Significance
We have engineered a light-activated caspase (Caspase-LOV) to better understand the roles of caspases in cells and to create a tool that would kill cells with temporal and spatial selectivity. This modified caspase is created based on the endogenous mechanism of caspase activation by adding a light-sensitive protein domain (LOV2) from oats into the intersubunit linker of the caspase. We tested variations of the Caspase-LOV construct in vitro and in Drosophila and found that it will kill many types of cells with light specificity. We were also able to use the Caspase-LOV to determine the time course of degeneration in two different neuronal cell types after caspase activation.
Keywords: protein engineering, optogenetics, Drosophila, apoptosis, neurodegeneration
Abstract
The circuitry of the brain is characterized by cell heterogeneity, sprawling cellular anatomy, and astonishingly complex patterns of connectivity. Determining how complex neural circuits control behavior is a major challenge that is often approached using surgical, chemical, or transgenic approaches to ablate neurons. However, all these approaches suffer from a lack of precise spatial and temporal control. This drawback would be overcome if cellular ablation could be controlled with light. Cells are naturally and cleanly ablated through apoptosis due to the terminal activation of caspases. Here, we describe the engineering of a light-activated human caspase-3 (Caspase-LOV) by exploiting its natural spring-loaded activation mechanism through rational insertion of the light-sensitive LOV2 domain that expands upon illumination. We apply the light-activated caspase (Caspase-LOV) to study neurodegeneration in larval and adult Drosophila. Using the tissue-specific expression system (UAS)-GAL4, we express Caspase-LOV specifically in three neuronal cell types: retinal, sensory, and motor neurons. Illumination of whole flies or specific tissues containing Caspase-LOV–induced cell death and allowed us to follow the time course and sequence of neurodegenerative events. For example, we find that global synchronous activation of caspase-3 drives degeneration with a different time-course and extent in sensory versus motor neurons. We believe the Caspase-LOV tool we engineered will have many other uses for neurobiologists and others for specific temporal and spatial ablation of cells in complex organisms.
Classical surgical ablation of neuronal tissues has had a major impact on our understanding of the brain in vertebrate animals. More precise surgical methods have been developed using laser ablation (1, 2) but are still invasive and produce collateral damage. Important progress has been made with chemo-optic approaches as induction of Mito-KillerRed or miniSOG with light activation of reactive oxygen species (3, 4), but these can induce collateral damage and/or complex forms of cell death induced through free-radical generation (5). By contrast, direct activation of caspases, the proteases that cleave thousands of proteins in their execution of apoptosis (for recent review, see ref. 6) prevents alternative pathway activations and leads to a natural form of cell death in metazoans without undesirable inflammatory reactions (7). Recently, it has been possible to induce selective cell death in small groups of neurons in Cre mice by adenoviral injection of a Cre-conditional, TEV-sensitive pro–caspase-3 transgene into the brain region of interest (8). However, injecting a virus is invasive and limits the approach to vertebrates. Additionally, inducing apoptosis by transgene expression lacks precise temporal control due to the transcriptional requirement of the system.
Here, we describe the engineering of a light-activated pro–caspase-3, Caspase-LOV, that can be expressed under tissue-specific promoters and precisely induce apoptosis under light control. We apply this genetically encoded tool for spatial and temporal activation of caspase-3 with light in Drosophila larvae and adult animals. In addition to cellular ablation, Caspase-LOV allows us to explore the effect of caspase activation in different cell types and in different cellular compartments. We selectively induce caspase activity in retinal, motor, and sensory neurons in Drosophila and demonstrate phenotypic consequences for each cell type. Optogenetic approaches have been extremely powerful means to rapidly activate specific cellular processes (9–12), and we believe Caspase-LOV may be broadly useful for specific cellular ablation and in increasing our understanding of the roles of caspases in a variety of organisms.
Results
Design and Characterization of Light-Activated Caspase-3 in Vitro.
We began by examining the activation mechanism of the wild-type human pro–caspase-3. Pro–caspase-3 is a constitutive dimer activated by proteolysis at Asp174 between the large and small subunit, typically by an initiator caspase or through autoactivation (Fig. 1A; for review, see ref. 13). Crystal structures comparing the proenzyme and mature enzyme (14–17) and mutational studies expanding the intersubunit linker (18) suggest a spring-loaded mechanism where cutting the intersubunit linker releases strain and allows the enzyme to reorganize into the active conformation (19). To test the degree to which expansion of the intersubunit linker would activate pro–caspase-3, we inserted noncleavable, flexible linkers of 30 and 50 residues (Methods and Fig. 1B). Kinetic analysis of these modified versions of caspase-3, containing 0, 30, or 50 amino acid extensions, showed systematic and dramatic increases in activity with linker length extension (Fig. 1C). The caspase missing the cleavage site and no amino acid extension had no detectable activity, while adding 30 and 50 amino acid residue extensions increased the catalytic efficiency to 1.6 and 3.8 (μM−1·s−1), respectively. These results further support the spring-loaded mechanism for caspase activation and suggested a strategy that could be exploited to generate a light-activated caspase.
Fig. 1.
Extending the intersubunit linker of caspase-3 creates a constitutively active caspase. (A) Schematic diagram of human caspase-3 activation. Wild-type caspase-3 is activated through the cleavage of the intersubunit linker (black) at D174 of the IETD cleavage site (red triangle). The 0-linker caspase is capsase-3 with D174N and DDD(179-181)NNN, mutations preventing the cleavage of the intersubunit linker, making the caspase inactive. The caspases with extended linkers have additional amino acids, primarily glycine and serine (see Methods for sequence), added in the place of D174 and also have the DDD(179-181)NNN mutation. (B) Gene diagrams of the constructs. (C) Kinetics of the modified caspases (50 nM) using DEVD-afc substrate. Each caspase was tested with two trials of four replicates. The error bars reflect the SEM.
The LOV2 (Light-Oxygen-Voltage-sensing) protein domain from Avena sativa (20, 21) is light sensitive and has been used extensively as an optogenetic protein modifier (22–24). When LOV2 is exposed to blue light, a Cys450 in its core domain binds the flavin mononucleotide (FMN) cofactor, causing the C-terminal Jα helix to undock and assume a flexible conformation (25–29). We hypothesized it may be possible to insert the LOV2 into the intersubunit linker of the procaspase so that in the dark the enzyme would be constrained, but in the light, the intersubunit linker would expand and allow the active site to fold into the active conformation (Fig. 2 A and B).
Fig. 2.
Computational modeling using Rosetta to design a light-activated caspase by extending the intersubunit linker with LOV2. (A) A schematic diagram of Caspase-LOV. Caspase-3 (blue) forms a homodimer, and LOV2 (green and orange) is inserted into the intersubunit linker of caspase-3. The Jα helix (orange) of LOV2 unfolds and extends when the protein is exposed to blue light. This extension allows the caspase to fold into the active conformation. (B) Gene diagrams of Caspase-LOV with alanine residue extensions added N-terminally and C-terminally to the pro–caspase-3/dark LOV2 and active caspase-3/light LOV2. (C) Representative models of pro–caspase-3/dark LOV2 and caspase-3/light LOV2 showing successful linker connections (gray) between the caspase and the LOV2 when seven alanine residues are added N-terminally. (D) Representative models of pro–caspase-3/dark LOV2 and caspase-3/light LOV2 showing successful linker connections (gray) between the caspase and the LOV2 when seven residues are removed from the caspase intersubunit linker C-terminally. The caspase-3/light LOV2 version includes the flexible Jα helix as part of the intersubunit linker. (E) Plot of the percent successful connections of the N-terminal caspase to LOV2 linker testing the extension of the intersubunit linker with 0–19 alanines. The black dashed (pro–caspase-3/dark-state LOV2) and blue solid (caspase- 3/light-state LOV2) lines represent the averages of the percent of successful connections across the three initial models. (F) Plot of the percent successful connections of the C-terminal caspase to LOV2 linker testing an intersubunit linker missing seven amino acids (−7) to the addition of 15 alanines. The black dashed (pro–caspase-3/dark-state LOV2) and blue solid (caspase-3/light-state LOV2) lines represent the averages of the percent of successful connections across the three initial models.
We considered several design criteria for inserting the LOV2 domain into the intersubunit linker including: (i) insertion of the LOV2 needs to be structurally compatible with the proform in the dark state and compatible with the active form in the light state, (ii) the insertion should keep the intersubunit linker conformationally restrained in the dark state to reduce activity, and (iii) the light-extended state should avoid blocking the access to the active site once activated. We used modeling tools, including the Rosetta software suite (30, 31), to assist in developing initial designs (Fig. 2).
We optimized the placement of LOV in relation to caspase-3 by manually orienting it, then using cluster analysis in Rosetta to select six starting point models (three for the pro–caspase-3 with dark LOV2 and three for the active caspase-3 with light LOV2) (Methods). To predict the optimal linker length for inserting the LOV2 domain within the intersubunit linker, we modeled linker connections between light LOV2 and the active caspase-3 and dark LOV2 and the pro–caspase-3. For the procaspase/dark LOV2 version, we split the intersubunit linker at the cleavage site and kept the Jα helix of the LOV2 rigid. We then added different numbers of alanine residues between the intersubunit linker and the LOV2 domain to form connections between them, N- and C-terminally (sample models Fig. 2 C and D). The linker connections for the active caspase versions were modeled identically except we allowed the Jα helix to extend in random conformations that would mimic the flexibility of the LOV2 domain in the light state. We ran 100 simulations per alanine linker length addition for each of the six starting structures to attempt linker connections between the caspase and LOV2 for both the light and dark states. For each linker length, we then quantified the number of successful connections across the three relaxed starting structures (Fig. 2 E and F). We assume that a linker that has many successful connections will be the most likely to form a functional protein when expressed. Additionally, we looked at the minimum Rosetta energies for each of the linkers to estimate which would be the most likely to fold properly. We preferred the shortest possible compatible linker to reduce flexibility between the caspase and the LOV when the protein is in the dark to keep the caspase in the inactive (zymogen) state. Based on these considerations, the Rosetta modeling results predicted that adding 5 to 12 residues to the N-terminal side (N5 to N12) and deleting up to seven residues of the caspase-3 intersubunit linker from the C-terminal side (C0 to C-7) should lead to successful fusions, with the N7/C-7 combination as the likely most optimal candidate.
We expressed the N7/C-7 Caspase-LOV protein in Escherichia coli (with two mutations in the LOV2 domain, G528A and N538E, to increase the dark stability; ref. 25). We were encouraged that the protein was intact, and expression levels were near those we found for wild-type caspase-3. We purified the enzyme and assayed it with the fluorogenic DEVD-afc substrate in the light (after 10 min of LED exposure) and found it to have comparable activity to wild-type caspase-3, and a two- to threefold lower activity when assayed in the dark (after 30-min incubation under foil) (Fig. 3A). Additionally, we tested the kinetics of the N7/C-7 Caspase-LOV to cleave the caspase substrate PARP from Jurkat cell lysate and found that it did cleave the substrate faster in the light than the dark although not as fast as active caspase-3 (SI Appendix, Fig. S1).
Fig. 3.
Testing light and dark activity and kinetics of activation for Caspase-LOV variants in vitro. (A) Kinetic assay with DEVD-afc substrate of the Caspase-LOV variant N7/C-7 compared with a dark-locked and light-insensitive mutation (C450A) in the light and the dark. (B) Absorbance reads over time at 450 nm of Caspase-LOV N7C-7 after a 3-min LED incubation and Caspase-LOV N7/C-7 kept in the dark. (C) Caspase-LOV gene diagram showing where deletions and insertions were made to make the Caspase-LOV variants. Different variants have different amounts of Gly-Ser amino acid additions to the N-terminal region of LOV2 where it connects to the large subunit of caspase (designated as Nx where x is the number of amino acids). C-terminal to the LOV2, there is either the addition of Gly-Ser amino acids (designated as Cx where x is the number of amino acids added) or the removal of endogenous amino acids of the intersubunit linker (designated as C-x where x is the number of amino acids removed). (D) Michaelis–Menten calculated kcat/KM of recombinant Caspase-LOV constructs (5 nM) using a DEVD-afc substrate in the light and in the dark (each variant and condition was tested with 3–15 trials of four replicates). The constructs are plotted in order of smallest to largest total linker length. (E) Light-to-dark ratio of Caspase-LOV constructs that have a consistent C-terminal linker (C-7) and changing N-terminal linkers. (F) Light-to-dark ratio of Caspase-LOV constructs that have a consistent N-terminal linker (N7) and changing C-terminal linkers. (G) Half-life of absorbance reads at 450 nm of different Caspase-LOV constructs. (All error bars reflect the SEM.)
To determine if the light-to-dark difference of the Caspase-LOV was due to activation of the LOV2 domain, we introduced the C450A mutation into the N7/C-7 enzyme which prevents LOV activation by locking it into the dark conformation (32). As expected, the LOV-inactive enzyme showed virtually no difference in activity in the light or dark (Fig. 3A). Interestingly, it showed threefold lower basal activity compared with the dark condition for N7/C-7 Caspase-LOV, suggesting that N7/C-7 Caspase-LOV was partially active in the dark condition during the assay. Additionally, an inhibitor active-site labeling assay showed reduced active site labeling for the dark condition compared with the light, but still more than the C450A dark-locked Caspase-LOV (SI Appendix, Fig. S2).
To understand the kinetics of the light-to-dark transition for N7/C-7 Caspase-LOV, we took advantage of the absorbance changes of LOV2. LOV2’s flavin mononucleotide chromophore has an absorbance peak at 450 nm in the dark that is extinguished in the light state. We found the enzyme rapidly folded into the dark state with a half-life of about 42 s, about the same rate as reported for the isolated LOV2 domain (28) (Fig. 3B). Thus, the N7/C-7 Caspase-LOV insertion does not appear to impede the ability of the LOV2 domain to undergo the light-induced transition. We also tested the kinetics of the Caspase-LOV in different light cycling states by exposing it to light, then letting it recover in the dark, then exposing it to light again. We have found that the Caspase-LOV has a comparable activity after light-dark-light cycling as light alone and a 30-min incubation in the dark after light exposure does reduce its catalytic activity to a level comparable to dark alone (SI Appendix, Fig. S3). These data support that the light-dark state changes are reversible.
To further test the effect the LOV2 location has on the light-dark Caspase-LOV kinetics, we generated a panel of nine linker variants by systematically adding and subtracting amino acids from each side linker (N-5/C-7, N-2/C-7, N0/C-9, N7/C-9, N5/C-7, N7/C-7, N9/C-7, N7/C-2, and N12/C-7) (Fig. 3C). All of the Caspase-LOV variants had greater kcat/KM in the light than the dark except the N-5/C-7 variant, which was completely inactive (Fig. 3D and SI Appendix, Fig. S4). (It is likely the N-5/C-7 variant was inactive due to the removal of the five amino acids before the cleavage site of caspase-3.) We then calculated the light-to-dark ratio for each variant to determine which had the greatest dynamic range (Fig. 3 E and F). We found that the ratios were roughly the same for the variants when the N-terminal connection was varied from −5 to +12 and C-terminal extension fixed at −7 (Fig. 3E). In contrast, the ratio increased when the C-terminal linker was increased and the N-terminal linker insertion was kept at +7 (Fig. 3F). This suggests that changing the C-terminal linker has a greater impact on the dark state of the caspase than varying the N-terminal linker. To test if the location of the LOV2 within the caspase impacted the light-to-dark transition rates, we tested several of these variants and found that the transition half-life was about the same (t1/2 = 42–45 s) (Fig. 3G).
Caspase-LOV Causes Light-Dependent Degeneration in Adult Drosophila Retinas.
Having demonstrated clear light activation of caspase-3 activity in vitro, we then explored its impact when expressed in specific neuronal tissues in Drosophila melanogaster. We exploited the GAL4/UAS expression system for tissue-specific gene expression in Drosophila (33). The UAS > N7/C-7 Caspase-LOV was transformed into the Drosophila genome and combined with cell type-specific expression of GAL4 according to standard methods. We used this system to test the Caspase-LOV in a variety of neuronal cell types including: retinal cells (GMR-GAL4), motor neurons (OK371-GAL4), individual motor neurons (MN1-GAL4), and dendritic arborizing sensory neurons (PPK-GAL4) (Fig. 4A).
Fig. 4.
Light-specific Caspase-LOV–induced cell death in retinas of Drosophila. (A) Diagram of the mechanism of Caspase-LOV expression in specific tissues using the UAS-GAL4 system. Genomic enhancers are used to express GAL4 in specific tissues or subsets of cells. The GAL4 in these cells can then bind to the UAS, and the Caspase-LOV will be expressed. (B) Light-raised flies are grown in the light from the embryo stage, while dark-raised flies are grown in the dark from embryo. For other assays, flies were raised in the dark and then put in the light or kept in the dark upon eclosion. (C and D) Representative images of Drosophila eyes expressing N7/C-7 Caspase-LOV with GMR-GAL4 and raised in the light (C) and the dark (D). (E) Representative images of Drosophila eyes without Caspase-LOV expression raised in the light. (F–H) Cryosectioned whole heads of Drosophila stained with anti-chaoptin (green) for retinal marking, anti-dlg (red), and dapi (blue).
We first expressed UAS > N7/C-7 Caspase-LOV using the GMR-GAL4 driver in retinal cells because retinal degeneration leads to obvious morphological changes (34). We raised animals in the light or dark from the embryo stage and then evaluated them once they eclosed to adults (Fig. 4B). The adult animals that were raised in the light have a very strong degenerative eye phenotype on the first day of eclosion compared with the animals raised in the dark (Fig. 4 C and D). This effect is due to light activation of Caspase-LOV and not light-raising alone as there was no defect in the light-raised animals that expressed the GMR-GAL4 driver without UAS > Caspase-LOV (Fig. 4E).
To determine whether this degeneration extends through the neurons, we cryosectioned 1-d-old adult heads and stained them for a retinal marker (anti-chaoptin) as well as a cell surface marker (anti-dlg) and a nuclear marker (DAPI). We found that the light-raised flies expressing N7/C-7 Caspase-LOV had lost all of the chaoptin-marked photoreceptor cells in the brain (Fig. 4F), while the dark-raised flies retained a layer of chaoptin-positive cells (Fig. 4G). Dark-raised flies had fewer chaoptin positive cells than the control flies (Fig. 4H), suggesting some amount of Caspase-LOV activity in the dark.
Caspase-LOV Causes Light-Dependent Degeneration in Adult Drosophila Motor Neurons.
To test Caspase-LOV in other neuronal systems, we expressed it with the motor neuron-specific driver OK371-GAL4. We raised animals in the dark and then divided adult animals, after eclosion, into either light-exposed or dark (foil-wrapped) vials (Fig. 4B). Caspase-LOV–expressing flies that were kept in the light had a much lower survival rate than flies kept in the dark (Fig. 5A). The median survival of the N7/C-7 Caspase-LOV flies kept in the light was 8 d, while those kept in the dark had a median survival of 54.5 d. In comparison, wild-type (w1118 was used as a genotypic control for transgenic insertion lines) flies kept in the light have a median survival of 56 d and those kept in the dark have a median survival of 91 d (SI Appendix, Fig. S5A). The Caspase-LOV–expressing flies became paralyzed before dying, suggesting a strong defect in motor function. We tested two different Caspase-LOV variants using this system: the N7/C-7 variant and the N7/C-2 variant, which had a higher light-to-dark activation in vitro than N7/C-7. The N7/C-2 Caspase-LOV showed a more dramatic effect in the rate of death in vivo with a median survival in the light of 4 d and in the dark of 20 d. (Fig. 5A). Both of these conditions are significantly faster than the N7/C-7, although it is unclear why the N7/C-2 rate in the dark would be faster from the in vitro data and suggests that the in vitro results cannot entirely predict the effects in vivo. Additionally, these two fly lines were made using a random genomic p-element insertion method. To confirm that the difference in survival rate was not from the genomic loci insertion and to expand the comparison of in vitro to in vivo variants, we made new fly lines using a site-specific integrase mediated repeated targeting (SIRT) strategy (35, 36). We compared the survival in the light and the dark of SIRT-inserted N7/C-7, N5/C-7, and N7/C-2 Caspase-LOV variants. We found that the SIRT-inserted N7/C-2 still died significantly faster than the N7/C-7 (median light survival of 10 d and >40 d, respectively), confirming our p-element insertion survival results were due to variant differences, rather than genomic loci differences. We did find the effects in vivo roughly reflected the degree of dark-to-light activation seen in vitro with the N7/C-2 variant being the strongest (SI Appendix, Fig. S5C). Interestingly, the maximum kcat values of each variant (SI Appendix, Fig. S4A) correlated better to the degree of phenotypic effects than the maximum catalytic efficiency. Since the animals with the SIRT insertions had only five copies of UAS rather than the 10 copies of UAS from the p-element insertion, the SIRT animals had a lower level of the Caspase-LOV expression. Thus, for the rest of the experiments, we used only the 10× UAS p-element versions of N7/C-7 and N7/C-2.
Fig. 5.
Survival and behavior of adult Drosophila with Caspase-LOV expression in motor neurons. (A) Survival of flies with N7/C-7 Caspase-LOV (blue) and N7/C-2 Caspase-LOV (red) expression in motor neurons. The light fly condition (solid lines, n = 87) were exposed to light upon eclosion, and the dark condition flies were kept in the dark their entire life (dashed lines, n = 40). (B) Motor behavior assay of flies expressing N7/C-7 Caspase-LOV in the light (solid line, n = 100) and dark (dashed line, n = 110). (C) Motor behavior assay of flies expressing N7/C-2 Caspase-LOV in the light (solid line, n = 20) and dark (dashed line, n = 20). (All error bars reflect the SEM.)
As a more sensitive phenotypic assay for early effects of the Caspase-LOV activation in motor neurons, we measured motor behavioral defects. To quantify the rate of degeneration of the motor neurons for the different groups, 10 flies were placed in a vial and we counted the number of flies that could climb 3 cm up the vial in 90 s. After 3 d in the light, about 50% of the flies containing the N7/C-7 Caspase-LOV variant were able to climb 3 cm in 90 s compared with 90% in the dark (Fig. 5B). The stronger N7/C-2 Caspase-LOV showed a more pronounced effect; in the light, by 20 h, only 50% were able to climb compared with 95% in the dark (Fig. 5C).
Evidence Caspase-LOV Activity Can Be Halted in Vivo.
As shown in the in vitro tests, the Caspase-LOV is reversible and will turn off within minutes in the dark after exposure to light (Fig. 3B). We wanted to test if neurons could recover after varying the amount of caspase activation using the Caspase-LOV. We expressed N7/C-7 Caspase-LOV (since these flies died less quickly than the N7/C-2 flies and we were more likely to see an effect over a longer time scale) in the motor neurons and found that the survival of the flies mirrored the amount of light they were exposed to; flies with 1 d of light exposure and then put into the dark did nearly as well as flies kept in the dark for the first 18 d of their life (Fig. 6A). Flies with 2 or 3 d of light exposure and then kept in constant darkness had an improvement in survival compared with flies that were kept in the light constantly. With more than 3 d of light exposure, the effect appeared irreversible.
Fig. 6.
Delay of survival and behavioral degradation when flies are put into the dark after being exposed to light. (A) Survival of adult flies expressing N7/C-7 Caspase-LOV in motor neurons with the OK371 GAL4. Flies were kept in the dark (black line, n = 40) or constant light (blue line, n = 87) or light for varying days and then put into the dark (1 d light-purple line n = 120, 2 d light-orange line n = 134, 3 d light-green line n = 76). (B) Behavioral assay tracking the percentage of flies able to crawl 3 cm in 90 s of flies kept in the dark (black line n = 110), the light (blue light n = 40), and light and then dark (1-d purple n = 90, 2-d orange n = 100 and 3-d green line n = 70). (All error bars reflect the SEM.)
When we look at the motor behavior of the flies exposed to light for 2 or 3 d, we see a plateau where degeneration stops 1 to 2 d after being put into the dark (Fig. 6B). At an individual level, some of the flies actually improved their motor function in this time period, which was never seen in flies constantly exposed to light.
Caspase-LOV Activity Can Be Blocked with an Irreversible Caspase-3 Inhibitor p35, and Partially by Endogenous Diap1, but Does Not Require Endogenous Drosophila Caspases To Induce Cell Death.
We next asked if Caspase-LOV engaged the endogenous apoptotic machinery in Drosophila cells. We first tested if it could be inhibited by genetically expressed caspase inhibitors such as the exogenously expressed baculoviral-derived p35 (37) or endogenous Drosophila Diap1. When UAS-p35 is expressed in motor neurons with N7/C-7 Caspase-LOV, there was a dramatic increase in survival in the light (Fig. 7A). The rescue by p35 confirms that the cell death phenotypes are due to the specific activity of the caspase. Overexpressing Diap1 had a small but significant increase in survival rates that were most pronounced in the dark (Fig. 7B). Diap1 may have had a weaker effect because it is not an irreversible caspase inhibitor and Diap1 is subject to caspase-cleavage–mediated degradation as well as full-length ubiquitin-proteasome–dependent degradation (38, 39). In the dark, we expect the Diap1 will be more able to inhibit any active Caspase-LOV because the Diap1 will be less likely to be cleaved and degraded by the smaller amount of active Caspase-LOV.
Fig. 7.
Caspase-LOV can be inhibited by the baculovrial caspase inhibitor p35, but does not need endogenous Drosophila caspases to degenerate motor neurons. (A and B) Survival of flies with motor neurons (OK371-GAL4) expressing N7/C-7 Caspase-LOV (blue lines, light: n = 166, dark: n = 50) and flies with N7/C-7 Caspase-LOV and overexpression of caspase inhibitors: p35 (irreversible baculoviral, light: n = 79, dark: n = 44) and diap1 (Drosophila, light: n = 31, dark: n = 28) (green lines). (C and D) Survival of flies with motor neurons (OK371-GAL4) expressing N7/C-7 Caspase-LOV with Drosophila initiator caspase droncRNAi (light: n = 34, dark: n = 28) and executioner caspase dcp1RNAi (light: n = 35, dark: n = 23). (E) Survival of flies expressing N7/C-7 Caspase-LOV and UAS-Wlds (Wallerian degeneration slowed, light: n = 30, dark: n = 23).
Once activated, executioner caspases are able to cleave and activate other executioner caspases in a feed-forward process. We were curious if Caspase-LOV activated endogenous fly caspases and cause the phenotypic effects we see. To explore this possibility, we expressed the UAS > Caspase-LOV in motor neurons with UAS > dronc-RNAi (initiator caspase) or UAS > dcp1-RNAi (executioner caspase and the fly homolog for caspase-3). Both transgenes have been validated in other systems (40, 41). We found that a reduction in either Dcp1 or Dronc had no effect on light-activated Caspase-LOV–induced degeneration (Fig. 7 C and D).
Neurodegeneration can occur during injury, development, or disease. It is thought that injury has a more distinct pathway for neurite removal compared with either development or disease, which have similar mechanisms. Wallerian degeneration is caused when an axon is severed from the cell body and the distal fragment is removed. This process has been thought to involve a pathway distinct from the caspase pathway (42, 43). To test this, we activated Caspase-LOV in animals expressing a protein fusion (Wlds) that significantly slows Wallerian degeneration in flies and mammals (44–46). We found flies with Wlds are not protected from Caspase-LOV–induced degeneration, which is consistent with the previous findings that Wallerian degeneration is not executed by proteins in the apoptotic pathway (Fig. 7E). Thus, the Caspase-LOV acts independent of the Wlds cellular machinery.
The Synchronous Activation of Caspase-LOV Allows Time-Course Measurements of Degeneration Across All Cellular Compartments in Motor and Sensory Neurons.
With the Caspase-LOV, we can activate caspases synchronously throughout the entire neuron. Previously, this has not been possible because caspases are expressed in an inactive state in overexpression systems, and the time and location of their activation cannot be controlled. With light activation, this is not a problem. We first tested the precise time of onset of neuronal cell death and damage following synchronous caspase activation in all compartments of motor neurons. We expressed N7/C-2 Caspase-LOV and a membrane-tethered GFP (UAS > CD8-GFP) to visualize the neuronal plasma membrane in a subset of motor neurons using the MN1-GAL4 driver. This driver selectively expresses in segmentally repeated pairs of motor neurons, allowing visualization of the individual dendritic tree, axon, and nerve terminal. For these experiments, eggs were laid in the dark and larva were moved to an agar-based apple juice plate for different amounts of timed light exposure pre-third instar stage. Larvae began to show signs of degeneration and apoptosis after 12–14 h of light exposure, which became more severe following 24 h of light (Fig. 8 A and B). Degeneration of axons, dendrites, and apoptosis of the cell body was quantified with a scoring system, blind to genotype and light treatment. There was not a substantive difference in the rates of degeneration among the axon, dendrite, or cell body (Fig. 8C). However, the neuromuscular synapse persists even at time points when severe dendritic degeneration is observed (Fig. 8D). To verify that this effect was not caused by a lack of Caspase-LOV at the synapse, we stained a cherry tagged version of the Caspase-LOV in larvae and found protein throughout the neurons including the synapse (SI Appendix, Fig. S6).
Fig. 8.
Time course of degeneration of motor neurons in Drosophila larvae. (A) Representative images of larval ventral nerve cords. N7/C-2 Caspase-LOV and CD8GFP are expressed in a subset of motor neurons using the MN1-GAL4 driver and the larvae were grown on apple plates for different amounts of time, as specified, prethird instar stage. Colored arrows represent differing degrees of degeneration scored in C. (B) Representative images of axons from time points in A. (C) Quantification of degeneration of cell bodies, axons, and dendrites. “Healthy” (black) signifies a morphologically healthy cell part, weak damage (blue) signifies a cell part that is starting to fragment or bleb, severe damage (green) signifies a cell part that has significant fragmentation or blebbing but has not been cleared, and removed (orange) signifies a cell part that has been cleared. The control fly is a fly with MN1-GAL4 driving CD8GFP, but not expressing Caspase-LOV. (Numbers of neurons counted for each condition: control n = 198, dark n = 379, 6 h n = 54, 12 h n = 46, 18 h n = 29, 24 h n = 29, and 28 h n = 28.) (D) Synapses at muscle one neuromuscular junction expressing N7/C-2 Caspase-LOV and CD8GFP with the MN1-GAL4 driver. Green is anti-GFP staining the neurons containing Caspase-LOV, and magenta is anti-dlg marking the postsynapse. (Scale bars: 10 μm.)
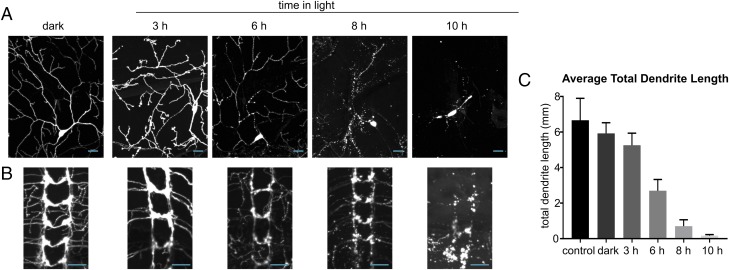
To see if the timing of degeneration and the rate of loss between compartments was consistent across neuronal cell types, we examined larvae expressing N7/C-2 Caspase-LOV in class IV dendritic arborizing (da) sensory neurons, using the PPK-GAL4 driver. Light exposure also induces degeneration in da neurons, but degeneration onset occurred more than 6 h earlier than in motor neurons and the rate of degeneration was much faster across all compartments (Fig. 9A). Blebbing of the dendrites and axons was observed 4–6 h after illumination. By 8 h, most of the neurites were significantly fragmented, severed, or absent. By 10 h, most of the neurites had been cleared, and by 12 h, all of the neurites were cleared. To quantify the rate of degeneration, we traced the dendrites of individual neurons to track their clearance (Methods and Fig. 9C). This is a striking difference, but we cannot rule out that this is due to potentially different strengths of the GAL4 drivers leading to higher Caspase-LOV expression. In contrast to the motor neurons, the synapses of the sensory neurons degenerated at roughly the same rate as the dendrites and axons (Fig. 9B).
Fig. 9.
Time course of degeneration of dendritic arborizing sensory neurons in Drosophila larva. (A) Representative images of larva expressing GFP and N7/C-2 Caspase-LOV in class iv da sensory neurons using PPKtdGFP and PPKGAL4 in the dark and grown on apple plates in the light for different amounts of time prethird instar stage when they were dissected and stained. (B) Representative images of sensory neuron synapses into the ventral nerve cord of larva expressing GFP and N7/C-2 Caspase-LOV in class iv da sensory neurons using PPKtdGFP and PPKGAL4 in the dark and light. (Scale bars: 20 μm.) (C) The average total dendrite branch length of neurons from larva with no caspase expression (control), kept in the dark, or in varying amounts of light, calculated using simple neurite tracer in FIJI with tracing through early blebbing branches unless the branch was severed or completely disrupted. The number of neurons traced for each: control n = 4, dark n = 4, 3 h light n = 7, 6 h light n = 7, 8 h light n = 12, and 10 h light n = 11. All error bars reflect the SEM, and a multiple comparisons test revealed no significant difference between control, dark, and 3 h (P = 0.9725 for control vs. dark, 0.6079 for control vs. 3 h, and 0.9724 for dark vs. 3 h). All other time points were significantly different from dark (P = 0.0068 for dark vs. 6 h, and P < 0.0001 for dark vs. 8 h and 10 h).
Discussion
Rational Design of a Light-Activated Caspase.
Our primary goal was to create a light-activated caspase (Caspase-LOV) especially for use in small model systems like Drosophila. We deliberately chose human caspase-3 because we had the most knowledge of its active and inactive structure, and human caspase-3 is likely to be effective in multiple organisms including human cells, rodents, and Drosophila. Although we cannot be sure the Caspase-LOV cleaves the same endogenous substrates, the specificities of the human and Drosophila caspases are very similar, and many of the substrates are conserved in apoptosis (47, 48). Additionally, the Caspase-LOV is small enough to be virally transduced in mammals and cell culture.
In designing the light-activated caspase-3, we focused on exploiting the natural zymogen activation mechanism. Our linker insertion experiments in a noncleavable form of pro–caspase-3 support previous studies (18) that activation occurs by releasing tension in the intersubunit linker, allowing it to fold into the active form. The LOV2 domain seemed an ideal candidate to mimic this activation since it is known to expand upon illumination; however, positioning this domain into the intersubunit linker was not obvious. The Rosetta design simulations we performed helped to triage a number of constructs to focus on the successful N7/C-7 linker insertion and variants thereof.
The in vitro tests showed a modest two- to fourfold activation in the light compared with the dark for most of our Caspase-LOV variants, and the catalytic efficiency in the light-activated state was virtually the same as that of the mature caspase-3. Most of the light-activation effect was seen as a gain in the kcat rather than a change in the KM, demonstrating that the light-sensitive conformational changes allowed the caspase to cleave the substrate more efficiently, rather than impacting the substrate affinity (SI Appendix, Fig. S4). Adjusting the positioning of the LOV2 domain in the linker had the most pronounced effects when the C-terminal side that connected the LOV2 to caspase-3 was altered. The in vitro results suggest that a longer linker is needed C-terminally to correctly associate the Jα helix to the LOV2 core domain in the dark state. Potentially, having a shorter linker does not allow complete association and folding of the Jα helix, leading to a Caspase-LOV that is more likely to be active in the dark.
Although altering the placement of the LOV2 within the caspase-3 intersubunit linker helped decrease the activity of the Caspase-LOV in the dark, the fold change between the on- and off-state is still orders of magnitude smaller than the fold change between active caspase-3 and inactive zymogen (107-fold difference) (49). The dark-locked C450A mutated Caspase-LOV does cause a threefold further drop in activity compared with the same Caspase-LOV variant without the mutation. This suggests that there could be some ambient light activation during the spectrophotometric enzyme activity assay so that the light-to-dark ratio is probably larger than measured. The remaining activity of the Caspase-LOV in the dark is likely due to either the LOV2 dark state not being tight enough to prevent the caspase from folding into the active state, or could be caused by dynamic movement of the LOV2 domain between on and off conformations in the dark. We attempted to preemptively minimize the later issue by introducing known stabilizing mutations in the LOV2 domain (G528A and N538E) (25). We believe further improvements could be made to the Caspase-LOV to further reduce the dark activity by additional stabilization of the LOV2 and, perhaps, engineering the interface between the LOV2 and caspase-3 domains.
Despite some caspase activity in the dark state, the Caspase-LOV was remarkably well tolerated in larvae and adult flies in the dark. This could reflect greater suppression of low levels of the caspase in tissues and less ambient activation. The dark activity we did see was the greatest in the N7/C-2 variant, which is not what we predicted from our in vitro results. This suggests that the in vitro results can not completely predict the biological response to the proteins and highlights the value of further exploration of the effects of different levels of caspase activity in vivo. We will note, that the levels of caspase activity in the dark could impact experiments, especially on older animals where we saw the greatest effects of caspase activity in the dark. Depending on the experiment, this could be alleviated by using a different Caspase-LOV variant or by expressing the Caspase-LOV in a Diap1 overexpression background, which reduces the rate of death in the dark while maintaining death in the light (Fig. 7B).
In designing the light-activated caspase-3, we studied previous work that constructed a light-activated caspase-7 (LOV-casp7) by an entirely different rationale (50). The investigators replaced the prodomain in caspase-7 with the LOV2 protein and observed about a twofold light activation in HeLa cells that were transiently transfected with the construct. The investigators proposed activation occurs by release of steric inhibition allowing exposure of the intersubunit linker, autoprocessing, and activation. Interestingly, Bcl2 overexpression in cells inhibited the photoactivation, suggesting activation required support from the intrinsic pathway, presumably by caspase-9 cleaving the intersubunit linker of LOV-casp7. This would also be consistent with their observation that cellular activation of apoptosis occurred within an hour, whereas cleavage of a large synthetic protein substrate in vitro occurred much slower over 24 h. Given the mild activation effects, the high dark apoptotic activity, and the dependence on the intrinsic apoptotic machinery, we felt the system was not an optimal starting point to develop a Caspase-LOV for animal experiments that we sought here.
Physiological Effects of Caspase-LOV in Vivo.
Caspase-LOV lets us control the amount, location, and length of time of caspase-3 activation in cells in an organism in vivo through light. Additionally, since the Caspase-LOV activation does not depend on the endogenous fly caspases or apoptotic machinery, virtually all of the phenotypic response is under illumination control and not positive feedback from the fly’s apoptotic system, which can vary by cell type.
Recent studies on anastasis, the recovery of a cell from significant caspase expression, suggest that many cells can survive transient caspase expression and proliferate in an animal (51, 52). Indeed, we find when flies expressing Caspase-LOV in their motor neurons were transiently exposed to light (24 h) and then returned to the dark, they showed a transient recovery of motor behavior (Fig. 6). Additionally, flies are able to handle the small amount of caspase activity in the dark for an extended period.
Caspases have also been shown to become activated transiently in noncell death processes in neurobiology including: selective pruning of dendritic branches in postmitotic neurons, the modulation of axon guidance in developing neurons, a role during long-term synapse depression and elimination, and other developmental roles (52–57). A number of studies have proposed that the apoptotic action of caspases is controlled either through local or temporal activation of caspases. A major hurdle in the study of nonapoptotic caspase signaling, particularly in postmitotic neurons, is the ability to precisely manipulate caspase activity. The light-activated control of caspase-3 activity in an animal allows us to control the temporal effect of caspase activation in vivo. We can express the caspase in an inactive state in cells so it can reach all compartments of a neuron and then activate it synchronously across compartments. When we look at two different neuronal cell types, sensory and motor neurons, we see that the sensory neurons degenerate much faster than the motor neurons. Since two different GAL4 drivers were used to express the UAS > Caspase-LOV in each of these cell types, we cannot conclusively say that the rates of degeneration were due to the response of caspase expression in each of the cell types. However, since the expression of Caspase-LOV was throughout the neurons (SI Appendix, Fig. S6), we can compare the effects of caspase expression in different compartments between these two cells. When Caspase-LOV is expressed globally in all compartments of the da sensory neurons, the axons, synapses, and dendrites all degenerate at the same rate. When the Caspase-LOV is expressed in motor neurons in all compartments, the axons, dendrites, and cell bodies all degenerate at roughly the same rate, but the synapses remain. During neuromuscular degenerative disease, the synapse is one of the first locations to undergo degeneration (for review, see ref. 58). If degeneration of the neuromuscular junction during disease is driven by caspase activation (59), then it seems likely that there must be localized activation of the caspase at that site. Since we saw a slower degeneration at synapses of the motor neuron with synchronous global activity, this suggests that the degenerative effects of neuromuscular diseases are due to local rather than global caspase expression.
Although we believe additional improvements are worthy to suppress the dark activity, there are many potential uses for the Caspase-LOV beyond those shown here. It may be applied broadly to different cell types to determine what level of activation is required to impact their cell fate both in the adult and in development. One could observe how normal cells respond when a neighboring cell undergoes apoptosis. One could also precisely ablate specific cells that do not yet have cell type-specific GAL4 expression lines. In addition, having a caspase that is controllable temporally and spatially not only allows us to study the role of caspase activation within an individual cell or cell type, but also allows us to define the sequence of cell biological events that occur, in vivo, following caspase activation. It may also be possible to produce light-activated derivatives of specific endogenous caspases to better understand their roles. Finally, this technique could allow one to study the role of individual neurons or groups of neurons within a circuit in a way that should not cause collateral damage to surrounding cells.
Methods
Cloning.
All cloning was performed with standard cloning procedures. See SI Appendix for details and plasmid sequences.
Rosetta Modeling.
All simulations were done using Rosetta revision 51511 (30, 31). See SI Appendix for details.
Recombinant Expression and Purification.
The expression and purification procedure was adapted from ref. 60. See SI Appendix for details.
LED Plate.
A 48-LED array was designed to illuminate one-half of a 96-well plate for the in vitro kinetic assays. Super Bright Blue 5-mm LEDs (Adafruit) were arranged in 12 series of four parallel LEDs on a Grid-Style PC board (RadioShack) with a BuckPuck DC LED Driver (LED Supply) and a low-pass filter. To connect to AC power, a Universal AC Adapter (Hausbell) was used. The LED plate was then attached to a black-sided clear bottom 96-well plate to direct the light of one LED into each 96-well and prevent side-scattered light. The power of the plate 1 cm away from LED is 5 mW. SI Appendix for circuit diagram.
In Vitro Kinetics Assay.
Assay adapted from ref. 40. Caspases with extended intersubunit linkers were diluted to 20 nM in caspase activity buffer (50 mM Hepes 7.4, 50 mM KCl, 0.1 mM EDTA, 10 mM DTT, 0.1% CHAPS) to 45 uL in a 96-well clear bottom plate (COSTAR 3915). The lower 48 wells were covered with a foil seal, and the whole plate was left in the dark at room temperature for 30 min. The following steps are all done in very dim light. After incubation, the plate was placed on a plate shaker and the LED plate array was placed to illuminate the top 48 wells for 10 min. After 10 min, 5 μL of DEVD-afc substrate was added to each well (piercing the foil for the dark condition), agitated briefly on the plate shaker, and then immediately placed into the Spectramax M5 to measure afc absorbance. Readings were taken every 20 s for 10 min. The DEVD-afc substrate concentration was varied across the 12 columns in 1:2 dilutions starting from a concentration of 400 μM. The fluorescence readings were analyzed by determining the initial rate. The Caspase-LOV variants were assayed identically except the initial protein concentration was 5 nM.
Absorbance Assay.
Caspases were diluted to 5 uM in caspase activity buffer without DTT or CHAPS (50 mM Hepes 7.4, 50 mM KCl, 0.1 mM EDTA) to a total volume of 200 μL and put into a clear UV 96-well plate (COSTAR 3635). The proteins were incubated at room temperature for 10–15 min in the dark and then illuminated for 2 min under the LED plate. They were then immediately placed into the TECAN plate reader, and 450-nm absorbance was read every 7 s for 5 min.
Larval Immunohistochemistry.
Wandering third-instar larvae were dissected and stained according to standard procedures (61). Larvae exposed to light were exposed 9 inches below a fluorescent lamp (power 1.6 mW, 488 nm; 1.8 mW, 470 nm) on an agar plate with a small amount of thin yeast paste at room temperature (21–22 °C). The dark controls were also kept on apple plates, but wrapped in foil at room temperature (21–22 °C). Larvae were mounted on slides with Vectashield (VectorLabs) and kept at 4 °C until imaging. For antibody information and imaging, see SI Appendix.
Cryosectioning.
Cryosectioning procedure was adapted from ref. 62. See SI Appendix.
Survival and Motor Behavior Assay.
For both assays, flies were raised in a vial wrapped in foil and kept in a cabinet in the dark at room temperature (21–22 °C). Flies were collected within 24 h of eclosion and put into fresh vials for either the light or dark condition. For the light condition, fly vials were kept 18 inches below a fluorescent lamp (power 1.2 mW, 488 nm; 1.2 mW, 470 nm), the dark condition flies were also kept under the lamp but in foil-wrapped vials and covered with a box lid to keep a consistent temperature but prevent light from entering the vials. Fly survival was recorded every 1–3 d, and flies were flipped into fresh vials every 3–4 d. For the motor behavior assay, flies were kept in identical conditions, but exactly 10 flies were put into every vial. On the day or hour of testing, the vial was marked with a line 3 cm above the food line. The vial was then banged with mild force on a padded surface, and the number of flies to cross the 3-cm line in 90 s was recorded. This was converted into a ratio of flies to successfully cross the line. Each vial of 10 flies was tested three to five times at each time period with at least a 2-min rest between trials. In the case that a fly would die in a vial before the experiment time was finished, that fly was considered to not cross the line and included in the total ratio.
MN1 Scoring.
Larval pelts were mounted on glass slides with vectashield. Each slide was blinded. Each compartment (cell body, dendrite, axon) of each neuron was scored and recorded using the following guidelines: A cell body was considered to have “weak damage” if it was starting to bleb, “severe damage” if it had a lot of blebbing or was breaking down, and “removed” if it had been removed. An axon was considered to have weak damage if it was starting to bead and fragment, severe damage if it had significant fragmentation, and removed if it had been removed or was mostly removed. Dendrites were considered to have weak damage if they were beginning to bead, severe damage if they were fragmented, and removed if they had been removed or were mostly removed. The analysis was done under a fluorescent microscope rather than with images to more easily see Z layers.
Da Neurite Tracing.
Dendrites were traced using the FIJI simple neurite tracer. Each dendrite was started at the cell body, and each branch was traced to the terminal. If the branches were slightly beaded but not broken, they were traced through and counted as the total dendrite length. If the branch was severed then it was not traced through.
Supplementary Material
Acknowledgments
We thank Laura DeVault, Shan Meltzer, and Yuh Nung Jan for flies and microscope use; the Nikon Imaging Center, Kurt Thorn, and Delaine Larsen for microscope use and assistance; Bryan Seybold for assistance with LED array design and construction; May Tran for assistance cryosectioning Drosophila retinas; Scott Gradia (California Institute for Quantitative Biosciences MacroLab) for expression plasmids; and Laura DeVault, Bryan Seybold, G.W.D. laboratory members, and J.A.W. laboratory members for helpful discussions and advice. Research was supported by NIH Grants 1R01GM097316-01 Regulating proteolysis to affect apoptosis and 2R01 GM081051 Global analysis of apoptosis (to J.A.W.) and National Institute of Neurological Disorders and Stroke 1R35NS097212 (to G.W.D.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705064114/-/DCSupplemental.
References
- 1.Difato F, et al. Combined optical tweezers and laser dissector for controlled ablation of functional connections in neural networks. J Biomed Opt. 2011;16:051306. doi: 10.1117/1.3560268. [DOI] [PubMed] [Google Scholar]
- 2.Roeser T, Baier H. Visuomotor behaviors in larval zebrafish after GFP-guided laser ablation of the optic tectum. J Neurosci. 2003;23:3726–3734. doi: 10.1523/JNEUROSCI.23-09-03726.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ertürk A, Wang Y, Sheng M. Local pruning of dendrites and spines by caspase-3-dependent and proteasome-limited mechanisms. J Neurosci. 2014;34:1672–1688. doi: 10.1523/JNEUROSCI.3121-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makhijani K, et al. Precision optogenetic tool for selective single- and multiple-cell ablation in a live animal model system. Cell Chem Biol. 2017;24:110–119. doi: 10.1016/j.chembiol.2016.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Uttara B, Singh AV, Zamboni P, Mahajan RT. Oxidative stress and neurodegenerative diseases: A review of upstream and downstream antioxidant therapeutic options. Curr Neuropharmacol. 2009;7:65–74. doi: 10.2174/157015909787602823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Julien O, Wells JA. Caspases in cell death and remodeling. Cell Death Differ. 2017;24:1380–1389. doi: 10.1038/cdd.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol. 2007;35:495–516. doi: 10.1080/01926230701320337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang CF, et al. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell. 2013;153:896–909. doi: 10.1016/j.cell.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 10.Laprell L, et al. Optical control of NMDA receptors with a diffusible photoswitch. Nat Commun. 2015;6:8076. doi: 10.1038/ncomms9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim B, Lin MZ. Optobiology: Optical control of biological processes via protein engineering. Biochem Soc Trans. 2013;41:1183–1188. doi: 10.1042/BST20130150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riggsbee CW, Deiters A. Recent advances in the photochemical control of protein function. Trends Biotechnol. 2010;28:468–475. doi: 10.1016/j.tibtech.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walters J, et al. A constitutively active and uninhibitable caspase-3 zymogen efficiently induces apoptosis. Biochem J. 2009;424:335–345. doi: 10.1042/BJ20090825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donepudi M, Grütter MG. Structure and zymogen activation of caspases. Biophys Chem. 2002;101-102:145–153. doi: 10.1016/s0301-4622(02)00151-5. [DOI] [PubMed] [Google Scholar]
- 16.Chai J, et al. Crystal structure of a procaspase-7 zymogen: Mechanisms of activation and substrate binding. Cell. 2001;107:399–407. doi: 10.1016/s0092-8674(01)00544-x. [DOI] [PubMed] [Google Scholar]
- 17.Thomsen ND, Koerber JT, Wells JA. Structural snapshots reveal distinct mechanisms of procaspase-3 and -7 activation. Proc Natl Acad Sci USA. 2013;110:8477–8482. doi: 10.1073/pnas.1306759110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacKenzie SH, et al. Lengthening the intersubunit linker of procaspase 3 leads to constitutive activation. Biochemistry. 2013;52:6219–6231. doi: 10.1021/bi400793s. [DOI] [PubMed] [Google Scholar]
- 19.Shi Y. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell. 2002;9:459–470. doi: 10.1016/s1097-2765(02)00482-3. [DOI] [PubMed] [Google Scholar]
- 20.Renicke C, Schuster D, Usherenko S, Essen L-O, Taxis C. A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem Biol. 2013;20:619–626. doi: 10.1016/j.chembiol.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 21.Strickland D, et al. TULIPs: Tunable, light-controlled interacting protein tags for cell biology. Nat Methods. 2012;9:379–384. doi: 10.1038/nmeth.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pudasaini A, El-Arab KK, Zoltowski BD. LOV-based optogenetic devices: Light-driven modules to impart photoregulated control of cellular signaling. Front Mol Biosci. 2015;2:18. doi: 10.3389/fmolb.2015.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu YI, et al. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strickland D, Moffat K, Sosnick TR. Light-activated DNA binding in a designed allosteric protein. Proc Natl Acad Sci USA. 2008;105:10709–10714. doi: 10.1073/pnas.0709610105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strickland D, et al. Rationally improving LOV domain-based photoswitches. Nat Methods. 2010;7:623–626. doi: 10.1038/nmeth.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoersch D, Bolourchian F, Otto H, Heyn MP, Bogomolni RA. Dynamics of light-induced activation in the PAS domain proteins LOV2 and PYP probed by time-resolved tryptophan fluorescence. Biochemistry. 2010;49:10811–10817. doi: 10.1021/bi101413v. [DOI] [PubMed] [Google Scholar]
- 27.Kennis JTM, et al. Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry. 2003;42:3385–3392. doi: 10.1021/bi034022k. [DOI] [PubMed] [Google Scholar]
- 28.Zayner JP, Antoniou C, Sosnick TR. The amino-terminal helix modulates light-activated conformational changes in AsLOV2. J Mol Biol. 2012;419:61–74. doi: 10.1016/j.jmb.2012.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harper SM, Neil LC, Gardner KH. Structural basis of a phototropin light switch. Science. 2003;301:1541–1544. doi: 10.1126/science.1086810. [DOI] [PubMed] [Google Scholar]
- 30.Leaver-Fay A, et al. Scientific benchmarks for guiding macromolecular energy function improvement. Methods Enzymol. 2013;523:109–143. doi: 10.1016/B978-0-12-394292-0.00006-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leaver-Fay A, et al. ROSETTA3: An object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krauss U, Lee J, Benkovic SJ, Jaeger K-E. LOVely enzymes–Towards engineering light-controllable biocatalysts. Microb Biotechnol. 2010;3:15–23. doi: 10.1111/j.1751-7915.2009.00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 34.Hawkins CJ, et al. The Drosophila caspase DRONC cleaves following glutamate or aspartate and is regulated by DIAP1, HID, and GRIM. J Biol Chem. 2000;275:27084–27093. doi: 10.1074/jbc.M000869200. [DOI] [PubMed] [Google Scholar]
- 35.Gao G, McMahon C, Chen J, Rong YS. A powerful method combining homologous recombination and site-specific recombination for targeted mutagenesis in Drosophila. Proc Natl Acad Sci USA. 2008;105:13999–14004. doi: 10.1073/pnas.0805843105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venken KJT, Simpson JH, Bellen HJ. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011;72:202–230. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay BA, Wolff T, Rubin GM. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- 38.Herman-Bachinsky Y, Ryoo H-D, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14:861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- 39.Ferreira KS, et al. Caspase-3 feeds back on caspase-8, Bid and XIAP in type I Fas signaling in primary mouse hepatocytes. Apoptosis. 2012;17:503–515. doi: 10.1007/s10495-011-0691-0. [DOI] [PubMed] [Google Scholar]
- 40.Kale A, Li W, Lee C-H, Baker NE. Apoptotic mechanisms during competition of ribosomal protein mutant cells: Roles of the initiator caspases Dronc and Dream/Strica. Cell Death Differ. 2015;22:1300–1312. doi: 10.1038/cdd.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, et al. Mitochondria and caspases tune Nmnat-mediated stabilization to promote axon regeneration. PLoS Genet. 2016;12:e1006503. doi: 10.1371/journal.pgen.1006503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn JT, et al. Evidence that Wallerian degeneration and localized axon degeneration induced by local neurotrophin deprivation do not involve caspases. J Neurosci. 2000;20:1333–1341. doi: 10.1523/JNEUROSCI.20-04-01333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon DJ, et al. A caspase cascade regulating developmental axon degeneration. J Neurosci. 2012;32:17540–17553. doi: 10.1523/JNEUROSCI.3012-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perry VH, Lunn ER, Brown MC, Cahusac S, Gordon S. Evidence that the rate of Wallerian degeneration is controlled by a single autosomal dominant gene. Eur J Neurosci. 1990;2:408–413. doi: 10.1111/j.1460-9568.1990.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 45.Conforti L, et al. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc Natl Acad Sci USA. 2000;97:11377–11382. doi: 10.1073/pnas.97.21.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoopfer ED, et al. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning. Neuron. 2006;50:883–895. doi: 10.1016/j.neuron.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 47.Song Z, et al. Biochemical and genetic interactions between Drosophila caspases and the proapoptotic genes rpr, hid, and grim. Mol Cell Biol. 2000;20:2907–2914. doi: 10.1128/mcb.20.8.2907-2914.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Crawford ED, et al. Conservation of caspase substrates across metazoans suggests hierarchical importance of signaling pathways over specific targets and cleavage site motifs in apoptosis. Cell Death Differ. 2012;19:2040–2048. doi: 10.1038/cdd.2012.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zorn JA, Wolan DW, Agard NJ, Wells JA. Fibrils colocalize caspase-3 with procaspase-3 to foster maturation. J Biol Chem. 2012;287:33781–33795. doi: 10.1074/jbc.M112.386128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mills E, Chen X, Pham E, Wong S, Truong K. Engineering a photoactivated caspase-7 for rapid induction of apoptosis. ACS Synth Biol. 2012;1:75–82. doi: 10.1021/sb200008j. [DOI] [PubMed] [Google Scholar]
- 51.Tang HL, Tang HM, Fung MC, Hardwick JM. In vivo CaspaseTracker biosensor system for detecting anastasis and non-apoptotic caspase activity. Sci Rep. 2015;5:9015. doi: 10.1038/srep09015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding AX, et al. CasExpress reveals widespread and diverse patterns of cell survival of caspase-3 activation during development in vivo. Elife. 2016;5:e10936. doi: 10.7554/eLife.10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Campbell DS, Okamoto H. Local caspase activation interacts with Slit-Robo signaling to restrict axonal arborization. J Cell Biol. 2013;203:657–672. doi: 10.1083/jcb.201303072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuo CT, Zhu S, Younger S, Jan LY, Jan YN. Identification of E2/E3 ubiquitinating enzymes and caspase activity regulating Drosophila sensory neuron dendrite pruning. Neuron. 2006;51:283–290. doi: 10.1016/j.neuron.2006.07.014. [DOI] [PubMed] [Google Scholar]
- 55.Li Z, et al. Caspase-3 activation via mitochondria is required for long-term depression and AMPA receptor internalization. Cell. 2010;141:859–871. doi: 10.1016/j.cell.2010.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schoenmann Z, et al. Axonal degeneration is regulated by the apoptotic machinery or a NAD+-sensitive pathway in insects and mammals. J Neurosci. 2010;30:6375–6386. doi: 10.1523/JNEUROSCI.0922-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Williams DW, Kondo S, Krzyzanowska A, Hiromi Y, Truman JW. Local caspase activity directs engulfment of dendrites during pruning. Nat Neurosci. 2006;9:1234–1236. doi: 10.1038/nn1774. [DOI] [PubMed] [Google Scholar]
- 58.Gillingwater TH, Ribchester RR. The relationship of neuromuscular synapse elimination to synaptic degeneration and pathology: Insights from WldS and other mutant mice. J Neurocytol. 2003;32:863–881. doi: 10.1023/B:NEUR.0000020629.51673.f5. [DOI] [PubMed] [Google Scholar]
- 59.Keller LC, et al. Glial-derived prodegenerative signaling in the Drosophila neuromuscular system. Neuron. 2011;72:760–775. doi: 10.1016/j.neuron.2011.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolan DW, Zorn JA, Gray DC, Wells JA. Small-molecule activators of a proenzyme. Science. 2009;326:853–858. doi: 10.1126/science.1177585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Massaro CM, Pielage J, Davis GW. Molecular mechanisms that enhance synapse stability despite persistent disruption of the spectrin/ankyrin/microtubule cytoskeleton. J Cell Biol. 2009;187:101–117. doi: 10.1083/jcb.200903166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolff T. Beta-galactosidase activity staining of frozen adult Drosophila retinas. Cold Spring Harb Protoc. 2010;2010:pdb.prot5418. doi: 10.1101/pdb.prot5418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.