Significance
The proliferation of multidrug-resistant organisms (MDROs) is one of the world’s most pressing public health problems. Despite great efforts to prevent MDRO spread in healthcare facilities, antibiotic-resistant infections remain a significant threat. We hypothesized that current infection prevention initiatives are hindered by their targeting of individual MDROs; neglecting the fact that healthcare institutions have multiple circulating MDRO species whose propagation is driven by competing risk factors. We tested this hypothesis by identifying risk factors for the spread of multiple MDROs among nursing home residents and unraveled a complex network of interactions between antibiotic exposure and microbial factors. Our results indicate that interactions among MDROs and underlying drivers must be considered simultaneously to design optimal strategies to reduce MDRO propagation.
Keywords: multidrug-resistant organisms, nursing homes, long-term care facilities, antibiotics, catheter-associated urinary tract infection
Abstract
The emergence and spread of multidrug-resistant organisms (MDROs) across global healthcare networks poses a serious threat to hospitalized individuals. Strategies to limit the emergence and spread of MDROs include oversight to decrease selective pressure for MDROs by promoting appropriate antibiotic use via antibiotic stewardship programs. However, restricting the use of one antibiotic often requires a compensatory increase in the use of other antibiotics, which in turn selects for the emergence of different MDRO species. Further, the downstream effects of antibiotic treatment decisions may also be influenced by functional interactions among different MDRO species, with the potential clinical implications of such interactions remaining largely unexplored. Here, we attempt to decipher the influence network between antibiotic treatment, MDRO colonization, and infection by leveraging active surveillance and antibiotic treatment data for 234 nursing home residents. Our analysis revealed a complex network of interactions: antibiotic use was a risk factor for primary MDRO colonization, which in turn increased the likelihood of colonization and infection by other MDROs. When we focused on the risk of catheter-associated urinary tract infections (CAUTI) caused by Escherichia coli, Enterococcus, and Staphylococcus aureus we observed that cocolonization with specific pairs of MDROs increased the risk of CAUTI, signifying the involvement of microbial interactions in CAUTI pathogenesis. In summary, our work demonstrates the existence of an underappreciated healthcare-associated ecosystem and strongly suggests that effective control of overall MDRO burden will require stewardship interventions that take into account both primary and secondary impacts of antibiotic treatments.
The successful treatment of bacterial infections has been significantly undermined by the emergence and spread of multidrug-resistant organisms (MDROs) (1). Antibiotic-resistant healthcare-acquired infections are of particular concern, as MDROs complicate treatment of infections, hinder effective treatment of comorbid conditions, and increase risk of morbidity and mortality (2–5). While the threats of MDROs in acute-care hospitals are well known, MDROs are not limited to those settings. In particular, residents in long-term care facilities (LTCFs) often have underlying medical conditions that make them particularly vulnerable to infections. Moreover, the high rates of MDRO colonization (6) and antibiotic use (7–9) among LTCF residents further put this population at risk for contracting infections. Furthermore, the frequent transfers between LTCFs and other healthcare facilities leads to the dissemination of MDROs throughout the healthcare network, creating a substantial economic and clinical burden that permeates healthcare systems (10, 11).
Given the central role of antibiotic use in both the emergence and proliferation of MDROs (12), antibiotic stewardship programs that enforce more judicious use of antibiotics have become a cornerstone of modern infection prevention (13–15). In addition to efforts to reduce total antibiotic consumption, stewardship programs also emphasize reductions in the use of specific antibiotics based on studies linking these antibiotics to increased rates of transmission and infection with high-priority MDROs (16–19). However, the studies used to inform stewardship decisions generally evaluate the risk of antibiotic resistance associated with single-bacterial species. This single-species view of MDRO epidemiology does not reflect the complex reality; most healthcare facilities have multiple circulating MDRO species (20, 21), and individuals are often colonized by more than one MDRO (22–24). Thus, interventions based on a single-species perspective can result in unintended consequences. For example, in one study, reducing cephalosporin use successfully decreased prevalence of cephalosporin-resistant Klebsiella (25), but—because carbapenems were used instead of cephalosporin—resulted in increased imipenem resistance in Pseudomonas aeruginosa (25). This example illustrates the inherent interdependencies between antibiotic use and MDROs and emphasizes that an optimal stewardship program must take into account circulating MDRO populations and the network of antibiotic-mediated interactions among them.
Here, we take an ecologic approach and describe how interactions among antibiotics and different MDRO species influence the dynamics of MDRO colonization and infection in LTCFs. To accomplish this, we leveraged longitudinal surveillance data and diagnostic records collected in the Targeted Infection Prevention study during 2010 and 2013 (26, 27). Briefly, the prevalence of MDROs in the groin and perirectum and associated metadata for 234 nursing home (NH) residents who were catheterized >72 h were included in this study. Our analysis revealed a complex network of associations between MDROs and antibiotics that drives colonization and infection. Intriguingly, in many cases, a primary event such as antibiotic administration or MDRO colonization increased risk of new MDRO acquisition or clinical infection. Our work strongly argues for a shift from the current single-species stewardship paradigm toward the design of interventions that account for the downstream impacts of treatment decisions mediated by the ecology of MDROs in healthcare facilities.
Results
NH Residents Are Heavily Colonized with Antibiotic-Resistant Bacteria.
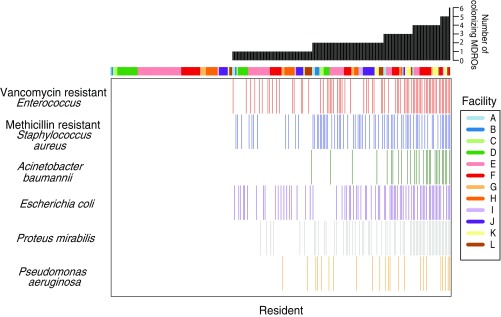
The characteristics of the study population have been published elsewhere (26, 27). Briefly, 122 (52%) males and 112 (48%) females were followed at baseline, day 15, and monthly for up to a year. Excluding participants that only had one visit, follow-up days ranged from 12 to 379 d, with a median of 57 d. The mean age was 73.9 ± 12.8 y, with an average physical self-maintenance score of 21.8 ± 3.9 and Charlson’s comorbidity score of 2.8 ± 1.9. MDROs most commonly colonizing the groin and perirectal area were VRE (n = 71; 30.3%); MRSA (n = 70; 29.9%); and resistant Gram-negative bacteria, including Escherichia coli (n = 71; 30.3%), Proteus mirabilis (n = 62; 26.5%), Acinetobacter baumannii (n = 28; 12.0%), and P. aeruginosa (n = 23; 9.8%) (Fig. 1). The distribution of various MDROs across facilities is shown in SI Appendix, Fig. S1. Only considering these species, 55 (23.5%) residents were colonized with one species, 49 (20.9%) were colonized with two species, and 46 (20%) were colonized with more than two species during their stay. Over the course of the study, 146 residents (62%) received at least one course of treatment with one of 50 different antibiotics (SI Appendix, Tables S1 and S2a). The high cocolonization and antibiotic use rates observed in these NHs prompted us to ask how antibiotic use and preexisting MDRO colonization influence risk for subsequent acquisition of new MDROs.
Fig. 1.
MDRO colonization patterns for 234 NH residents. The prevalence of most commonly observed multidrug-resistant organisms (MDROs) in the groin or perirectum of 234 nursing home residents who had a urinary catheter in place for >72 h is shown. Each column in the heatmap represents a catheterized resident, and each colored cell indicates colonization by the corresponding MDRO during their stay. The facility each resident resided in is color-coded on the top of the heatmap, with legend provided on the right (A–L). The total number of different species detected in each resident over time is summarized in the black barplot above.
Existing Bacterial Colonization Increases the Risk of Secondary Acquisition.
To discern if functional interactions were likely to be mediating MDRO spread, we first tested whether the observed high frequency of multi-MDRO colonization is due to the chance accumulation of MDROs in susceptible individuals, or if particular sets of MDROs preferentially cocolonized individuals. Applying a permutation analysis that controls for the observed burden of multi-MDRO colonization (28), we observed that colonization patterns are highly nonrandom, with pairs of MDROs having varying degrees of positive and negative association with each other (SI Appendix, Fig. S2). To more effectively leverage our longitudinal sampling, we next considered colonization as a time-varying variable and tested whether preexisting MDRO colonization predicted new colonization with a different MDRO species (29). In particular, we constructed models that only considered colonization with other MDROs and assessed the hazard ratio (HR) of acquiring a secondary organism in the presence of another organism. In this analysis, P. mirabilis showed high connectivity, where preexisting P. mirabilis colonization increased the risk of acquiring A. baumannii, MRSA, and VRE (unadjusted HR > 2 and P < 0.05 in all cases), and preexisting colonization with A. baumannii, E. coli, and P. aeruginosa increased the risk of acquiring P. mirabilis (HR > 2 and P < 0.05 in all cases) (SI Appendix, Table S3). Note that observed associations were not always bidirectional, as exemplified by E. coli predicting MRSA acquisition, but MRSA not being associated with E. coli acquisition (unadjusted HR = 2.06 and 0.37; P = 0.03 and 0.33, respectively) (SI Appendix, Table S3). Together, these results indicate that cocolonization is not simply a stochastic process, but is shaped by underlying microbial and host factors.
Many-to-Many Relationships Between Antibiotic Exposure and MDRO Colonization.
We next explored associations between antibiotic use and risk of subsequent MDRO acquisition (30, 31). We grouped the 50 different antibiotics used during the study into 18 classes; residents were most often exposed to cephalosporins, quinolones, penicillin combinations, and glycopeptides (SI Appendix, Table S2a). The spectrum of activity of each class is provided in SI Appendix, Table S2b. As different antibiotics have different modes of action and target specificity, we investigated whether individual antibiotic classes were associated with the acquisition of specific MDRO species. We treated antibiotic exposure as a time-varying variable, adding 30 d after the end of antibiotic administration to account for prolonged disruption of microbiome-mediated colonization resistance. We observed that multiple antibiotics could be associated with acquisition of a single MDRO; for example, aminoglycosides, cephalosporins, and glycopeptides exposures were all associated with VRE acquisition. Conversely, a single antibiotic could be associated with acquiring multiple MDRO species; for example, aminoglycosides increased risk of acquiring VRE, A. baumannii, E. coli, P. mirabilis, and P. aeruginosa (unadjusted HR > 2 and P < 0.05 in all cases) (SI Appendix, Table S4). These observations highlight the many-to-many relationship between antibiotics and MDROs, offering a perspective that could be overlooked in data analyzed with a focus on single MDRO species or individual antibiotics.
Microbial and Antibiotic Factors Jointly Influence Bacterial Colonization.
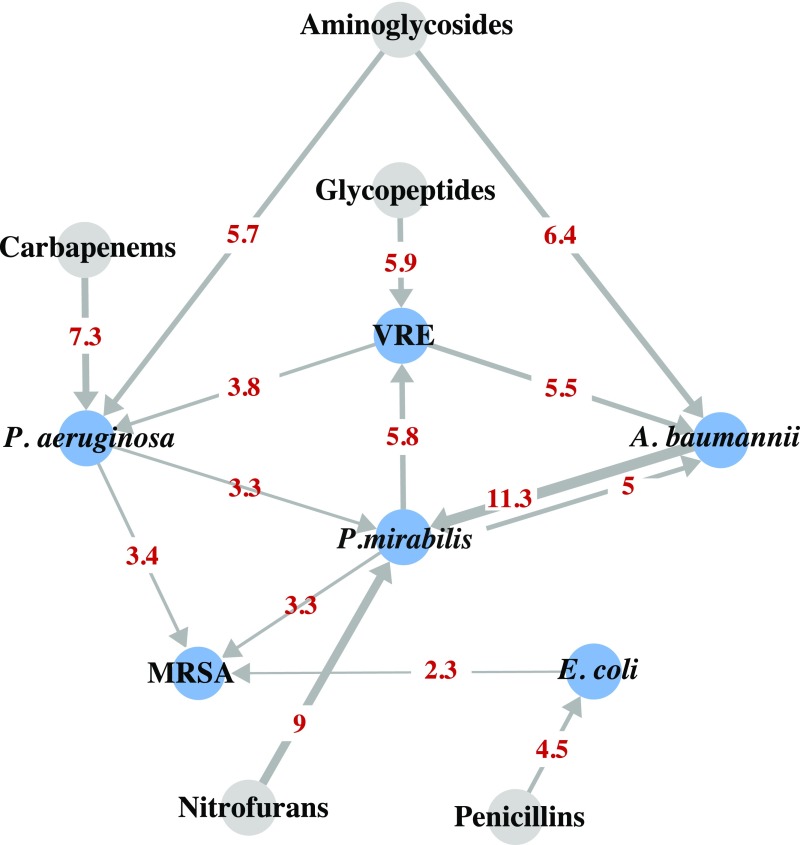
Having shown that the acquisition of MDRO species is influenced by microbial and antibiotic factors, we next fit multivariate Cox regression models to estimate individual and joint effects. We included in the models all microbial and antibiotic factors with at least modestly significant associations (P < 0.1) and strongly associated resident characteristics (HR of >2 and P value <0.05, namely, sex) and accounted for clustering of measures by resident and facility. Using associations that were statistically significant in the multivariate Cox regression model (P < 0.05) (Fig. 2 and SI Appendix, Table S5), we constructed a network model demonstrating how risk of acquiring each MDRO was influenced by a distinct set of microbial and antibiotic factors. Strikingly, except for MRSA, all MDRO colonization was found to be positively associated with at least one other MDRO and one antibiotic. Most MDROs emerged in response to a single antibiotic class, such as VRE to glycopeptides, E. coli to penicillin, P. mirabilis to nitrofurans, and P. aeruginosa to carbapenems. While several MDROs were associated with the acquisition of other MDROs, P. mirabilis appeared to be the “hub” species, given its central role in connecting many MDRO species. This network of interactions suggests that antibiotic exposure increased the risk of acquiring MDROs, which in turn altered the risk of colonization with an additional MDRO.
Fig. 2.
Risk network for MDRO colonization. Blue nodes represent MDRO colonization, and gray nodes represent antibiotic exposure. Each directed arrow indicates that the source node is predicative of the recipient node. Here antibiotic exposure is assumed to be risk factors for colonization, whereas bacterial colonization could either be the risk factor for subsequent colonization, or a result of antibiotic exposure and/or previous colonization. The magnitude of the hazard ratio is reflected in edge thickness and shown in numbers. All associations shown are statistically significant with P < 0.05.
Cocolonization Is Associated with Increased Risk of Having Specific Species in CAUTI Urine Culture.
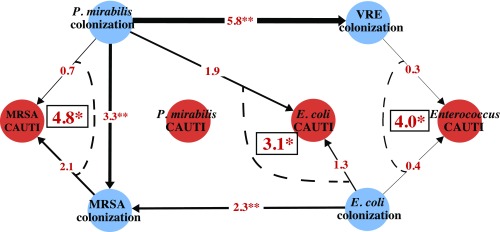
Colonization is a first step toward infection; therefore we wondered whether microbial interactions in colonized residents influence clinical trajectories. One of the most common hospital-acquired infections is urinary tract infection, especially among those with an indwelling catheter (32). For this analysis, we included the 234 residents with an indwelling urinary catheter for longer than 72 h. In total, 70 residents had a clinically diagnosed catheter-associated urinary tract infection (CAUTI) during the course of the study (26). Excluding CAUTIs that were present before enrollment, P. mirabilis (n = 38), Enterococcus spp. (n = 22), E. coli (n = 20), and Staphylococcus spp. (n = 13) were the most commonly isolated bacteria in their urine. The distribution of species-specific CAUTI across facilities is shown in SI Appendix, Fig. S1. After adjustment for functional status and sex, risk of CAUTI was not associated with preexisting groin or perirectum monocolonization by the species detected in the urine (SI Appendix, Table S6). However, for CAUTI associated with Enterococcus, E. coli, and MRSA, we observed that the presence of a specific cocolonizing species in the groin or perirectum increased the risk of infection. In particular, the cooccurrence of VRE and E. coli increased risk of Enterococcus in urine, and the cooccurrence of E. coli or MRSA with P. mirabilis increased the risk of E. coli or MRSA in the urine, respectively (HR > 2, P < 0.1 in all cases; Fig. 3). This bacterial network shows that the effects of antibiotic exposure contribute to colonization as well as disease, potentially mediated by synergistic interactions between colonizing MDROs.
Fig. 3.
Risk network for catheter-associated urinary tract infection (CAUTI) events. CAUTI events were divided into four subsets based on the species found in the urine (Enterococcus, P. mirabilis, MRSA, and E. coli). Orange nodes represent species-specific CAUTI events, and blue nodes represent MDRO colonization. Figure includes only colonizing species that were also found in CAUTI urine. Directed arrows indicate the risk of a subsequent colonization in the presence of another MDRO colonization (**P < 0.05), or the risk of having a species-specific CAUTI when colonized by an MDRO (*P < 0.1). Red numbers indicate the hazard ratio of having a species-specific CAUTI in cocolonized residents; the hazard ratio of having such outcome in residents colonized by only one MDRO is not significant.
Discussion
Antibiotic use invariably selects for antibiotic resistance. Our task is to preserve the effectiveness of existing antibiotics by minimizing the emergence and spread of MDROs to maximize the time until existing antibiotics become ineffective. As we show using longitudinal data from nursing homes, this task is complicated by the interdependencies among MDROs and antibiotics. We uncovered a complex set of relationships, where colonization and infection with different MDRO species were associated with distinct sets of microbial and antibiotic risk factors. This work demonstrates the importance of designing strategies that account for the complex set of interdependencies among different MDROs and the antibiotics that influence their spread.
Among the MDRO species included in our study, P. mirabilis was located most centrally. P. mirabilis increased risk of acquiring other MDROs, and risk of acquiring P. mirabilis was influenced by the presence of other colonizing MDROs. P. mirabilis is known for its propensity to produce both monospecies and polymicrobial biofilms, commonly with urease-producing species (33, 34). Although MRSA is the only species in our culture collection that has been characterized as a urease producer (35), a recent study showed that P. mirabilis urease production is enhanced in the presence of urease nonproducers, including Enterococcus, A. baumannii, and E. coli (36). Thus, communication among MDROs, whether dependent on chemical signaling or physical contact, may underlie the associations among these organisms.
The colonization of bacterial pairs, including E. coli/Enterococcus, P. mirabilis/E. coli, and P. mirabilis/MRSA, synergistically increased risk of developing a CAUTI. In animal and in vitro systems, E. coli and Enterococcus together alter biofilm formation and modulate the metabolic milieu, potentially promoting polymicrobial infections (37). Similarly, P. mirabilis and E. coli promote one another’s growth by using complementary metabolic pathways, with coinfection promoting the colonization and persistence of both species (38). These interspecies interactions are consistent with our observation that P. mirabilis/E. coli cocolonization increased the risk of subsequent movement into the urine and suggests enhanced pathogenic potential when both species were present. Lastly, Proteus species and Staphylococcus aureus increase each other’s virulence in mouse and Caenorhabditis elegans models, although the molecular mechanism remains to be investigated (39–41). It is also noteworthy that while P. mirabilis increased risk of CAUTI with other organisms, no colonizing partners were identified as specifically increasing the risk of P. mirabilis CAUTI. We hypothesize that the lack of observed association may be a consequence of Proteus’ diverse interaction partners precluding the detection of specific interactions with our small number of CAUTI cases (SI Appendix, Fig. S5). Together, these observations suggest that interspecies interactions may enhance microbial pathogenic potential. That we found supporting evidence from model systems for each of our associations validates our approach of using surveillance data analysis to identify clinically significant polymicrobial interactions. These interactions can then be studied mechanistically using model systems.
A limitation in our study is that only high-priority MDROs were collected; thus we cannot know the role of other members of host microbial communities in mediating risk of acquisition and infection with MDROs. Future studies should collect both resistant and susceptible pathogens and, potentially, specimens for microbiome analysis to provide a complete picture of the microbial ecology at both intra- and interresident levels (42–46). Despite this limitation, our study represents an important proof-of-concept for the application of surveillance data and standard epidemiological tools to dissect how host, microbial, and treatment factors influence colonization and infection with MDROs. The success of our approach, as indicated by the concordance of our findings with prior epidemiologic and experimental studies, supports its potential role in studying the ecology of MDROs in a variety of healthcare settings. The complex network of interactions among MDROs and antibiotic treatments found in the current population indicates that additional studies are necessary to better inform the design and evaluation of stewardship interventions aimed at preventing emergence and spread of MDROs.
Methods
Study Population and Design.
We performed a post hoc analysis of data collected through the Targeted Infection Prevention (TIP) study. Written informed consent to collect microbiological and resident-level data was obtained from each resident or his or her durable power of attorney, and the study was approved by the University of Michigan Institutional Review Board. The TIP study, a cluster-randomized intervention trial aiming to reduce the prevalence of drug-resistant organisms in nursing homes, was conducted in Michigan between May 2010 and April 2013 (27). Details of the trial design and outcomes have been reported previously (27, 47–49). In short, 12 community-based nursing homes (NHs) were randomly assigned as control or intervention sites, with 215 and 203 enrolled residents, respectively. Longitudinal data regarding indwelling device use, functional status, comorbidities, urine culture results, and antibiotic administration were collected from each resident at the time of enrollment up to 1 y, until discharge, device removal, or death. Microbiological samples were collected from multiple body sites to assess antibiotic-resistant organism (MDRO) colonization at baseline, day 15, and monthly for up to a year. The body sites included the nares, oropharynx, enteral feeding tube insertion site, suprapubic catheter site, groin, perirectal area, and wounds. Standard microbiological methods were used to isolate and identify methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus (VRE), and Gram-negative bacteria resistant to ciprofloxacin or ceftazidime.
For the present study, we limited our population to 234 residents who had urinary catheter for longer than 72 h, as catheterized residents are known to experience different clinical trajectories from noncatheterized residents (50). We included in our analysis only MDROs colonizing the groin and perirectum. The analysis of catheter-associated urinary tract infection (CAUTI) risk included only residents who had full baseline demographic and CAUTI symptom information (n = 234) (26).
Data Organization.
Based on the positive correlation found between rectal colonization and infecting strains present in CAUTI (51, 52), we specifically considered MDROs colonizing two body sites, the groin and perirectal region, in the analysis. We further defined our bacteria of interest to those that were detected microbiologically in more than 20 residents across visits during the study to focus on the more prevalent colonizers, namely, VRE, MRSA, Acinetobacter baumannii, Escherichia coli, Proteus mirabilis, and Pseudomonas aeruginosa. As samples were collected monthly, the exact time when bacterial colonization started and/or ended was unknown. Therefore, when a positive culture was identified, we arbitrarily assigned each day until the next visit as colonized. Colonization status was treated as a time-dependent variable and was coded dichotomously, 1 if detected, and 0 if undetected, on a day-to-day basis (example scenarios provided in SI Appendix, Fig. S3). Antibiotics were categorized into classes (SI Appendix, Table S1). Antibiotic exposure has long-lasting impact on the intestinal microbiota, such as community structure and development of MDROs, for years (53, 54). To assess the prolonged effects of antibiotics on the acquisition of MDROs, we extended antibiotic exposure by 30 d past the end of antibiotic administration (example scenarios provided in SI Appendix, Fig. S4).
Statistical Analysis and Data Visualization.
All analyses were conducted using RStudio R version 3.3.2 (55). We used Cox proportional hazard models to test for the effects of (i) presence of an earlier bacterial colonizer on acquiring secondary colonization; (ii) antibiotic exposure on MDRO colonization; and, (iii) bacterial colonization on CAUTI urine culture results (MRSA, Enterococcus, E. coli, and P. mirabilis). The “coxph” function from the “survival” package in R was used to perform the analysis (56). Without adjusting for covariates, our preliminary results indicated that both primary colonization and antibiotic exposure increased the risk of acquiring specific MDROs. Subsequently we entered all variables significantly associated (P < 0.1) with secondary colonization into a single multivariate analysis, adjusting for age, sex, comorbidity score, functional status, intervention, and clustering by resident and facilities. Here we used the “coxme” function (57). Resident-level characteristics, namely, age, comorbidity score, functional status, and intervention that did not have a significant HR (P < 0.05) were removed from the model. The final model for colonization was adjusted for sex, and the CAUTI model was adjusted for functional status and sex. Statistically significant variables (P < 0.05) associated with each outcome were then organized into a matrix for directed network visualization using the “igraph” package in R (58).
Supplementary Material
Acknowledgments
We thank members of the L.M. laboratory and Dr. C. Armbruster for data collection and analysis; Sophie Yu-Pu Chen at the Center for Statistical Consultation and Research (CSCAR, University of Michigan), Dr. A. Galecki, and M. Kabeto (University of Michigan Older Americans Independence Centers Pepper Center) for statistical consultation; and Dr. Mary Hayden (Rush University Medical Center, Chicago) for antibiotic classification. This work was supported by the Pepper Center Pilot Grant AG-024824 (to E.S.S.) and the Centers for Disease Control (CDC) Contract 2016-N-17812 (to E.S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710235114/-/DCSupplemental.
References
- 1.Rossolini GM, Arena F, Pecile P, Pollini S. Update on the antibiotic resistance crisis. Curr Opin Pharmacol. 2014;18:56–60. doi: 10.1016/j.coph.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Giske CG, Monnet DL, Cars O, Carmeli Y. ReAct-Action on Antibiotic Resistance Clinical and economic impact of common multidrug-resistant gram-negative bacilli. Antimicrob Agents Chemother. 2008;52:813–821. doi: 10.1128/AAC.01169-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strausbaugh LJ, Joseph CL. The burden of infection in long-term care. Infect Control Hosp Epidemiol. 2000;21:674–679. doi: 10.1086/501712. [DOI] [PubMed] [Google Scholar]
- 4.Friedman ND, Temkin E, Carmeli Y. The negative impact of antibiotic resistance. Clin Microbiol Infect. 2016;22:416–422. doi: 10.1016/j.cmi.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Lambert ML, et al. Clinical outcomes of health-care-associated infections and antimicrobial resistance in patients admitted to European intensive-care units: A cohort study. Lancet Infect Dis. 2011;11:30–38. doi: 10.1016/S1473-3099(10)70258-9. [DOI] [PubMed] [Google Scholar]
- 6.Cassone M, Mody L. Colonization with multi-drug resistant organisms in nursing homes: Scope, importance, and management. Curr Geriatr Rep. 2015;4:87–95. doi: 10.1007/s13670-015-0120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren JW, Palumbo FB, Fitterman L, Speedie SM. Incidence and characteristics of antibiotic use in aged nursing home patients. J Am Geriatr Soc. 1991;39:963–972. doi: 10.1111/j.1532-5415.1991.tb04042.x. [DOI] [PubMed] [Google Scholar]
- 8.Han JH, et al. Risk factors for the development of gastrointestinal colonization with fluoroquinolone-resistant Escherichia coli in residents of long-term care facilities. J Infect Dis. 2014;209:420–425. doi: 10.1093/infdis/jit471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loeb MB, et al. Risk factors for resistance to antimicrobial agents among nursing home residents. Am J Epidemiol. 2003;157:40–47. doi: 10.1093/aje/kwf173. [DOI] [PubMed] [Google Scholar]
- 10.Bonomo RA, Rice LB. Emerging issues in antibiotic resistant infections in long-term care facilities. J Gerontol A Biol Sci Med Sci. 1999;54:B260–B267. doi: 10.1093/gerona/54.6.b260. [DOI] [PubMed] [Google Scholar]
- 11.Kahvecioglu D, et al. Multidrug-resistant organism infections in US nursing homes: A national study of prevalence, onset, and transmission across care settings, October 1, 2010-December 31, 2011. Infect Control Hosp Epidemiol. 2014;35(Suppl 3):S48–S55. doi: 10.1086/677835. [DOI] [PubMed] [Google Scholar]
- 12.Holmes AH, et al. Understanding the mechanisms and drivers of antimicrobial resistance. Lancet. 2016;387:176–187. doi: 10.1016/S0140-6736(15)00473-0. [DOI] [PubMed] [Google Scholar]
- 13.Malani AN, et al. Clinical and economic outcomes from a community hospital’s antimicrobial stewardship program. Am J Infect Control. 2013;41:145–148. doi: 10.1016/j.ajic.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Davey P, et al. Interventions to improve antibiotic prescribing practices for hospital inpatients. Cochrane Database Syst Rev. 2017;2:CD003543. doi: 10.1002/14651858.CD003543.pub4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feazel LM, et al. Effect of antibiotic stewardship programmes on Clostridium difficile incidence: A systematic review and meta-analysis. J Antimicrob Chemother. 2014;69:1748–1754. doi: 10.1093/jac/dku046. [DOI] [PubMed] [Google Scholar]
- 16.Wiener J, et al. Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA. 1999;281:517–523. doi: 10.1001/jama.281.6.517. [DOI] [PubMed] [Google Scholar]
- 17.Won SY, et al. Centers for Disease Control and Prevention Epicenter Program Emergence and rapid regional spread of Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae. Clin Infect Dis. 2011;53:532–540. doi: 10.1093/cid/cir482. [DOI] [PubMed] [Google Scholar]
- 18.Fisch J, et al. New acquisition of antibiotic-resistant organisms in skilled nursing facilities. J Clin Microbiol. 2012;50:1698–1703. doi: 10.1128/JCM.06469-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pop-Vicas A, Mitchell SL, Kandel R, Schreiber R, D’Agata EM. Multidrug-resistant gram-negative bacteria in a long-term care facility: Prevalence and risk factors. J Am Geriatr Soc. 2008;56:1276–1280. doi: 10.1111/j.1532-5415.2008.01787.x. [DOI] [PubMed] [Google Scholar]
- 20.Sheppard AE, et al. Modernising Medical Microbiology (MMM) Informatics Group Nested Russian doll-like genetic mobility drives rapid dissemination of the carbapenem resistance gene blaKPC. Antimicrob Agents Chemother. 2016;60:3767–3778. doi: 10.1128/AAC.00464-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sanon MA, Watkins S. Nurses’ uniforms: How many bacteria do they carry after one shift? J Public Health Epidemiol. 2012;4:311–315. doi: 10.5897/JPHE12.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snyder GM, O’Fallon E, D’Agata EM. Co-colonization with multiple different species of multidrug-resistant gram-negative bacteria. Am J Infect Control. 2011;39:506–510. doi: 10.1016/j.ajic.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D’Agata EM, Habtemariam D, Mitchell S. Multidrug-resistant gram-negative bacteria: Inter- and intradissemination among nursing homes of residents with advanced dementia. Infect Control Hosp Epidemiol. 2015;36:930–935. doi: 10.1017/ice.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buke C, et al. Epidemiology of multidrug-resistant bacteria in patients with long hospital stays. Infect Control Hosp Epidemiol. 2007;28:1255–1260. doi: 10.1086/522678. [DOI] [PubMed] [Google Scholar]
- 25.Rahal JJ, et al. Class restriction of cephalosporin use to control total cephalosporin resistance in nosocomial Klebsiella. JAMA. 1998;280:1233–1237. doi: 10.1001/jama.280.14.1233. [DOI] [PubMed] [Google Scholar]
- 26.Armbruster CE, Prenovost K, Mobley HL, Mody L. How often do clinically diagnosed catheter-associated urinary tract infections in nursing homes meet standardized criteria? J Am Geriatr Soc. 2017;65:395–401. doi: 10.1111/jgs.14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mody L, et al. A targeted infection prevention intervention in nursing home residents with indwelling devices: A randomized clinical trial. JAMA Intern Med. 2015;175:714–723. doi: 10.1001/jamainternmed.2015.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gotelli N, Hart EM, Ellison AM. 2015 EcoSimR: Null model analysis for ecological data. R Package Version 0.1.0. Available at github.com/gotellilab/EcoSimR. Accessed August 29, 2017.
- 29.Shaffer ML, D’Agata EM, Habtemariam D, Mitchell SL. Examining the relationship between multidrug-resistant organism acquisition and exposure to antimicrobials in long-term care populations: A review. Ann Epidemiol. 2016;26:810–815. doi: 10.1016/j.annepidem.2016.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiébaut AC, et al. ColoRea Study Group Variability of intestinal colonization with third-generation cephalosporin-resistant Enterobacteriaceae and antibiotic use in intensive care units. J Antimicrob Chemother. 2012;67:1525–1536. doi: 10.1093/jac/dks072. [DOI] [PubMed] [Google Scholar]
- 31.Tacconelli E, et al. Antibiotic usage and risk of colonization and infection with antibiotic-resistant bacteria: A hospital population-based study. Antimicrob Agents Chemother. 2009;53:4264–4269. doi: 10.1128/AAC.00431-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foxman B. The epidemiology of urinary tract infection. Nat Rev Urol. 2010;7:653–660. doi: 10.1038/nrurol.2010.190. [DOI] [PubMed] [Google Scholar]
- 33.Stickler D, Ganderton L, King J, Nettleton J, Winters C. Proteus mirabilis biofilms and the encrustation of urethral catheters. Urol Res. 1993;21:407–411. doi: 10.1007/BF00300077. [DOI] [PubMed] [Google Scholar]
- 34.Jacobsen SM, Stickler DJ, Mobley HL, Shirtliff ME. Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin Microbiol Rev. 2008;21:26–59. doi: 10.1128/CMR.00019-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murchan S, Aucken HM, O’Neill GL, Ganner M, Cookson BD. Emergence, spread, and characterization of phage variants of epidemic methicillin-resistant Staphylococcus aureus 16 in England and Wales. J Clin Microbiol. 2004;42:5154–5160. doi: 10.1128/JCM.42.11.5154-5160.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Armbruster CE, et al. The pathogenic potential of Proteus mirabilis is enhanced by other uropathogens during polymicrobial urinary tract infection. Infect Immun. 2017;85:e00808-16. doi: 10.1128/IAI.00808-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Keogh D, et al. Enterococcal metabolite cues facilitate interspecies niche modulation and polymicrobial infection. Cell Host Microbe. 2016;20:493–503. doi: 10.1016/j.chom.2016.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Alteri CJ, Himpsl SD, Mobley HL. Preferential use of central metabolism in vivo reveals a nutritional basis for polymicrobial infection. PLoS Pathog. 2015;11:e1004601. doi: 10.1371/journal.ppat.1004601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arndt WF, Ritts RE. Synergism between staphylococci and proteus in mixed infection. Proc Soc Exp Biol Med. 1961;108:166–169. doi: 10.3181/00379727-108-26881. [DOI] [PubMed] [Google Scholar]
- 40.JebaMercy G, Balamurugan K. Effects of sequential infections of Caenorhabditis elegans with Staphylococcus aureus and Proteus mirabilis. Microbiol Immunol. 2012;56:825–835. doi: 10.1111/j.1348-0421.2012.00509.x. [DOI] [PubMed] [Google Scholar]
- 41.Arndt WF, Young EJ, Ritts RE. Staphylococcal enhancement of susceptibility to bacterial infection in the mouse. J Infect Dis. 1963;112:255–263. [Google Scholar]
- 42.Stein RR, et al. Ecological modeling from time-series inference: Insight into dynamics and stability of intestinal microbiota. PLOS Comput Biol. 2013;9:e1003388. doi: 10.1371/journal.pcbi.1003388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher CK, Mehta P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS One. 2014;9:e102451. doi: 10.1371/journal.pone.0102451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.David LA, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15:R89. doi: 10.1186/gb-2014-15-7-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Andersen H, et al. Use of shotgun metagenome sequencing to detect fecal colonization with multidrug-resistant bacteria in children. J Clin Microbiol. 2016;54:1804–1813. doi: 10.1128/JCM.02638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Araos R, Tai AK, Snyder GM, Blaser MJ, D’Agata EMC. Predominance of Lactobacillus spp. among patients who do not acquire multidrug-resistant organisms. Clin Infect Dis. 2016;63:937–943. doi: 10.1093/cid/ciw426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cao J, Min L, Lansing B, Foxman B, Mody L. Multidrug-resistant organisms on patients’ hands: A missed opportunity. JAMA Intern Med. 2016;176:705–706. doi: 10.1001/jamainternmed.2016.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cassone M, McNamara SE, Perri MB, Zervos M, Mody L. Impact of intervention measures on MRSA clonal type and carriage site prevalence. MBio. 2016;7:e00218. doi: 10.1128/mBio.00218-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ismail MD, et al. Long-term carriage of ciprofloxacin-resistant Escherichia coli isolates in high-risk nursing home residents. Infect Control Hosp Epidemiol. 2016;37:440–447. doi: 10.1017/ice.2015.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kunin CM, Douthitt S, Dancing J, Anderson J, Moeschberger M. The association between the use of urinary catheters and morbidity and mortality among elderly patients in nursing homes. Am J Epidemiol. 1992;135:291–301. doi: 10.1093/oxfordjournals.aje.a116283. [DOI] [PubMed] [Google Scholar]
- 51.Daifuku R, Stamm WE. Association of rectal and urethral colonization with urinary tract infection in patients with indwelling catheters. JAMA. 1984;252:2028–2030. [PubMed] [Google Scholar]
- 52.Mathur S, Sabbuba NA, Suller MT, Stickler DJ, Feneley RC. Genotyping of urinary and fecal Proteus mirabilis isolates from individuals with long-term urinary catheters. Eur J Clin Microbiol Infect Dis. 2005;24:643–644. doi: 10.1007/s10096-005-0003-0. [DOI] [PubMed] [Google Scholar]
- 53.Jernberg C, Löfmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 54.Dethlefsen L, Relman DA. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci USA. 2011;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.R Development Core Team 2016. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna)
- 56.Therneau TM, Grambsch PM. 2015 A package for survival analysis in S, Version 2.38 Available at https://CRAN.R-project.org/package=survival. Accessed August 29, 2017.
- 57.Therneau TM. 2015 coxme: Mixed Effects Cox Models. Version 2.2-5. Available at https://CRAN.R-project.org/package=coxme. Accessed August 29, 2017.
- 58.Csardi G, Nepusz T. The igraph software package for complex network research. Int J Complex Syst. 2006;1695:1–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.