Significance
Elevated downstream signals of androgen receptor (AR) and its variants are important for prostate cancer progression. We show that an RNA-binding transcriptional and splicing factor, splicing factor proline and glutamine-rich (PSF/SFPQ), predicts worse prognosis of prostate cancer patients. Inhibition of PSF expression repressed treatment-resistant prostate tumor growth in our animal model. Our global analysis of PSF-binding RNAs revealed that PSF enhances AR-regulated genes and noncoding RNAs associated with prostate cancer progression. Interestingly, various splicing factors, which are primary targets of PSF, are upregulated in metastatic prostate tumors. These enhanced factors form complexes with PSF to promote AR expression and splicing. Our findings suggest a role of RNA-binding protein for AR activation for prostate cancer progression.
Keywords: androgen receptor, RNA-binding protein, PSF, NONO, prostate cancer
Abstract
Developing therapeutic approaches are necessary for treating hormone-refractory prostate cancer. Activation of androgen receptor (AR) and its variants’ expression along with the downstream signals are mostly important for disease progression. However, the mechanism for marked increases of AR signals and its expression is still unclear. Here, we revealed that various spliceosome genes are aberrantly induced by RNA-binding protein PSF, leading to enhancement of the splicing activities for AR expression. Our high-speed sequence analyses identified global PSF-binding transcripts. PSF was shown to stabilize and activate key long noncoding RNAs and AR-regulated gene expressions in prostate cancer cells. Interestingly, mRNAs of spliceosome-related genes are putative primary targets of PSF. Their gene expressions are up-regulated by PSF in hormone-refractory prostate cancer. Moreover, PSF coordinated these spliceosome proteins to form a complex to promote AR splicing and expression. Thus, targeting PSF and its related pathways implicates the therapeutic possibility for hormone-refractory prostate cancer.
Androgen receptor (AR) regulates many genes central to the identity and behavior of prostate cancer cells (1). AR functions in a ligand-dependent manner in many prostate cancers, and androgen deprivation therapy is effective in inhibiting tumor growth. However, most of the patients acquire resistance to this therapy and eventually suffer from castration-resistant prostate cancer (CRPC) (2, 3). Previous studies have discovered the importance of enhanced AR downstream signaling (3, 4), expression of AR and its splice variants lacking the ligand binding domain (ARVs) in CRPC (5, 6). The variant AR-V7 was shown to regulate distinct and androgen-independent activation of its downstream signals, which contributes to the development of CRPC (7). Thus, targeting AR, AR-V7, and its downstream signals could have efficacy in treating CRPC.
Long noncoding (lnc) RNAs function through interaction with epigenetic factors in cancer (8–11). Previous reports highlighted the lncRNA-mediated association of RNA-binding proteins or transcription factors with specific genomic regions for prostate cancer progression (10, 11). We have reported that androgen-induced lncRNA (named CTBP1-AS) in the antisense region of carboxyl terminal binding protein 1 (CTBP1) promotes castration-resistant tumor growth. CTBP1-AS interacts with an RNA-binding transcriptional and splicing factor, splicing factor proline and glutamine-rich (PSF/SFPQ), and represses cell cycle regulators by epigenetic mechanism (11). We also found that PSF and its interacting factor, non–POU-domain–containing octamer-binding protein (NONO), have an important role in prostate cancer cell proliferation, suggesting the possibility that these RNA-binding proteins are also implicated in the disease progression (11).
PSF is a ubiquitous nuclear protein essential for neural development by regulating axon viability (12, 13). PSF has a unique structure possessing both DNA- and RNA-binding domains, implicated in transcription and nuclear RNA processing (14–16). However, the transcriptional and posttranscriptional specific targets and the clinical significance of these proteins in prostate cancer progression still remain to be elucidated. Here, we conducted global systematic analyses to determine both transcriptional targets and PSF-binding RNAs using CRPC model cells. Notably, we found that a broad range of spliceosome genes are primary targets of PSF, and they are induced for activation of splicing and mRNA production in CRPC. Thus, our findings shed light on the regulatory mechanism of various splicing factors by RNA-binding proteins in prostate cancer progression.
Results
PSF Promotes Castration-Resistant Prostate Tumor Growth.
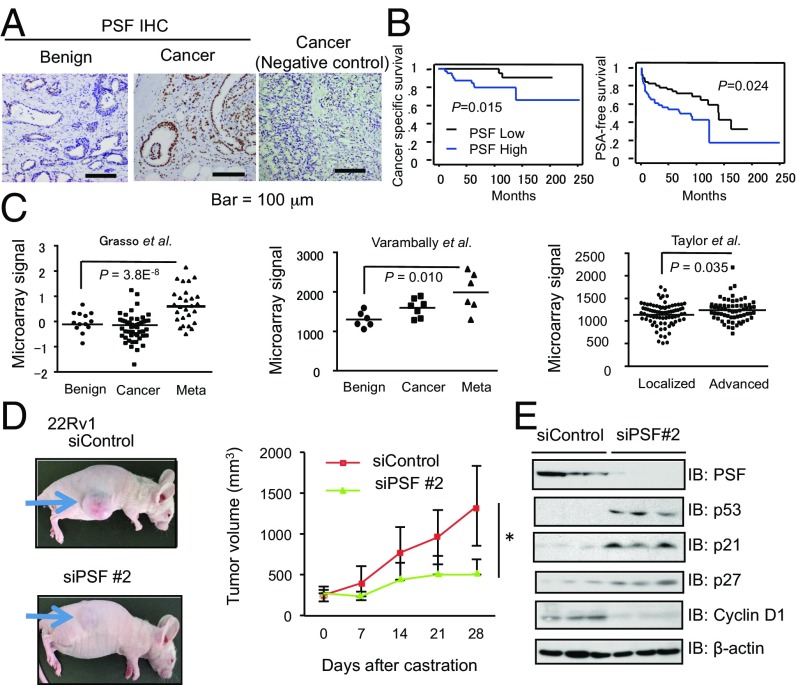
To investigate the role of PSF in prostate cancer, we analyzed the expression level of PSF. In prostate cancer cell lines, we observed increased expression of PSF in several prostate cancer cells such as DU145, LNCaP, and LNCaP-derived CRPC model cells, long-term androgen deprivation (LTAD) cells compared with normal prostate epithelial cells (PrEC) (SI Appendix, Fig. S1A). We evaluated the PSF protein expression in clinical tumor samples by immunohistochemical (IHC) analysis (Fig. 1A) using anti-PSF specific antibody (SI Appendix, Fig. S1B). Low PSF staining was observed in benign prostate tissues, whereas high PSF staining was observed in a subset of tumor samples, and higher expression of PSF correlated with the patients’ cancer-specific survival after surgery and the PSA-free survival after hormone therapy (Fig. 1B and SI Appendix, Fig. S1C). According to public databases of expression profiles in prostate cancer clinical samples (such as Oncomine) (17–19), PSF mRNA expression is significantly elevated in metastatic or advanced prostate cancer samples, suggesting that PSF expression is associated with prostate cancer progression (Fig. 1C and SI Appendix, Fig. S1 D–F).
Fig. 1.
Clinical significance of PSF expression in prostate cancer progression and CRPC tumor growth. (A) PSF is up-regulated in a subset of prostate cancer samples. Immunohistochemistry of PSF in prostate cancer and benign prostate tissues (n = 102) was performed. A negative control using normal rabbit IgG as a primary antibody in the case with PSF positive staining is shown. (B) Higher expression of PSF (n = 51) is a prognostic factor for prostate cancer patients. Kaplan–Meier analysis using the log-rank test was performed. (C) PSF expression levels in metastatic prostate cancer tissues were analyzed using public database (GSE35988, GSE3325, GSE21034). Meta, metastatic cancer; Localized, stage < T1; Advanced, stage > T2. (D) Nude mice were inoculated with 22Rv1 cells. After tumor development, we performed castration and divided into two groups randomly. Tumor growth of xenografted 22Rv1 cells in nude mice treated with siControl or siPSF is shown (n = 8). Representative views of tumors in nude mice are shown. (E) Western blot analysis was performed to evaluate PSF and its downstream signals in tumors. Values represent the mean ± SD. *P < 0.05, **P < 0.01.
We previously reported that PSF-inhibited cell cycle-associated gene such as p53 and SMAD3 expression by transcriptional mechanism in AR-dependent prostate cancer cell lines (LNCaP, VCaP) and their CRPC models, LTAD cells (11). In this study, by using short-interference (si) RNAs targeting PSF, we efficiently depleted the expression of PSF in other hormone-refractory prostate cancer cells such as DU145 and 22Rv1 cells (SI Appendix, Fig. S2A). We then observed the inhibited cell growth of these prostate cancer cells by siPSF treatment (SI Appendix, Fig. S2B). Next, we examined the roles of PSF using in vivo xenograft assays. Injection of PSF siRNA into the tumors significantly inhibited tumor growth derived from LNCaP cells (SI Appendix, Fig. S2 C and D). As for in vivo model of CRPC, we injected 22Rv1 cells into nude mice s.c., castrated the mice after the tumors formed, followed by injection of siPSF or siControl into the tumors. Interestingly, siPSF treatment reduced the growth of tumors derived from 22Rv1 cells (Fig. 1D). In tumors with reduced PSF expression, induced cell cycle regulators such as p53 and p21 proteins were also observed, suggesting that PSF could be a potential target for the treatment of CRPC (Fig. 1E). These data highlight a potential role of increased PSF in the CRPC tumor growth. (Additional results are shown in SI Appendix, Text S1 and Figs. S3–S5.)
Global Mapping of PSF-Binding Transcripts Revealed That AR-Regulated Genes and Prostate Cancer-Associated lncRNAs Are Specific Targets of PSF.
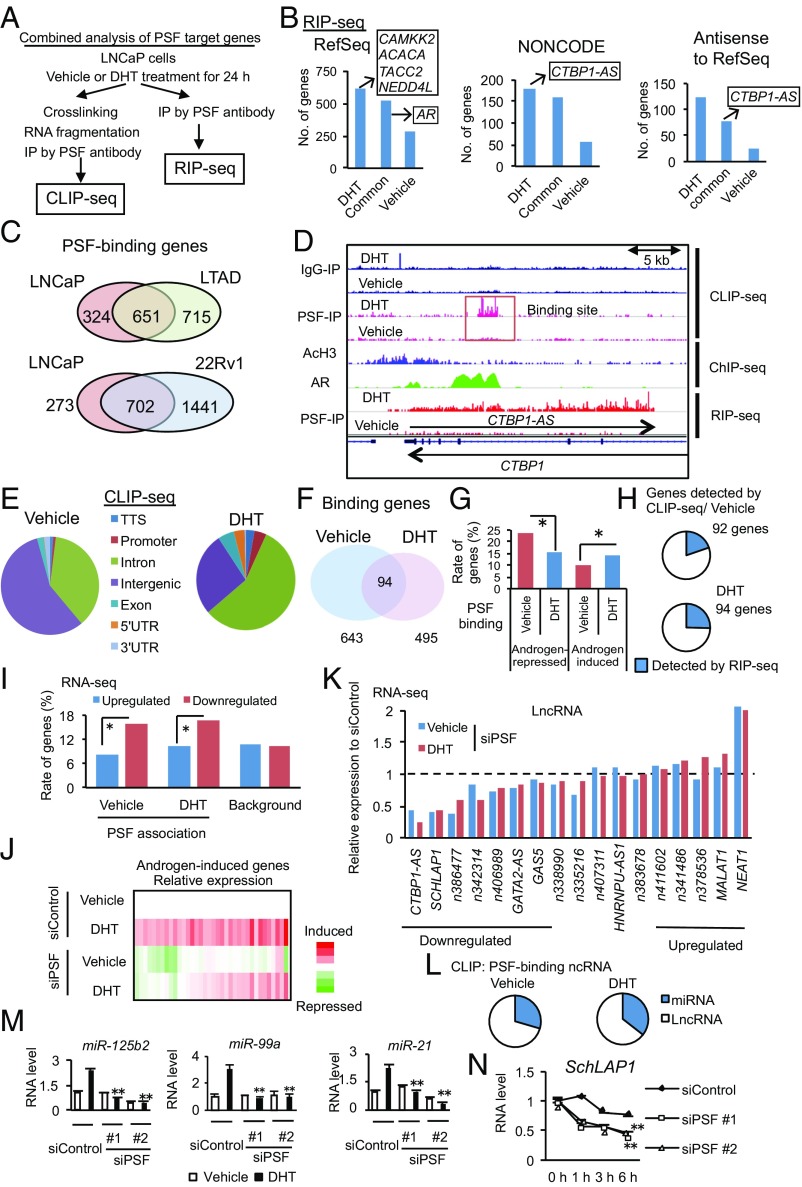
To explore other functions of PSF in prostate cancer, we next analyzed RNAs bound with PSF using deep-sequence analysis (Fig. 2A). First, we performed RNA immunoprecipitation coupled with deep sequencing (RIP-seq) in LNCaP cells treated with vehicle or DHT. By mapping sequence tags to RefSeq, GENCODE, and NONCODE, we identified putative PSF target genes (Fold > 2, P < 10−5) including lncRNAs (Fig. 2B and SI Appendix, Fig. S6 A–C). We further performed RIP-seq in CRPC model cells (LTAD and 22Rv1) (Fig. 2C). In LTAD and 22Rv1 cells, we found more binding genes, which significantly overlapped with those of LNCaP cells. Interestingly, CTBP1-AS was identified as one of the most significant lncRNAs in this comprehensive study among NONCODE and RefSeq antisense transcripts (SI Appendix, Fig. S6B), suggesting the specific interaction between PSF and CTBP1-AS in LNCaP cells treated with DHT. In addition, we observed that DHT treatment changed the binding transcripts of PSF. We found that androgen-regulated genes such as CAMKK2 (20) are enriched among DHT-specific target genes of PSF (Fig. 2B). In contrast to the PSF targets identified by ChIP-seq (SI Appendix, Fig. S4A), pathway analysis showed that spliceosome genes are assumed to be the primary targets of PSF at RNA level in LNCaP cells (SI Appendix, Fig. S6D), suggesting this gene cluster is specifically regulated by PSF as an RNA-binding protein. Prostate cancer-associated signals were also significantly included in these pathways. We next performed crosslinking immunoprecipitation (CLIP)-seq (21, 22) in LNCaP cells to identify the peak positions of PSF bindings (Fig. 2D). PSF-binding sites were mainly distributed in the intron regions (Fig. 2E) of either protein-coding or noncoding genes (SI Appendix, Fig. S6E). Motif analysis (MEME) (23) revealed that GA-rich motifs were evident in the binding peaks of PSF (SI Appendix, Fig. S6F). We then identified putative PSF-binding genes including annotated RefSeq genes (Vehicle: 643 DHT: 495 genes) and noncoding genes among them (Vehicle: 173 DHT: 125) (Fig. 2F and SI Appendix, Fig. S6G). Different lists of PSF-binding genes in the absence and presence of DHT treatment implicated the involvement of AR in the PSF bindings to RNA. In fact, we found that transcripts bound with PSF in the presence of DHT were mainly derived from DHT-induced genes (Fig. 2G). Conversely, transcripts bound with PSF in the absence of DHT were derived from repressed genes by DHT. The list of PSF-binding genes obtained by CLIP-seq showed significant but partial overlap with those by RIP-seq, probably due to the different techniques (Fig. 2H). Those PSF-binding genes were validated by RIP assays (SI Appendix, Fig. S6H). Moreover, we found that PSF-binding genes in the presence of DHT were up-regulated in prostate cancer according to The Cancer Genome Atlas (TCGA) database (SI Appendix, Fig. S6I), suggesting the importance of PSF interaction with transcripts for prostate cancer progression.
Fig. 2.
Global analysis of PSF-binding RNAs revealed roles of PSF in the spliceosome and lncRNA maturation. (A) Schematic diagram of global analysis of PSF-binding RNAs. (B) Identification of RefSeq and NONCODE transcripts bound with PSF. (C) Global mapping of PSF-binding transcripts in CRPC cells. RIP-seq was performed in LNCaP, LTAD, and 22Rv1 cells. The number of significant PSF-binding genes annotated in RefSeq are shown. (D) Representative mapping of PSF-binding RNAs in the CTBP1-AS locus. We present the results of RIP-seq, CLIP-seq, and ChIP-seq (AR, AcH3) in LNCaP cells. Arrows indicate the direction of CTBP1 and CTBP1-AS. (E) Genomic location of the identified peaks in the presence and absence of androgen. TTS, terminal transcription site; 5′UTR, 5′ untranslated region; 3′UTR, 3′ untranslated region. (F) The difference of PSF-binding genes identified by CLIP in the absence and presence of DHT. (G) PSF binds to androgen-induced genes significantly in the presence of DHT. The number of PSF-binding genes was counted, and χ2 test was performed. (H) Overlap of identified genes by CLIP-seq with RIP-seq. (I) Regulation of PSF-binding genes in prostate cancer cells. LNCaP cells were treated with siControl or siPSF#1. After 48 h incubation, cells were treated with vehicle or DHT for 24 h. PSF-binding genes identified by RIP-seq were selected, and changes in their expression levels by PSF knockdown are summarized. (J) PSF regulates androgen-dependent gene activation. Heatmap results of RNA-seq in androgen-regulated genes are shown. (K) Regulation of lncRNAs by PSF binding. PSF-binding lncRNAs identified by CLIP-seq and RIP-seq are selected. (L) The ratio of miRNAs and lncRNAs in PSF-binding lncRNAs identified by CLIP. (M) Expressions of androgen-induced miRNAs (miR-125b2, miR-99a, and miR-21) were repressed by knockdown of PSF. LNCaP cells were treated with siControl, siPSF#1, or siPSF#2 for 48 h. We then measured miRNA expression levels by qRT-PCR in cells after treatment of vehicle or DHT for 24 h (n =3). (N) mRNA stability of SchLAP1 was decreased by knockdown of PSF. LNCaP cells are treated with siControl, siPSF #1, or siPSF #2 for 48 h. To inhibit transcription, actinomycin-D (1 nM) was added. After incubation for indicated times, mRNA expression levels were measured by qRT-PCR (n = 3). Values represent the mean ± SD. *P < 0.05, **P < 0.01.
To understand how PSF affects the binding transcripts, we performed directional RNA-seq to investigate the gene regulation by PSF. These results revealed that PSF mainly increases the expression of its binding transcripts (Fig. 2I). In addition, knockdown of PSF resulted in a marked decrease in DHT-induced expression of AR target genes (Fig. 2J). For lncRNAs, we observed both negative [NEAT1, MALAT1 (9, 24, 25)] and positive [CTBP1-AS, SchLAP1 (26)] effects on expression in LNCaP cells (Fig. 2K). These effects were also confirmed by quantitative reverse transcription PCR (qRT-PCR) in both LNCaP and VCaP cells (SI Appendix, Fig. S7 A and B). Interestingly, we found that lncRNAs overlapping with miRNAs (such as miR-99AHG) were also included in the list of genes with PSF CLIP peaks (Fig. 2L). We then performed miRNA-qPCR, showing that the AR-regulated miRNAs (27) were repressed by PSF knockdown. This suggests that production of androgen-mediated miRNAs could also be modulated by PSF binding to its primary transcripts (Fig. 2M and SI Appendix, Fig. S7C). To investigate the mechanism of PSF for up-regulation of its target genes, we analyzed transcript stability by incubating cells with actinomycin-D. Consistent with the result of RNA-seq, we observed reduced half times in these PSF-binding genes in VCap and LNCaP cells by PSF knockdown (Fig. 2N and SI Appendix, Fig. S8). Thus, PSF was assumed to control the gene expression of coding and noncoding signals associated with prostate cancer, spliceosome genes, and AR-regulated genes by binding to these gene transcripts to increase stability. (Additional results are shown in SI Appendix, Text S2 and Fig. S9.)
PSF Binding to AR Transcripts Is Enhanced in Hormone-Refractory Prostate Cancer Cells for Activating Their Expression and Splicing.
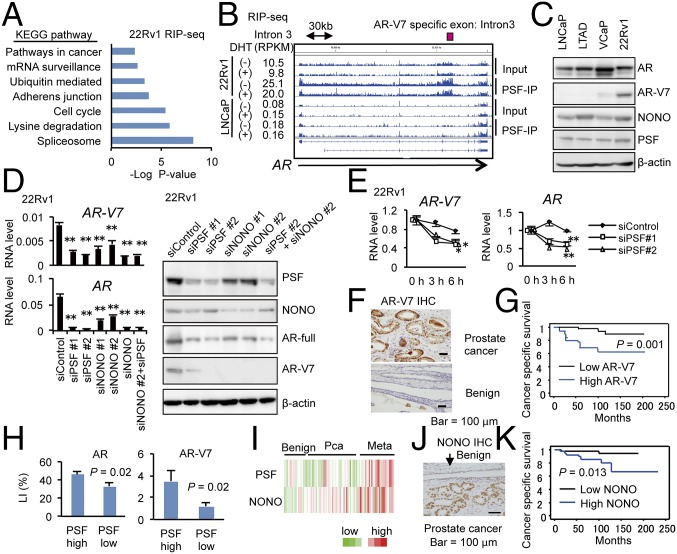
We next investigated the PSF function in CRPC model cells. Importantly, the pathway analysis of PSF-binding genes in 22Rv1 cells indicated the importance of PSF in induction of spliceosome genes, as observed in LNCaP cells (Fig. 3A and SI Appendix, Figs. S6D and S10A). In addition, the interaction of PSF with prostate cancer-associated signals was also enhanced. For example, the binding of PSF to the introns of SchLAP1 and AR (Fig. 3B and SI Appendix, Fig. S10 B–D) was enhanced in LTAD and 22Rv1 cells compared with LNCaP cells, suggesting the role of PSF in expressing these important signals in CRPC (SI Appendix, Fig. S10E).
Fig. 3.
PSF bindings to the target transcripts are enhanced in CRPC. (A) Pathway analysis of PSF-binding genes in 22Rv1 cells is shown. (B) Representative mapping of PSF-binding RNAs in AR locus. (C) Expression levels of PSF, NONO, AR, and AR-V7 in prostate cancer cells. Lysates from LNCaP, LTAD, VCaP, and 22Rv1 cells were used for immunoblots with indicated antibodies. (D) Regulation of AR and AR-V7 expression by PSF and NONO in CRPC model cells. (Left) The 22Rv1 cells were treated with siPSF or siNONO for 72 h. AR and AR-V7 mRNA levels were measured by qRT-PCR (n = 3). (Right) Lysates from 22Rv1 cells transfected with siPSF or siNONO are used for immunoblots to detect AR and AR-V7 protein levels. (E) mRNA stability of AR and AR-V7 was decreased by knockdown of PSF. The 22Rv1 cells are treated with siControl, siPSF #1, or siPSF #2 for 48 h. To inhibit transcription, actinomycin-D (1 nM) was added. After incubation for indicated times, mRNA expression levels were measured by qRT-PCR (n = 3). (F) Immunohistochemistry of AR-V7 in prostate cancer tissues (n = 102) was performed. (G) AR-V7 expression (high, n = 18; low, n = 86) is a strong prognostic factor for prostate cancer patients. Cases with labeling index (LI) > 10% were determined to be AR-V7 high expression. (H) Positive correlation of PSF expression with AR and AR-V7 levels in prostate cancer tissues. LI in each group is shown. (I) NONO as well as PSF was up-regulated in metastatic prostate cancer tissues. Heatmap results using microarray database (GSE35988) is shown. (J) Immunohistochemistry of NONO in prostate cancer tissues (n = 102). (K) NONO expression (high, n = 51; low, n = 51) is a prognostic factor of poor outcome of prostate cancer patients. Values represent the mean ± SD. *P < 0.05, **P < 0.01.
We then investigated the role of PSF in AR splicing and posttranscriptional regulation. We have detected CLIP signals in the intron and 3′-UTR regions of AR gene, suggesting the involvement of PSF in AR mRNA stability and splicing (SI Appendix, Fig. S10F). First, we confirmed the binding of PSF with AR and AR-V7, an AR-splicing variant associated with CRPC development, by RIP assays (SI Appendix, Fig. S10 B and G). In addition, Western blot analyses showed that AR-V7 protein expression was detected in VCaP and 22Rv1 cells (Fig. 3C). Interestingly, PSF expression was also up-regulated in these cell lines compared with LNCaP and LTAD, in which AR-V7 protein could not be detected. To determine whether PSF induces AR-V7 protein production or not, we performed qRT-PCR and Western blot analysis (Fig. 3D) by using siRNAs targeting PSF and its associated factor, NONO. Surprisingly, PSF knockdown in 22Rv1 cells inhibited full-length AR and AR-V7 mRNA and protein dramatically, whereas AR mRNA (SI Appendix, Fig. S11A) and protein (11) levels were not affected by PSF knockdown in LNCaP cells. Knockdown of NONO also effectively reduced the expression of AR-V7 as well as AR at mRNA and protein levels in 22Rv1 cells (Fig. 3D). Analysis of mRNA stability of AR and AR-V7 by inhibiting transcription indicated that PSF and NONO are involved in the posttranscriptional regulation of these mRNAs (Fig. 3E and SI Appendix, Fig. S11B). We also observed similar results in VCaP cells (SI Appendix, Fig. S11 C–E). As expected, our AR-V7 IHC analysis in clinical samples has shown that AR-V7 expression predicts poor prognosis of prostate cancer patients (Fig. 3 F and G). Importantly, AR-V7 immunoreactivity score is correlated with that of PSF expression level, suggesting the role of PSF in the production of AR-V7 protein (Fig. 3H). Microarray dataset (Fig. 3I) and our IHC analysis (Fig. 3J) revealed that expression of NONO was also elevated in the metastatic prostate cancer tissues and that high expression of NONO was associated with AR-V7 expression (SI Appendix, Fig. S11F) and poor prognosis of patients (Fig. 3K), suggesting the potential role of NONO in prostate cancer progression and AR-V7 expression by interaction with PSF.
Spliceosome Genes Are Primary Targets of PSF and Aberrantly Expressed in Hormone-Refractory Prostate Cancer.
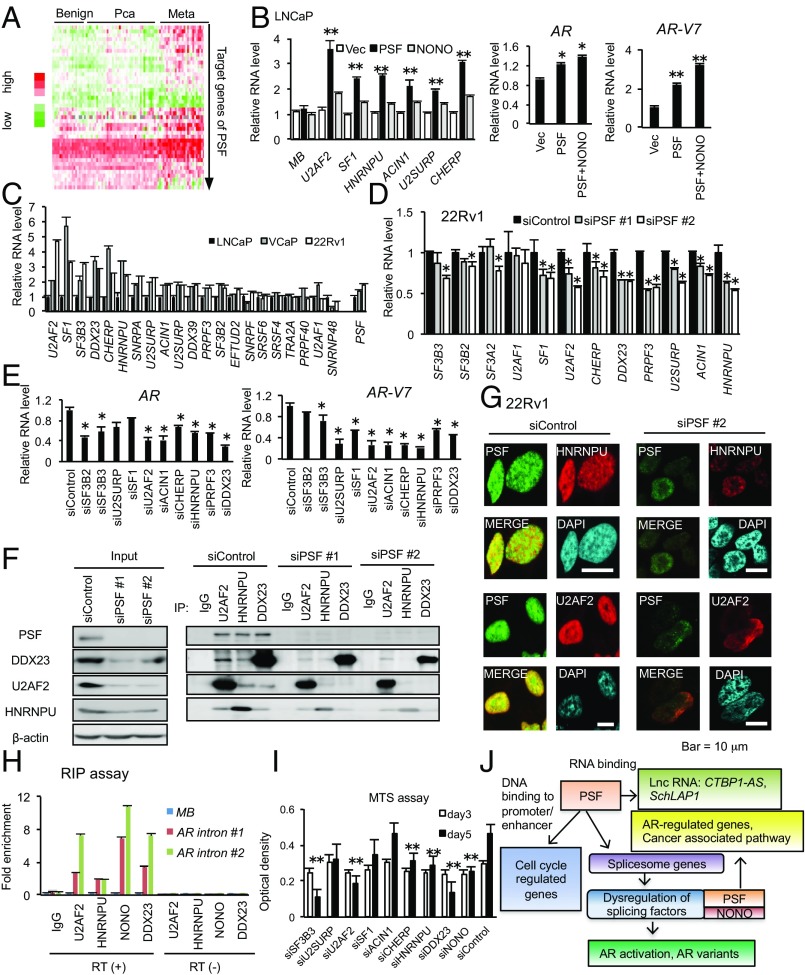
We next hypothesized that elevated expression of spliceosome genes by PSF binding might enhance splicing activity to induce oncogenic effects. Clinically, most of PSF-targeted spliceosome genes are up-regulated in metastatic prostate cancer tissues (Fig. 4A). To test the hypothesis that this up-regulation is mediated by PSF, we selected a set of spliceosome genes up-regulated in the metastatic cancer tissues for further analysis. First, we observed up-regulation of spliceosome genes in addition to the up-regulation of AR and AR-V7 in LNCaP cells overexpressing PSF and NONO that we established previously (13) (Fig. 4B), indicating the role of PSF and NONO in these elevated expressions. We confirmed the up-regulation of these spliceosome genes in AR-V7 positive, VCaP, and 22Rv1 compared with LNCaP cells (Fig. 4C), while PSF and NONO knockdown repressed the expression of these spliceosome genes (Fig. 4D). Knockdown of these spliceosome genes inhibited AR and AR-V7 expression (Fig. 4E and SI Appendix, Fig. S12 A–C). Thus, these results revealed that PSF could increase the expression of spliceosome genes to regulate for AR/AR-V7 production.
Fig. 4.
Spliceosome genes targeted by PSF are up-regulated in CRPC and promote AR splicing by forming a complex with PSF. (A) Expression levels of spliceosome genes (42 genes) targeted by PSF. Microarray expression data were downloaded from GEO database (GSE35988). Meta, metastatic cancer. (B) Overexpression of PSF and NONO enhances the expression level of spliceosome genes. We measured mRNA expression levels of spliceosome genes by qRT-PCR in LNCaP cells stably expressing PSF or NONO and control cells (n =3). For analyzing AR and AR-V7 expression levels, we transfected transiently control vector or NONO to LNCaP cells overexpressing PSF. (C) Expression level of spliceosome genes in AR-V7 positive prostate cancer cells. In LNCaP, VCaP, and 22Rv1 cells, mRNA levels of spliceosome genes targeted by PSF were measured by qRT-PCR (n = 3). (D) Negative regulation of spliceosome genes by knockdown of PSF in CRPC cells. We treated 22Rv1 cells with siPSF #1, #2, or siControl for 72 h. mRNA level of each spliceosome gene was measured by qRT-PCR (n = 3). (E) Regulation of AR and AR-V7 mRNA expression by splicing factors regulated by PSF in CRPC model cells. The 22Rv1 cells were treated with siRNA targeting spliceosome genes for 72 h. AR and AR-V7 mRNA levels were measured by qRT-PCR (n = 3). (F) Forming complex of PSF with splicing factors for modulating AR expression. Lysates of 22Rv1 transfected with siControl or siPSF for 72 h are used for immunoprecipitation. (G) Immunofluorescence analysis of splicing factors and PSF in 22Rv1 cells. (H) RIP analysis of splicing factors in 22Rv1 cells (n = 3). (I) Spliceosome genes regulate CRPC cell growth. Growth of 22Rv1 prostate cancer cells after transfection of siControl or siRNA targeting splicing factors (n = 4). (J) Schematic model of PSF via DNA binding and RNA binding ability. Values represent the mean ± SD. *P < 0.05, **P < 0.01. MB, myoglobin; Vec, vector.
We further investigated whether up-regulation of spliceosome genes could enhance splicing activity cooperating with PSF and NONO in CRPC. Our immunoprecipitation and Western-blotting analysis indicated that PSF interacts with these spliceosome components and was responsible for the complex formation of spliceosome (Fig. 4F). In addition, immunofluorescence analysis showed that this interaction occurred in the nucleus (Fig. 4G). RIP assay also indicated that these factors bind to AR transcripts and act as an integrator for PSF-mediated splicing of AR transcripts (Fig. 4H). Taken together, these results revealed that PSF also orchestrated its target splicing factors at protein levels to form a complex for splicing and protein expression in CRPC.
Finally, we evaluated the clinical significance of these PSF target genes in CRPC development and progression. By qRT-PCR analysis of PSF, AR-V7, and spliceosome genes in prostate cancer tissues and tissues obtained from CRPC patients including metastatic tissues (bone, lymph node, and liver), we found that all tested spliceosome genes were up-regulated in prostate cancer compared with the benign regions. Interestingly, several spliceosome genes are up-regulated in CRPC tissues (SI Appendix, Fig. S12D). Among those, we found expression of U2AF2, DDX23, CHERP, and HNRNPU was elevated in CRPC. AR-V7 expression was also correlated with PSF, consistent with the result of IHC (Fig. 3H). In addition, knockdown of these factors showed effective growth inhibition of 22Rv1 cells, particularly in cells where CHERP, HNRNPU, DDX23, SF3B3, and U2AF2 (28–32) were knocked down (Fig. 4I and SI Appendix, Fig. S12E). Overall, our findings suggest that aberrant overexpression of spliceosome genes in prostate cancer by PSF contributes to the progression from hormone-naïve prostate cancer to CRPC (Fig. 4J).
Discussion
In the present study, we demonstrate the importance of PSF in the progression of prostate cancer using deep-sequence–based approach, clinicopathological analysis, and public database. We explored a mechanistic insight by identifying global RNAs bound with PSF. We found that some lncRNAs and miRNAs are important targets of PSF. Our analysis revealed that CTBP1-AS and SchLAP1 are positively regulated lncRNAs by PSF in prostate cancer cells. Interestingly, SchLAP1 has been known to be overexpressed in metastatic prostate tumors (26). The most unique finding of this study is that spliceosome genes are almost uniformly up-regulated in metastatic CRPC tissues. Importantly, our cell model-based analyses indicate that PSF is responsible for the expressional regulation of these spliceosome genes. In addition, our experimental data indicate that PSF coordinates these factors to regulate PSF target genes.
It is notable that one of the most important targets of PSF in CRPC would be AR. Our RIP-seq analysis demonstrated that enhanced association of AR transcript with PSF in CRPC model, 22Rv1 cells, compared with LNCaP cells (Fig. 3B). This might be caused by upregulated PSF expression as shown by Western blot and qRT-PCR analyses (Fig. 3C). Thus, impact of PSF knockdown on AR expression level may be evident in 22Rv1 cells that express PSF abundantly. Since the AR and AR-V7 have been assumed as the driver for the hormone-refractory state (5–8), the up-regulation of PSF would be responsible for the up-regulation of AR and AR-V7 by aberrant splicing activity for CRPC development. We observed PSF forms a complex with other splicing factors and NONO in the intronic region of AR transcripts by RIP and Western blotting following immunoprecipitation analysis. In addition, AR mRNA stability was decreased by PSF knockdown (Fig. 3E), although PSF recruitment to the promoter region of AR was not detected in ChIP-seq analysis. Therefore, these findings suggest that PSF regulates AR-splicing process and promotes production of AR and its variants at mRNA level. Moreover, our comprehensive analysis unveiled the variety of PSF-binding transcripts associated with cancer development in addition to AR. By regulating spliceosome gene expressions and cooperating with those factors, PSF could activate such a broad range of oncogenic pathways as an RNA-binding protein. In the future study, it will be interesting to further analyze specific targets of each splicing factor for revealing the functions of PSF complexes in cancer progression.
Our clinicopathological analyses also demonstrated the importance of NONO, showing the higher expression of NONO correlated with poor prognosis of patients. Our cell model study showed that knockdown of NONO in 22Rv1 and VCaP cells inhibits the expression of AR-V7 more effectively than full-length AR, suggesting the cooperative function of RNA-binding protein complex including PSF and NONO in prostate cancer. These functions of RNA-binding proteins could be effective to enhance the expressions of prostate cancer-associated genes as well as AR.
We indicated the importance of PSF in the regulation of splicing machinery in the progression of aggressive prostate cancer. Our finding revealed a mechanism in which a broad range of splicing components were aberrantly regulated for cancer progression. The wide-ranging up-regulation of the splicing pathway in metastatic prostate cancer could affect the splicing complexes in cancer. RNA splicing is mediated by an assembly, rearrangement, and disengagement of a set of small nuclear ribonucleoprotein (snRNP) complexes (U1, U2, and either U4/5/6 or U11/U12) or other proteins onto the pre-mRNAs (33, 34). The broad coverage of the wide spectrum of these components in our study suggests the involvement of PSF in enhancing this machinery. Interestingly, frequent mutations of splicing pathway have been reported in several malignancies by whole-exome sequencing (35, 36). These clinical analyses provide an intriguing insight into the mechanism of cancer progression by splicing machinery, since RNA splicing system has essential cellular roles for the diversity of protein species using a limited number of genes. Loss of splicing activity by mutations would induce severe developmental abnormality and tumorigenesis (37). Conversely, such mutation has been rarely identified in prostate cancer (18, 38). Enhanced splicing machinery could be indispensable for the cancer progression in some types of malignancies (39). Moreover, the present study would illustrate the specificity of PSF function and splicing machinery in prostate cancer.
In summary, our global analysis of PSF functions revealed its target signals based on the RNA-binding ability in addition to cell cycle regulator control by epigenetic silencing of the transcription with histone modification. Importantly, we proposed a mechanistic link between RNA-binding proteins and AR as well as prostate cancer-associated signals, indicating the clinical and biological impact of PSF in the development of abnormal splicing machinery in hormone-refractory prostate cancer. Considering the potential function of AR and AR variants in the development of metastatic CRPC, PSF and its associated factors could have the potential for therapy targets.
Materials and Methods
All animal experimental protocols were performed in accordance with the guidelines of the Animal Ethics Committee of the University of Tokyo. Further details are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank RIKEN for sequencing our samples. We are grateful to E. Sakamoto, N. Sasaki, and T. Oishi for technical assistance. This work was supported by grants of the Cell Innovation Program, the P-DIRECT, and the P-CREATE from Ministry of Education, Culture, Sports, Science and Technology, Japan (to S.I.); Japan Society for the Promotion of Science, Japan Grant 15K15581 (to K.T.) and Grant 15K15353 (to S.I.); a grant of the Program for Promotion of Fundamental Studies in Health Sciences from the National Institute of Biomedical Innovation, Japan (to S.I.); Grants-in-Aid from the Ministry of Health, Labor and Welfare, Japan (to S.I.); and grants from the Terumo Foundation for Life Sciences and Arts (to K.T.), the Princess Takamatsu Cancer Research Fund (to K.T.), and Uehara Memorial Foundation, Japan [201520122j (to S.I.)].
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.-J.K. is a guest editor invited by the Editorial Board.
Data deposition: Sequences have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession nos. GSE94577, GSE94028, GSE94243, and GSE100239).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706076114/-/DCSupplemental.
References
- 1.Yuan X, et al. Androgen receptor functions in castration-resistant prostate cancer and mechanisms of resistance to new agents targeting the androgen axis. Oncogene. 2014;33:2815–2825. doi: 10.1038/onc.2013.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen CD, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10:33–39. doi: 10.1038/nm972. [DOI] [PubMed] [Google Scholar]
- 3.Wang Q, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–256. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Waltering KK, Urbanucci A, Visakorpi T. Androgen receptor (AR) aberrations in castration-resistant prostate cancer. Mol Cell Endocrinol. 2012;360:38–43. doi: 10.1016/j.mce.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 5.Lu J, Van der Steen T, Tindall DJ. Are androgen receptor variants a substitute for the full-length receptor? Nat Rev Urol. 2015;12:137–144. doi: 10.1038/nrurol.2015.13. [DOI] [PubMed] [Google Scholar]
- 6.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715–2730. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahu A, Singhal U, Chinnaiyan AM. Long noncoding RNAs in cancer: From function to translation. Trends Cancer. 2015;1:93–109. doi: 10.1016/j.trecan.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarty D, et al. The oestrogen receptor alpha-regulated lncRNA NEAT1 is a critical modulator of prostate cancer. Nat Commun. 2014;5:5383. doi: 10.1038/ncomms6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung CL, et al. A long noncoding RNA connects c-Myc to tumor metabolism. Proc Natl Acad Sci USA. 2014;111:18697–18702. doi: 10.1073/pnas.1415669112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takayama K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–1680. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowery LA, Rubin J, Sive H. Whitesnake/sfpq is required for cell survival and neuronal development in the zebrafish. Dev Dyn. 2007;236:1347–1357. doi: 10.1002/dvdy.21132. [DOI] [PubMed] [Google Scholar]
- 13.Cosker KE, Fenstermacher SJ, Pazyra-Murphy MF, Elliott HL, Segal RA. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat Neurosci. 2016;19:690–696. doi: 10.1038/nn.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee M, et al. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 2015;43:3826–3840. doi: 10.1093/nar/gkv156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patton JG, Potro EB, Galceran J, Tempst P, Nadal-Ginard B. Cloning and characterization of PSF, a novel pre-mRNA splicing factor. Genes Dev. 1993;7:393–406. doi: 10.1101/gad.7.3.393. [DOI] [PubMed] [Google Scholar]
- 16.Dong B, Horowitz DS, Kobayashi R, Krainer AR. Purification and cDNA cloning of HeLa cell p54nrb, a nuclear protein with two RNA recognition motifs and extensive homology to human splicing factor PSF and Drosophila NONA/BJ6. Nucleic Acids Res. 1993;21:4085–4092. doi: 10.1093/nar/21.17.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taylor BS, et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18:11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grasso CS, et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature. 2012;487:239–243. doi: 10.1038/nature11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varambally S, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Massie CE, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719–2733. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen B, Yun J, Kim MS, Mendell JT, Xie Y. PIPE-CLIP: A comprehensive online tool for CLIP-seq data analysis. Genome Biol. 2014;15:R18. doi: 10.1186/gb-2014-15-1-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, et al. Design and bioinformatics analysis of genome-wide CLIP experiments. Nucleic Acids Res. 2015;43:5263–5274. doi: 10.1093/nar/gkv439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hirose T, et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol Biol Cell. 2014;25:169–183. doi: 10.1091/mbc.E13-09-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji Q, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111:736–748. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–1398. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takayama K, Inoue S. The emerging role of noncoding RNA in prostate cancer progression and its implication on diagnosis and treatment. Brief Funct Genomics. 2016;15:257–265. doi: 10.1093/bfgp/elv057. [DOI] [PubMed] [Google Scholar]
- 28.Crisci A, et al. Mammalian splicing factor SF1 interacts with SURP domains of U2 snRNP-associated proteins. Nucleic Acids Res. 2015;43:10456–10473. doi: 10.1093/nar/gkv952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye J, et al. hnRNP U protein is required for normal pre-mRNA splicing and postnatal heart development and function. Proc Natl Acad Sci USA. 2015;112:E3020–E3029. doi: 10.1073/pnas.1508461112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teigelkamp S, Mundt C, Achsel T, Will CL, Lührmann R. The human U5 snRNP-specific 100-kD protein is an RS domain-containing, putative RNA helicase with significant homology to the yeast splicing factor Prp28p. RNA. 1997;3:1313–1326. [PMC free article] [PubMed] [Google Scholar]
- 31.Golas MM, Sander B, Will CL, Lührmann R, Stark H. Molecular architecture of the multiprotein splicing factor SF3b. Science. 2003;300:980–984. doi: 10.1126/science.1084155. [DOI] [PubMed] [Google Scholar]
- 32.Voith von Voithenberg L, et al. Recognition of the 3′ splice site RNA by the U2AF heterodimer involves a dynamic population shift. Proc Natl Acad Sci USA. 2016;113:E7169–E7175. doi: 10.1073/pnas.1605873113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wahl MC, Will CL, Lührmann R. The spliceosome: Design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 34.Lee Y, Rio DC. Mechanisms and regulation of alternative pre-mRNA splicing. Annu Rev Biochem. 2015;84:291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida K, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 36.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell. 2014;156:1324–1335. doi: 10.1016/j.cell.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 37.Daguenet E, Dujardin G, Valcárcel J. The pathogenicity of splicing defects: Mechanistic insights into pre-mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015;16:1640–1655. doi: 10.15252/embr.201541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–1228. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.