Significance
Human cytomegalovirus (HCMV) causes birth defects and serious disease in immunocompromised patients. We do not fully understand the cellular processes that HCMV manipulates during infection. A holistic understanding of the cellular response to HCMV will help clarify mechanisms that underlie its replication and spread. This work uses systems virology to globally map the host response to HCMV infection. Our method identifies unappreciated pathways modulated by HCMV, including mesenchymal-to-epithelial transition (MET), an important developmental pathway involved in epithelial tissue formation, wound healing, and cancer metastasis. Our findings demonstrate that HCMV induces MET and raise the possibility that the transition influences not only viral pathogenesis but also the behavior of normal and diseased cells within an infected human host.
Keywords: cytomegalovirus, RNA-seq, GSEA, EMT, MET
Abstract
Human cytomegalovirus (HCMV) is the prototypical human β-herpes virus. Here we perform a systems analysis of the HCMV host-cell transcriptome, using gene set enrichment analysis (GSEA) as an engine to globally map the host–pathogen interaction across two cell types. Our analysis identified several previously unknown signatures of infection, such as induction of potassium channels and amino acid transporters, derepression of genes marked with histone H3 lysine 27 trimethylation (H3K27me3), and inhibition of genes related to epithelial-to-mesenchymal transition (EMT). The repression of EMT genes was dependent on early viral gene expression and correlated with induction E-cadherin (CDH1) and mesenchymal-to-epithelial transition (MET) genes. Infection of transformed breast carcinoma and glioma stem cells similarly inhibited EMT and induced MET, arguing that HCMV induces an epithelium-like cellular environment during infection.
Human cytomegalovirus (HCMV) is a β-herpes virus that latently infects a large proportion of the human population (1). The HCMV DNA genome encodes ∼200 genes (2), and may express >700 RNAs and proteins (3). Despite its large genetic endowment (for a virus), HCMV relies on numerous cellular functions for efficient replication and spread. A comprehensive understanding of the fundamental cellular networks modulated by HCMV will advance our understanding of HCMV infection, persistence, and pathogenesis.
Theoretically, gene-expression profiling experiments can reveal the global structure of cellular changes induced by HCMV infection. Unlike many other viruses, HCMV does not cause host shutoff, a mechanism whereby viral gene products impede cellular translation. Rather, changes to the cellular transcriptome transfer to the proteome in HCMV-infected cells, as the majority of cellular transcripts are translated with equal efficiency in uninfected and infected cells (4). Although posttranscriptional mechanisms likely disconnect a proportion of gene-expression signatures from their phenotypes, analysis of the infected-cell transcriptome should provide a large portion of the information necessary to globally model HCMV infection.
Cellular functions required for HCMV replication continue to be discovered, suggesting that our understanding of the pathways manipulated by infection has not been saturated. For example, recently HCMV has been shown to require fatty acid synthesis (5) and histone methylation (6) to replicate efficiently.
We aimed to derive biological insights into HCMV infection by creating global maps of gene expression structure. Our method used gene set enrichment analysis (GSEA) and gene set overlap networks to reduce dimensionality of host-cell transcriptome data generated by RNA-sequencing (RNA-seq). This analysis identified several thousand gene sets concordantly altered by HCMV in two different cell types, at three times following infection. These gene sets self-organized into <20 functionally related communities at each time point; defining the core cellular functions modulated during infection. We identified multiple, previously unappreciated, gene expression signatures induced during infection. Most notably, we discovered that HCMV inhibits epithelial-to-mesenchymal transition (EMT) gene expression. EMT inhibition was accompanied by mesenchymal-to-epithelial transition (MET) gene induction in primary cells. Infection of mesenchymal cancer cells with HCMV similarly inhibited EMT gene expression and induced MET. These data suggest that HCMV induces an epithelial cellular state as part of its replication program. Further, since HCMV has been found in multiple tumor samples (7), our data raise the possibility that HCMV might alter the EMT/MET status of tumor cells, and thereby influence the progression of human cancers.
Results
A Systems Approach to Identify Pathways Modulated by HCMV Infection.
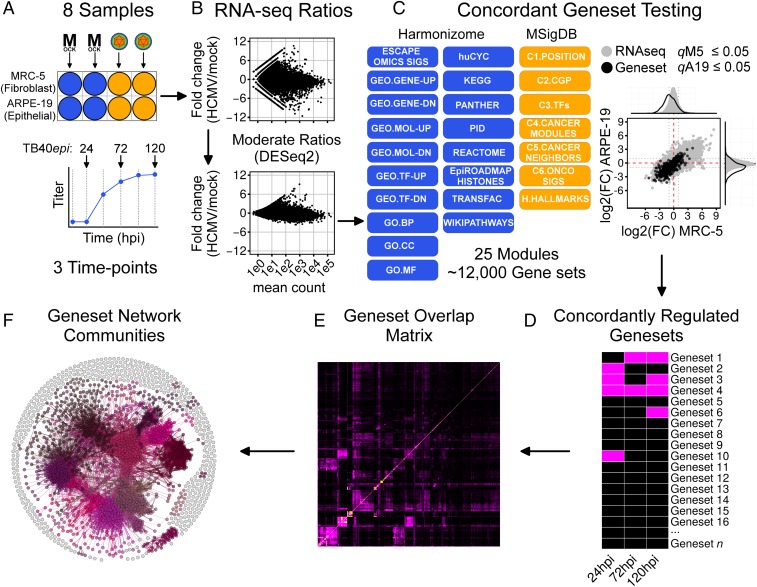
To filter cell-type-specific responses from general responses to infection, we collected gene-expression data from MRC-5 fibroblasts and ARPE-19 retinal epithelial cells; both commonly used HCMV infection models. Clinical isolates of HCMV readily accumulate mutations during growth in fibroblasts that limit infection of epithelial cells (8). Therefore, to infect both cell types with the same virus stock, we propagated the clinical isolate TB40/E (9) in ARPE-19 cells, which preserved epithelial tropism (SI Appendix, Fig. S1). We refer to stocks grown in fibroblasts as “TB40fibro” and those grown in epithelial cells as “TB40epi.” We then performed RNA-seq analysis of infected MRC-5 and ARPE-19 cells and devised a systems workflow to analyze the data (Fig. 1). Both cell types were infected with TB40epi in biological duplicate, and RNA samples were extracted at 24, 72, and 120 h postinfection (hpi) (Fig. 1A). To place these times into context, 24 hpi marks the “early” phase of infection, before viral DNA replication has begun; 72 and 120 hpi mark the “late” phase of infection with active viral DNA replication and the production of progeny. Mock-infected cell samples were collected at each time, as mock RNAs can fluctuate over the extended HCMV infection time course. Fold-change ratios (HCMV/mock) and significance values were then determined at each time, for each cell type, using DESeq2 (10), which provides shrunken ratios by modeling both gene level and biological variance (Fig. 1B). To identify coregulated pathways and cellular functions, we used a simplified GSEA procedure allowing for multidimensional, or “concordant,” gene set testing (Fig. 1C). Our procedure was based on the observation (11) that simple distribution tests perform similarly to the original GSEA method (12). For each cell type, each gene set distribution was compared with the parent RNA-seq distribution using a Wilcoxon rank-sum test. Gene sets with adjusted P values (q-values) ≤0.05 in both cell types and showing the same directional component were selected as concordant gene sets (Fig. 1D). To reduce dimensionality, gene set overlap matrices were constructed (Fig. 1E) and converted into force-directed networks. Lastly, gene set networks were partitioned into functionally related communities (Fig. 1F) and annotated as described in SI Appendix, SI Materials and Methods. We hypothesized that these networks might reveal unknown processes modulated by HCMV.
Fig. 1.
Concordant gene set and network analysis workflow. (A) Experimental design. MRC-5 or ARPE-19 cells (two independent cultures) were mock infected or infected with TB40epi at a multiplicity of 3 IU/cell. RNA samples were prepared at 24, 72, and 120 hpi. (B) RNA-seq ratio scatterplots demonstrating moderation of transcript fold-change ratios using DESeq2. (C) Gene set testing using GSEA. Gene sets scored as significant in both cell types were considered concordantly regulated. (D) Concordantly regulated gene sets can be analyzed temporally or at individual time points. For the time point analysis conducted here, overlap matrices (E) were used to construct gene set overlap networks, which were analyzed for functionally related communities (F). The communities represent the core processes modulated during HCMV infection.
To test our ability to rank enriched gene sets, we applied our procedure to the chemical and genetic perturbations (cgp; C2) module of the Molecular Signatures Database (MSigDB) (13). This module contains ∼3,400 gene sets of which 28 are from a microarray time course of HCMV-infected fibroblasts (14). We reasoned that the majority of these gene sets should be enriched at the top of the ranked list of regulated gene sets, and that microarray signatures obtained at times coinciding with those of the RNA-seq dataset (24, 72, and 120 hpi) should rank higher than signatures obtained at times very early after infection, which were analyzed only in the microarray study. Of the 2,312 gene sets that passed size filtering (SI Appendix, SI Materials and Methods), 54%, 43%, and 36% of the HCMV gene sets ranked within the top 5% of all gene sets at 24, 72, and 120 hpi, respectively (SI Appendix, Table S1). Considering only microarray time points after 12 hpi, 100%, 86%, and 71% were contained within the top 5% of all ranked gene sets (SI Appendix, Table S1), representing enrichment factors of ∼165×, 141×, and 94×, respectively. Microarray gene sets showed expected trends within the RNA-seq results (i.e., up-regulated microarray gene sets were up-regulated by RNA-seq assay; SI Appendix, Fig. S2). These results validated that our approach could effectively identify and rank concordantly enriched gene sets.
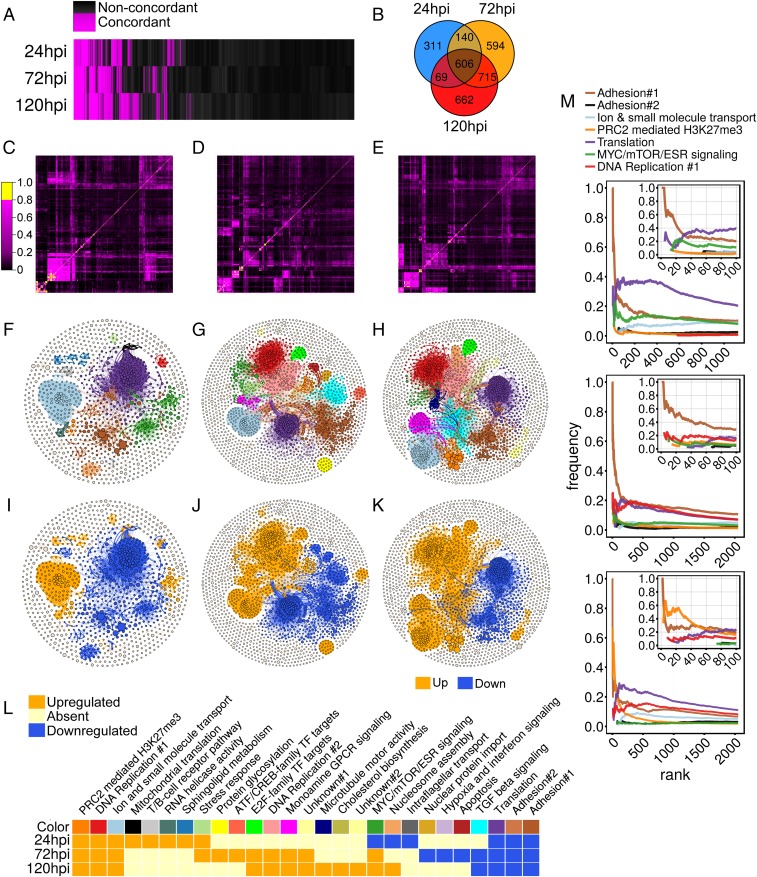
We then performed gene set tests using a larger gene set library containing 12,293 size-filtered gene sets from MSigDB (13) and Harmonizome (15) (Fig. 1C). We identified 1,126, 2,055, and 2,052 concordantly regulated gene sets at 24, 72, and 120 hpi, respectively (Fig. 2A and Dataset S1). More than 50% of the concordant gene sets at 24 hpi were also regulated at 72 and 120 hpi, suggesting that signaling trends initiated early are sustained throughout the infection cycle (Fig. 2B). Overlap adjacency matrices showed considerable substructure (Fig. 2 C–E), suggesting the presence of interconnected communities. Therefore, gene set networks were rendered, partitioned into communities, and annotated (Fig. 2 F–L). Considering communities containing more than 0.5% of total nodes per time point, the networks self-organized into 14, 18, and 16 communities at 24, 72, and 120 hpi, respectively (Fig. 2 F–H). Most communities showed polarized directionality, with most nodes either exclusively up-regulated or down-regulated (Fig. 2 I–K and SI Appendix, Tables S2–S4). Given this polarization, an overall community trend was assigned by scoring the most frequent gene set direction in each community.
Fig. 2.
Concordant gene set analysis of TB40epi infected MRC-5 and ARPE-19 cells at 24, 72, and 120 hpi. (A) Binary heatmap showing all significant gene set tests (q-values ≤ 0.05 in both cell types). (B) Venn diagram of concordantly regulated gene sets at each time point indicating overlap of detected gene sets. (C–E) Heatmaps of overlap matrices used to create gene set networks at 24 (Left), 72 (Middle), and 120 (Right) hpi. (F–H) Gene set networks and detected functional communities at 24, 72, and 120 h. (I–K) Gene set networks and communities colored by trend. (L) Gene set community heatmap with annotations, trends, and color key corresponding to F–H. (M) Frequency-rank plots of concordantly regulated communities at all time points showing enrichment of adhesion-related gene sets at high ranks. Insets show the top 100 gene sets at each time point. Colors are consistent with the key in L, with the exception that the adhesion#2 community is colored black to increase contrast.
Temporal Communities Modulated by HCMV.
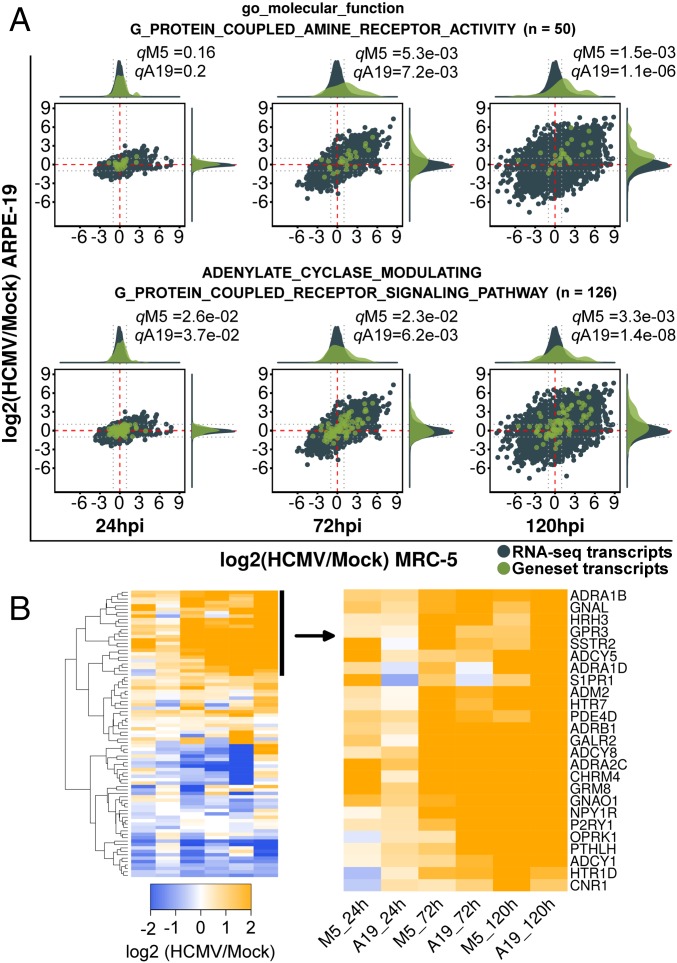
Approximately half of the communities at each time point showed “temporal” regulation, with concordant enrichment at one or two, but not all, times assayed (Fig. 2L). These temporal communities were small and represented cellular functions known to be modulated by HCMV, such as apoptosis, stress response, cholesterol biosynthesis, ATF/CREB or E2F-mediated transcription, sphingolipid metabolism, and nucleosome assembly (1). Three temporal communities, “RNA helicase activity,” “intraflagellar transport,” and “monoamine G-protein-coupled receptor (GPCR) signaling,” represented functions not yet identified as pathways modulated by HCMV. Of the three, the monoamine GPCR signaling community showed the most significant regulation at the RNA level. Gene sets in this community were concordantly up-regulated (Fig. 3A). Further examination of these gene sets using hierarchical clustering revealed up-regulation of GPCR signaling pathway components like ADCY-1, -5, -8, GNAL, and GNAO1, as well as a set of upstream initiating receptors, including seven biogenic amine activated GPCRs (ADRA1B, ADRA1D, ADRA2C, ADRB1, CHRM4, HTR1D, and HTR7) (Fig. 3B).
Fig. 3.
GSEA analysis of the monoamine GPCR temporal community. (A) RNA-seq ratio scatterplots of representative gene sets partitioned into the monoamine GPCR community. Distribution density plots used for significance testing are displayed Above and to the Right of each scatterplot. Dotted gray lines are twofold reference guides. RNA-seq transcripts and distributions are shown in blue, while those from each gene set are shown in green. (B) Heatmap of the union of gene sets shown in A showing up-regulated and down-regulated transcripts grouped by hierarchical clustering.
Core Communities Modulated by HCMV.
Seven of the ∼15 annotated communities at each time point were concordantly regulated at all three times (Fig. 2L). They tended to be the largest communities and represented 86%, 61%, and 62% of all community gene sets, at 24, 72, and 120 hpi, respectively, suggesting that the changes seen during infection are dominated by a few “core” functions/pathways. We analyzed each core community in detail. Three of the seven core communities represented pathways known to be regulated by HCMV infection: DNA replication and cell-cycle modulation, translation, and MYC/mTOR/ESR signaling (1, 16). Two core communities were related to cell adhesion and were enriched for gene sets related to EMT, a pathway not known to be modulated by HCMV. The remaining core communities, “PRC2-mediated H3K27Me3” and “ion and small molecule transport,” were additional previously unidentified pathways modulated during infection. Gene sets from these communities were enriched in the top 100 ranked gene sets (Fig. 2M). Below we describe these communities, referring to each by its network color as shown in Fig. 2.
Ion and Small Molecule Transport (Light Blue).
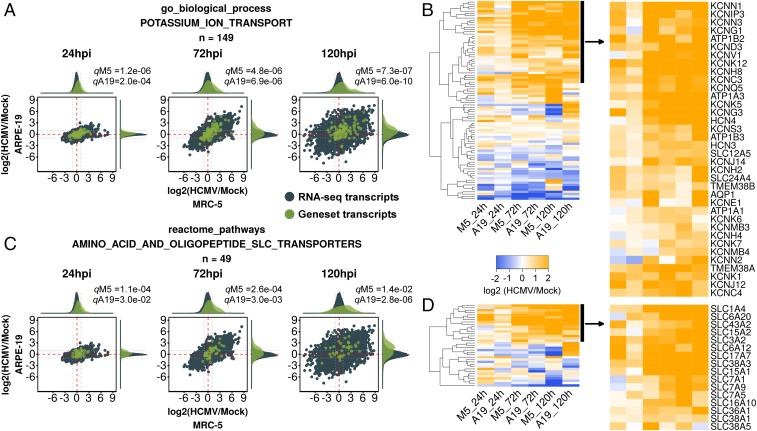
A large community related to ion and small molecule transport was up-regulated by infection at all times tested. The most significantly regulated gene sets in the community were related to Gene Ontology (GO) terms such as “transporter activity” and “ion transport.” Gene sets related to potassium ion channels and amino acid transporters were enriched in this community (Datasets S2–S4). Inspection of the potassium channel gene sets revealed 10 potassium transporter RNAs that were up-regulated at 72 and 120 hpi, including three subunits of the sodium/potassium ATPase (ATP1A1, ATP1A3, and ATP1B3), three outward rectifying voltage-gated potassium channels (KCNC4, KCNH8, and KCNQ5), several two-pore-domain leak channels (KCNK1 and KCNK12), one calcium-activated potassium channel (KCNN1), and one phosphatidylinositol 4,5-bisphosphate (PIP2) activated, inwardly rectifying, potassium channel (KCNJ12) (Fig. 4 A and B). Eleven additional potassium channels were up-regulated at 120 hpi (Fig. 4B), highlighting dramatic, coordinated, potassium channel expression late during infection. Inspection of the amino acid transport gene sets revealed up-regulation of two surface oligopeptide transporters (SLC15A1 and SLC15A2) and eight solute-carrier family amino acid transporters (SLC1A4, SLC6A20, SLC7A1, SLC3A2/SLC7A5, SLC17A7, SLC36A1, SLC38A3, and SLC43A2) during infection (Fig. 4 C and D). As with potassium channels, up-regulation of RNAs encoding amino acid transporters could be detected at 24 hpi and increased dramatically at 72 and 120 hpi, suggesting that induction of ion and small molecule transporters is a feature of the early and late phases of infection. Gene sets for iron/transferrin, sodium, carbohydrate, calcium, and zinc transport were also partitioned into the ion and small molecule transport community (Datasets S2–S4).
Fig. 4.
GSEA analysis of the ion and small molecule transport community. (A and B) RNA-seq ratio scatterplots and up-regulated transcripts (as in Fig. 3) of GO biological process gene set “potassium ion transport.” This gene set is a representative potassium transport gene set from the ion and small molecule transport community. (C and D) RNA-seq ratio scatterplots and up-regulated transcripts (as in Fig. 3) of reactome gene set “amino acid and oligopeptide SLC transporters.” This gene set is a representative amino acid transport gene set from the ion and small molecule transport community.
PRC2-Mediated H3K27Me3 (Orange).
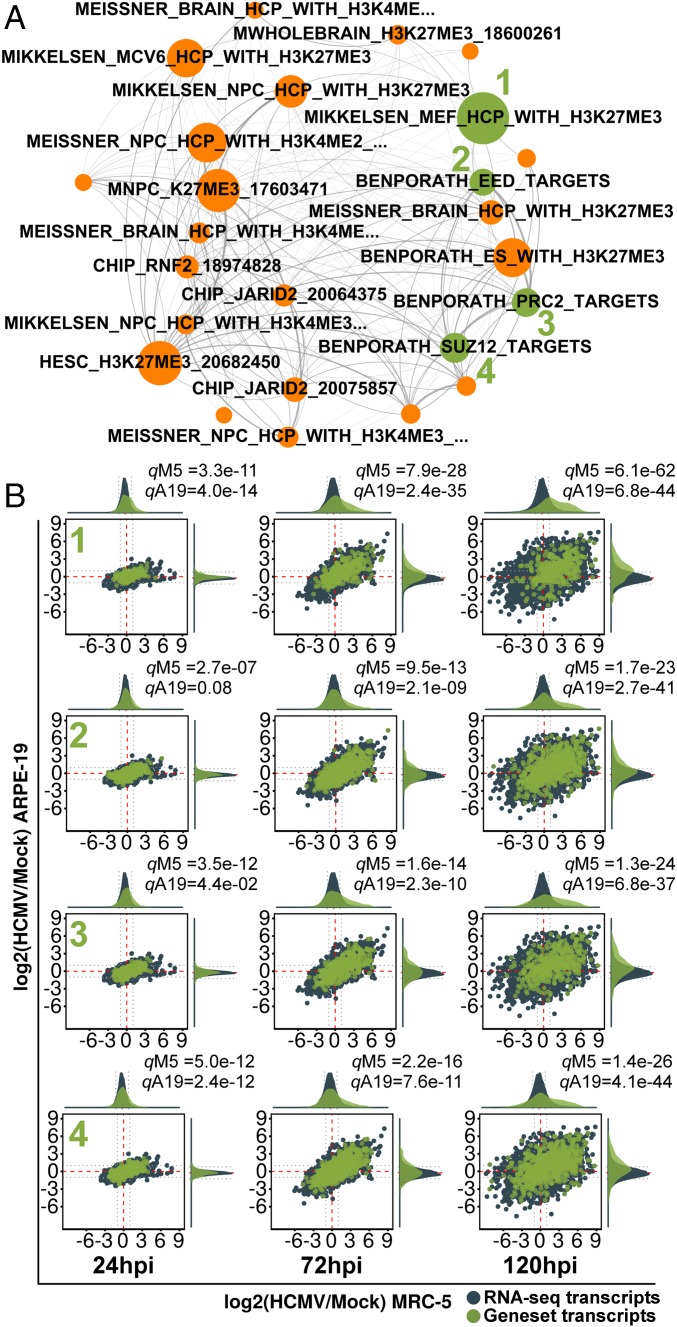
A small cluster of gene sets related to polycomb repressive complex 2 (PRC2) composed the orange core community. The 38%, 36%, and 46% of the gene sets in this community were present in the top 10% of all concordant gene sets, at 24, 72, and 120 hpi, respectively; indicating an approximately fourfold enrichment in the highest ranked gene sets. This community expanded as infection proceeded, constituting ∼50% of the top 20 ranked gene sets at 120 hpi (Fig. 2M). At 24 hpi, the orange community exclusively partitioned gene sets from chromatin immunoprecipitation sequencing (ChIP-seq) experiments defining genes marked with H3K27me3 or H3K27me3 + H3K4me3 (Dataset S2). These gene sets persisted at 72 and 120 hpi, when additional gene sets composed of targets of the PRC2 enzyme complex, the individual subunits SUZ12 and EED1, and the PRC2-recruitment protein JARID2 were partitioned into the community (Fig. 5A and Datasets S3 and S4). PRC2 is responsible for methylating H3K27, a repressive histone mark (17). These gene sets were concordantly up-regulated (Fig. 5B), suggesting that HCMV infection globally derepresses PRC2 target genes.
Fig. 5.
Derepression of PRC2 target genes during HCMV infection. (A) Subnetwork displaying all concordantly regulated gene sets in the PRC2-mediated H3K27Me3 community at 72 hpi (colored orange as in Fig. 2). Node size is proportional to its significance or inverse product of MRC-5 and ARPE-19 q values. (B) RNA-seq ratio scatterplots of select PRC2-related gene sets (green nodes in A) showing derepression of PRC2 target genes.
Adhesion (Brown and Light Brown).
A large community of gene sets involved in cell adhesion and extracellular matrix composition (“Adhesion#1”; brown) was observed at all time points (Fig. 2L). A related, smaller, community containing a mixture of cell adhesion and vesicle transport gene sets (adhesion#2, light brown) was also observed at 24 and 72 hpi and considered an extension of adhesion#1. These gene sets were globally down-regulated (Fig. 2 I–L) and functionally related to production of matrix components such as collagen, fibronectin, and fibrillin, and cell–matrix attachment (i.e., “focal adhesion”). HCMV infection decreases focal adhesion (4, 18, 19). However, further analysis (described below) suggested that loss of cell–matrix adhesion was part of a larger cellular response centered around induction of epithelial characteristics.
HCMV Infection Inhibits EMT and Induces an MET Signature.
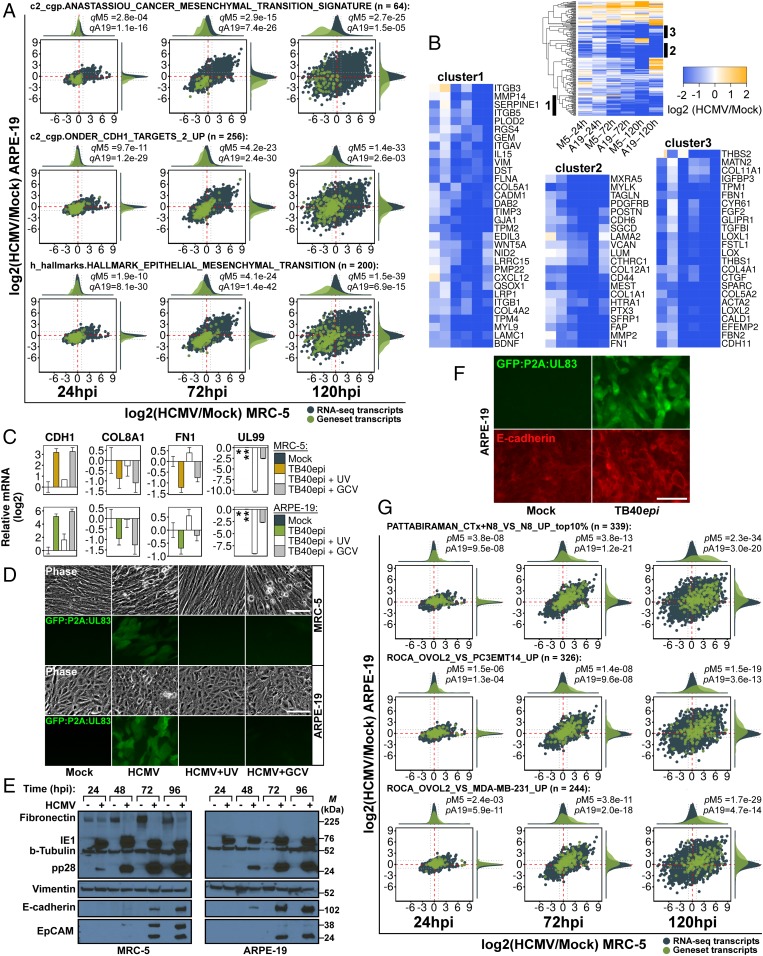
A disproportionate number of highly ranked gene sets were partitioned into the brown community, representing 45%, 50%, and 25% of the top 20 gene sets at 24, 72, and 120 hpi, respectively (Fig. 2M). This suggested that they played a dominant role during infection. To our surprise, the majority of highly ranked gene sets in this community were related to epithelial-to-mesenchymal transition (EMT), such as gene sets relating to breast cancer aggressiveness, matrix protein synthesis, and cadherin signaling (Datasets S2–S4). Several highly ranked gene sets implicated EMT as a pathway inhibited during infection. For example, the MSigDB gene sets “hallmark epithelial mesenchymal transition” and “Anastassiou cancer mesenchymal transition signature” were ranked numbers 12 and 51, out of 12,293 at 72 hpi. These gene sets were inhibited in a coordinated manner (Fig. 6A) and revealed the majority of EMT markers were down-regulated during infection (Fig. 6B). For example, the mesenchymal markers vimentin (VIM) and fibronectin (FN1), as well as several collagens (e.g., COL1A1, COL4A1, COL5A1, and COL12A1), fibrillins (FBN1 and FBN2), matrix metalloproteinases (MMP2 and MMP14), and thrombospondins (THBS1 and THBS2) were all concordantly down-regulated. Additionally, several gene sets in the adhesion community suggested activation of E-cadherin signaling, such as the down-regulation of genes from “Onder CDH1 targets 2 up” (Fig. 6A), a set of genes up-regulated upon knockdown of CDH1. Using quantitative real-time PCR (qRT-PCR) we confirmed EMT gene inhibition and found that CDH1 was induced during infection (SI Appendix, Fig. S3). We also confirmed EMT down-regulation by performing GSEA on a previously analyzed RNA-seq dataset from fibroblasts infected with HCMV (4) (SI Appendix, Fig. S4). The induction of CDH1, as well as the inhibition of several EMT marker genes (FN1 and COL8A1), was dependent on de novo immediate-early or early viral gene expression, as it was abrogated upon infection with UV-inactivated HCMV, but not treatment with the viral polymerase inhibitor, ganciclovir (GCV) (Fig. 6C), which blocks progression to the late phase of infection. Both treatments were shown to be efficacious by monitoring HCMV late gene expression using qRT-PCR (Fig. 6C, UL99) and monitoring GFP expression from a TB40epi strain marked at the UL83 locus (Fig. 6D). Steady-state E-cadherin protein expression was induced by HCMV in both MRC-5 and ARPE-19 cells starting at 48 hpi (Fig. 6E), precisely when its RNA levels increased (SI Appendix, Fig. S3). Vimentin and fibronectin showed little change at the protein level, with the exception of fibronectin in MRC-5 cells, which was repressed during infection. HCMV-induced E-cadherin showed plasma membrane localization (Fig. 6F), suggesting it might be incorporated into adherens junctions, a hallmark of the epithelial state.
Fig. 6.
Inhibition of EMT and induction of MET during HCMV infection. (A) RNA-seq ratio scatterplots of representative EMT-related gene sets from the adhesion#1 community. (B) Heatmap and down-regulated clusters of hallmark epithelial mesenchymal transition gene set. (C) Effect of UV and ganciclovir (GCV; 100 μM) treatment on CDH1, COL8A1, FN1, and UL99 at 72 hpi. Samples were calibrated to mock, except for UL99, which was calibrated to TB40epi (**), since the mock (*) sample was not assayed. Data are mean ± SD from three separate infections, each assayed in technical triplicate. (D) Phase-contrast and fluorescence images of cell populations from C, showing inhibition of late gene expression due to UV and GCV treatment. (E) Western analysis of EMT and MET markers during HCMV infection in MRC-5 and ARPE-19 cells. M, marker (kDa). (F) Immunostaining of E-cadherin (red) in mock and infected ARPE-19 cells. Infected cells are marked with GFP expressed from the virus-encoded GFP:P2A:UL83 reporter. (G) RNA-seq ratio scatterplots of MET gene sets during HCMV infection. (Scale bars, 100 μm.)
Mesenchymal gene expression is coupled to opposing changes in epithelial gene expression during development, wound repair, and carcinogenesis (20). Given the observed inhibition of EMT and induction of E-cadherin, we hypothesized that infection might cause an MET phenotype. RNA-seq ratios showed that levels of additional epithelial genes were induced late during infection, such as CLDN6, EPCAM (confirmed at the protein level; Fig. 6E), PKP1, CRB3, and the gap junction proteins GJB2 and GJB3. However, others, such as KRT8, KRT18, TJP1, PARD6A, and CTNNB1, were not (SI Appendix, Table S5). Therefore, some, but not all, canonical epithelial features were induced during infection. Therefore, we sought a broader metric for determining whether an MET was induced during infection. Three MET gene expression signatures (gene sets) were generated by extracting up-regulated genes from recent MET studies (SI Appendix, SI Materials and Methods), one examining cholera toxin (CTx)-induced MET in NAMEC8 (N8) mammary epithelial cells (21), and two comparing OVOL2-induced MET in PC3EMT14 prostate cancer cells (22) and MDA-MB-231 breast cancer cells (23). We then used GSEA to test whether these signatures were modulated during HCMV infection. All three gene sets were strongly (P << 0.01) up-regulated at 72 and 120 hpi (Fig. 6G), precisely when EMT was inhibited and E-cadherin was expressed. Thus, HCMV infection induces a gene expression profile similar to experimental METs observed in cancer cell models.
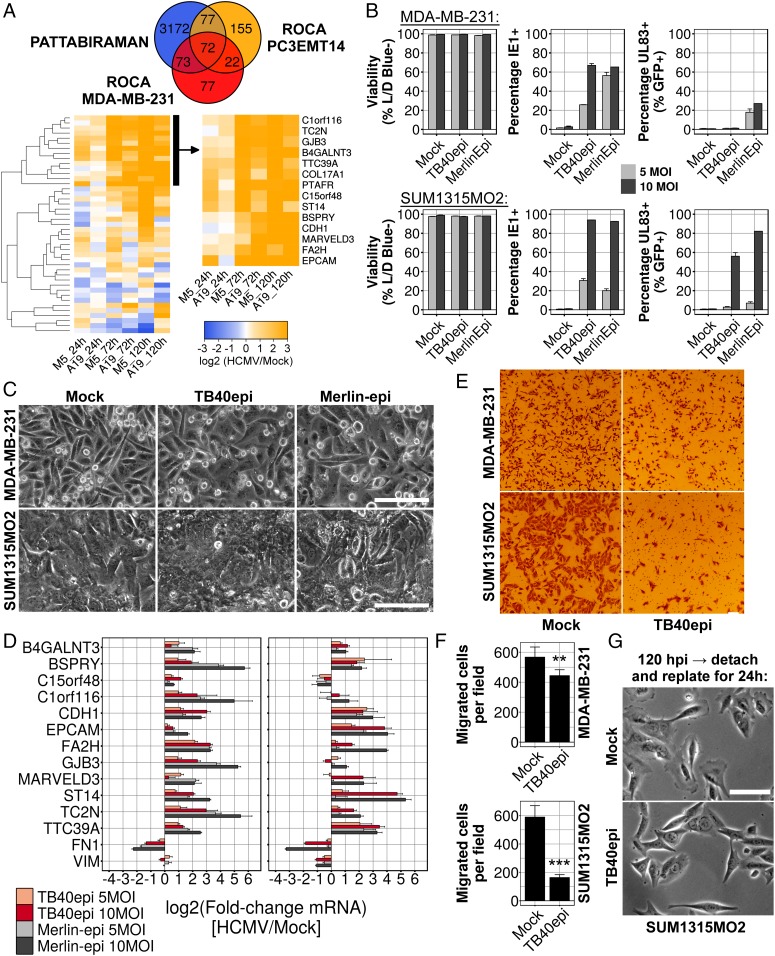
To facilitate monitoring of HCMV-induced MET, we generated a 14-gene, HCMV-specific, MET signature from our RNA-seq data by extracting genes that intersected the three cancer MET gene sets (Fig. 7A, Top), and clustering the resulting 72 genes based upon their expression during infection (Fig. 7A, Bottom). This signature was primarily composed of epithelial genes such as CDH1, EPCAM, and GJB3; the tight-junction component MARVELD3; the hemidesmosomal structural protein, COL17A1; and the epithelial serine protease ST14. We monitored 12 of the 14 genes in this signature by qRT-PCR late during infection and all 12 correlated perfectly with infection in both MRC-5 and ARPE-19 cells (SI Appendix, Fig. S5A).
Fig. 7.
Induction of MET in HCMV-infected mesenchymal breast cancer cell lines. (A) Venn diagram and heatmaps showing intersection of MET gene sets and extraction of an HCMV-specific gene expression signature by hierarchical clustering. (B) Flow-cytometry analysis of infected MDA-231 and SUM1315 cells at 120 hpi. Viability was assessed using a fixable live/dead (L/D) blue stain, infection efficiency by intracellular staining for viral IE1 protein and late gene expression using the virus-encoded GFP:P2A:UL83 reporter. (C) Phase-contrast images of infected breast cancer cells at 120 hpi. (D) qRT-PCR analysis of HCMV-specific MET signature genes in infected breast cancer cells at 120 hpi. (E) Representative images from Transwell migration assays using mock- and TB40epi-infected [10 multiplicity of infection (MOI)] breast cancer cells. (F) Quantification of Transwell migration assays shown in E. Data are mean ± SD of 15 fields taken at 40× magnification, from three separate infections. Significance was determined by two-tailed t test (**P = 1.63e-04; ***P = 1.37e-08). (G) Morphology of SUM1315 cells replated for an additional 16 h after an initial 120-h infection period. (Scale bars, 100 μm.)
HCMV Induction of MET in Mesenchymal Cancer Cells.
METs are most well characterized in cancer cells, such as metastatic breast cancer cells, which transition between mesenchymal and epithelial states (24). Therefore, we tested whether HCMV could induce MET in two mesenchymal breast cancer lines, MDA-MB-231 (MDA-231) and SUM1315MO2 (SUM1315). Both lines were infected at low confluency with either TB40epi or an epithelial cell-grown stock of the HCMV clinical isolate Merlin (25). At 120 hpi, cells were analyzed for viability and viral immediate-early (IE1) protein expression. Nearly all SUM1315 cells, and ∼65% of MDA-231 cells, were infected with HCMV, as judged by the number of cells expressing IE1 at 120 hpi (Fig. 7B). However, only a fraction of the infected cells expressed a late gene reporter and infection did not induce cell death at 120 hpi (Fig. 7B), suggestive of a stalled or significantly delayed replication cycle. Both HCMV strains induced a flattened, cobblestone morphology in MDA-231 cells, which typically appear elongated and spindle-like (Fig. 7C). Infected SUM1315 cells grew to confluency over the 120-h infection period and their morphology appeared similar to mock-infected cells (Fig. 7C). Nonetheless, nearly all genes from the HCMV-MET signature were induced in both cell types in a multiplicity-dependent manner, and the mesenchymal markers FN1 and VIM (SUM1315 only) were down-regulated (Fig. 7D). Since induction of MET in mesenchymal breast cancer cells inhibits migration (21), we next assayed their migratory capacity after infection. Cells were infected for 120 h and then equal numbers were seeded into Transwell dishes, or standard tissue culture plates, for another 16 h. Infection with HCMV decreased Transwell migration in MDA-231 cells modestly, but had a dramatic inhibitory effect on SUM1315 migration (Fig. 7 E and F). Examination of the infected SUM1315 cultures reseeded onto standard plates revealed an altered morphology. Mock-infected cells showed prominent lamellipodia with phase-dense membrane ruffles at the leading edge of the majority of cells. In contrast, infected SUM1315 cells displayed straight, triangular edges, with few, if any, lamellipodia (Fig. 7G), consistent with reduced migratory capacity. These data provide functional evidence that HCMV can induce a bona fide MET in mesenchymal breast cancer cells. We also observed MET gene expression upon infection of two glioma stem cell lines (SI Appendix, Fig. S5B). Our observations in fibroblasts, epithelial cells, breast cancer cells, and glioma stem cells provide strong evidence that MET induction is a general feature of HCMV infection.
Discussion
Gene set databases are inherently biased toward known pathways, and experimentally derived gene sets can be biased toward more “popular” pathways or cell types. Nonetheless, GSEA enabled us to identify at least four unidentified pathways modulated during productive HCMV infection: biogenic amine GPCR signaling, potassium and amino acid transport, PRC2-mediated histone methylation, and EMT/MET.
Biogenic Amine GPCR, Potassium Channel and Amino Acid Transporter Signatures.
The amine GPCRs we observed to be up-regulated during infection typically function in neurons or skeletal muscle, such as the acetylcholine receptor CHRM4; the serotonin receptors HTR1D and HTR7; or adrenergic receptors ADRA1B, ADRA2C, and ADRB1. Further experiments will be required to clarify the role of biogenic amine signaling during infection and whether their activities converge on MET, which can be modulated by cyclic-AMP production (21).
Most up-regulated potassium transport proteins, were outward rectifying channels, which repolarize neurons after an action potential. Patch clamp experiments have shown increases in outward rectifying potassium currents late during HCMV infection (26). The function of these currents, potentially caused by the up-regulated channels we identified, is not presently clear. However, some viruses require potassium channel function for virus release (27). Given the late kinetics of the HCMV-induced potassium channels, a role in virion release is feasible.
The increase in amino acid transporters suggests that HCMV requires exogenous amino acids to maintain protein synthesis. Metabolomic studies of HCMV infection did not observe changes in steady-state amino acid pools, with the exception of an increase in alanine. However, this work monitored only 5 of the 20 amino acids. We observed up-regulation of the transporters SLC1A4/ASCT-1, SLC38A3, and SLC3A2/4F2 during infection, starting between 24 and 72 hpi, a time aligned with peak metabolic changes induced by HCMV (∼48 hpi) (16). SLC1A4 has specificity for alanine, SLC38A3 for glutamine, and SLC3A2 modulates surface expression of LAT-1, which can exchange intracellular arginine for extracellular glutamine (28). These transporters could be responsible for the increased alanine levels and glutamine anaplerosis observed during infection (16).
EMT/MET and PRC2 Signatures.
The MET signature was one of the most dominant cellular responses we observed. MET succinctly explains the modulation of focal adhesion, extracellular organization, and adhesion junction complexes previously noted during HCMV infection (4, 18, 19, 29).
Why does HCMV induce a MET phenotype? EMT and MET are essential for development, wound healing, and cancer metastasis (20). Little is known about these transitions during viral replication, so the functional relevance of MET to HCMV pathogenesis is not presently clear. An important question to be resolved is whether the activation of MET is beneficial to the virus, part of the cellular antiviral response, or a collateral disruption induced by infection that is of no consequence to the virus. Conceivably, the MET environment causes expression of a constellation of cellular genes that modulate viral growth in a cell autonomous manner. Alternatively, MET induction might control the choice between active HCMV growth versus latency. In Epstein-Barr virus (EBV)-transformed B cells, expression of miR-200 RNAs, which directly induces MET in cancer models (30), causes a switch from latency to lytic replication (31). Similarly, during Kaposi’s sarcoma-associated herpesvirus (KSHV) infection of B cells, the latency-associated nuclear antigen (LANA) induces mesenchymal features by up-regulating SNAIL (32). Suppression of LANA breaks latency (33), inhibits SNAIL, and induces epithelial genes including CDH1 (32), features consistent with MET. Thus, there is a correlation between the mesenchymal state and latency and the epithelial state and productive replication in this family of viruses. It will be interesting to see if the EMT/MET switch is functionally coupled to the HCMV lytic/latency switch. It is also possible that the MET supports viral spread by enforcing an epithelial phenotype where cells establish zones of tight cell–cell contact. This could allow the virus to spread from cell to cell without being exposed to the external environment and possible neutralization by antiviral antibodies. It has been proposed that herpes simplex virus and measles virus use epithelial adherens junctions to pass directly from cell to cell (34).
How does HCMV induce an MET phenotype? The PRC2 signature may provide a clue to the mechanism. A group of PRC2 targets, including CDH1, was derepressed during infection. The EMT transcription factor SNAIL1 can recruit PRC2 to the CDH1 promoter and mediate its silencing, and inhibition of PRC2 derepresses CDH1 (35). During development, displacement of PRC2 from promoters is mediated by transcriptional activator binding and/or demethylase recruitment (17). Therefore, a putative mechanism driving MET during HCMV infection might involve the displacement of PRC2 or recruitment of a demethylase by cellular or viral transcription factors at the CDH1 promoter. The HCMV immediate-early transcription factors IE1 and IE2, which activate viral and cellular promoters (36), are prime candidates for this activity. IE1/IE2-mediated derepression at one or more master MET genes, perhaps CDH1, would provide a simple mechanism for MET induction during infection. In addition to its role in the HCMV-induced MET, many concordantly regulated genes in other novel infection signatures, appear to be putative PRC2 derepressed genes. For example, nine monoamine GPCR signaling components, all 10 potassium transporters mentioned above, and five amino acid transporters were found in PRC2-related gene sets. This suggests that epigenetic alteration is an apical event driving remodeling of the host cell during infection.
HCMV-Induced MET as a Potential Oncomodulatory Mechanism.
Our identification of an HCMV-induced MET has important implications regarding the possible role of HCMV in oncogenesis. Numerous studies have confirmed the presence of HCMV nucleic acids and in some cases proteins in multiple cancers, including glioblastoma (GBM) (7), breast (37–39), and ovarian (40) cancers. However, HCMV infection does not transform cells, leading many to suspect that HCMV does not contribute to oncogenesis. However, enforced expression of the viral GPCR US28 can transform cells (41), as can coexpression of IE1 and IE2 with adenovirus E1A (42). Furthermore, autologous dendritic cells pulsed with HCMV RNAs can recognize and kill GBM tumors in vivo (43), indicating that GBM cells are exposed to HCMV during their development. Although MET can inhibit the migratory phase of metastasis, it encourages metastatic colonization during secondary tumor formation (44). Primary cells infected with HCMV induce MET with early-late kinetics, but the virus eventually kills the cells. However, HCMV can induce a functional MET in at least some mesenchymal tumor cells without affecting their viability (Fig. 7). This scenario might encourage tumor colonization.
Conclusion.
Our results demonstrate that a virus can induce MET, possibly by relief of PRC2-mediated repression. Further delineation of the mechanism by which HCMV represses EMT and induces MET may reveal new insights into the normal and aberrant activation cellular MET, the role of cellularity transitions in the HCMV latency switch, and the potential impact of HCMV infection on tumor cell behavior. Additionally, this work provides a framework for further investigation into the roles of monoamine GPCR signaling, potassium transport, and amino acid transport during HCMV replication.
Materials and Methods
Systems Analysis.
RNA-seq reads were aligned to the human and HCMV genomes using HISAT2 (45) and converted to gene counts using featureCounts (46). HCMV/mock ratios were determined using DESeq2 (10). The modified GSEA procedure was performed using gene sets from the MSigDB (13) and Harmonizome (15) databases, and comparing each gene set’s ratio distribution to its parental RNA-seq ratio distribution using the Wilcoxon rank-sum test. Concordantly regulated gene sets (MRC-5 q-value ≤0.05 and ARPE-19 q-values ≤0.05) were organized into gene set overlap networks essentially as describe in the “enrichment map” technique (47); by representing each gene set as a network node and connecting all node pairs with edges weighted by the overlap of their underlying gene memberships. Networks were rendered and analyzed using igraph (48) and Gephi (49). Network communities were detected using the Louvain modularity maximization algorithm (50). Additional methods and computational details are provided in SI Appendix, SI Materials and Methods.
Data Availability.
RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession no. GSE99454.
Supplementary Material
Acknowledgments
We thank Ben Greenbaum, Anastasia Baryshnikova, and Andres Blanco for critical reading of the manuscript; Yuka Imamura for technical assistance with RNA-sequencing; and members of the T.S. laboratory for suggestions. This work was supported by NIH Grant AI112951 (to T.S.) and Ruth L. Kirschstein National Research Service Award AI106175 (to A.O.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE99454).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710799114/-/DCSupplemental.
References
- 1.Mocarski E, Shenk T, Griffiths P, Pass RF. Cytomegaloviruses. In: Knipe DM, Howley PM, editors. Fields Virology. 6th Ed. Wolters Kluwer Lippincott Williams & Wilkins; Philadelphia: 2013. pp. 1960–2014. [Google Scholar]
- 2.Murphy E, et al. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc Natl Acad Sci USA. 2003;100:14976–14981. doi: 10.1073/pnas.2136652100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stern-Ginossar N, et al. Decoding human cytomegalovirus. Science. 2012;338:1088–1093. doi: 10.1126/science.1227919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tirosh O, et al. The transcription and translation landscapes during human cytomegalovirus infection reveal novel host-pathogen interactions. PLoS Pathog. 2015;11:e1005288. doi: 10.1371/journal.ppat.1005288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koyuncu E, Purdy JG, Rabinowitz JD, Shenk T. Saturated very long chain fatty acids are required for the production of infectious human cytomegalovirus progeny. PLoS Pathog. 2013;9:e1003333. doi: 10.1371/journal.ppat.1003333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Connor CM, DiMaggio PA, Jr, Shenk T, Garcia BA. Quantitative proteomic discovery of dynamic epigenome changes that control human cytomegalovirus (HCMV) infection. Mol Cell Proteomics. 2014;13:2399–2410. doi: 10.1074/mcp.M114.039792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2011;157:193–203. doi: 10.1016/j.virusres.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol. 2005;79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinzger C, et al. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J Gen Virol. 2008;89:359–368. doi: 10.1099/vir.0.83286-0. [DOI] [PubMed] [Google Scholar]
- 10.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Irizarry RA, Wang C, Zhou Y, Speed TP. Gene set enrichment analysis made simple. Stat Methods Med Res. 2009;18:565–575. doi: 10.1177/0962280209351908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Browne EP, Wing B, Coleman D, Shenk T. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: Viral block to the accumulation of antiviral mRNAs. J Virol. 2001;75:12319–12330. doi: 10.1128/JVI.75.24.12319-12330.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rouillard AD, et al. The harmonizome: A collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (Oxford) 2016;2016:baw100. doi: 10.1093/database/baw100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shenk T, Alwine JC. Human cytomegalovirus: Coordinating cellular stress, signaling, and metabolic pathways. Annu Rev Virol. 2014;1:355–374. doi: 10.1146/annurev-virology-031413-085425. [DOI] [PubMed] [Google Scholar]
- 17.Voigt P, Tee W-W, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318–1338. doi: 10.1101/gad.219626.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stanton RJ, et al. Cytomegalovirus destruction of focal adhesions revealed in a high-throughput Western blot analysis of cellular protein expression. J Virol. 2007;81:7860–7872. doi: 10.1128/JVI.02247-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weekes MP, et al. Quantitative temporal viromics: An approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattabiraman DR, et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016;351:aad3680. doi: 10.1126/science.aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roca H, et al. Transcription factors OVOL1 and OVOL2 induce the mesenchymal to epithelial transition in human cancer. PLoS One. 2013;8:e76773. doi: 10.1371/journal.pone.0076773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roca H, et al. A bioinformatics approach reveals novel interactions of the OVOL transcription factors in the regulation of epithelial–Mesenchymal cell reprogramming and cancer progression. BMC Syst Biol. 2014;8:29. doi: 10.1186/1752-0509-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tam WL, Weinberg RA. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med. 2013;19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanton RJ, et al. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J Clin Invest. 2010;120:3191–3208. doi: 10.1172/JCI42955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakhramov A, Boriskin YS, Booth JC, Bolton TB. Activation and deactivation of membrane currents in human fibroblasts following infection with human cytomegalovirus. Biochim Biophys Acta. 1995;1265:143–151. doi: 10.1016/0167-4889(94)00230-c. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, Xie S, Sun B. Viral proteins function as ion channels. Biochim Biophys Acta. 2011;1808:510–515. doi: 10.1016/j.bbamem.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wise DR, Thompson CB. Glutamine addiction: A new therapeutic target in cancer. Trends Biochem Sci. 2010;35:427–433. doi: 10.1016/j.tibs.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reinhardt B, et al. Human cytomegalovirus-induced reduction of extracellular matrix proteins in vascular smooth muscle cell cultures: A pathomechanism in vasculopathies? J Gen Virol. 2006;87:2849–2858. doi: 10.1099/vir.0.81955-0. [DOI] [PubMed] [Google Scholar]
- 30.Gregory PA, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 31.Ellis-Connell AL, Iempridee T, Xu I, Mertz JE. Cellular microRNAs 200b and 429 regulate the Epstein-Barr virus switch between latency and lytic replication. J Virol. 2010;84:10329–10343. doi: 10.1128/JVI.00923-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jha HC, et al. KSHV-mediated regulation of Par3 and SNAIL contributes to B-cell proliferation. PLoS Pathog. 2016;12:e1005801. doi: 10.1371/journal.ppat.1005801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uppal T, Banerjee S, Sun Z, Verma SC, Robertson ES. KSHV LANA: The master regulator of KSHV latency. Viruses. 2014;6:4961–4998. doi: 10.3390/v6124961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mateo M, Generous A, Sinn PL, Cattaneo R. Connections matter: How viruses use cell–cell adhesion components. J Cell Sci. 2015;128:431–439. doi: 10.1242/jcs.159400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herranz N, et al. Polycomb complex 2 is required for E-cadherin repression by the Snail1 transcription factor. Mol Cell Biol. 2008;28:4772–4781. doi: 10.1128/MCB.00323-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paulus C, Nevels M. The human cytomegalovirus major immediate-early proteins as antagonists of intrinsic and innate antiviral host responses. Viruses. 2009;1:760–779. doi: 10.3390/v1030760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banerjee S, et al. Distinct microbiological signatures associated with triple negative breast cancer. Sci Rep. 2015;5:15162. doi: 10.1038/srep15162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harkins LE, et al. Detection of human cytomegalovirus in normal and neoplastic breast epithelium. Herpesviridae. 2010;1:8. doi: 10.1186/2042-4280-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taher C, et al. High prevalence of human cytomegalovirus proteins and nucleic acids in primary breast cancer and metastatic sentinel lymph nodes. PLoS One. 2013;8:e56795. doi: 10.1371/journal.pone.0056795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Banerjee S, et al. The ovarian cancer oncobiome. Oncotarget. 2017;8:36225–36245. doi: 10.18632/oncotarget.16717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maussang D, et al. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc Natl Acad Sci USA. 2006;103:13068–13073. doi: 10.1073/pnas.0604433103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen Y, Zhu H, Shenk T. Human cytomagalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitchell DA, et al. Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature. 2015;519:366–369. doi: 10.1038/nature14320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168:670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, Langmead B, Salzberg SL. HISAT: A fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao Y, Smyth GK, Shi W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 47.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal Complex Syst. 2006;1695:1–9. [Google Scholar]
- 49.Bastian M, Heymann S, Jacomy M. Gephi: An open source software for exploring and manipulating networks. ICWSM. 2009;8:361–362. [Google Scholar]
- 50.Blondel VD, Guillaume J-L, Lambiotte R, Lefebvre E. Fast unfolding of communities in large networks. J Stat Mech Theory Exp. 2008;2008 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) under accession no. GSE99454.