Significance
Age-related macular degeneration (AMD) and related macular dystrophies (MDs) are a major cause of vision loss. However, pharmacological treatments in these diseases are limited due to the lack of knowledge of underlying disease mechanisms, partly because appropriate human models to study AMD and related MDs are lacking. Furthermore, in the living human eye, the entire retina acts as a functional unit, making it difficult to investigate the specific contribution of a particular retinal cell type in the disease. Here, we established human models of multiple MDs, which demonstrated similar molecular and phenotypic manifestations within these diseases. Furthermore, we showed that dysfunction of an individual cell type, retinal pigment epithelium cells in the retina, is sufficient for the development of key pathological features in these MDs.
Keywords: human induced pluripotent stem cells, retinal pigment epithelium, macular dystrophies, drusen, sub-RPE deposits
Abstract
Age-related macular degeneration (AMD) and related macular dystrophies (MDs) are a major cause of vision loss. However, the mechanisms underlying their progression remain ill-defined. This is partly due to the lack of disease models recapitulating the human pathology. Furthermore, in vivo studies have yielded limited understanding of the role of specific cell types in the eye vs. systemic influences (e.g., serum) on the disease pathology. Here, we use human induced pluripotent stem cell-retinal pigment epithelium (hiPSC-RPE) derived from patients with three dominant MDs, Sorsby’s fundus dystrophy (SFD), Doyne honeycomb retinal dystrophy/malattia Leventinese (DHRD), and autosomal dominant radial drusen (ADRD), and demonstrate that dysfunction of RPE cells alone is sufficient for the initiation of sub-RPE lipoproteinaceous deposit (drusen) formation and extracellular matrix (ECM) alteration in these diseases. Consistent with clinical studies, sub-RPE basal deposits were present beneath both control (unaffected) and patient hiPSC-RPE cells. Importantly basal deposits in patient hiPSC-RPE cultures were more abundant and displayed a lipid- and protein-rich “drusen-like” composition. Furthermore, increased accumulation of COL4 was observed in ECM isolated from control vs. patient hiPSC-RPE cultures. Interestingly, RPE-specific up-regulation in the expression of several complement genes was also seen in patient hiPSC-RPE cultures of all three MDs (SFD, DHRD, and ADRD). Finally, although serum exposure was not necessary for drusen formation, COL4 accumulation in ECM, and complement pathway gene alteration, it impacted the composition of drusen-like deposits in patient hiPSC-RPE cultures. Together, the drusen model(s) of MDs described here provide fundamental insights into the unique biology of maculopathies affecting the RPE–ECM interface.
Maculopathies are a major cause of blindness, with age-related macular degeneration (AMD) being the leading cause of irreversible vision loss in adults in the United States. Histopathological and clinical studies have shown that AMD and a subset of inherited macular dystrophies (MDs) share extensive phenotypic and clinical similarities (1–4). Specifically, AMD and related MDs have a cumulative etiology with adult onset of signs and symptoms and similar disease presentation including drusen formation, extracellular matrix (ECM) protein accumulation, thickening of Bruch’s membrane, development of choroidal neovascularization, retinal pigment epithelium (RPE) atrophy, and ultimately the loss of vision (1–5). Although, the major pathological manifestations in these maculopathies are localized to the RPE–ECM interface in the retina, the multifactorial nature of these diseases, including the involvement of multiple retinal cell layers (photoreceptors, RPE, and choriocapillaris) (3, 6–9) and environmental risk factors (e.g., cigarette smoke) (10), has complicated the pursuit of the underlying disease mechanisms(s). The animal models of macular degeneration, including MDs, while significantly contributing to our understanding of specific cellular process(es) in retinal physiology, have often failed to recapitulate crucial aspects of the human disease pathology. Furthermore, isolating the contribution of individual cell types to the pathophysiology of macular degeneration has proven difficult in these in vivo models.
RPE cells play an essential role in vision by performing several important tasks necessary for retinal homeostasis. For instance, RPE cells support the visual cycle, phagocytose and degrade the shed photoreceptor outer segments (POS), absorb damaging light, and control the flux of ions to and from the choroidal vasculature. Impaired RPE function thus could be sufficient for the development of key pathological manifestations of specific maculopathies, including drusen formation and aberrant thickening of underlying ECM. Dysregulated expression/activity of RPE-secreted matrix metalloproteases (MMPs), for example, should be sufficient for the increased accumulation of ECM proteins and consequent thickening of the underlying ECM (Bruch’s membrane) (11). ECM thickening could then potentially inhibit the flux of metabolic waste products by RPE cells and lead to development of sub-RPE deposits (drusen) (12). In fact, recent studies using both mouse- (13) and porcine-derived (14) RPE have demonstrated that RPE cells in culture are capable of forming basal deposits in a cell-autonomous fashion. Furthermore, human fetal RPE (hfRPE) cultures, when exposed to exogenous stressors such as serum (15) or POS (16), have been shown to develop drusen-like basal deposits. However, the ability of cultured human RPE obtained from a macular degeneration patient, in the absence of any exogenous stressors, to develop drusen, has not been investigated. This is partly due to the lack of access to RPE samples from a living patient’s eyes for cellular studies. Furthermore, the scope of investigation is limited in postmortem eyes that are typically available at the end stage of the disease and thus are not conducive to studying early pathophysiological changes of the disease.
The advent of human induced pluripotent stem cell (hiPSC) technology has provided a unique platform to interrogate disease-associated pathology and physiology in an individual cell type using a patient’s own cells. Importantly, hiPSCs are also conducive to genome editing by the CRISPR-Cas system (17). The correction of disease-causing mutations in hiPSCs is a powerful tool that provides access to isogenic controls that are necessary to clearly correlate an identified disease phenotype to a mutation. In fact, hiPSC technology in conjunction with CRISPR technology has previously been used to gain important insights into the underlying mechanism of several diseases (e.g., Huntington’s disease, Duchenne muscular dystrophy) (18, 19). Using hiPSCs as a platform to study human ocular diseases is particularly relevant to RPE-based disorders, as hiPSC-RPE in culture displays morphological, transcriptional, and functional characteristics similar to adult human RPE in vivo (20–24). Furthermore, hiPSC-RPEs have already been used to gain significant insight into the disease mechanisms of retinal diseases including AMD, Best disease, and retinitis pigmentosa (25–28). Thus, patient-derived hiPSC-RPE cells offer a suitable platform to investigate the RPE-autonomous molecular events that contribute to the development of specific-disease–relevant pathology.
The overarching goal of this study was to determine the specific role of RPE-autonomous dysfunction in drusen biogenesis and ECM alterations in maculopathies affecting the RPE–ECM complex. To address this question, we used hiPSCs from patients with the monogenetic MDs, Sorsby’s fundus dystrophy (SFD) and Doyne honeycomb retinal dystrophy/malattia Leventinese (DHRD). SFD and DHRD arise from point mutation(s) within the tissue inhibitor of metalloproteinase 3 (TIMP3) (29) and EGF-containing fibulin-like extracellular matrix protein 1 (EFEMP1) genes (30), respectively. Importantly, SFD and DHRD share a very similar disease presentation, and both display the two key pathologic manifestations central to our disease-modeling efforts, the presence of sub-RPE deposits (drusen) and the accumulation of ECM proteins (1, 2, 5). To further confirm the sensitivity of patient-derived hiPSC-RPE to recapitulate MD phenotype(s), we also included patients with autosomal dominant radial drusen (ADRD) from a pedigree phenotypically similar to DHRD in the absence of EFEMP1 gene mutations. Control hiPSCs used in this study included (i) an unaffected family member of the SFD patients, (ii) a gene-corrected DHRD hiPSC line, (iii) an unaffected family member of the ADRD patients, and (iv) two individuals from the general population with no history of macular degeneration.
Using data from 11 different hiPSC-RPE lines (two from SFD patients harboring mutation in TIMP3 gene, two from DHRD patients harboring mutation in EFEMP1 gene, two from ADRD patients with a yet-unidentified gene defect, and five control hiPSC lines), we demonstrate that (i) RPE-autonomous dysfunction is sufficient for both drusen biogenesis and ECM alterations; (ii) drusen composition varies in control vs. patient hiPSC-RPE cell models; (iii) systemic influences are not required for development of the drusen phenotype in specific maculopathies; and (iv) complement pathway gene alteration occurs locally in the RPE within three distinct inherited MDs. Together, the patient-derived hiPSC-RPE models described here demonstrate the singular contribution of RPE cells to the development of drusen in specific MDs.
Results
“Aged” SFD and DHRD hiPSC-RPE Cultures Display a Larger Number of Sub-RPE Basal Deposits than Control hiPSC-RPE Cultures.
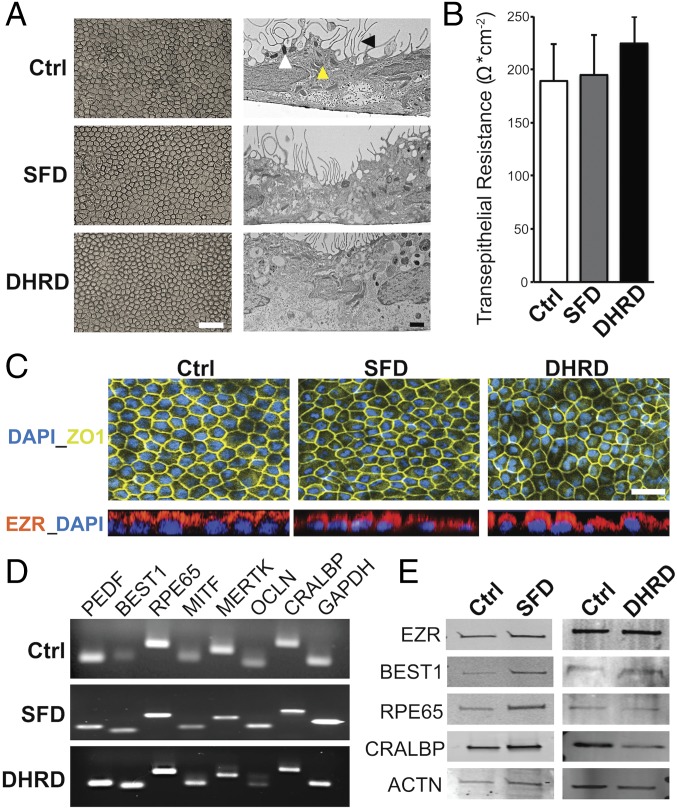
SFD and DHRD are monogenic diseases caused by mutations in TIMP3 and EFEMP1 genes, respectively. Histopathologic studies on donor eyes of both SFD and DHRD patients have shown abnormal lipid- and protein-rich sub-RPE deposits (drusen) (1, 2). The fact that both TIMP3 and EFEMP1 are expressed by RPE cells increases the likelihood that RPE-specific cellular defects in SFD and DHRD patients contribute to the development of drusen. Therefore, in an attempt to recapitulate this central phenotype and isolate the contribution of RPE cells to drusen development within these diseases, SFD, DHRD, and control (Ctrl) hiPSC-RPE were cultured in parallel over an extended period (≥90 d, D90) and analyzed. hiPSC-RPE differentiated from SFD, DHRD, and Ctrl hiPSC lines displayed typical RPE characteristics (Fig. 1). Specifically, light microscopy and electron microscopy analyses revealed similar cobblestone morphology and the presence of apical microvilli, melanosomes, and mitochondria in Ctrl, SFD, and DHRD hiPSC-RPE cultures (Fig. 1A). Consistent with a polarized epithelium, Ctrl, SFD, and DHRD hiPSC-RPE cell monolayers on a nonpermeable plastic support formed fluid domes (SI Appendix, Fig. S1). Similarly, Ctrl, SFD, and DHRD hiPSC-RPE cells plated on Transwells (Corning) developed functional tight junctions with transepithelial resistance similar to that formed by human RPE cells in vivo (150 Ω/cm2) (Fig. 1B) (31). Immunocytochemical analyses of Ctrl, SFD, and DHRD hiPSC-RPE cells revealed uniform expression of the tight junction marker ZO-1 and apical localization of EZR (Fig. 1C). Furthermore, in agreement with an overall normal RPE phenotype, gene and protein expression analyses by RT-PCR and Western blot, respectively, demonstrated similar and robust expression of RPE signature genes and/or proteins PEDF, BEST1, RPE65, MITF, MERTK, OCLN, and CRALBP in Ctrl, SFD, and DHRD hiPSC-RPE cultures (Fig. 1 D and E). Taken together, the baseline evaluation of Ctrl, SFD, and DHRD hiPSC-RPE showed similar physical and functional attributes that are characteristic of in vivo adult RPE.
Fig. 1.
No difference in baseline RPE characteristics were seen in Ctrl, SFD, and DHRD hiPSC-RPE at D90 in culture. (A) Light and electron microscopy images of D90 SFD, DHRD, and Ctrl hiPSC-RPE cultures grown on Transwell inserts (Corning) showed the characteristic cobblestone RPE morphology, apical microvilli (black arrowhead), melanosomes (white arrowhead), and mitochondria (yellow arrowhead). (Scale bars: 50 μm, Left; 1 μm, Right.) (B) Transepithelial resistance measurement of D90 SFD, DHRD, and Ctrl hiPSC-RPE cultures was comparable to the proposed in vivo threshold, 150 Ω·cm−2 (31). Data are expressed as mean + SEM. (C) Immunocytochemical analyses demonstrated RPE-specific morphology, i.e., tight junction formation (ZO-1) and proper polarization (EZR: Apical), of cultured SFD, DHRD, and Ctrl hiPSC-RPE at D90. (Scale bar: 50 μm.) (D and E) RT-PCR (D) and Western blot (E) analyses showed robust expression of RPE characteristic genes and proteins in D90 hiPSC-RPE derived from SFD, DHRD, and Ctrl hiPSCs. GAPDH and ACTN served as loading controls in RT-PCR and Western blotting analysis, respectively. Note: The color palette in confocal images throughout the article has been altered to accommodate colorblind readers.
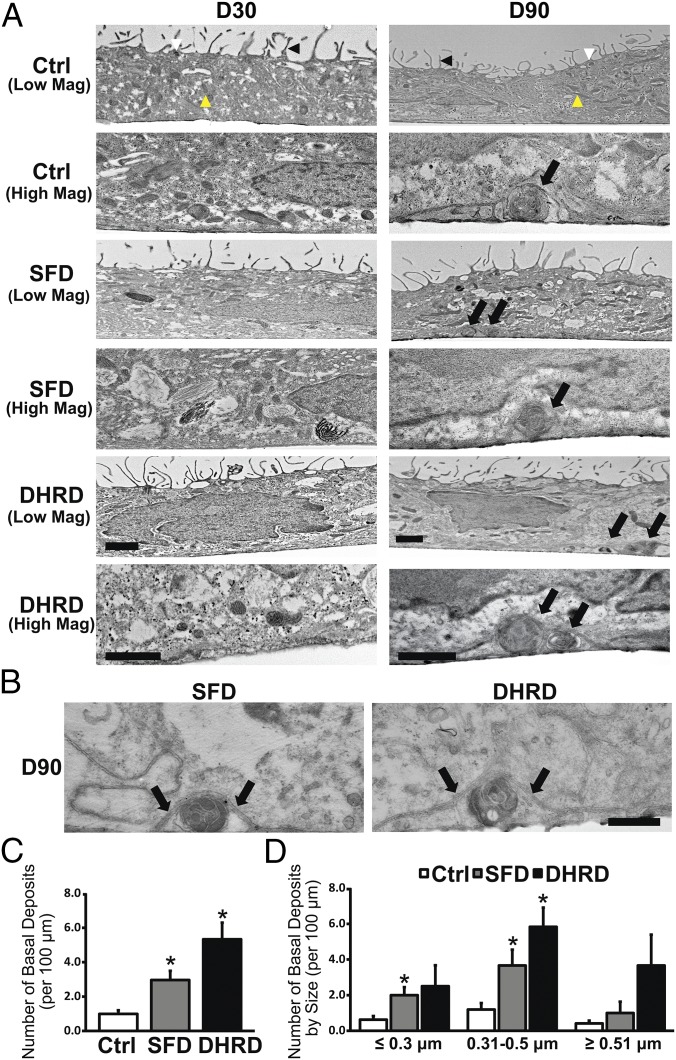
Although hiPSC-RPE in culture lacked a Bruch’s membrane, electron microscopy and immunocytochemical analyses of hiPSC-RPE monolayers (D30–D90) revealed (i) a basement membrane containing basal in-foldings (SI Appendix, Fig. S2A) and (ii) COL4, LAM, and TIMP3 localization consistent with the presence of a defined basement membrane (SI Appendix, Fig. S2 B–D). Electron microscopy analyses of “young” (∼D30) Ctrl, SFD, and DHRD hiPSC-RPE on Transwells were indistinguishable and displayed similar ultrastructure with apical microvilli, mitochondria, and melanosomes (Fig. 2A). Of note, although no basal deposits were seen in any hiPSC-RPE cultures at D30 (Fig. 2A), accumulation of electron-dense material lacking drusen morphology was sporadically observed in DHRD hiPSC-RPE cultures. In contrast to D30 hiPSC-RPE cultures, aged (D90) hiPSC-RPE cells on Transwells displayed basal deposits that were localized in a position consistent with that of drusen in patient eyes (Fig. 2A). Specifically, basal deposits in Ctrl and patient-derived SFD and DHRD hiPSC-RPE cultures were circular, electron-dense structures present extracellularly and underneath the hiPSC-RPE cell membrane (Fig. 2 A and B). In fact, basal deposits in Ctrl, SFD, and DHRD hiPSC-RPE cultures often displaced the overlying hiPSC-RPE basement membrane (Fig. 2 A and B). Quantitative analyses of basal deposits in Ctrl vs. SFD and DHRD hiPSC-RPE cultures revealed increased number of basal deposits in SFD and DHRD hiPSC-RPE cultures compared with that in Ctrl hiPSC-RPE cultures (Fig. 2C). The characterization of basal deposits by diameter showed deposits in three distinct size ranges: ≤0.3 μm, 0.31–0.5 μm, and ≥0.51 μm in Ctrl, DHRD, and SFD hiPSC-RPE cultures. Of note, the largest number of deposits in all cultures ranged between 0.31 and 0.5 μm (Fig. 2D). The comparison of basal deposits by size in Ctrl, SFD, and DHRD hiPSC-RPE cultures further demonstrated significantly more deposits in Ctrl vs. SFD hiPSC-RPE culture in both the ≤0.3 μm and the 0.31–0.5 μm size range and in Ctrl vs. DHRD cultures in the 0.31–0.5 μm size range (Fig. 2D). It is noteworthy that in addition to an increased number of basal deposits in Ctrl vs. SFD and Ctrl vs. DHRD hiPSC-RPE cultures, a greater number of basal deposits were observed in DHRD hiPSC-RPE cultures than in SFD hiPSC-RPE cultures (P = 0.07). Coupled with the aforementioned early initiation of sparse electron-dense material accumulation beneath D30 DHRD hiPSC-RPE cultures but not Ctrl and SFD hiPSC-RPE cultures, these data are consistent with an earlier onset of DHRD pathophysiology compared with SFD.
Fig. 2.
Increased number of basal deposits underlying aged (D90) Ctrl vs. SFD and DHRD hiPSC-RPE cultures. (A) Transmission electron microscopy (TEM) analyses showed cellular features consistent with RPE cells, i.e., apical microvilli (black arrowhead), melanosomes (white arrowhead), and mitochondria (yellow arrowhead), in both D30 (Left) and D90 (Right) Ctrl, SFD, and DHRD hiPSC-RPE cultures. No sub-RPE basal deposits were seen in Ctrl, SFD, and DHRD hiPSC-RPE cells at D30 (Left). In contrast, Ctrl, SFD, and DHRD hiPSC-RPE cultures at D90 in culture exhibited basal deposits (black arrows, Right). (Scale bars: 1 μm.) (B) Higher-magnification TEM images further demonstrated membrane-displacing basal deposits in SFD and DHRD hiPSC-RPE cultures (the basement membrane is indicated by black arrows). (Scale bar: 200 nm.) (C) Quantification of the total number of basal deposits confirmed an increased number of basal deposits in D90 SFD and DHRD hiPSC-RPE cultures compared with age-matched Ctrl hiPSC-RPE cultures. (D) Quantification and characterization of basal deposits by diameter demonstrated basal deposits in three distinct size ranges: ≤0.3 μm, 0.31–0.5 μm, and ≥0.51 μm in Ctrl, DHRD, and SFD hiPSC-RPE cultures. Furthermore, deposits ≤0.3 μm and/or 0.31–0.5 μm were more abundant in SFD and DHRD hiPSC-RPE cultures compared with Ctrl hiPSC-RPE cultures. Data are presented as mean + SEM. *P < 0.05.
Taken together, our results show that hiPSC-RPE cells when aged in culture and in the absence of any exogenous stressor can form sub-RPE basal deposits. Furthermore, consistent with existing clinical data, basal deposits are more abundant in hiPSC-RPE derived from SFD and DHRD patients than in those from control individuals.
The Composition of Basal Deposits Varies in SFD and DHRD hiPSC-RPE Cultures Compared with Ctrl hiPSC-RPE Cultures.
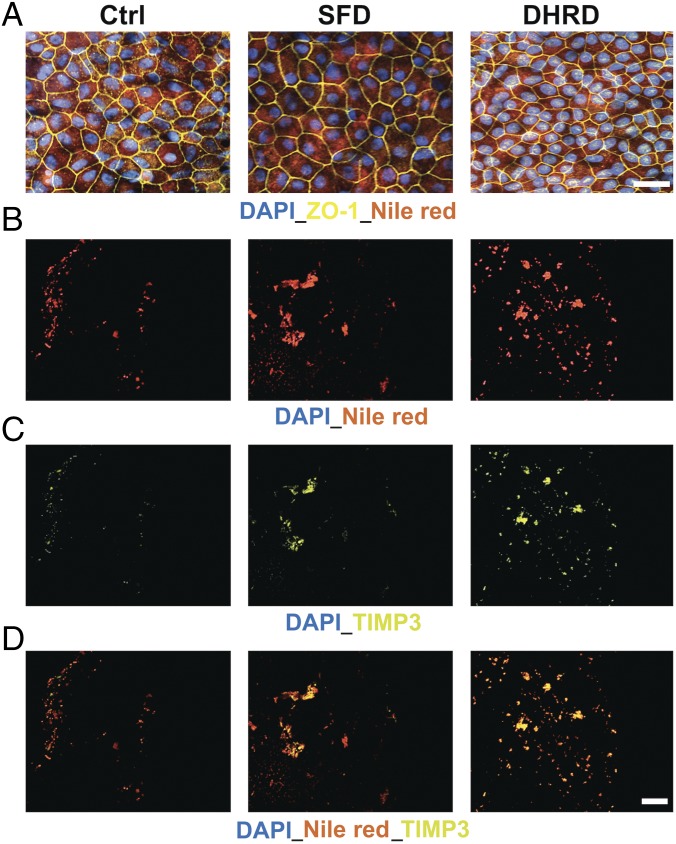
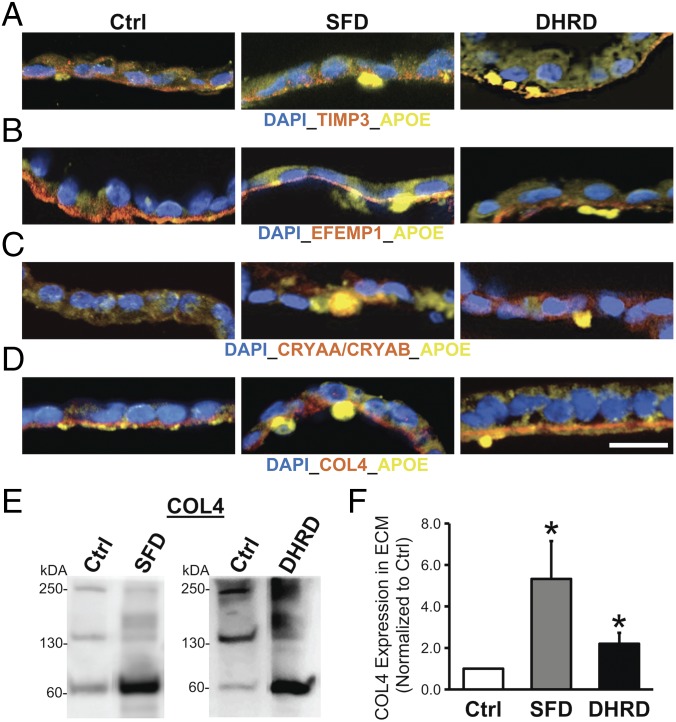
Given that drusen in macular degeneration patient eyes are lipoproteinaceous deposits underlying RPE cells, we next evaluated if sub-RPE deposits in SFD and DHRD hiPSC-RPE in culture were also rich in neutral lipids and the RPE-synthesized drusen proteins APOE, CRYAA/CRYAB, and TIMP3 (32–35). Consistent with published studies on mammalian RPE (14, 36), staining of hiPSC-RPE cultures with Nile red demonstrated the presence of neutral lipids in Ctrl, SFD, and DHRD hiPSC-RPE cultures (Fig. 3A). To probe for the presence of drusen-like deposits underneath the hiPSC-RPE monolayer, we used trypsin and removed the polarized monolayer of aged (D90) hiPSC-RPE from Transwell membranes and subsequently performed immunostaining on the surface of the Transwell membrane for drusen-characteristic lipids and proteins (Nile red and TIMP3). Of note, DAPI staining in these experiments was used to confirm the lack of any residual RPE cells on the surface of Transwell membranes. Nile red staining of Transwell membranes underlying Ctrl vs. SFD and Ctrl vs. DHRD hiPSC-RPE cultures revealed an increased amount of neutral lipid deposition on Transwell membranes underlying SFD and DHRD hiPSC-RPE cultures (Fig. 3B). Similarly, immunolabeling of the Transwell membrane after hiPSC-RPE removal demonstrated increased deposition of a drusen-associated protein, TIMP3, in SFD and DHRD hiPSC-RPE cultures compared with Ctrl hiPSC-RPE cultures (Fig. 3C). Importantly, consistent with the prominent presence of both neutral lipids and TIMP3 in drusen underlying patient eyes, TIMP3 and Nile red immunostaining almost completely colocalized in the basal deposits formed on the surface of the Transwell membrane underneath the SFD and DHRD hiPSC-RPE monolayers (Fig. 3D). To further analyze the composition and localization of basal deposits underlying Ctrl vs. SFD and DHRD hiPSC-RPE cells, we also performed immunocytochemical analyses of RPE cross-sections (D90) for the presence and localization of the basement membrane proteins TIMP3, EFEMP1, and COL4 alongside known drusen constituents APOE, TIMP3, and CRYAA/CRYAB. Of note, the confocal images shown in Fig. 4 A–D have been selected to show the entire range of the immunostaining pattern observed in our analyses of basement membrane proteins (TIMP3, EFEMP1, and COL4) and drusen constituents (APOE, TIMP3, and CRYAA/CRYAB). Importantly, Ctrl, DHRD, and SFD hiPSC-RPE cultures showed the expected basement membrane localization of TIMP3, EFEMP1, and COL4 proteins (Fig. 4 A–D and SI Appendix, Fig. S2 B–D) (32, 37–40). Although APOE-positive sub-RPE deposits were occasionally observed in Ctrl hiPSC-RPE cultures (Fig. 4 A–D and SI Appendix, Figs. S3 and S4), sub-RPE deposits that showed immunoreactivity to TIMP3, CRYAA/CRYAB, and EFEMP1 in conjunction with APOE were present only underneath SFD and DHRD hiPSC-RPE cultures (Fig. 4 A–D and SI Appendix, Figs. S3 and S4). It is also noteworthy that APOE-positive basal deposits in SFD and DHRD hiPSC-RPE cultures were extracellular and were present underneath the basement membrane stained by COL4 antibody (Fig. 4D and SI Appendix, Fig. S4A). The genetic basis of sub-RPE drusen-like deposits was further confirmed by parallel experiments on hiPSC-RPE cultures from the DHRD patient and its corresponding isogenic hiPSC line in which APOE-, CRYAA/CRYAB-, EFEMP1-, and TIMP3-positive sub-RPE drusen-like deposits were exclusively present in DHRD hiPSC-RPE cultures (SI Appendix, Fig. S4).
Fig. 3.
Transwell membranes underneath aged (D90) SFD and DHRD hiPSC-RPE show deposition of Nile red-stained TIMP3 containing lipid–protein complexes. (A) Nile red staining demonstrated uniform intracellular expression of neutral lipids in D90 Ctrl, DHRD, and SFD hiPSC-RPE. (Scale bar: 50 μm.) (B–D) Immunostaining of Transwell membranes after removal of the D90 hiPSC-RPE monolayer showed increased sub-RPE deposition of neutral lipids (B), TIMP3 (C), and colocalized neutral lipid-TIMP3 complexes (D) on the surface of Transwell membranes underlying patient SFD and DHRD hiPSC-RPE cultures compared with Ctrl hiPSC-RPE cultures. (Scale bar: 50 μm.) Of note, confocal images from the same experiment showing the same Transwell membrane are shown in B–D to emphasize the almost complete colocalization of TIMP3 and Nile red staining in sub-RPE deposits on Transwell membranes underlying SFD and DHRD hiPSC-RPE cultures.
Fig. 4.
Presence of sub-RPE deposits with drusen-like composition underneath aged (D90) SFD and DHRD hiPSC-RPE cultures. (A–D) Confocal images of age-matched (D90) Ctrl vs. SFD and DHRD hiPSC-RPE cross-sections displayed the presence of TIMP3-APOE–positive (A), EFEMP1-APOE–positive (B), CRYAA/CRYAB-APOE–positive (C), and APOE-positive deposits underlying basement membrane marked by COL4 (D) in SFD and DHRD hiPSC-RPE cultures. (Scale bar: 25 µm.) Of note, sporadic APOE-positive sub-RPE basal deposits (A and D) were observed underneath Ctrl hiPSC-RPE cultures. (E and F) Quantitative Western blot analyses revealed increased amount of COL4 protein in the ECM underlying SFD and DHRD hiPSC-RPE cultures compared with Ctrl hiPSC-RPE cultures at D90. Of note, data are presented as mean + SEM, and COL4 bands at ∼250, 150, and 70 kDa are consistent with multimeric and monomeric α1 and α2 subunits and fragments of human COL4 protein. *P ≤ 0.05.
Taken together, in addition to confirming that hiPSC-RPE in culture is capable of forming basal deposits in a cell-autonomous fashion, these results demonstrate that the composition of sub-RPE deposits varies between Ctrl vs. patient-derived (SFD, DHRD) hiPSC-RPE cultures. Specifically, colocalization of neutral lipids and several RPE-synthesized drusen proteins (APOE, CRYAA/CRYAB, EFEMP1, and TIMP3) is a pathologic phenotype present only in patient-derived (SFD and DHRD) hiPSC-RPE cultures.
COL4 Levels Are Higher in ECM Isolated from SFD and DHRD hiPSC-RPE Cultures than in Ctrl hiPSC-RPE Cultures.
Increased deposition of ECM constituents underneath RPE cells has previously been shown in both SFD and DHRD (2, 5, 41) and has been implicated in the Bruch’s membrane thickening identified in vivo (1, 2). To investigate whether a similar phenotype of increased deposition of specific ECM proteins could be mimicked in SFD and DHRD hiPSC-RPE monocultures, we isolated and analyzed ECM underlying Ctrl vs. SFD and DHRD hiPSC-RPE cultures. Quantitative Western blot analyses of ECM extracts from Ctrl, DHRD, and SFD hiPSC-RPE revealed increased abundance of COL4, a major component of basement membrane and specifically of the Bruch’s membrane (39), in the ECM extracted from DHRD and SFD hiPSC-RPE cells relative to Ctrl hiPSC-RPE cells (Fig. 4 E and F). Of note, Western analyses of COL4 in ECM underlying hiPSC-RPE cells showed bands of ∼250, 150, and 60 kDa consistent with multi- and monomeric α1, α2 subunits and fragments of human COL4 protein (42, 43). In addition to COL4, we quantified the levels of LAM, another RPE basement membrane protein, in the ECM underlying hiPSC-RPE cells using a LAM antibody that recognizes all subunits of human LAM protein, which range in size from 400 kDa for the alpha chain to 210 and 200 kDa for the β1 and β2 chains, respectively (44). No difference in the levels of LAM was seen in Ctrl, SFD, and DHRD hiPSC-RPE cultures (SI Appendix, Fig. S5). Overall, these results demonstrate increased levels of a specific RPE basement membrane protein, COL4, in the ECM underlying hiPSC-RPE derived from patients with two distinct MDs, SFD and DHRD.
ADRD hiPSC-RPEs Mimic the Phenotypic Alterations Observed in SFD and DHRD hiPSC-RPE Cultures.
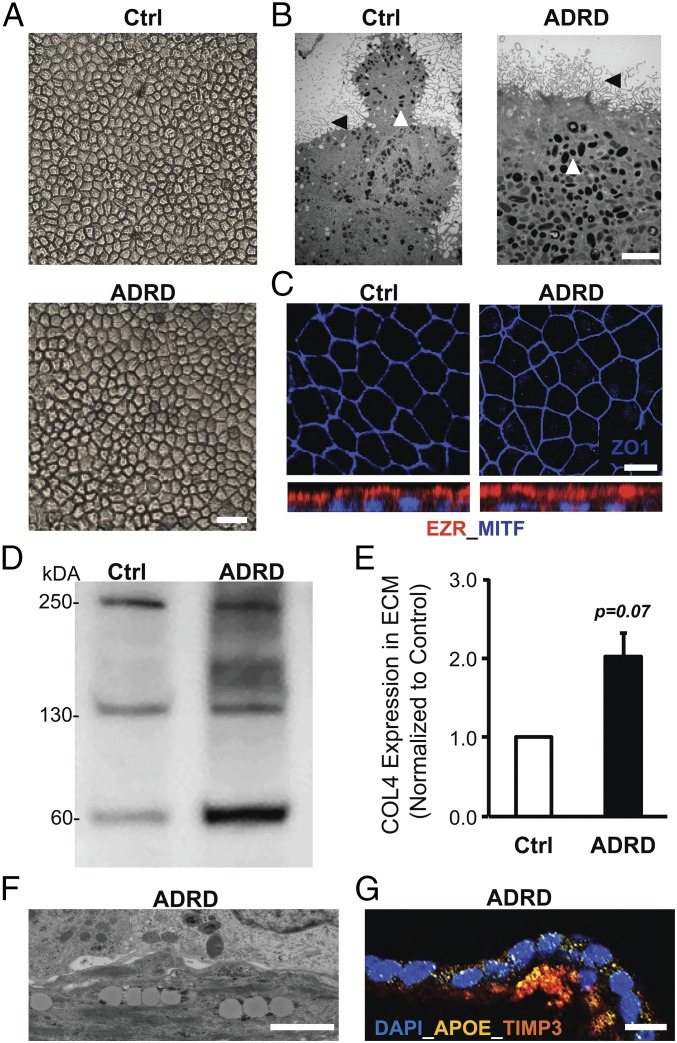
Experiments on SFD and DHRD hiPSC-RPE cultures had thus far demonstrated that molecular alterations limited to RPE cells alone were sufficient for drusen formation in monogenic maculopathies in which the affected gene (TIMP3, EFEMP1) is expressed predominantly by RPE cells (2, 32). To further assess the sensitivity of hiPSC-RPE to capture a maculopathy-relevant phenotype, we next chose to investigate the origin of drusen development in a pedigree with the same disease phenotype as DHRD but with an unidentified genetic defect. Similar to SFD and DHRD hiPSC-RPE, ADRD hiPSC-RPE in culture displayed baseline characteristics comparable to Ctrl hiPSC-RPE cultures. Specifically, light and electron microscopy analyses demonstrated that ADRD hiPSC-RPE cells, like Ctrl hiPSC-RPE cells, display the RPE-characteristic cobblestone morphology as well as apical microvilli and melanosomes (Fig. 5 A and B). Furthermore, ADRD hiPSC-RPE, like Ctrl hiPSC-RPE, formed a polarized epithelium in culture as evident from the formation of fluid domes (SI Appendix, Fig. S6) and localization of ZO-1 and EZR (Fig. 5C). Interestingly, similar to SFD and DHRD hiPSC-RPE, quantitative Western blot analyses of ECM underlying RPE cells revealed an increase in the amount of COL4 in Ctrl vs. ADRD hiPSC-RPE cultures (Fig. 5 D and E). Importantly and consistent with the disease pathology and our previous characterization of SFD and DHRD hiPSC-RPE culture (Figs. 2–4), electron microscopy and confocal microscopy analyses of aged (D90) ADRD hiPSC-RPE in culture revealed lipid deposition and sub-RPE deposits, with strong colocalization of the drusen-resident proteins APOE and TIMP3 (Fig. 5 F and G).
Fig. 5.
Increased accumulation of COL4 in ECM and the presence of TIMP3-APOE–positive deposits in aged (D90) ADRD hiPSC-RPE cultures. (A and B) Light microscopy (A) and electron microscopy (B) analysis at D90 showed similar cobblestone morphology in Ctrl vs. ADRD hiPSC-RPE cultures. Apical microvilli (black arrowhead), and melanosomes (white arrowhead) are seen in Ctrl vs. ADRD hiPSC-RPE cultures. (Scale bars: 100 µm in A; 5 µm in B.) (C) Immunocytochemical analyses demonstrated similar localization of the tight junction protein ZO-1 and the apical RPE cell marker EZR in Ctrl and ADRD hiPSC-RPE at D90. (Scale bar: 10 μm.) (D and E) Quantitative Western blot analyses demonstrated higher levels of COL4 protein in the ECM underlying Ctrl vs. ADRD hiPSC-RPE cultures at D90. Of note, data are presented as mean + SEM in the bar graph. Furthermore, COL4 bands at ∼250, 150, and 70 kDa are consistent with multi- and monomeric α1, α2 subunits and fragments of the human COL4 protein. (F and G) Electron microscopy (F) and immunocytochemical (G) analyses revealed lipid droplet accumulation and TIMP3-APOE–positive sub-RPE deposits in D90 ADRD hiPSC-RPE cultures. (Scale bars: 1 µm in F and 50 µm in G.)
The analysis of ADRD cultures demonstrates that RPE-autonomous cellular alterations are sufficient for drusen biogenesis and ECM alterations in multiple maculopathies affecting the RPE-ECM complex and highlight the sensitivity of hiPSC-RPE as a model system to recapitulate specific disease phenotype(s) of macular degeneration in the absence of a known genetic defect.
Increased Expression of Complement Pathway Genes in SFD, DHRD, and ADRD hiPSC-RPE Monocultures.
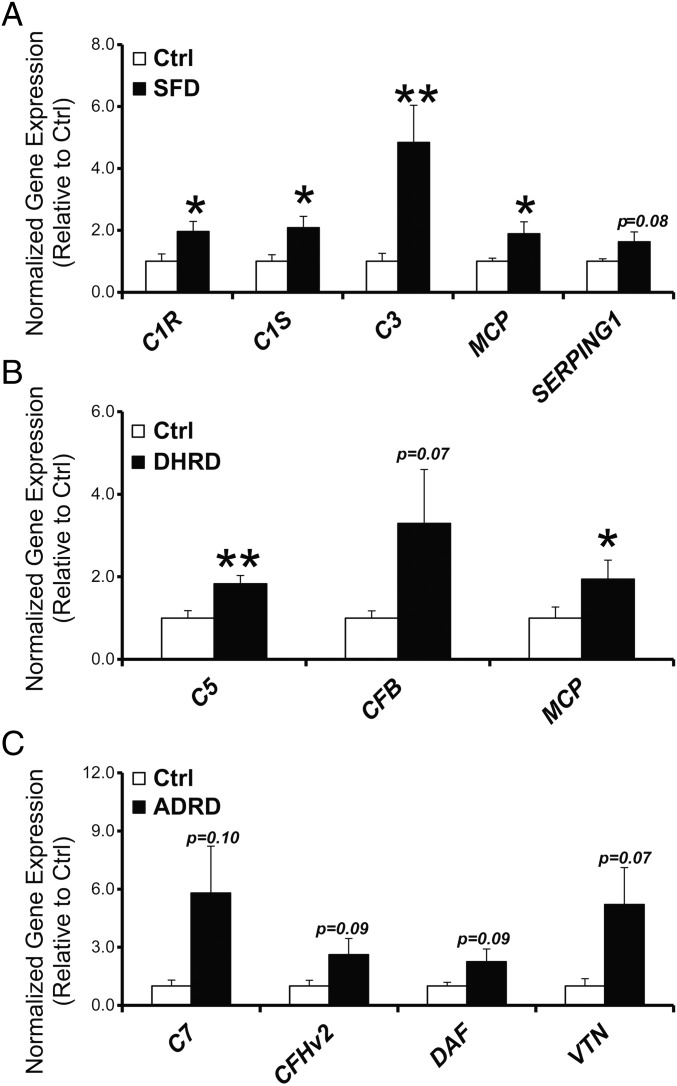
Our results comparing pathological manifestations of three distinct MDs in patient-derived hiPSC-RPE cultures had thus far established common phenotypic alterations, i.e., formation of drusen-like deposits, and accumulation of COL4 in the ECM beneath RPE cells in SFD, DHRD, and ADRD hiPSC-RPE cultures. These phenotypic similarities led us to investigate whether SFD, DHRD, and ADRD hiPSC-RPE also shared similar molecular alterations. We chose to investigate the complement pathway based on studies implicating the complement cascade in drusen formation in specific maculopathies localized to the RPE-ECM complex. Previously published data on the DHRD mouse model and cultured mouse RPE cells from the DHRD mouse model shows a possible role of local complement alteration in the disease pathology (13, 45). A recent paper further showed increased expression of several complement pathway genes in hiPSC-RPE monocultures derived from AMD patients (46). Activation of the classical complement pathway has also been shown to be necessary for drusen formation in hfRPE cultures (15). Furthermore, broad suppression of the complement response using the C3 inhibitor compstatin has previously been shown to regress drusen in a cellular and nonhuman primate model of macular degeneration (47, 48). These data together support a link between complement pathway alteration/activation at the local level of RPE cells and drusen formation in multiple macular degeneration models. Although evaluating a direct link between complement pathway alterations was beyond the scope of this study, we were interested in evaluating whether hiPSC-RPE from three distinct inherited maculopathies, SFD, DHRD, and ADRD, that display drusen-like deposits will also show alterations in complement pathway genes similar to that observed in hiPSC-RPE from AMD patients (46). We limited our analysis to complement pathway genes that have previously been shown to be expressed by human RPE cells (49). Of note, hiPSC-RPE cultures aged to D90, a time point consistent with the formation of drusen-like deposits in SFD, DHRD, and ADRD hiPSC-RPE cultures, were used in these experiments. Furthermore, Ctrl, SFD, DHRD, and ADRD hiPSC-RPE cultures in these experiments were not exposed to any exogenous stressors (e.g., serum, POS). Quantitative real-time PCR analyses demonstrated increased expression of several distinct complement pathway genes in Ctrl vs. SFD (C1R, C1S, C3, MCP, and SERPING1), in Ctrl vs. DHRD (C5, CFB, and MCP), and in Ctrl vs. ADRD (C7, CFHv2, DAF, and VTN) hiPSC-RPE monocultures (Fig. 6 and SI Appendix, Table S1). Taken together, our results show that aged D90 SFD, DHRD, and ADRD hiPSC-RPE monocultures that display drusen-like sub-RPE deposits (Figs. 2–5) concurrently demonstrate increased expression of several complement pathway genes (Fig. 6).
Fig. 6.
Increased expression of complement pathway genes in ADRD, SFD, and DHRD compared with Ctrl hiPSC-RPE monoculture at D90. (A–C) Quantitative real-time PCR analysis demonstrated increased expression of several complement pathway genes in SFD (A), DHRD (B), and ADRD (C) hiPSC-RPE cultures at D90 compared with age-matched Ctrl hiPSC-RPE. Data are presented as mean + SEM. *P < 0.05, **P < 0.005.
Serum Supplementation Affects Composition of Drusen-Like Basal Deposits in SFD and DHRD hiPSC-RPE Cultures.
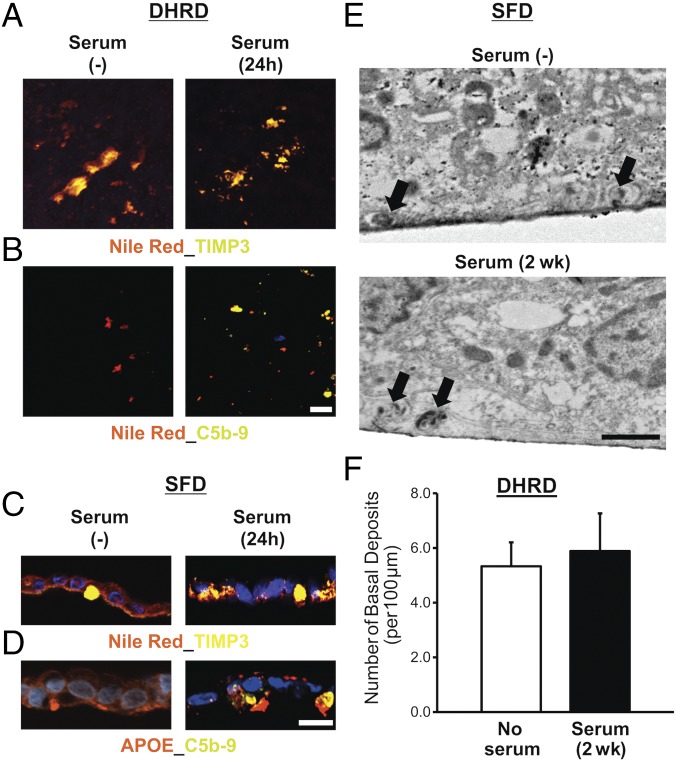
It has previously been shown that serum supplementation is sufficient to promote drusen formation in hfRPE cultures (15). Furthermore, drusen deposits in serum-exposed hfRPE cultures displayed the deposition of complement proteins, i.e., the C5b-9 complex and VTN (15). Although, our previous data showed that patient-derived SFD, DHRD, and ADRD hiPSC-RPE in culture display increased expression of several complement pathway genes (Fig. 6) and cell-autonomously developed drusen-like deposits (Figs. 2–5), sub-RPE deposits in SFD, DHRD, and ADRD hiPSC-RPE monocultures did not contain the complement proteins (Fig. 7 A–D). The lack of complement proteins in sub-RPE deposits underlying SFD, DHRD, and ADRD hiPSC-RPE cultures in conjunction with the documented presence of complement proteins in (i) drusen in AMD patient eyes (50, 51), (ii) sub-RPE deposits in the DHRD mouse model (13, 45, 52), and (iii) sub-RPE deposits/drusen underlying hfRPE in culture after serum exposure (15) suggested that complement proteins in drusen are derived from an extra-RPE source, such as serum. To validate and further confirm these findings in patient-derived drusen models of MDs, we evaluated the impact of acute (24-h) and chronic (2-wk) serum exposure on drusen composition in Ctrl and SFD, DHRD, and ADRD hiPSC-RPE cultures. Important for these studies, live-cell staining with Calcein-AM showed no adverse effect of chronic (2-wk) serum supplementation on hiPSC-RPE cell viability (SI Appendix, Fig. S7). Consistent with the published study on hfRPE (15), Ctrl hiPSC-RPE exposed to exogenous serum displayed sub-RPE deposits immunoreactive for both APOE and C5b-9 complex (SI Appendix, Fig. S8). Furthermore, immunocytochemical analyses showed that both acute (24-h) and chronic (2-wk) serum supplementation leads to the deposition of the complement proteins C3, C5b-9, and VTN in drusen-like deposits underneath the SFD, DHRD, and ADRD hiPSC-RPE cultures (Fig. 7 A–D and SI Appendix, Fig. S9). Specifically, immunostaining of Transwell membranes after removal of the overlying hiPSC-RPE monolayer with antibodies against TIMP3 and C5b-9 and Nile red staining showed (i) Nile red- and TIMP3-containing deposits in both untreated and serum-treated (10%, 24 h) DHRD hiPSC-RPE cultures (Fig. 7 A and B) and (ii) the selective presence of C5b-9–containing deposits in serum-treated (10%, 24 h) DHRD hiPSC-RPE cultures (Fig. 7B). Similarly, immunocytochemical analyses of SFD hiPSC-RPE sections demonstrated the presence of Nile red-, TIMP3-, and APOE-positive sub-RPE deposits in both untreated and serum-treated (10%, 24 h) cultures (Fig. 7C). In contrast, C5b-9 and VTN immunoreactive basal deposits were seen only underneath serum-treated (10%, 24 h) SFD and DHRD hiPSC-RPE cells (Fig. 7 B and D and SI Appendix, Fig. S9A). In a subset of experiments, we further confirmed the extracellular localization of complement protein C3 containing drusen-like deposits after serum exposure. Specifically, when immunocytochemical analyses were performed after the removal of the ADRD hiPSC-RPE monolayer in a portion of the culture dish, C3- and APOE-immunoreactive deposits were found only in the area void of cells (SI Appendix, Fig. S9B). Of note, DAPI staining in these experiments was used to differentiate between RPE cell-containing vs. RPE-cell void areas. It is also noteworthy that the serum-derived drusen proteins C3, C5b-9 complex, and VTN colocalized with the other drusen constituents TIMP3 and APOE in sub-RPE deposits underneath SFD, DHRD, and ADRD hiPSC-RPE cultures (Fig. 7 B and D and SI Appendix, Fig. S9). In contrast to the effect of serum supplementation on drusen composition, electron microscopy analyses revealed no difference in the number of drusen deposits before and after serum exposure (Fig. 7 E and F).
Fig. 7.
Serum supplementation affects the composition of sub-RPE deposits in aged (D90) patient-derived hiPSC-RPE cultures. (A) Confocal microscopy after immunocytochemical analyses showed similar sub-RPE deposition of colocalized neutral lipid–TIMP3 complexes on the surface of Transwell membranes underlying the DHRD hiPSC-RPE (D90) monolayer in untreated vs. serum-treated (10%, 24-h) cultures. (B) Confocal microscopy demonstrated deposition of complement proteins C5b-9 in conjunction with neutral lipids after supplementation of D90 DHRD hiPSC-RPE cultures with 10% serum for 24 h. [Scale bar (applies also to A): 50 µm.] (C) Immunocytochemical analyses of D90 SFD hiPSC-RPE cross-sections after serum supplementation (10%, 24 h) in culture medium demonstrated the presence of Nile red-, TIMP3-positive deposits in both untreated and serum-treated SFD hiPSC-RPE cultures. (D) Immunostaining of D90 SFD hiPSC-RPE cross-sections showed selective deposition of C5b-9 complex with APOE-positive sub-RPE deposits in serum-treated (10%, 24 h) compared with untreated SFD hiPSC-RPE cultures. (E and F) Electron microscopy analyses showed similar numbers of basal deposits (black arrows) in both untreated and serum-treated SFD (E) and DHRD (F) hiPSC-RPE cultures after chronic serum supplementation (10%, 2 wk). Data are presented as mean + SEM. (Scale bar in E: 1 µm.)
Overall, our results show that although serum is not necessary for drusen formation, it can affect the composition of drusen in patient-derived SFD, DHRD, and ADRD hiPSC-RPE cultures. The selective deposition of complement proteins (C3, C5b-9) and VTN in sub-RPE deposits after serum exposure suggests that systemic influence can modulate drusen composition in maculopathies such as SFD, DHRD, and ADRD.
Discussion
Maculopathies, a major cause of blindness, are complex heterogeneous disorders that lead to the loss of central vision. In AMD and related MDs, the primary site of disease pathology in the retina is the RPE-ECM complex. However, because multiple cell layers in the eye (photoreceptor, RPE, and choroid) are affected in these maculopathies and because in vivo the retina acts as a functional unit, the singular contribution of RPE cells in the pathology of these diseases has not been established. Here, using patient-derived drusen models of three distinct MDs, SFD, DHRD, and ADRD, we address fundamental questions about the role of RPE cells in specific maculopathies. The major findings of our study are concisely summarized in the following points. (i) Consistent with previously published studies (13–15, 45, 52) and human clinical data, we show that, although drusen can be present underneath both control (unaffected) and macular degeneration patient RPE, these deposits are more abundant in patient-derived hiPSC-RPE cultures (Fig. 2). Interestingly, our data also show that the composition of drusen-like sub-RPE deposits varies in control vs. patient hiPSC-RPE samples (Figs. 3–5 and SI Appendix, Figs. S3 and S4). This has significant implications for macular degeneration research and suggests that a patient-derived model is necessary for understanding the pathogenesis of drusen deposits in maculopathies such as SFD, DHRD, and ADRD. (ii) It has been hypothesized that photoreceptors and choriocapillaris are required for the development of macular degeneration phenotypes within maculopathies such as AMD and SFD. Here, using the patient-derived hiPSC maculopathy models, we show that development of drusen is an RPE-autonomous cellular pathology in multiple maculopathies (Figs. 2–5). Specifically, in the absence of photoreceptors and choriocapillaris, patient-derived hiPSCs form drusen, the key pathological hallmark of MDs. (iii) Consistent with previous studies (15), we show that serum-derived factors contribute to drusen composition in patient-derived hiPSC-RPE models of MDs (Fig. 7 and SI Appendix, Fig. S9). Importantly, we extend these findings and demonstrate that serum-derived factors are not required for drusen biogenesis in specific MDs such as SFD, DHRD, and ADRD (Figs. 2–5). (iv) It is well established that complement gene variants, including CFH, C3, CFB, and CFI, confer a risk for AMD. Complement activation has also been associated with drusen development in cellular and mouse models of AMD and DHRD. However, complement pathway alteration in patients with specific MDs has not been established. Here we show that the expression of multiple complement pathway genes is increased in patient-derived hiPSC-RPE models of SFD, DHRD, and ADRD (Fig. 6). This implicates complement pathway dysregulation at the local level, within the RPE cell of the eye, in the pathology of several inherited MDs. Taken together, the patient-derived drusen models of MDs described here provide important insights into the cell biology underlying specific maculopathies affecting the RPE–ECM complex and provide a suitable platform to further investigate the RPE-autonomous molecular and pathological alterations that contribute to the development and progression of these diseases.
A pathologic hallmark of AMD and similar maculopathies, such as SFD and DHRD, is the accumulation of drusen, lipoproteinaceous aggregates localized between the RPE and its underlying ECM, the Bruch’s membrane in the eye. Although RPE cells in culture do not lay a complex ECM like the in vivo Bruch’s membrane, previous studies using primary RPE cells of human, mouse, and porcine origin (13–15) have demonstrated the formation of sub-RPE deposits in cell culture. Consistent with the aforementioned studies, we show that Ctrl hiPSC-RPE cells in culture are capable of forming sub-RPE deposits (Fig. 2 A, C, and D and SI Appendix, Figs. S3 and S4). In fact, similar to data published on porcine RPE in culture (14), our results demonstrate spontaneous development of APOE-positive sub-RPE deposits in aged (D90) Ctrl hiPSC-RPE cultures (Figs. 2 A, C, and D and 4 A and D and SI Appendix, Fig. S4D). It is plausible that, as in the other cell culture models of sub-RPE basal deposits (14, 15), the presence of a physical barrier in the form of the 10-µm-thick polyester Transwell support (14) or nonpermeable plastic support underlying the hiPSC-RPE monolayer contributed to the development of the sub-RPE deposits in our cultures. Interestingly, young (D30) Ctrl hiPSC-RPE cultures did not contain basal deposits (Fig. 2A), highlighting the role of physiologically stressing or aging RPE cells in culture to promote the development of drusen-like deposits. Importantly and in agreement with the ability of hiPSC-derived RPE cells to recapitulate features of specific hiPSC-donor human RPE cells in vivo, our results demonstrate a higher abundance of sub-RPE basal deposits in aged (D90) patient-derived cultures than in aged (D90) Ctrl hiPSC-RPE cultures (Fig. 2). Furthermore, consistent with histological and proteomic characterization of drusen in human cadaver eyes (12), our data show that the composition of drusen-like deposits varies between Ctrl vs. patient hiPSC-RPE cultures (Figs. 3–5 and SI Appendix, Figs. S3 and S4). In addition, the impact of serum supplementation on drusen composition in hfRPE cultures (15) and patient-derived hiPSC-RPE cultures (Fig. 7 and SI Appendix, Fig. S9) emphasizes the plausible role of RPE-extrinsic factors in the modulation of the drusen phenotype in specific maculopathies. In this context, it would be important and relevant to further interrogate the impact of other physiologically relevant stressors (e.g., POS and RPE–vascular interaction) on the drusen phenotype in Ctrl vs. patient-derived hiPSC-RPE cultures.
Previous clinical studies have described striking similarities between symptoms, pathology, and functional loss in maculopathies affecting the RPE-ECM complex, including the inherited maculopathies SFD and DHRD and AMD. Specifically, these maculopathies lead to the bilateral loss of central vision due to choroidal neovascularization and/or RPE atrophy (6, 53, 54). Pathologically these diseases are characterized by common alterations, including drusen formation and thickening of Bruch’s membrane (1–5). These similarities suggest that similar mechanistic defects may underlie the shared pathological manifestations in these distinct maculopathies. In support of this hypothesis, we demonstrate increased accumulation of a RPE-secreted ECM protein, COL4, in the ECM beneath hiPSC-RPE cultures derived from three distinct MDs, i.e., SFD, DHRD, and ADRD (Figs. 4 E and F and 5 D and E). Interestingly, impaired MMP2 activity and consequently COL4 accumulation has previously been linked to AMD pathology (55). In this regard, it is also noteworthy that MMP2 activity is regulated by both TIMP3 and EFEMP1 (56–59), the protein product of genes mutated in SFD and DHRD, respectively. Extrapolation of our findings with the known regulatory roles of TIMP3 and EFEMP1 on MMP2 activity and the published linkage of MMP2 activity to COL4 accumulation in Bruch’s membrane leads us to hypothesize that impaired MMP2 activity contributes to Bruch’s membrane thickening in multiple maculopathies (e.g., SFD, DHRD, and AMD) by affecting COL4 turnover in the ECM underlying RPE cells. Our data showing increased expression of several complement pathway genes in SFD, DHRD, and ADRD hiPSC-RPE monocultures (Fig. 6) and a recent paper showing elevated expression of several complement pathway genes in hiPSC-RPE cells derived from AMD patients further highlight similar molecular alteration locally in RPE cells in distinct maculopathies affecting the RPE–ECM interface (46). Of note, in our studies, distinct complement pathway genes were up-regulated in SFD, DHRD, and ADRD hiPSC-RPE cultures, potentially highlighting similar (but not the same) molecular change in distinct maculopathies affecting the RPE–ECM complex. On the same note, although Saini et al. (46) did not interrogate the formation and composition of sub-RPE deposits underlying hiPSC-RPE cultures from patients with AMD, they also show, in agreement with our findings, increased expression of several drusen-related proteins in AMD patient hiPSC-RPE cultures compared with Ctrl hiPSC-RPE cultures. Overall, our data showing the similar pathological phenotype (drusen-like deposits) and molecular change (COL4 accumulation in the ECM, complement gene alteration) in the SFD, DHRD, and ADRD hiPSC-RPE models support a shared mechanistic defect at the local level of RPE cells in the eye in multiple maculopathies.
Methods
Ethics.
Experimental work performed was approved by the Institutional Regulatory Board of the University of Rochester (RSRB00056538) and the Human Research Ethics Committees of the Royal Victorian Eye and Ear Hospital (11/1031H) in accordance with the requirements of the National Institutes of Health and the National Health and Medical Research Council of Australia and conformed to the Declarations of Helsinki.
Patient Samples.
Skin biopsies of two genotype- and phenotype-confirmed patients each with SFD (S204C in TIMP3), DHRD (R345W in EFEMP1), and ADRD (no mutation in EFEMP1) were obtained following informed consent (SI Appendix, Fig. S10). In addition, skin biopsy/fibroblast samples from four control individuals (one unaffected family member of an SFD and an ADRD patient and two unaffected individuals with no history of macular degeneration) were obtained following informed consent (SI Appendix, Fig. S10). All patient identification was removed, and de-identified patient samples were used for subsequent experimentation/analysis. The pedigree, patient age, age at diagnosis, visual acuity at the time of diagnosis, and representative fundus and/or autofluorescence images from SFD, DHRD, and ADRD families are presented in SI Appendix, Figs. S11–S13, respectively.
Fibroblast Culture.
Human fibroblasts were cultured in DMEM with high glucose supplemented with 10% FBS, 2 mM glutamine, and 100 U/mL penicillin-streptomycin (Life Technologies). Before reprogramming was performed, the presence or absence of disease-specific gene mutations (S204C in TIMP3 in SFD; R345W in EFEMP1 in DHRD) in patient and Ctrl fibroblasts, respectively, was confirmed by PCR amplification of relevant gene fragments and consequent DNA sequencing. The primer pairs used for mutation confirmation included TIMP3_S204C_F1 (5′-CCTGCTACTACCTGCCTTGC-3′), TIMP3_S204C_R1 (5′-AGTGTCCAAGGGAAGCTCAG-3′), and EFEMP1_R345W_F1 (5′-TGTCTCAGCTCTGTCTGTCC-3′), EFEMP1_R345_R1 (5′-CGGCTGCAGAGACAAACAAA-3′).
Generation of hiPSCs.
Fibroblasts were reprogrammed to hiPSCs with nonintegrating episomal vectors containing six reprogramming factors (OCT4, SOX2, KLF4, l-MYC, LIN28, and shRNA for p53) (60). More detailed information is presented in SI Appendix, SI Methods. Of note, multiple hiPSC clones for each patient and Ctrl fibroblast sample were isolated, expanded, and characterized as described below (25, 61).
Characterization of hiPSCs.
All Ctrl and patient hiPSC lines used in this study (SI Appendix, Fig. S10A) were analyzed for the expression of the pluripotency markers OCT4 and NANOG by immunocytochemistry (SI Appendix, Fig. S10 B–L). Sequencing analyses were used to confirm the absence or presence of the expected point mutations in TIMP3 (S204C) and EFEMP1 (R345W) in Ctrl vs. SFD and DHRD hiPSC lines (SI Appendix, Fig. S10 M–Q). Furthermore, because a subset of ADRD patients has previously been shown to harbor a mutation in the EFEMP1 gene (30), we confirmed that the ADRD patient fibroblast and subsequently ADRD hiPSC samples did not contain a mutation in the EFEMP1 gene (SI Appendix, Fig. S10R).
CRISPR Correction of DHRD Patient hiPSCs.
Guide RNAs against the target region were designed over the EFEMP1 Arg345Trp (rs121434491; NM_001039348.2:c.1033C > T) mutation site (SI Appendix, Fig. S14A) using the CRISPR design tool (crispr.mit.edu/). Guide RNA oligos (Integrated DNA Technologies) were cloned into the pSpCas9(BB)purov2.0 vector (Addgene plasmid no. 62988) as described by Ran et al. (62). Two million cells were nucleofected with 2.5 µg pSpCas9(BB)purov2.0 DHRD single-guide RNA, and 2.5 µL of 10 µM single-stranded oligo DNA correction templates (Integrated DNA Technologies) were cotransfected using the Human Stem Cell Nucleofector Kit 2 (Lonza), program A-23. Nucleofected cells were plated onto irradiated mouse embryo fibroblasts (MEFs) in one well of a six-well plate in KSR medium supplemented with FGF2 (10 ng/mL) and Y27632 (10 µM) (Selleck Chemicals). L755507 (5 µM) (Tocris Bioscience) was added to the medium for two consecutive days following transfection to enhance homology-dependent repair efficiency. Puromycin (150 ng/µL) (Life Technologies) was added to the culture medium on the second day post nucleofection and was removed after 5 d. Nucleofected hiPSCs were replated at low density onto MEFs on 10-cm dishes (4,000 cells per 10-cm plate), and single-cell colonies were picked after 2 wk of culture maintenance before being expanded for clone screening and characterization.
Screening and Characterization of CRISPR-Corrected hiPSCs.
Genomic DNA from the hiPSC clones was isolated using QuickExtract DNA extraction solution 1.0 (EpicentreBio) with the following thermal cycle program: 65 °C for 20 min, 68 °C for 20 min, 98 °C for 20 min. The Taqman SNP genotyping assay (Life Technologies) was used as the primary screening method to detect the presence of the corrected cytosine allele versus the mutant thymine allele. Secondary screening using restriction diagnostic digest of the PCR product was performed with enzyme HpyCH4III (New England Biolabs) and resolved on 1.8% Tris/boric acid/EDTA buffer agarose gel. Furthermore, mutation correction was verified by Sanger sequencing (SI Appendix, Fig. S14C). To confirm that the corrected clones originated from DHRD patient hiPSCs, genomic DNA samples were sent for 12-marker human microsatellite analysis, and chromosomal aberrations were excluded using virtual karyotyping (Australian Genome Research Facility) (SI Appendix, Fig. S14 D–F). Verification of chromosomal integrity in the CRISPR-corrected DHRD hiPSC line was further performed by karyotyping analysis (SI Appendix, Fig. S14G).
hiPSC Culture and Differentiation to RPE.
hiPSCs were maintained on either irradiated MEF feeders in hiPSC basal medium or on Matrigel in mTeSR medium (Stem Cell Technologies). Differentiation of hiPSCs toward the retinal fate and consequently RPE was conducted as previously described (22, 25). More detailed information is presented in SI Appendix, SI Methods. RPE cells, characterized by their distinct cobblestone morphology, appeared in the adherent cultures, at approximately day 40 (63, 64) and were allowed to mature for a total of 60–90 d before subsequent passaging (25). Thereafter, pure patches of RPE monolayer on these adherent cultures (passage 0, P0) were isolated by manual dissection and passaged onto nonpermeable plastic support and Transwell inserts (6.5-mm diameter, 0.4-µm pore size) (Costar; Corning) in accordance with our previously described protocol (22). Of note, passaging of RPE cells was limited to P3 to avoid loss of RPE characteristics (22). Furthermore, unless stated otherwise, age-matched (≥90 d in culture) Ctrl and patient (SFD, DHRD, ADRD) hiPSC-RPE monolayers were used in all experiments (25).
Electron Microscopy Analysis.
Mature monolayers of hiPSC-RPE grown on transwells were fixed in 2.5% gluteraldehyde and 4% paraformaldehyde in 0.1 M sodium cacodylate and were supplied to the University of Rochester electron microscopy core for further processing and imaging. Briefly, fixed hiPSC-RPE samples were embedded in epoxy resin, and 60-nm-thick sections were cut at 10-μm depth advancements into the sample. Subsequently, hiPSC-RPE sections were imaged using a transmission electron microscope (7650 Analytical transmission electron microscope; Hitachi). For basal deposit quantification, sequential images at 12,000× magnification were taken across ∼250 μm of a continuous hiPSC-RPE section at three different depths. For quantification of basal deposit, number, and diameter, images were further analyzed using Image J software (NIH).
Extracellular Matrix Isolation.
Mature monolayers of hiPSC-RPE overlying the ECM were removed nonenzymatically from Transwells or the nonpermeable plastic support as previously described (65, 66). A more in-depth description is provided in SI Appendix, SI Methods.
Transepithelial Resistance Measurement.
The transepithelial resistance of hiPSC-RPE was recorded as previously described (22). A more detailed description is provided in SI Appendix, SI Methods.
RPE Culture Treatments with Serum.
Human sera used in this study were either obtained commercially (Complement Technologies) or isolated at the University of Rochester from a subject in the sixth decade of life with no clinical history of macular degeneration in accordance with an approved institutional regulatory board or IRB protocol. For acute (24-h) and chronic (2-wk) serum treatments, serum was diluted to 10% and/or 25% in the RPE cell culture medium, in accordance with previously published studies (15), and was applied apically to the hiPSC-RPE culture in either Transwell or nonpermeable plastic supports. The medium was changed, and fresh serum supplementation was provided to hiPSC-RPE on a daily basis (every 24 h) until the end of the experiment and ensuing sample processing/analysis.
Western Blotting.
ECM protein was extracted as described above, and total cellular protein was isolated in RIPA buffer (Thermo Fisher Scientific) in accordance with the manufacturer’s protocol. Total cellular protein levels were quantified using the DC protein assay (Bio-Rad), and equal amounts of Ctrl and patient (SFD, DHRD, ADRD) protein samples were used for Western blotting. ECM samples were normalized to an approximately equal number of RPE cells, and both cellular protein and ECM samples were resolved on 4–20% gradient gels by SDS/PAGE, transferred to PVDF membrane, and processed according to our previously described protocol (22, 25). More detailed information on Western blot analysis, including antibody description/concentration, is provided in SI Appendix, SI Methods.
Immunocytochemical Analysis.
hiPSC-RPE for immunocytochemical analysis were prepared and processed as described in SI Appendix. Immunocytochemistry was performed in accordance with our previously published protocol (22, 25). More detailed information on immunocytochemical analysis, including antibody description/concentration, is also provided in SI Appendix, SI Methods.
Quantitative Real-Time RT-PCR Analysis.
RNA was isolated, processed, and analyzed in accordance with a previously published protocol (22, 25). More detailed information is presented in SI Appendix, SI Methods.
Experimental Set-Up and Data Analyses.
All hiPSC-RPE experiments were performed on age-matched (days in culture) Ctrl and patient (SFD, DHRD, ADRD) hiPSC-RPE cells grown on the same substrate (Transwell or nonpermeable plastic support). Of note, a subset of all experiments was performed on a polarized monolayer of hiPSC-RPE grown on Transwells. Furthermore, all experiments, with the exception of electron microscopy analyses, used three to five distinct Ctrl lines and two distinct patient lines per disease (SFD, DHRD, and ADRD). Electron microscopy analyses data constitute basal deposit quantification at three different depths of at least two distinct hiPSC-RPE samples differentiated from two different Ctrl and patient hiPSC-RPE lines. In each experiment, data from all the Ctrl lines/clones and patient lines/clones for each individual disease (SFD, DHRD, and ADRD) were grouped together and used in Ctrl vs. SFD, DHRD, and ADRD analyses. For quantitative analyses, data are expressed as mean + SEM throughout the article and are compared using the two-tailed unpaired Students t test and/or Mann–Whitney u test. Significance was assigned as a P value ≤0.05. Of note, the color palette of all of the figures, including individual confocal images, has been altered to accommodate colorblind readers. Furthermore, confocal images showing drusen localization and composition have been selected to illustrate the entire range of the immunostaining pattern observed in our analyses of Ctrl vs. patient-derived (SFD, DHRD, ADRD) hiPSC-RPE cultures.
Supplementary Material
Acknowledgments
We thank Dr. Helena Hai Liang [Centre for Eye Research Australia (CERA)] for fibroblast culturing; Dr. Robert Buttery, Melinda Cain (CERA), and Lisa S. Kearns (CERA) for assistance with clinical phenotyping; Stacey Jackson (CERA) for help with CRISPR screening; Stefanie Volland (University of California, Los Angeles) for her assistance and advice with electron microscopy; Gavin Jenkins (University of Rochester) for helping with qPCR analyses; and Dr. Bela-Anand Apte (Cleveland Clinic) for help with writing and editing the manuscript. This work was supported by funding from BrightFocus Foundation (R.S.), David Bryant Trust (R.S.), Foundation Fighting Blindness (R.S.), Knights Templar Eye Foundation (R.S.), Research to Prevent Blindness (R.S. and Department of Ophthalmology, University of Rochester), Retina Research Foundation (R.S.), University of Rochester University Research Award (R.S.), Retina Research Foundation E.A. Humble Directorship of the McPherson Eye Research Institute (D.M.G.), Sandra Lemke Trout Chair in Eye Research (D.M.G.), NIH Grant EY013048 (to D.S.W.), an Australian Research Council Future Fellowship FT140100047 (to A.P.), National Health and Medical Research Council (NHMRC) Project Grant 1059369 (to R.H.G. and A.P.), NHMRC Practitioner Fellowship 1103329 (to A.W.H.), NHMRC Elizabeth Blackburn Fellowship 1103013 (to R.H.G.), the Ophthalmic Research Institute of Australia (A.P. and A.W.H.), Retina Australia (S.S.H., A.P., and A.W.H.), and Clinical and Translational Science Institute Grant UL1TR000042 (University of Rochester).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.H.T. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710430114/-/DCSupplemental.
References
- 1.Capon MR, et al. Sorsby’s fundus dystrophy. A light and electron microscopic study. Ophthalmology. 1989;96:1769–1777. doi: 10.1016/s0161-6420(89)32664-9. [DOI] [PubMed] [Google Scholar]
- 2.Marmorstein LY, et al. Aberrant accumulation of EFEMP1 underlies drusen formation in Malattia Leventinese and age-related macular degeneration. Proc Natl Acad Sci USA. 2002;99:13067–13072. doi: 10.1073/pnas.202491599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green WR. Histopathology of age-related macular degeneration. Mol Vis. 1999;5:27. [PubMed] [Google Scholar]
- 4.Young RW. Pathophysiology of age-related macular degeneration. Surv Ophthalmol. 1987;31:291–306. doi: 10.1016/0039-6257(87)90115-9. [DOI] [PubMed] [Google Scholar]
- 5.Fariss RN, Apte SS, Luthert PJ, Bird AC, Milam AH. Accumulation of tissue inhibitor of metalloproteinases-3 in human eyes with Sorsby's fundus dystrophy or retinitis pigmentosa. Br J Ophthalmol. 1998;82:1329–1334. doi: 10.1136/bjo.82.11.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis MD, et al. Age-Related Eye Disease Study Group The Age-Related Eye Disease Study severity scale for age-related macular degeneration: AREDS report no. 17. Arch Ophthalmol. 2005;123:1484–1498. doi: 10.1001/archopht.123.11.1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Curcio CA, Medeiros NE, Millican CL. Photoreceptor loss in age-related macular degeneration. Invest Ophthalmol Vis Sci. 1996;37:1236–1249. [PubMed] [Google Scholar]
- 8.Dorey CK, Wu G, Ebenstein D, Garsd A, Weiter JJ. Cell loss in the aging retina. Relationship to lipofuscin accumulation and macular degeneration. Invest Ophthalmol Vis Sci. 1989;30:1691–1699. [PubMed] [Google Scholar]
- 9.Bhutto I, Lutty G. Understanding age-related macular degeneration (AMD): Relationships between the photoreceptor/retinal pigment epithelium/Bruch’s membrane/choriocapillaris complex. Mol Aspects Med. 2012;33:295–317. doi: 10.1016/j.mam.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan JC, et al. Genetic Factors in AMD Study Smoking and age related macular degeneration: The number of pack years of cigarette smoking is a major determinant of risk for both geographic atrophy and choroidal neovascularisation. Br J Ophthalmol. 2006;90:75–80. doi: 10.1136/bjo.2005.073643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hussain AA, Lee Y, Zhang JJ, Marshall J. Disturbed matrix metalloproteinase activity of Bruch’s membrane in age-related macular degeneration. Invest Ophthalmol Vis Sci. 2011;52:4459–4466. doi: 10.1167/iovs.10-6678. [DOI] [PubMed] [Google Scholar]
- 12.Crabb JW, et al. Drusen proteome analysis: An approach to the etiology of age-related macular degeneration. Proc Natl Acad Sci USA. 2002;99:14682–14687. doi: 10.1073/pnas.222551899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez-Godino R, Garland DL, Pierce EA. A local complement response by RPE causes early-stage macular degeneration. Hum Mol Genet. 2015;24:5555–5569. doi: 10.1093/hmg/ddv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pilgrim MG, et al. Subretinal pigment epithelial deposition of drusen components including hydroxyapatite in a primary cell culture model. Invest Ophthalmol Vis Sci. 2017;58:708–719. doi: 10.1167/iovs.16-21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson LV, et al. Cell culture model that mimics drusen formation and triggers complement activation associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2011;108:18277–18282. doi: 10.1073/pnas.1109703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radu RA, Hu J, Jiang Z, Bok D. Bisretinoid-mediated complement activation on retinal pigment epithelial cells is dependent on complement factor H haplotype. J Biol Chem. 2014;289:9113–9120. doi: 10.1074/jbc.M114.548669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockemeyer D, Jaenisch R. Induced pluripotent stem cells meet genome editing. Cell Stem Cell. 2016;18:573–586. doi: 10.1016/j.stem.2016.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young CS, et al. A single CRISPR-Cas9 deletion strategy that targets the majority of DMD patients restores dystrophin function in hiPSC-derived muscle cells. Cell Stem Cell. 2016;18:533–540. doi: 10.1016/j.stem.2016.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu X, et al. Reversal of phenotypic abnormalities by CRISPR/Cas9-mediated gene correction in Huntington disease patient-derived induced pluripotent stem cells. Stem Cell Reports. 2017;8:619–633. doi: 10.1016/j.stemcr.2017.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kokkinaki M, Sahibzada N, Golestaneh N. Human induced pluripotent stem-derived retinal pigment epithelium (RPE) cells exhibit ion transport, membrane potential, polarized vascular endothelial growth factor secretion, and gene expression pattern similar to native RPE. Stem Cells. 2011;29:825–835. doi: 10.1002/stem.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buchholz DE, et al. Derivation of functional retinal pigmented epithelium from induced pluripotent stem cells. Stem Cells. 2009;27:2427–2434. doi: 10.1002/stem.189. [DOI] [PubMed] [Google Scholar]
- 22.Singh R, et al. Functional analysis of serially expanded human iPS cell-derived RPE cultures. Invest Ophthalmol Vis Sci. 2013;54:6767–6778. doi: 10.1167/iovs.13-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, et al. Long-term safety and efficacy of human-induced pluripotent stem cell (iPS) grafts in a preclinical model of retinitis pigmentosa. Mol Med. 2012;18:1312–1319. doi: 10.2119/molmed.2012.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davidson KC, Guymer RH, Pera MF, Pébay A. Human pluripotent stem cell strategies for age-related macular degeneration. Optom Vis Sci. 2014;91:887–893. doi: 10.1097/OPX.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 25.Singh R, et al. iPS cell modeling of best disease: Insights into the pathophysiology of an inherited macular degeneration. Hum Mol Genet. 2013;22:593–607. doi: 10.1093/hmg/dds469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang J, et al. Validation of genome-wide association study (GWAS)-identified disease risk alleles with patient-specific stem cell lines. Hum Mol Genet. 2014;23:3445–3455. doi: 10.1093/hmg/ddu053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, et al. Gene therapy in patient-specific stem cell lines and a preclinical model of retinitis pigmentosa with membrane frizzled-related protein defects. Mol Ther. 2014;22:1688–1697. doi: 10.1038/mt.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsden CM, et al. Rescue of the MERTK phagocytic defect in a human iPSC disease model using translational read-through inducing drugs. Sci Rep. 2017;7:51. doi: 10.1038/s41598-017-00142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber BH, Vogt G, Pruett RC, Stöhr H, Felbor U. Mutations in the tissue inhibitor of metalloproteinases-3 (TIMP3) in patients with Sorsby’s fundus dystrophy. Nat Genet. 1994;8:352–356. doi: 10.1038/ng1294-352. [DOI] [PubMed] [Google Scholar]
- 30.Stone EM, et al. A single EFEMP1 mutation associated with both Malattia Leventinese and Doyne honeycomb retinal dystrophy. Nat Genet. 1999;22:199–202. doi: 10.1038/9722. [DOI] [PubMed] [Google Scholar]
- 31.Sonoda S, et al. Attainment of polarity promotes growth factor secretion by retinal pigment epithelial cells: Relevance to age-related macular degeneration. Aging (Albany NY) 2009;2:28–42. doi: 10.18632/aging.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz A, Brett P, Bok D. TIMP-3 is expressed in the human retinal pigment epithelium. Biochem Biophys Res Commun. 1996;226:467–474. doi: 10.1006/bbrc.1996.1379. [DOI] [PubMed] [Google Scholar]
- 33.Strunnikova NV, et al. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Hum Mol Genet. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pikuleva IA, Curcio CA. Cholesterol in the retina: The best is yet to come. Prog Retin Eye Res. 2014;41:64–89. doi: 10.1016/j.preteyeres.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson DH, et al. Local cellular sources of apolipoprotein E in the human retina and retinal pigmented epithelium: Implications for the process of drusen formation. Am J Ophthalmol. 2001;131:767–781. doi: 10.1016/s0002-9394(00)00961-2. [DOI] [PubMed] [Google Scholar]
- 36.Curcio CA, Johnson M, Huang JD, Rudolf M. Apolipoprotein B-containing lipoproteins in retinal aging and age-related macular degeneration. J Lipid Res. 2010;51:451–467. doi: 10.1194/jlr.R002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishida BY, et al. Regulated expression of apolipoprotein E by human retinal pigment epithelial cells. J Lipid Res. 2004;45:263–271. doi: 10.1194/jlr.M300306-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Fariss RN, Apte SS, Olsen BR, Iwata K, Milam AH. Tissue inhibitor of metalloproteinases-3 is a component of Bruch’s membrane of the eye. Am J Pathol. 1997;150:323–328. [PMC free article] [PubMed] [Google Scholar]
- 39.Marshall GE, Konstas AG, Reid GG, Edwards JG, Lee WR. Type IV collagen and laminin in Bruch's membrane and basal linear deposit in the human macula. Br J Ophthalmol. 1992;76:607–614. doi: 10.1136/bjo.76.10.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marmorstein LY, McLaughlin PJ, Peachey NS, Sasaki T, Marmorstein AD. Formation and progression of sub-retinal pigment epithelium deposits in Efemp1 mutation knock-in mice: A model for the early pathogenic course of macular degeneration. Hum Mol Genet. 2007;16:2423–2432. doi: 10.1093/hmg/ddm199. [DOI] [PubMed] [Google Scholar]
- 41.Klenotic PA, Munier FL, Marmorstein LY, Anand-Apte B. Tissue inhibitor of metalloproteinases-3 (TIMP-3) is a binding partner of epithelial growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1). Implications for macular degenerations. J Biol Chem. 2004;279:30469–30473. doi: 10.1074/jbc.M403026200. [DOI] [PubMed] [Google Scholar]
- 42.Rautavuoma K, et al. Premature aggregation of type IV collagen and early lethality in lysyl hydroxylase 3 null mice. Proc Natl Acad Sci USA. 2004;101:14120–14125. doi: 10.1073/pnas.0404966101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grosfeld A, et al. Interaction of hydroxylated collagen IV with the von hippel-lindau tumor suppressor. J Biol Chem. 2007;282:13264–13269. doi: 10.1074/jbc.M611648200. [DOI] [PubMed] [Google Scholar]
- 44.Sitterley G. Laminin. BioFiles. 2008;3:11. [Google Scholar]
- 45.Fu L, et al. The R345W mutation in EFEMP1 is pathogenic and causes AMD-like deposits in mice. Hum Mol Genet. 2007;16:2411–2422. doi: 10.1093/hmg/ddm198. [DOI] [PubMed] [Google Scholar]
- 46.Saini JS, et al. Nicotinamide ameliorates disease phenotypes in a human iPSC model of age-related macular degeneration. Cell Stem Cell. 2017;20:635–647.e7. doi: 10.1016/j.stem.2016.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chi ZL, Yoshida T, Lambris JD, Iwata T. Suppression of drusen formation by compstatin, a peptide inhibitor of complement C3 activation, on cynomolgus monkey with early-onset macular degeneration. Adv Exp Med Biol. 2010;703:127–135. doi: 10.1007/978-1-4419-5635-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gorham RD, Jr, et al. Novel compstatin family peptides inhibit complement activation by drusen-like deposits in human retinal pigmented epithelial cell cultures. Exp Eye Res. 2013;116:96–108. doi: 10.1016/j.exer.2013.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Anderson DH, et al. The pivotal role of the complement system in aging and age-related macular degeneration: Hypothesis re-visited. Prog Retin Eye Res. 2010;29:95–112. doi: 10.1016/j.preteyeres.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anderson DH, Mullins RF, Hageman GS, Johnson LV. A role for local inflammation in the formation of drusen in the aging eye. Am J Ophthalmol. 2002;134:411–431. doi: 10.1016/s0002-9394(02)01624-0. [DOI] [PubMed] [Google Scholar]
- 51.Mullins RF, Russell SR, Anderson DH, Hageman GS. Drusen associated with aging and age-related macular degeneration contain proteins common to extracellular deposits associated with atherosclerosis, elastosis, amyloidosis, and dense deposit disease. FASEB J. 2000;14:835–846. [PubMed] [Google Scholar]
- 52.Garland DL, et al. Mouse genetics and proteomic analyses demonstrate a critical role for complement in a model of DHRD/ML, an inherited macular degeneration. Hum Mol Genet. 2014;23:52–68. doi: 10.1093/hmg/ddt395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans K, et al. Assessment of the phenotypic range seen in Doyne honeycomb retinal dystrophy. Arch Ophthalmol. 1997;115:904–910. doi: 10.1001/archopht.1997.01100160074012. [DOI] [PubMed] [Google Scholar]
- 54.Sivaprasad S, Webster AR, Egan CA, Bird AC, Tufail A. Clinical course and treatment outcomes of Sorsby fundus dystrophy. Am J Ophthalmol. 2008;146:228–234. doi: 10.1016/j.ajo.2008.03.024. [DOI] [PubMed] [Google Scholar]
- 55.Nita M, Strzałka-Mrozik B, Grzybowski A, Mazurek U, Romaniuk W. Age-related macular degeneration and changes in the extracellular matrix. Med Sci Monit. 2014;20:1003–1016. doi: 10.12659/MSM.889887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butler GS, Apte SS, Willenbrock F, Murphy G. Human tissue inhibitor of metalloproteinases 3 interacts with both the N- and C-terminal domains of gelatinases A and B. Regulation by polyanions. J Biol Chem. 1999;274:10846–10851. doi: 10.1074/jbc.274.16.10846. [DOI] [PubMed] [Google Scholar]
- 57.Hu B, Thirtamara-Rajamani KK, Sim H, Viapiano MS. Fibulin-3 is uniquely upregulated in malignant gliomas and promotes tumor cell motility and invasion. Mol Cancer Res. 2009;7:1756–1770. doi: 10.1158/1541-7786.MCR-09-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gill SE, Pape MC, Leco KJ. Tissue inhibitor of metalloproteinases 3 regulates extracellular matrix–Cell signaling during bronchiole branching morphogenesis. Dev Biol. 2006;298:540–554. doi: 10.1016/j.ydbio.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 59.Apte SS, Olsen BR, Murphy G. The gene structure of tissue inhibitor of metalloproteinases (TIMP)-3 and its inhibitory activities define the distinct TIMP gene family. J Biol Chem. 1995;270:14313–14318. doi: 10.1074/jbc.270.24.14313. [DOI] [PubMed] [Google Scholar]
- 60.Crombie DE, et al. Friedreich's ataxia induced pluripotent stem cell-derived cardiomyocytes display electrophysiological abnormalities and calcium handling deficiency. Aging (Albany NY) 2017;5:1440–1452. doi: 10.18632/aging.101247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips MJ, et al. Blood-derived human iPS cells generate optic vesicle-like structures with the capacity to form retinal laminae and develop synapses. Invest Ophthalmol Vis Sci. 2012;53:2007–2019. doi: 10.1167/iovs.11-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meyer JS, et al. Optic vesicle-like structures derived from human pluripotent stem cells facilitate a customized approach to retinal disease treatment. Stem Cells. 2011;29:1206–1218. doi: 10.1002/stem.674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer JS, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci USA. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weber BH, et al. A mouse model for Sorsby fundus dystrophy. Invest Ophthalmol Vis Sci. 2002;43:2732–2740. [PubMed] [Google Scholar]
- 66.Qi JH, et al. S156C mutation in tissue inhibitor of metalloproteinases-3 induces increased angiogenesis. J Biol Chem. 2009;284:19927–19936. doi: 10.1074/jbc.M109.013763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.