobleaching. Streptavidin-functionalized 1.09-m beads were coated with biotinylated pMHC (Fig. 1B). Methods of quantification of pMHC on the bead are detailed in SI Appendix, SI Materials and Methods and Fig. S2. Beads were subsequently saturated with biotin-labeled BSA (bBSA) to prevent nonspecific interaction. Vectoral piconewton force was generated by stepping the piezo stage to the desired distance to create a displacement X as shown in Fig. 1C. The trapped bead was calibrated before the attachment, and the trap force was calculated by the product of trap stiffness and X.
Fig. 1.
TCR triggering using an optical trap to apply vectoral piconewton forces over nanometer distances. (A) Mechanical force is applied using an optical trap. Individual T lymphocytes are immobilized on a coverslip surface. After a trapped ligand-coated polystyrene bead is guided into contact with a T cell, a vectoral one-step force is applied to the bead by moving the piezoelectric stage. X denotes displacement of the bead out of the trap center. The bead abutting the T-cell surface is shown in the DIC image indicated by a white arrow and the fluorescence image as a gray sphere and yellow circle. The optimal force along the T-cell surface triggers a rise in free Ca2+ shown in colorization as an increase in yellow intensity (state 2) compared with the initial state of the same T cell (state 1). EPF, epifluorescence. (B) Cartoon detailing the bead–cell contact interface as expanded from the yellow box in A. Optical traps are used for application of force to a streptavidin (SA)-coated polystyrene bead arrayed with specific biotinylated TCR ligand (pMHC; in this case VSV8/Kb) and then saturated with bBSA to prevent potential nonspecific streptavidin binding to cells. The trapped bead was calibrated before attachment, and the trap force was calculated as the product of trap stiffness and X. The molecular model depicts the TCR heterodimer from 1NFD and the pMHC from 1KPU. For simplicity, only the clonotypic heterodimer in the TCR complex is represented. (Inset) Side view of a bead-attached T cell. (C) Cartoon showing optimal shear and normal force directions of the bead relative to the cell.
Two different TCRs were used in our studies. The first is the N15 TCR specific for a vesicular stomatitis virus nuclear protein octapeptide (RGYVYQGL) bound to the H-2Kb molecule (VSV8/Kb). The second is the NP63 TCR specific for influenza A virus PR8 nucleoprotein nonapeptide (NP366; ASNENMETM) bound to the H-2Db molecule. These TCR epitopes are targets of protective adaptive T-cell immunity against the two viruses. Comparable results with these diverse specificities imply the broadness of the dynamic mechanosensing behaviors that we observe.
The Chemical Threshold for Triggering in the Absence of External Force Requires Numbers of pMHC Molecules Exceeding Physiological Levels.
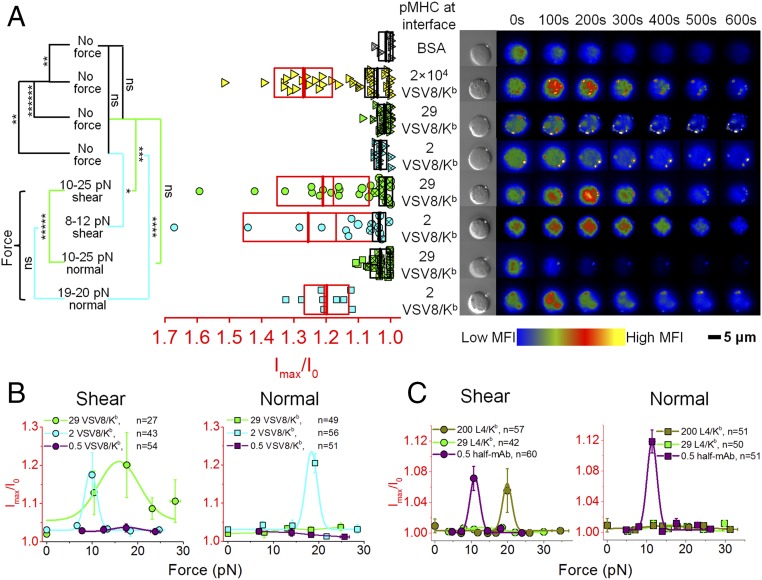
We first tested the sensitivity of triggering N15 T cells using beads displaying different VSV8/Kb copy numbers at the bead–cell interface in the absence of load, beginning with saturating ligand coverage and diluting until activation was no longer observed. Beads were trapped and gently placed on the cell to facilitate attachment. The trap was then immediately turned off. Bead position and fluorescence levels were monitored for 10 min. As seen in Fig. 2A and Movie S1, a saturated VSV8/Kb -coated bead with 2 × 104 molecules at the bead–cell interface generated robust TCR triggering (18 of 34 cells) (yellow triangles in Fig. 2A). With 200 molecules at the interface, triggering was reduced (4 of 11 cells) with minimal fluorescence increase (SI Appendix, Fig. S3B). However, beads with 29 VSV8/Kb at the interface, a pMHC density found physiologically on normal APCs (44), did not trigger (1 of 28 cells) (green triangles in Fig. 2A), and lower pMHC copies (2 VSV8/Kb) did not activate the T cells (0 of 10 cells) (cyan triangles in Fig. 2A). Measurements on T cells with other pMHC ligands were also tested, including the weak agonist L4 [vesicular stomatitis virus nuclear protein octapeptide (VSV8) with p4 V to L VSV8 mutation]/Kb and the unrelated Sendai virus SEV9/Kb complexes. With 200 L4/Kb or SEV9/Kb molecules at the interface, no triggering was observed (0 of 17 cells each) (SI Appendix, Fig. S3A). We also performed controls with BSA-only coated beads (gray triangles in Fig. 2A), with fluorescence signals that were used as background for the nontriggered cells without external trapping force. These control beads manifest no cell surface binding and failed to trigger T cells, despite repeated apposition.
Fig. 2.
Thresholds for TCR triggering are dependent on both pMHC ligand copy number and directional force. (A) Ca2+ flux was triggered in N15 T cells by using either a high VSV8/Kb copy number at the bead–cell interface (2) without external force or physiological numbers of VSV8/Kb (29 and 2) in conjunction with an optimal vectoral force. Ca2+ flux signal is indicated as the ratio of maximum fluorescence intensity (Imax) to the initial fluorescence intensity (I0) of the Ca2+-sensitive dye. Width of rectangles illustrates the SD. Mean value and median value are shown as thick lines and thin lines, respectively. On the Right, a rise in intracellular free Ca2+ is shown by colorization through a rise in red and yellow intensities at indicated times for representative cells. Boxes with red and black boundaries in the same column were statistically analyzed for triggered T cells (open symbols) and nontriggered T cells (symbols with ), respectively. P values refer to significance analysis obtained using one-way ANOVA for all of the data points. MFI, mean fluorescence intensity; ns, not significant. *P 0.05; **P 10−2; ***P 10−3; ****P 10−4; *****P 10−5; ******P 10−6. (B and C) Directional force vs. N15 T cell triggering of Ca2+ flux under different load and (B) VSV8/Kb, (C) L4/Kb, or (C) the anti-TCR V MR9.4 fragment copies at the interface. Curves represent fitting with Gaussian distribution. Error bars represent SEM. The duration of the increase in calcium for shear and normal directions for VSV8/Kb in B and half-mAb in C are 269 vs. 132 and 136 vs. 87 s, respectively.
External Force Enhances TCR Sensitivity to Achieve Triggering Thresholds with Limited Numbers of pMHC Molecules at the Interaction Interface.
To determine if external force can facilitate TCR triggering at lower pMHC copy number, a step force was applied by actuating the piezo stage to move the cell relative to the fixed trap, yielding a desired displacement and force. Measurements on T cells with 29 VSV8/Kb at the bead–cell interface showed robust triggering (16 of 26 cells) (green circles in Fig. 2B) under conditions detailed below using specified directional forces ranging from 10 to 25 pN compared with those without external load (Fig. 2 A and B, SI Appendix, Fig. S4, and Movie S2). Likewise, bead–cell interfaces containing 2 VSV8/Kb molecules yielded 8 of 15 triggered cells at triggering force of 8–12 pN (cyan circles in Fig. 2B). In contrast, with an average 0.5 pMHC copy number per interface, where we expect no more than one TCR–pMHC interaction in the bead–cell-contacting interface (33), no triggering was observed (Fig. 2B, purple circles and Movie S2). Plots of fluorescence increase vs. force show a narrow distribution of triggering forces at the 2 VSV8/Kb pMHC copy numbers dependent on force application as described below (Fig. 2B). L4/Kb shows triggering at 200 but not at 29 copies per interface. To determine whether the TCR can be triggered with a single sustained interaction, we used one-half anti-TCR V MR9.4 mAb fragment at 0.5 molecules per interface, yielding a robust 12 of 18 cell triggering responses (at 8- to 12-pN shear force) (Fig. 2C and SI Appendix, Fig. S3A, purple).
T-Cell Triggering Is Anisotropic, Preferring Shear over Normal Directionally Applied Loads.
We directly measured the directional dependence of the TCR–pMHC interaction with similar step force and identical pMHC numbers at the cell–bead interface. Whereas step displacements in the shear direction exhibited triggering with as few as two pMHCs at the interface and 10 pN (cyan circles in Fig. 2 A and B), forces applied in the normal direction required 18 pN (cyan squares in Fig. 2 A and B) to trigger (Fig. 2 A and B and Movie S3). Ca2+ flux durations triggered by 29 and 2 VSV8/Kb molecules at the interface along the shear direction are 250 ± 92 s (16 cells; ±SD) and 288 ± 119 s (8 cells), respectively, whereas that stimulated by 2 VSV8/Kb at the interface with optimal normal force is only 132 ± 43 s (9 cells). Interestingly, beads displaying 29 pMHCs at the interface failed to trigger Ca2+ flux with any force application along the normal direction (green squares in Fig. 2 A and B) but triggered along the shear direction (green circles in Fig. 2 A and B) using a range of forces examined (Fig. 2 A and B and SI Appendix, Fig. S4). As with shear (Fig. 2B, purple circles), single molecule-level pMHC (0.5 VSV8/Kb molecules at the interface) failed to trigger TCR with force applied normal to the cell surface (Fig. 2B, purple squares and Movie S3). In contrast, triggering with the very weak agonist L4/Kb was only achieved along the shear direction (Fig. 2C). Triggering with the one-half antibody was possible in both shear and normal directions.
T-Cell Triggering Is Associated with Rapid Relaxation and Large Displacements.
Our measurements in the absence of force simultaneously monitor bead trajectory and Ca2+ flux caused by T-cell triggering. In SI Appendix, Fig. S5A, cells with pMHC-saturated beads (2 VSV8/Kb at the interface) that triggered without force exhibited relatively large displacements (1,000 nm in 200 s) and rapid positional relaxation (peak velocities of 30 nm/s). Note that, during these measurements, the trap is off, only actuated to facilitate the initial bead to cell contact; thus, bead motions originate from the cell. Change in position is contemporaneous with Ca2+ rise, with peak flux occurring at the end of positional relaxation and within minutes of the initial bead–cell association. Statistically, unloaded pMHC-saturated beads with limited displacement (below 625 nm) typically did not trigger (SI Appendix, Fig. S5B). Cells which have fewer pMHC molecules at the interface (29 and 2 VSV8/Kb) show little responsiveness, with no correlation between displacement and Ca2+ flux, even for those showing large displacement.
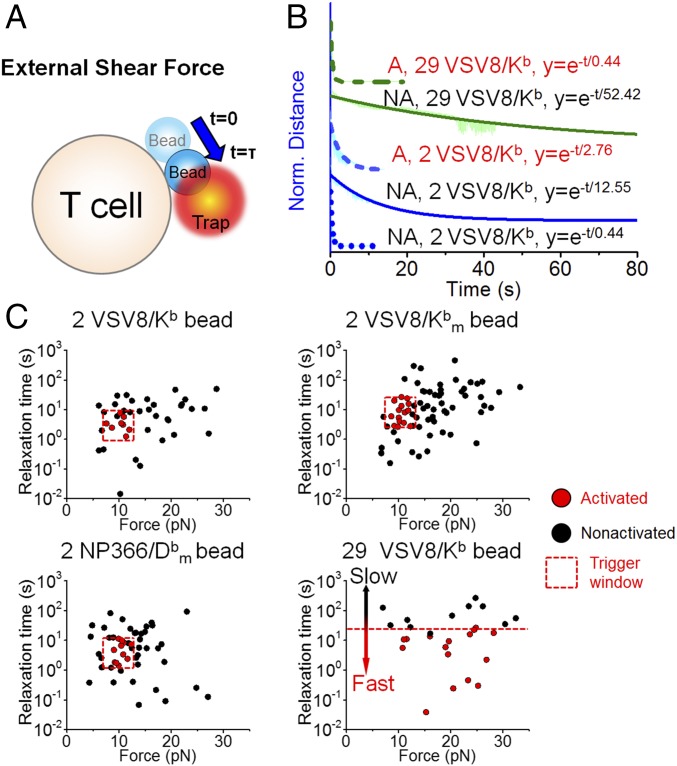
In the presence of shear force, cells exhibit complete positional relaxation as shown in Fig. 3A. Here, force increases as a step and steadily decreases as the bead moves back to the trap center. Triggered and nontriggered traces were pooled, normalized, and fit to a single exponential decay to extract representative time constants for the average force–displacement transients (Fig. 3B) (45). Force-triggered cells exhibit rapid positional relaxation (Fig. 3B, dashed lines) followed by a rise in Ca2+ signal (SI Appendix, Fig. S6, red lines). Traces were also fit individually to map the relation between trigger state, relaxation time, and force (Fig. 3C). The detailed fitting procedures are illustrated in SI Appendix, Fig. S7A. Our results illustrate that, for cells stimulated with 29 VSV8/Kb at the interface, triggered cells relaxed more quickly than nontriggered cells: 0.44 4.65 10−4 s compared with 52.42 8.82 10−3 s, respectively, for all measured forces. For T cells stimulated with cognate ligands at identical interfacial copy number (two pMHCs), triggered cells can be represented by a window centered around 10 pN exhibiting a relaxation width of 10 s compared with nontriggered cells neighboring this window. The subset of particularly fast relaxing, nontriggered cells (average 0.44 4.89 10−4 s) remained bound but was effectively only under load for a fraction of a second (Fig. 3B and SI Appendix, Fig. S7C). A few beads with single molecule-level array (0.5 VSV8/Kb) led to abrupt unbinding, where the bead snapped back to the trap center within 5 ms (n = 19) (SI Appendix, Fig. S7 B and C) and failed to trigger. Fig. 3C shows clearly that the triggered N15 T cells were centered in the same window irrespective of whether pMHC was competent to bind the CD8 coreceptor (VSV8/Kb) or mutated to abrogate coreceptor binding to the Kb 3 domain (VSV8/). The NP63 TCR-expressing T cells also yielded the same profile using NP366/. Results with triggered N15 and NP63 T cells using VSV8/ and NP336/ beads were pooled and illustrated in SI Appendix, Fig. S8 A and B. A possible triggering mechanism is illustrated in SI Appendix, Fig. S8C, highlighting that, because the pulling direction is shear, TCR–pMHC bonds at the back edge of the bead will sustain the load. TCR-triggered Ca2+ flux requires that the magnitude of pulling force on each single TCR–pMHC complex achieve a 10-pN physical threshold as well as an optimal relaxation rate. A greater force (i.e., 20 pN) is required for TCR triggering with 29 VSV8/Kb at the interface, because the load is distributed across multiple bonds.
Fig. 3.
Relationship between TCR triggering and external shear force relaxation. (A) Cartoon showing shear force relaxation (blue arrow) with characteristic time constant after a step displacement of the trap relative to the bead. (B) External force relaxation for different densities of VSV8/Kb bead on the cell surface interface with fits to a single-exponential decay profile. External forces were firstly normalized, shifted to the same time zero, and then averaged. Traces from top to bottom were averaged from 16 (olive dashed line), 11 (olive solid line), 8 (blue dashed line), 25 (blue solid line), and 10 (blue dotted line) separate experiments, respectively. The nonactivated (NA) blue dotted trace represents a rarely seen fast fluid-like relaxation. A, activated. (C) Scatter plots showing relationship among relaxation rate, external force magnitude, and TCR triggering at different pMHC densities for WT VSV8/Kb as well as the mutants VSV8/ and NP366/. For 29 VSV8/Kb, “Activated” is defined as an Imax/I0 higher than 1.04 (the upper boundary of 29 VSV8/Kb box without external force in Fig. 2A). For 2 VSV8/Kb, an Imax/I0 higher than 1.05 (upper boundary of 2 VSV8/Kb box without external force in Fig. 2A) is defined as activated. For 2 VSV8/ and NP366/, Imax/I0 higher than 1.00 (upper boundary of other forces box in SI Appendix, Fig. S8 A and B) is defined as activated. Note that the trigger window depicted by dashed lines is the same in VSV8 and NP366/. In all cases with two pMHCs at interface, the window spans a force range from 8 to 12 pN.
For pulls in the normal direction (SI Appendix, Fig. S9A), relaxation includes an initial exponential decay and then, a terminal position, where unlike pulling in shear, complete relaxation to the trap center is not achieved (SI Appendix, Fig. S9B). The initial relaxation portion exhibited exponential time constants (0.1–10 s) similar to shear pulls above. However, unlike pulls in the shear direction, large displacement relaxations in the normal direction were not possible. In SI Appendix, Fig. S9C, with 2 VSV8/Kb at the interface, the T cell can be triggered and exhibited partial relaxation (n = 9). Bead–cell interface with higher density (29 VSV8/Kb) did not trigger the TCR but exhibited frustrated motion caused by active intracellular processes with a sustained displacement similar to 2 VSV8/Kb. Single-molecule densities (0.5 VSV8/Kb) failed to trigger the TCR because of abrupt unbinding returning to the trap center within 5 ms (n = 16). A possible triggering mechanism along the normal direction is illustrated in SI Appendix, Fig. S9D. Force, shared evenly by the 2 VSV8/Kb molecules, requires 20 pN to reach the TCR triggering threshold for any single TCR. For 29 VSV8/Kb at the interface, however, force is distributed among many TCRs, where any single TCR is below the threshold force for triggering. These data and those in Fig. 3 also rationalize the distinctions among the vectoral pulling forces required to trigger T-cell activation in Fig. 2B. Similar optimal triggering windows were obtained for L4/Kb and one-half antibody shear triggering (SI Appendix, Fig. S10).
Regular Steps Linked to Actomyosin Machinery Rather than TCR Serial Engagement.
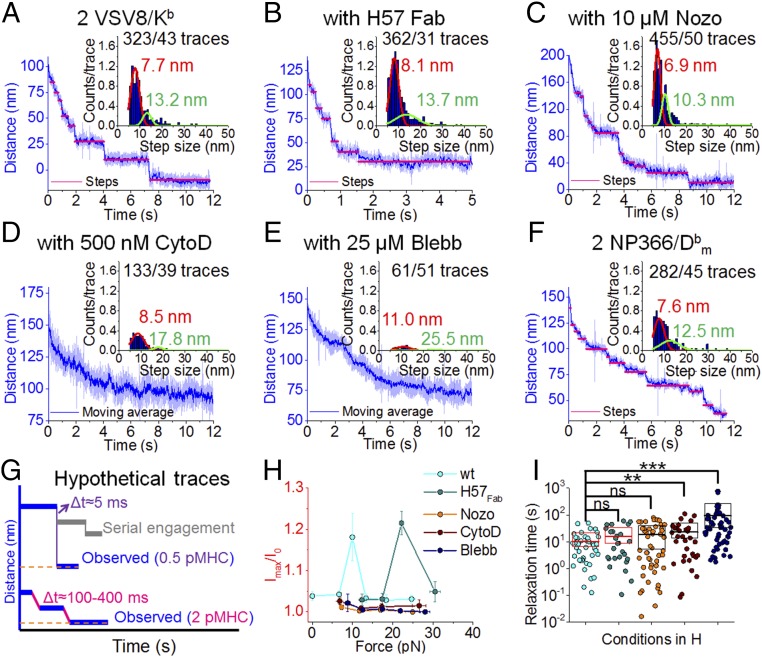
Given the prominence of T cell-induced bead motion on TCR–pMHC ligation, we studied these movements in greater detail. Bead motion reveals discrete steps after shear force application at threshold pMHC numbers (2 VSV8/Kb at the interface). The traces are characteristic of single-molecule motility of motor proteins on cytoskeletal filaments (46). Regular stepping occurs with average displacements and dwells of 7.7 nm for the primary peak (Fig. 4A). Additional step analysis is found in SI Appendix, Fig. S11A. To rule out steps being caused by sequentially unbinding and rebinding of TCRs, we stabilized the pMHC–TCR interaction with H57 Fab, with a single-TCR binding site on the FG loop of the C domain that leads to a 10-fold increase in bond lifetime (33). H57 Fab-ligated T cells exhibited similar step size and dwell times of 8.1 nm (Fig. 4B). To unveil the origin of the steps, we challenged the cells with cytoskeletal disrupting drugs, including Nocodazole (Nozo), Cytochalasin D (CytoD), and Blebbistatin (Blebb). Drug studies were conducted using concentrations that significantly disrupt the cytoskeleton but avoid cell toxicities that alter overall cell shape or ability to obtain measurements. Step probability and peak location were unaffected by 10 M Nozo, a microtubule depolymerizer (Fig. 4C) (47). Nozo blocks Lck phosphorylation (48) but enhances myosin regulatory light-chain phosphorylation, which induces activation of the motor (49). However, both CytoD, an actin depolymerizer at 500 nM, and Blebb, a myosin inhibitor at 25 M, showed clear reduction in step probability (Fig. 4 D and E and SI Appendix, Fig. S11B). Normally, we can extract 7.5 steps per trace, which were reduced to 3.4 and 1.2 steps for CytoD and Blebb, respectively. Blebb also shifted the step size distribution to 11.0 nm and reduced bead velocity returning to the trap center. Regular 8-nm steps were also visualized for another TCR–pMHC pair (NP63 TCR–NP366/) (Fig. 4F). Purified, naïve N15 transgenic CD8+ T cells (SI Appendix, Fig. S12A) exhibited step-linked triggering (SI Appendix, Fig. S12 B–D). All cells challenged with drugs did not trigger as expected given the significant role of the cytoskeleton in TCR triggering (Fig. 4H, SI Appendix, Fig. S13, and Movie S4) (50). Although H57 typically inhibits triggering (31), at high forces (23 pN), some cells showed an increase in Ca2+-mediated fluorescence but with reduced Ca2+ flux: 85 29 s (8 of 20) above 22 pN. Force relaxation time also shows significant difference when cells are treated with CytoD and Blebb (Fig. 4I).
Fig. 4.
High-avidity TCR triggering is a dynamic process requiring actomyosin rearrangement rather than serial engagement. (A–F) Typical stepwise traces for N15 T cells treated with H57 Fab, Nocodazole, Cytochalasin D, or Blebbistatin with 11- to 13-pN shear force and 2 VSV8/Kb at the interface. Stepping traces are common for NP63 T cells at 11 pN with two NP366/. Insets show relative overall step histograms fit to the sum of two Gaussian distributions with indicated numbers of steps. (G) Hypothetical trace under load of serial engagement (gray) compared with the empirically observed completely ruptured at 0.5 VSV8/Kb and stepwise trace at 2 VSV8/Kb conditions. t between adjacent dwells are averaged from 2 VSV8/Kb at the interface (steps) for the observed regular stepwise trace under shear force. For the hypothetical serial engagement trace, an upper bound of t = 5 ms is obtained from 0.5 VSV8/Kb beads (single rupture) measurements, where unbinding and complete snapback to the trap center are observed. Serial engagement, which is not observed, would either show a rebind to a random location or alternatively, return to baseline within this 5 ms. (H) H57 Fab or drug-treated N15TCR triggering under load. Mean fluorescence intensities of T cells triggering by external forces were monitored using 2 VSV8/Kb stimulation through a shear direction. Error bars represent SEM. (I) Comparison of relaxation times resulting from the indicated treatments. Box height illustrates SD. Red and black boxes illustrate the triggered and nontriggered conditions, respectively. Mean values are shown as thick lines. P values were determined using one-way ANOVA. ns, not significant. **P 10−2; ***P 10−3.
The concept of serial engagement was suggested, because very few pMHC ligands on an APC induced loss of hundreds of copies of surface TCRs on the relevant antigen-specific interacting T cell over a timeframe of many hours (17). However, rather than a direct serial single-pMHC ligation of many TCRs at initial TCR triggering, establishment of a network of cytoskeletal interactions might indirectly modulate TCR distribution and/or surface expression. We tested this possibility by developing a two-bead assay, with both beads opposed to a T cell in the absence of external force. One bead is coated with a low copy number of nonactivating anti-CD3 mAb 17A2 (31) to monitor pMHC-unligated TCRs on the cell, while a second bead of different size arrays high density-specific pMHC. As shown in Fig. 5 and Movie S5, only in activated T cells (i.e., fluxing Ca2+) does the 17A2 bead move a greater distance to become more proximal to the pMHC bead. This phenomenon may relate to activation of motors in the vicinity of the motor ligated to the triggered (Fig. 5). The convergence of TCRs is consistent with initiation of IS formation. Hypothetically, serial engagement in the early timeframe (seconds to minutes) would exhibit random dwell displacements caused by sequential bond rupture and rebinding with step time intervals no greater than 5 ms (SI Appendix, Figs. S7C, S9B, and S10B), the time measured for a ruptured bead to completely snap back to the trap center, whereas we observed a dwell time between steps ranging from 100- to 400-ms intervals (Fig. 4G). SI Appendix, Fig. S1F shows functional avidity plots of N15 for VSV8 and L4 as well as NP63 for NP366 depicting IL-2 production in response to graded peptide stimulation. Given that the functional avidities differ by 10,000-fold among these receptor–ligand pairs and no pattern of 5 ms t was observed, the phenomenon of serial engagement is not occurring in this timeframe.
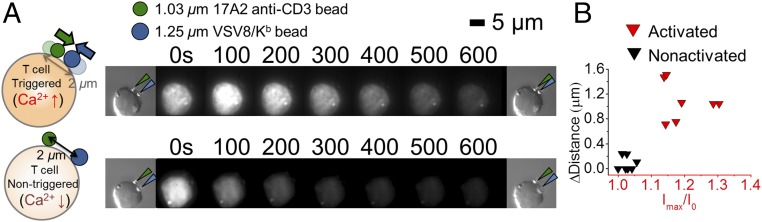
Fig. 5.
pMHC-triggered T cells cluster ligand-engaged as well as ligand-unengaged TCRs. (A) During triggering, more N15 TCRs (binding with a 1.03-m bead; olive arrow) translocate to the interface, where the pMHC-specific triggering initiates (1.25-m bead; n = 7 cells; dark blue arrow). By contrast, nonactivated cells manifest limited TCR motion (n = 10 cells). (B) Changes of distances between the two beads for activated and nonactivated T cells. The two clusters are significantly different at P 1 10−6.
TCR Triggering Impacted by Directional Tangential Force.
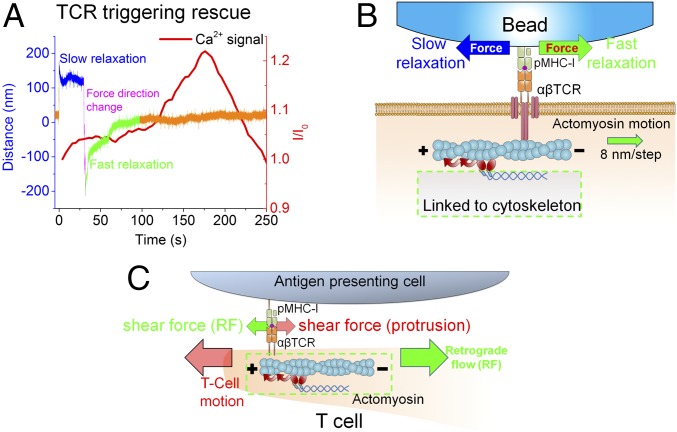
Failed TCR triggering associated with slow force relaxation could be rescued by rapidly reversing the pulling direction, resulting in a fast relaxation response. Representative studies were performed on beads presenting 29 VSV8/Kb at the interface with N15 T cells (Fig. 6A and SI Appendix, Fig. S14). Since TCRs are reported to directly link to actin filaments via CD3 chains (51), the fast- vs. slow-force relaxation could be explained by the assistance or hindrance, respectively, of the optical trap force as shown in Fig. 6B. Concordance of force direction resulting from actin motion and optical trap as applied to the TCR could facilitate release of the force-dependent structural transition of the TCR heterodimer–pMHC complex (33, 52). In contrast, the structural extension would persist in the case of a pMHC-arrayed bead, slowly relaxing back to trap center. This behavior is consistent with that of a T cell performing immune surveillance on an APC surface; force loaded on the TCR through one direction [e.g., cell moving/spreading direction or microvilli protrusion (53–55)] may be quickly released by the motion along the other direction (e.g., actin retrograde flow), as shown in Fig. 6C (41). Although N15 T cell-triggering rescue is hard to actuate for 2 VSV8/Kb molecules at the interface, hallmarks distinguishing fast from slow relaxation are evident when the position of the VSV8/Kb-coated bead is close to the trap center (SI Appendix, Fig. S15). There, bead traces exhibiting fast relaxation smoothly return to the trap center (SI Appendix, Fig. S15 A and B). However, slow trajectories exhibit fluctuations and even step against the trap at low displacements (SI Appendix, Fig. S15C). Collectively, these results reveal a clear force vector directionality linked to successful TCR triggering by pMHC at the single-molecule level.
Fig. 6.
External force in the direction of retrograde flow synergizes with actomyosin motion to facilitate TCR triggering. (A) Preference in external pulling force direction revealed by optical trap analysis. Failed TCR triggering by 29 VSV8/Kb beads after a slow relaxation (blue) lacking Ca2+ flux (red curve) can be rescued by quickly switching pulling force direction (magenta). This directional change induces fast relaxation (green) and attendant Ca2+ flux (red curve) without altering pulling force magnitude. The portion of the trace in orange represents the position of the trap center. (B) Possible actomyosin motion when rapid trapping force relaxation initiates along the tangential direction. Polarization of F-actin is indicated by plus and minus marks. Relative motion of F-actin is indicated by the green arrow. Different external force directions are indicated by blue and green arrows, which correspond to slow and fast relaxation, respectively. The actomyosin motion will facilitate force relaxation and steps when pulled in the green direction generated by myosin II pulling (gradient red arrows). In contrast, pulling in the blue direction will exhibit a tug of war that results in a slow relaxation, such that the TCR cannot be triggered, despite application of identical force magnitude. (C) Cartoon of synergistic shear forces-facilitated TCR triggering during T cell–APC contact; CD3 components were omitted for clarity. A T cell scans an APC surface in search of specific pMHC. Shear force could be generated either by cell motion (as drawn) or actin retrograde flow (RF, green arrow). Scanning force (red arrow) loaded on the TCR caused by T-cell motion (large red arrow) may be quickly released by the force generated by local actomyosin stepping (red curved arrows). Synergy between these two forces may foster TCR molecular recoil to facilitate T-cell activation and recruit more TCRs to the uropod. Note that the actin cortex is a much more complex array of actin and myosin filaments but simplified here for illustration.
Discussion
T cells executing surveillance motions orchestrate a complex process that operates on many length scales. The cell’s leading edge, microvilli, uropod, and trailing elements exhibit extensions and retractions as adhesions are formed and broken, exerting nanonewton forces to deform the underlying substratum (56). Within the cell, actin structures cycle through states of polymerization, depolymerization, and flow mediated by motors and actin binding proteins that couple filaments, sever filaments, generate branches, cap ends, and facilitate depolymerization (reviewed in ref. 57). T cells cluster their TCRs in the uropod, which is central to the cell and tied to actin structure undergoing retrograde flow (58, 59). T cells exhibit frequent periods of persistent motion but with intermittent stops and turns. The underlying adhesions result in pulling and dragging motions, where shear forces similar in magnitude and direction to those in our experiments can be generated (Fig. 6C). Our data show that the native environment of the TCR mechanosensor exploits direct coupling to the cell motility machinery for its performance. Cell motions provide the energy to activate the 10-nm 10-pN conformational change, where the equivalent of 100 pNnm work is done on the system (33).
At physiological pMHC numbers, triggering requires external load (Fig. 2 A and B). We observe a triggering preference for shear vs. normal-directed forces as occur when T cells perform their scanning functions (Fig. 3 and SI Appendix, Figs. S4 and S9) (59, 60). Stresses were applied experimentally via loading through our optical trap system, delivering, on average, 10 pN per TCR–pMHC interaction. The anisotropic response can be explained, in part, by geometrical factors. TCR triggering along the normal direction shares the load equally among each interfacial TCR–pMHC complex (SI Appendix, Fig. S9D) and thus, requires a greater overall pulling force for any one of them to breach the 10-pN conformational change threshold (33). On the contrary, shear motion distributes the force unevenly at the bead–cell interface, concentrating it where fewer TCRs engage (SI Appendix, Fig. S8C). Thus, for shear pulls, triggering is seen with both 29 and 2 pMHC interfacial molecules.
Without load, the TCR can be triggered when 2 104 VSV8/Kb molecules are present at the bead–cell interface (Fig. 2A). We expect that the high densities provide a stabilizing interface to create multiple points against which internal cytoskeletal forces can pull to activate TCR–pMHC mechanosensors (40, 61). Consistent with this notion, triggered cells are associated with large bead displacements along the shear direction (SI Appendix, Fig. S5). Such a high-density array is relevant to exogenously loaded APCs under in vitro conditions but rarely, if at all, to naturally processed viral antigens (62).
In addition to force, direction impacts coupling of the mechanosensor to the underlying actin machinery. Shear pulls permit motility of TCRs along actomyosin structures, even at low pMHC copy number (2 pMHC) at the interface (Fig. 4 A and F). Pulling in the normal direction directs the TCR away from membrane-associated actin. Our results are consistent with this anisotropic phenomenon, as a much stronger Ca2+ flux is seen with shear over normal pulls (SI Appendix, Figs. S6 and S9C).
Bead trajectories for cells that trigger reveal processive active transport in the direction of applied shear force. Although visually, the bead positional relaxation appears smooth, the high spatiotemporal resolution of our instrument allows discrete stepping of the TCR coupling to the actin machinery to be detected. Although the step size (8 nm) suggests microtubule-based transport, drug inhibition studies and stall force (3.8 pN) measured here (Fig. 4 A and F and SI Appendix, Fig. S15) are consistent with myosin-based transport (and steps sizes) on actin filaments (63–65). We cannot formally rule out that the transients originate from bulk contraction of actomyosin networks, release of actin cross-linkers or other attachments, or encountering of other obstacles in the cell. Notwithstanding, SI Appendix, Fig. S15 shows motion against the trap force, revealing that active transport contributes to bead behavior after an activating manipulation.
Unlike the rapid and relatively long-distance bead motion observed in triggered cells, bead trajectories of most nontriggered cells show a slow return to the trap center (Fig. 3B and SI Appendix, Fig. S6). The slow relaxation further suggests that a strong linkage connects the TCR to cytoskeleton. In contrast, a fluid-like microenvironment, with TCR uncoupled to the underlying actin structure, results in fast bead relaxation to zero displacement as the large trap force easily overcomes the relatively small drag force of the TCR complex within the membrane (Fig. 3B, dotted blue line). Fluid-like relaxation is directly seen in some traces, which exhibit distinctly faster timescales relative to observed active motility (Fig. 3B and SI Appendix, Fig. S7C). In fact, while naïve cells that triggered showed stepping, those that failed to trigger did not relax to the trap center (SI Appendix, Fig. S12D). Moreover, rapid fluid-like relaxation is not observed. This is consistent with the greater densities of TCR and associated actomyosin on the surface of the 4-m-diameter naïve cell.
Direct coupling to underlying cytoskeleton is a desirable feature underpinning our mechanosensor model, as TCR activation requires a sustained load. The direct connection to actin (i.e., a load-sustaining pathway) is likely through a single active motor, because discrete steps are observed. If multiple motors were transporting the TCR, then that discreteness would be blurred, and the single-molecule nature of the stepping would be lost because of averaging over an ensemble of motors. Our rescue experiments (Fig. 6A) also support the model that motility is linked to actomyosin and a single or small number of motors and only permitted in a direction aligned with the polarity of the underlying actin cytoskeleton. Triggered T cells manifest actomyosin motion in the direction of force application; thus, load is in a direction that assists motility (SI Appendix, Fig. S15A). Force in the opposite direction works against motility and opposes transport, setting up a tug of war between the trap and motor, leading to a stall. Myosin II stall forces have been reported in the range of 3.4 0.9 pN, consistent with lack of motion against the 10- to 20-pN trap forces applied here (SI Appendix, Fig. S15C) (65). If a handful of motors was engaged and activated, collectively, they would likely overcome the opposing trap force and displace the bead. Trajectories of nontriggered cells under load exhibit small amplitude displacements above the thermal noise distribution of the trapped bead, likely driven by TCR-linked actomyosin machinery.
Given that the TCR complex has a wide base, optical trap shear load applied parallel to actin structures, analogous to forces at work during immune surveillance motion, will torque the mechanosensor apparatus. The pushing and pulling on signaling structures drive T-cell activation. Although we observe some bead motion at low force, TCR activation correlates with activation of cytoskeleton motility (40). A number of mechanisms may drive the latter, including phosphorylation, Ca2+ increase, and mechanically triggering activation. Motors within the neighboring microenvironment may be able to receive these signals and activate their associated TCRs to amplify the signaling cascade (66). In two-bead experiments, a pMHC array-saturated bead was used to trigger the cell, while a second bead coated with a nonstimulatory anti-CD3 mAb (31) showed motion toward the triggered bead. This motion was not observed in cases where the pMHC-coated bead failed to trigger the cell, suggesting that the triggering event activates neighboring motors. Thus, during T-cell activation, the neighboring, nonengaged TCRs will accumulated at the contact site (67) by direct active transportation of F-actin–coupled TCRs and assembly of initial noncoupled TCRs to F-actin through ERM (ezrin/radixin/moesin) proteins (68).
In contrast, without direct connection to the cytoskeleton to sustain load and drive force generation, the TCR–pMHC bond simply unbinds or displaces within the fluid-like membrane, never achieving critical load thresholds required for mechanosensor-based activation, including bond strengthening, conformational change, and other push and pull motions (33, 52). A fluid-like microenvironment does not have the densely organized actin machinery of a motile cell, and thus, second wave signaling after TCR–pMHC triggering is muted. Similarly, load in the normal direction pulls the TCR away from membrane-associated actin. We underscore that mechanosensor activation requires a physical anchor, namely the underlying actin cytoskeleton. Fortunately, our pulling geometry capitalizes on cell surface attachment to the coverslip, where an organized actin structure is created (Fig. 1B, Inset) (69), similar to that of T cells scanning a cellular substratum. Beads are placed on the side of the cell adjacent to this structure, where TCRs are engaged with actin machinery.
Biomembrane force probe analysis has been used to investigate early T-cell activation events (19) by applying load through pMHC to the distal pole of the T cell, typically opposite the holding micropipette location, where organized actin structure is sparse or lacking (69). Repeat pulls are required to activate individual T cells with force direction normal to the cell surface (which we have shown here is less efficient than shear force). Ligated TCRs likely form a locus to organize actin machinery, permitting actomyosin-mediated motility and neighboring TCR signaling. Because signal buildup independent of those originating from a single TCR contributes to T-cell activation, the process is empirically conceptualized as cumulative bond lifetime (19).
The threshold studies on three different systems show a very similar window for triggering requirements centered at 8–12 pN (Fig. 3C), force ranges and relaxation times consistent with myosin stepping. Forces below this region do not permit catch bond formation necessary to create an energized TCR–pMHC bond (19, 33). On the other hand, higher force regimes also fail to trigger. Although we believe that these TCR–pMHC molecular interactions are capable of supporting conformational change, they may either rapidly rupture or not allow repetitive hopping back and forth between conformational states seen in our single-molecule experiments (52). Actomyosin-based transport acts as a force regulator, maintaining the TCR–pMHC system within this window optimal for conformational transitioning. Oscillating conformational changes in single TCRs likely foster signal transmission by altering membrane lipids, inducing transmembrane segment conformational change, or exposing interfacial cytoplasmic tail segments for downstream modifications (9). Our triggering of single cells is quite robust, with 53.3% of T cells triggered by the lowest pMHC density studied here using optimally applied force. This frequency is remarkable, assuming a 50:50 chance of applying the correct pulling direction relative to actin polarity.
Supported lipid bilayer (SLB) experiments investigating TCR activation are performed at high pMHC densities, where the T cell is able to elicit signals without an apparent external force and in geometries initially devoid of organized actin (70). T cells are then placed on the bilayer surface, establishing fluid-like areas in which TCRs–pMHC bonds move freely flanked by rigid supports, where their motion is blocked. Force across the TCR can only be sustained against those supports or within internal cellular actomyosin structures. The triggering chemical thresholds are very high for SLB experiments, 0.2 pMHC per 1 m2, which corresponds to 16 pMHC beneath the cell (28). At least four pMHCs within a single TCR cluster spanning 400 400 nm2 and containing 18–100 TCRs are sufficient to induce the Ca2+ flux (71). TCR–pMHC clusters form and begin to organize actin machinery, eliciting transport and IS formation with distal, peripheral, and central supramolecular activation clusters. Triggering is not immediate but requires sufficient ligand density and time (minutes) to generate motions and forces necessary for clustering and triggering. A mature IS contains all of the features of our mechanosensor-based activation, including shear motions and the ability to generate force across a TCR–pMHC bond, processive motility, and activation and transport of neighboring TCRs. These IS structures are reflective of uropod actin organization for mediating rapid Ca2+ flux and motor activation featured by our experiments.
Earlier studies have suggested that only a single or very few pMHC molecules per APC/target cell are required to stimulate T-cell activation (72–74). Although our lowest pMHC densities capable of triggering cells involve two pMHC molecules at the interface, the bead trajectories with nanometer-scale resolution suggest that a single interaction is sustaining the load. Perhaps the second molecule serves to stabilize the interaction along a line, defined by two points. This finding is consistent with adhesion activation studies requiring two ligands in related systems (75). While our assay geometry recreates many of the elements found in cells undergoing surveillance, it lacks APC adhesion molecules, such as CD58/48, that are known to be important for cognate recognition by pairing with CD2 on the T cell to enforce the relevant intermembrane spacing between cells forming conjugates (76). Thus, in the context of a T-cell–target cell or T-cell–APC conjugate, one TCR–pMHC bond is probably sufficient to trigger. Triggering with single molecules of the MR9.4 mAb fragment supports this notion.
Serial engagement is a leading model for T-cell activation based on a concept of sequential binding and unbinding of a single pMHC molecule to multiple TCRs (17). The model rests on evidence that many TCRs on a single T cell are down-regulated in the presence of small numbers of pMHC molecules. Serial engagement has morphed over years to incorporate arguments germane to 2D vs. 3D apparent binding kinetics (77), rationalizing the requirements for optimal TCR affinity to avoid interactions that are too strong or too weak to achieve repeat binding and sustained T-cell signaling (18). Liu et al. (19) suggested that serial engagement is occurring through early and rapidly accumulated bond lifetimes.
Multiple observations made here are in direct conflict with serial engagement as being critical for initial TCR activation over seconds to minutes as opposed to hours. First, we observe immediate and direct stepping in trajectories of beads triggering cells. These findings indicate a rapid, sustained, systematic, and strong connection to the actomyosin-based machinery (Fig. 4A). Second, in the presence of H57 Fab, which extends the N15 TCR-VSV8/Kb bond lifetime 10-fold, where any unbinding probability is further reduced (33), we observe a similar stepping signature, suggesting that TCR–pMHC maintains a single bond throughout the trajectory (Fig. 4B). Third, given known estimates for 2D densities of pMHC and TCRs at the bead–cell interface, the probability in our system of pMHC rupturing and binding a second TCR is less than 1% (77, 78) at the single-molecule level (0.5 VSV8/Kb at interface). Thus, we would expect to see only a single binding event during the trace. In fact, for experiments where we observe unbinding, beads return to the trap center within 5 ms (SI Appendix, Fig. S7C), a time window extremely short with respect to requirements for serially engaging a second receptor. If serial engagement were occurring, we would observe a single short dwell followed by a single rupture and snap back to the center of the trap position (Fig. 4G). Rather, we observe regular discrete steps (Fig. 4A). This is also consistent with single-pMHC tracking experiments, showing that reentrapment of the same pMHC with multiple TCRs nearby is unlikely after it is disengaged from the TCR (79). Fourth, conditions expected to severely reduce serial engagement, such as pulling in the normal force direction where pMHC is moved away from the cell surface, reducing the probability for repeat binding, still allow triggering (SI Appendix, Fig. S9). Fifth, a wide array of ligand affinities was investigated here, where weaker binding L4 showed reduced triggering over VSV8. In contrast, sustained binding through a single one-half antibody was capable of triggering cells at the single-molecule level. Sixth, we can directly show clustering of pMHC-triggered and pMHC-untriggered TCRs in two-bead experiments (Fig. 5). Our two-bead measurements suggest that serial engagement is not required for initiation of IS formation in this timeframe.
The translational implications of our findings are significant. T-cell activation requires recognition of but a handful of specific pMHC ligands on a given target cell. This sensitive TCR signal transduction apparatus is dependent on external mechanical force resulting from cell movement and coordinated intracellular retrograde cortical actin flow similarly directed. A correlate is that foreign antigens derived from infectious pathogens or nonsynonymous mutations in tumor exons need not be displayed at more than a few copies on the target, because the TCR functions as an exquisitely sensitive anisotropic mechanosensor. Highly potent CTL for adoptive immunotherapy may be most effectively stimulated in vitro on relevant pMHC molecule arrays using piconewton shear forces to train and expand CTL for subsequent in vivo use. Chimeric antigen receptor T cells (CAR-Ts) consisting of an antibody antigen binding fragment of predefined specificity as an ectodomain linked to an intracellular T-cell activation domain originally proposed by Eshhar and coworkers in 1989 (80) have shown great clinical promise recently (81). Molecular features of TCR mechanobiology revealed here should be exploited to optimize the synthetic biology of CAR-T design to confer heightened potency as well as intrinsic regulation features.
Materials and Methods
Various external mechanical force applications using optical traps. A single pMHC-functionalized bead (d = 1.09 m) was trapped with a 1,064-nm laser (stiffness 0.06–0.13 pN/nm) and placed alongside a surface-bound T cell with an automated piezoelectric stage. The bead position relative to the trap was monitored so that contact could be achieved with minimal pushing force (<5 pN). After T cell–bead contact was verified, differential interference contrast (DIC) images were taken to calculate the contact area between bead and cell membrane. The contact area was calculated as the surface area of a zone of a spherical cap with its height, which was controlled during the course of the experiment by viewing the position signal along the normal direction. A shear or normal force was then applied to the bead by displacing the bead-attached cell relative to the fixed trap using fine motions of the piezo stage. The force on the bead was determined by measuring the trap stiffness before cell–bead contact by the equipartition method and by measuring the displacement of the bead from the trapping center via postprocessing of experiment videos. Distances between bead and trap center were sampled at 3 kHz, antialias-filtered at 1.5 kHz, and recorded for 10 min. Simultaneously, T cells were excited using a 532-nm laser to monitor Ca2+-sensitive Quest Rhod-4 fluorescence every 4 s right after force application. Assays were performed at 37 °C. All of these experiments were done using an automated custom-built optical trap designed for simultaneous force and fluorescence experiments at the single-molecule and single-cell scales (42). Additional details can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Sonia K. Brady for helpful discussions and Rebecca E. Hussey for technical assistance. We also thank the NIH Tetramer Core Facility at Emory University for the VSV8/H2-K𝑏-3A2 and NP336/H2-D𝑏-3A2 monomers. Flow cytometry experiments were performed in the Vanderbilt University Medical Center Flow Cytometry Shared Resource supported by the Vanderbilt-Ingram Cancer Center (NIH Grant CA68485) and the Vanderbilt Digestive Disease Research Center (NIH Grant DK058404). This work was supported by NIH Grants R01AI100643, R01AI37581, P01GM047467, and SU2C-AACR-DT13-14.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.L.D. is a guest editor invited by the Editorial Board.
See Commentary on page 10303.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1703559114/-/DCSupplemental.
References
- 1.Rossjohn J, et al. T cell antigen receptor recognition of antigen-presenting molecules. Annu Rev Immunol. 2015;33:169–200. doi: 10.1146/annurev-immunol-032414-112334. [DOI] [PubMed] [Google Scholar]
- 2.Rudolph MG, Stanfield RL, Wilson IA. How TCRs bind MHCs, peptides, and coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 3.Wang Jh, Reinherz EL. The structural basis of T-lineage immune recognition: TCR docking topologies, mechanotransduction, and co-receptor function. Immunol Rev. 2012;250:102–119. doi: 10.1111/j.1600-065X.2012.01161.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 5.Reinherz EL. TCR-mediated recognition: Relevance to tumor-antigen discovery and cancer immunotherapy. Cancer Immunol Res. 2015;3:305–312. doi: 10.1158/2326-6066.CIR-15-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roche PA, Cresswell P. Antigen processing and presentation mechanisms in myeloid cells. Microbiol Spectr. 2016;4:MCHD-0008-2015. doi: 10.1128/microbiolspec.MCHD-0008-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stern LJ, Wiley DC. Antigenic peptide binding by class I and class II histocompatibility proteins. Structure. 1994;2:245–251. doi: 10.1016/s0969-2126(00)00026-5. [DOI] [PubMed] [Google Scholar]
- 8.Brazin KN, et al. Constitutively oxidized CXXC motifs within the CD3 heterodimeric ectodomains of the T cell receptor complex enforce the conformation of juxtaposed segments. J Biol Chem. 2014;289:18880–18892. doi: 10.1074/jbc.M114.574996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu C, et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3 cytoplasmic tyrosine-based motif. Cell. 2008;135:702–713. doi: 10.1016/j.cell.2008.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty AK, Weiss A. Insights into the initiation of TCR signaling. Nat Immunol. 2014;15:798–807. doi: 10.1038/ni.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith-Garvin JE, Koretzky GA, Jordan MS. T cell activation. Annu Rev Immunol. 2009;27:591–619. doi: 10.1146/annurev.immunol.021908.132706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, et al. STIM1 gates the store-operated calcium channel ORAI1 in vitro. Nat Struct Mol Biol. 2010;17:112–116. doi: 10.1038/nsmb.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards L, Zarnitsyna V, Hood J, Evavold B, Zhu C. Insights into T cell recognition of antigen: Significance of two-dimensional kinetic parameters. Front Immunol. 2012;3:86. doi: 10.3389/fimmu.2012.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hwang JK, Alt FW, Yeap LS. Related mechanisms of antibody somatic hypermutation and class switch recombination. Microbiol Spectr. 2015;3:MDNA3-0037-2014. doi: 10.1128/microbiolspec.MDNA3-0037-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasada T, Ghendler Y, Wang Jh, Reinherz EL. Thymic selection is influenced by subtle structural variation involving the p4 residue of an MHC class I-bound peptide. Eur J Immunol. 2000;30:1281–1289. doi: 10.1002/(SICI)1521-4141(200005)30:5<1281::AID-IMMU1281>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 17.Valitutti S, Müller S, Cella M, Padovan E, Lanzavecchia A. Serial triggering of many T-cell receptors by a few peptide MHC complexes. Nature. 1995;375:148–151. doi: 10.1038/375148a0. [DOI] [PubMed] [Google Scholar]
- 18.Valitutti S. The serial engagement model 17 years after: From TCR triggering to immunotherapy. Front Immunol. 2012;3:272. doi: 10.3389/fimmu.2012.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu B, Chen W, Evavold BD, Zhu C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell. 2014;157:357–368. doi: 10.1016/j.cell.2014.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henrickson SE, et al. In vivo imaging of T cell priming. Sci Signal. 2008;1:pt2. doi: 10.1126/stke.112pt2. [DOI] [PubMed] [Google Scholar]
- 21.Ananthakrishnan R, Ehrlicher A. The forces behind cell movement. Int J Biol Sci. 2007;3:303–317. doi: 10.7150/ijbs.3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trepat X, et al. Physical forces during collective cell migration. Nat Phys. 2009;5:426–430. [Google Scholar]
- 24.Lanzavecchia A, Iezzi G, Viola A. From TCR engagement to T cell activation: A kinetic view of T cell behavior. Cell. 1999;96:1–4. doi: 10.1016/s0092-8674(00)80952-6. [DOI] [PubMed] [Google Scholar]
- 25.Ma Z, Sharp KA, Janmey PA, Finkel TH. Surface-anchored monomeric agonist pMHCs alone trigger TCR with high sensitivity. PLoS Biol. 2008;6:e43. doi: 10.1371/journal.pbio.0060043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208–1212. doi: 10.1126/science.1175862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wülfing C, Davis MM. A receptor/cytoskeletal movement triggered by costimulation during T cell activation. Science. 1998;282:2266–2269. doi: 10.1126/science.282.5397.2266. [DOI] [PubMed] [Google Scholar]
- 28.Grakoui A, et al. The immunological synapse: A molecular machine controlling T cell activation. Science. 1999;285:221–227. doi: 10.1126/science.285.5425.221. [DOI] [PubMed] [Google Scholar]
- 29.Yi J, Wu XS, Crites T, Hammer JA. Actin retrograde flow and actomyosin II arc contraction drive receptor cluster dynamics at the immunological synapse in jurkat T cells. Mol Biol Cell. 2012;23:834–852. doi: 10.1091/mbc.E11-08-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu Ch, Wu HJ, Kaizuka Y, Vale RD, Groves JT. Altered actin centripetal retrograde flow in physically restricted immunological synapses. PLoS One. 2010;5:e11878. doi: 10.1371/journal.pone.0011878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim ST, et al. The T cell receptor is an anisotropic mechanosensor. J Biol Chem. 2009;284:31028–31037. doi: 10.1074/jbc.M109.052712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim ST, et al. TCR mechanobiology: Torques and tunable structures linked to early T cell signaling. Front Immunol. 2012;3:76. doi: 10.3389/fimmu.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Das DK, et al. Force-dependent transition in the T-cell receptor -subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci USA. 2015;112:1517–1522. doi: 10.1073/pnas.1424829112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bashour KT, et al. CD28 and CD3 have complementary roles in T-cell traction forces. Proc Natl Acad Sci USA. 2014;111:2241–2246. doi: 10.1073/pnas.1315606111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basu R, et al. Cytotoxic T cells use mechanical force to potentiate target cell killing. Cell. 2016;165:100–110. doi: 10.1016/j.cell.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu KH, Butte MJ. T cell activation requires force generation. J Cell Biol. 2016;213:535–542. doi: 10.1083/jcb.201511053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hui KL, Balagopalan L, Samelson LE, Upadhyaya A. Cytoskeletal forces during signaling activation in jurkat T-cells. Mol Biol Cell. 2015;26:685–695. doi: 10.1091/mbc.E14-03-0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Husson J, Chemin K, Bohineust A, Hivroz C, Henry N. Force generation upon T cell receptor engagement. PLoS One. 2011;6:e19680. doi: 10.1371/journal.pone.0019680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li YC, et al. Cutting edge: Mechanical forces acting on T cells immobilized via the TCR complex can trigger TCR signaling. J Immunol. 2010;184:5959–5963. doi: 10.4049/jimmunol.0900775. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, et al. DNA-based nanoparticle tension sensors reveal that T-cell receptors transmit defined pN forces to their antigens for enhanced fidelity. Proc Natl Acad Sci USA. 2016;113:5610–5615. doi: 10.1073/pnas.1600163113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tabdanov E, et al. Micropatterning of TCR and LFA-1 ligands reveals complementary effects on cytoskeleton mechanics in T cells. Integr Biol (Camb) 2015;7:1272–1284. doi: 10.1039/c5ib00032g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brau RR, Tarsa PB, Ferrer JM, Lee P, Lang MJ. Interlaced optical force-fluorescence measurements for single molecule biophysics. Biophys J. 2006;91:1069–1077. doi: 10.1529/biophysj.106.082602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lang MJ, Fordyce PM, Engh AM, Neuman KC, Block SM. Simultaneous, coincident optical trapping and single-molecule fluorescence. Nat Methods. 2004;1:133–139. doi: 10.1038/nmeth714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bernardeau K, et al. Assessment of CD8 involvement in T cell clone avidity by direct measurement of HLA-A2/Mage3 complex density using a high-affinity TCR like monoclonal antibody. Eur J Immunol. 2005;35:2864–2875. doi: 10.1002/eji.200526307. [DOI] [PubMed] [Google Scholar]
- 45.Ramesh P, et al. FBAR Syndapin 1 recognizes and stabilizes highly curved tubular membranes in a concentration dependent manner. Sci Rep. 2013;3:1565. doi: 10.1038/srep01565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfenson H, et al. Tropomyosin controls sarcomere-like contractions for rigidity sensing and suppressing growth on soft matrices. Nat Cell Biol. 2016;18:33–42. doi: 10.1038/ncb3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun D, Huang A, Sharma S, Koller A, Kaley G. Endothelial microtubule disruption blocks flow-dependent dilation of arterioles. Am J Physiol Heart Circ Physiol. 2001;280:H2087–H2093. doi: 10.1152/ajpheart.2001.280.5.H2087. [DOI] [PubMed] [Google Scholar]
- 48.Huby RDJ, Weiss A, Ley SC. Nocodazole inhibits signal transduction by the T cell antigen receptor. J Biol Chem. 1998;273:12024–12031. doi: 10.1074/jbc.273.20.12024. [DOI] [PubMed] [Google Scholar]
- 49.Kolodney MS, Elson EL. Contraction due to microtubule disruption is associated with increased phosphorylation of myosin regulatory light chain. Proc Natl Acad Sci USA. 1995;92:10252–10256. doi: 10.1073/pnas.92.22.10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joseph N, Reicher B, Barda-Saad M. The calcium feedback loop and T cell activation: How cytoskeleton networks control intracellular calcium flux. Biochim Biophys Acta. 2014;1838:557–568. doi: 10.1016/j.bbamem.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 51.Klieger Y, et al. Unique -chain motifs mediate a direct TCR-actin linkage critical for immunological synapse formation and T-cell activation. Eur J Immunol. 2014;44:58–68. doi: 10.1002/eji.201243099. [DOI] [PubMed] [Google Scholar]
- 52.Das DK, et al. Pre-T cell receptors (pre-TCRs) leverage V complementarity determining regions (CDRs) and hydrophobic patch in mechanosensing thymic self-ligands. J Biol Chem. 2016;291:25292–25305. doi: 10.1074/jbc.M116.752865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jung Y, et al. Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc Natl Acad Sci USA. 2016;113:E5916–E5924. doi: 10.1073/pnas.1605399113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guillou L, et al. T-lymphocyte passive deformation is controlled by unfolding of membrane surface reservoirs. Mol Biol Cell. 2016;27:3574–3582. doi: 10.1091/mbc.E16-06-0414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majstoravich S, et al. Lymphocyte microvilli are dynamic, actin-dependent structures that do not require wiskott-aldrich syndrome protein (WASp) for their morphology. Blood. 2004;104:1396–1403. doi: 10.1182/blood-2004-02-0437. [DOI] [PubMed] [Google Scholar]
- 56.Fu J, et al. Mechanical regulation of cell function with geometrically modulated elastomeric substrates. Nat Methods. 2010;7:733–736. doi: 10.1038/nmeth.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 58.Ritter AT, et al. Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity. 2015;42:864–876. doi: 10.1016/j.immuni.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tibaldi EV, Salgia R, Reinherz EL. CD2 molecules redistribute to the uropod during T cell scanning: Implications for cellular activation and immune surveillance. Proc Natl Acad Sci USA. 2002;99:7582–7587. doi: 10.1073/pnas.112212699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brazin KN, et al. Structural features of the TCR mechanotransduction apparatus that promote pMHC discrimination. Front Immunol. 2015;6:441. doi: 10.3389/fimmu.2015.00441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mizuno D, Tardin C, Schmidt CF, MacKintosh FC. Nonequilibrium mechanics of active cytoskeletal networks. Science. 2007;315:370–373. doi: 10.1126/science.1134404. [DOI] [PubMed] [Google Scholar]
- 62.Keskin DB, et al. Physical detection of influenza a epitopes identifies a stealth subset on human lung epithelium evading natural CD8 immunity. Proc Natl Acad Sci USA. 2015;112:2151–2156. doi: 10.1073/pnas.1423482112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finer JT, Simmons RM, Spudich JA. Single myosin molecule mechanics: Piconewton forces and nanometre steps. Nature. 1994;368:113–119. doi: 10.1038/368113a0. [DOI] [PubMed] [Google Scholar]
- 64.Murphy CT, Rock RS, Spudich JA. A myosin II mutation uncouples atpase activity from motility and shortens step size. Nat Cell Biol. 2001;3:311–315. doi: 10.1038/35060110. [DOI] [PubMed] [Google Scholar]
- 65.Hundt N, Steffen W, Pathan-Chhatbar S, Taft MH, Manstein DJ. Load-dependent modulation of non-muscle myosin-2A function by tropomyosin 4.2. Sci Rep. 2016;6:20554. doi: 10.1038/srep20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ilani T, Vasiliver-Shamis G, Vardhana S, Bretscher A, Dustin ML. T cell receptor signaling and immunological synapse stability require myosin IIA. Nat Immunol. 2009;10:531–539. doi: 10.1038/ni.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.San José E, Borroto A, Niedergang F, Alcover A, Alarcón B. Triggering the TCR complex causes the downregulation of nonengaged receptors by a signal transduction-dependent mechanism. Immunity. 2000;12:161–170. doi: 10.1016/s1074-7613(00)80169-7. [DOI] [PubMed] [Google Scholar]
- 68.Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: The role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Burnette DT, et al. A contractile and counterbalancing adhesion system controls the 3D shape of crawling cells. J Cell Biol. 2014;205:83–96. doi: 10.1083/jcb.201311104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gustafsson N, et al. Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat Commun. 2016;7:12471. doi: 10.1038/ncomms12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Manz BN, Jackson BL, Petit RS, Dustin ML, Groves J. T-cell triggering thresholds are modulated by the number of antigen within individual T-cell receptor clusters. Proc Natl Acad Sci USA. 2011;108:9089–9094. doi: 10.1073/pnas.1018771108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ebert PJ, Ehrlich LIR, Davis MM. Low ligand requirement for deletion and lack of synapses in positive selection enforce the gauntlet of thymic T cell maturation. Immunity. 2008;29:734–745. doi: 10.1016/j.immuni.2008.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Purbhoo MA, Irvine DJ, Huppa JB, Davis MM. T cell killing does not require the formation of a stable mature immunological synapse. Nat Immunol. 2004;5:524–530. doi: 10.1038/ni1058. [DOI] [PubMed] [Google Scholar]
- 74.Sykulev Y, Joo M, Vturina I, Tsomides TJ, Eisen HN. Evidence that a single peptide-MHC complex on a target cell can elicit a cytolytic T cell response. Immunity. 1996;4:565–571. doi: 10.1016/s1074-7613(00)80483-5. [DOI] [PubMed] [Google Scholar]
- 75.Roein-Peikar M, Xu Q, Wang X, Ha T. Ultrasensitivity of cell adhesion to the presence of mechanically strong ligands. Phys Rev X. 2016;6:011001. [Google Scholar]
- 76.Wang Jh, et al. Structure of a heterophilic adhesion complex between the human CD2 and CD58 (LFA-3) counterreceptors. Cell. 1999;97:791–803. doi: 10.1016/s0092-8674(00)80790-4. [DOI] [PubMed] [Google Scholar]
- 77.Huang J, et al. The kinetics of two dimensional TCR and pMHC interactions determine T cell responsiveness. Nature. 2010;464:932–936. doi: 10.1038/nature08944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mallis RJ, et al. Pre-TCR ligand binding impacts thymocyte development before TCR expression. Proc Natl Acad Sci USA. 2015;112:8373–8378. doi: 10.1073/pnas.1504971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O’Donoghue GP, Pielak RM, Smoligovets AA, Lin JJ, Groves JT. Direct single molecule measurement of TCR triggering by agonist pMHC in living primary T cells. Elife. 2013;2:e00778. doi: 10.7554/eLife.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gross G, Gorochov G, Waks T, Eshhar Z. Generation of effector T cells expressing chimeric T cell receptor with antibody type-specificity. Transplant Proc. 1989;21:127–130. [PubMed] [Google Scholar]
- 81.Sadelain M. Chimeric antigen receptors: Driving immunology towards synthetic biology. Curr Opin Immunol. 2016;41:68–76. doi: 10.1016/j.coi.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.