Significance
Rab GTPases are central regulators of vesicle-mediated cargo transport. They are switched from the GDP-bound inactive conformation to the GTP-bound active state by guanine nucleotide exchange factors (GEFs). Rab-interacting factor (RABIF)/mammalian suppressor of Sec4 (MSS4) and its yeast homolog DSS4 are the first putative GEFs isolated for Rabs, but their biological function and molecular mechanism remained unclear. In this work, we demonstrate that RABIF plays a crucial role in exocytosis. Unexpectedly, we discovered that RABIF does not function as a GEF, as previously assumed. Instead, RABIF is a Rab-stabilizing holdase chaperone, a previously unrecognized mode of Rab regulation independent of its GDP-releasing activity. We suggest that a requirement for holdase chaperones is likely a general feature of Rab GTPases.
Keywords: GLUT4, exocytosis, Rab GTPase, CRISPR screen, vesicle transport
Abstract
Rab GTPases are switched from their GDP-bound inactive conformation to a GTP-bound active state by guanine nucleotide exchange factors (GEFs). The first putative GEFs isolated for Rabs are RABIF (Rab-interacting factor)/MSS4 (mammalian suppressor of Sec4) and its yeast homolog DSS4 (dominant suppressor of Sec4). However, the biological function and molecular mechanism of these molecules remained unclear. In a genome-wide CRISPR genetic screen, we isolated RABIF as a positive regulator of exocytosis. Knockout of RABIF severely impaired insulin-stimulated GLUT4 exocytosis in adipocytes. Unexpectedly, we discovered that RABIF does not function as a GEF, as previously assumed. Instead, RABIF promotes the stability of Rab10, a key Rab in GLUT4 exocytosis. In the absence of RABIF, Rab10 can be efficiently synthesized but is rapidly degraded by the proteasome, leading to exocytosis defects. Strikingly, restoration of Rab10 expression rescues exocytosis defects, bypassing the requirement for RABIF. These findings reveal a crucial role of RABIF in vesicle transport and establish RABIF as a Rab-stabilizing holdase chaperone, a previously unrecognized mode of Rab regulation independent of its GDP-releasing activity. Besides Rab10, RABIF also regulates the stability of two other Rab GTPases, Rab8 and Rab13, suggesting that the requirement of holdase chaperones is likely a general feature of Rab GTPases.
A universal feature of eukaryotic cells is a compartmentalized cytoplasm filled with functionally specialized membrane-bound organelles (1, 2). Maintenance and propagation of the organelles require constant interorganelle transport of cargo proteins via membrane-enclosed vesicles (3–5). Vesicle-mediated cargo transport was first genetically dissected in yeasts, leading to the identification of vesicle-transport mediators conserved in all eukaryotes (3). Vesicle transport is substantially more complex in mammalian cells and is often tightly regulated by extracellular and intracellular stimuli so that the speed and direction of cargo flow can be adjusted according to physiological demands (1, 6). However, mammalian vesicle transport has not been systematically dissected due to a lack of robust genetic tools.
The recent advent of the CRISPR-Cas9 system has revolutionized mammalian cell genetics by enabling rapid and complete ablation of target genes (7–10). Pooled CRISPR libraries introduced into cultured cells generate mutant populations that can be subsequently selected based on cellular phenotypes (8, 11–14). Previous CRISPR genetic screens, however, were mainly based on straightforward cell-viability or growth-advantage assays (11, 12, 15–19), which cannot be directly used to dissect multifaceted membrane pathways such as vesicle transport.
In this work, we developed a fluorescent reporter-based CRISPR screening platform to identify regulators of insulin-stimulated glucose transporter type 4 (GLUT4) exocytosis, a classic but poorly understood vesicle-transport pathway. Insulin-stimulated GLUT4 exocytosis plays critical roles in nutrient homeostasis in adipocytes and skeletal muscles (6, 20–22). Upon binding to its receptor, the anabolic hormone insulin facilitates glucose uptake by acutely relocating GLUT4 from intracellular compartments to the cell surface (6, 20, 21, 23). Upon the termination of insulin signaling, GLUT4 is retrieved from the plasma membrane through endocytosis and returns to intracellular storage vesicles (6). Importantly, the components of GLUT4 exocytosis are also involved in the regulation of other exocytic pathways such as insulin secretion and T cell degranulation (6, 22, 24–27). Thus, findings from the GLUT4 exocytosis screens will serve as a springboard to understanding the principles of vesicle transport in general.
Our CRISPR screens identified several known regulators of insulin-stimulated GLUT4 exocytosis, but the great majority of the genes were not previously linked to the pathway. One of the regulators isolated in the screens is Rab-interacting factor (RABIF), also known as “mammalian suppressor of Sec4” (MSS4). When overexpressed, a dominant mutant form of DSS4 (Dominant suppressor of Sec4), the yeast homolog of RABIF, could partially rescue the secretion defects caused by mutations in the exocytic Rab (Ras-related protein in brain) Sec4p (28). DSS4 and RABIF stimulate GDP release from Rab GTPases such as Sec4p in vitro, leading to the hypothesis that they function as a guanine nucleotide exchange factor (GEF) (28, 29). However, no obvious loss-of-function phenotype was observed for DSS4 in yeast except its synthetic lethality with Sec4p mutations (28). Likewise, the biological function of RABIF in mammalian cells has remained unclear. We confirmed the findings of our CRISPR screens by demonstrating that deletion of RABIF severely impaired insulin-stimulated GLUT4 exocytosis. RABIF binds to Rab10, an exocytic Rab GTPase regulating GLUT4 exocytosis, and robustly stimulates GDP release from Rab10 in vitro. However, this GDP-releasing activity is dispensable for the stimulatory function of RABIF in GLUT4 exocytosis. Instead, our results suggest that RABIF regulates GLUT4 exocytosis by stabilizing Rab10 protein. In the absence of RABIF, newly synthesized Rab10 protein is rapidly degraded by the proteasome, leading to abrogation of exocytosis. Restoration of Rab10 expression through a transient overexpression system rescued exocytosis defects, bypassing the requirement of RABIF. Thus, although initially identified as a putative Rab GEF, RABIF regulates exocytosis as a Rab-stabilizing holdase chaperone, a function independent of its GDP-releasing activity.
Results
Genome-Wide CRISPR Screens Identify Regulators of Insulin-Stimulated GLUT4 Exocytosis.
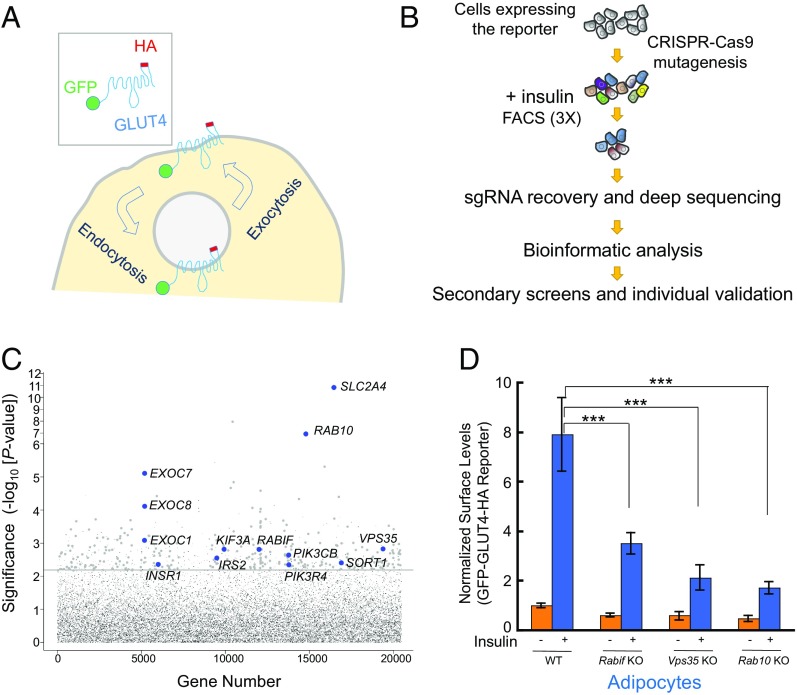
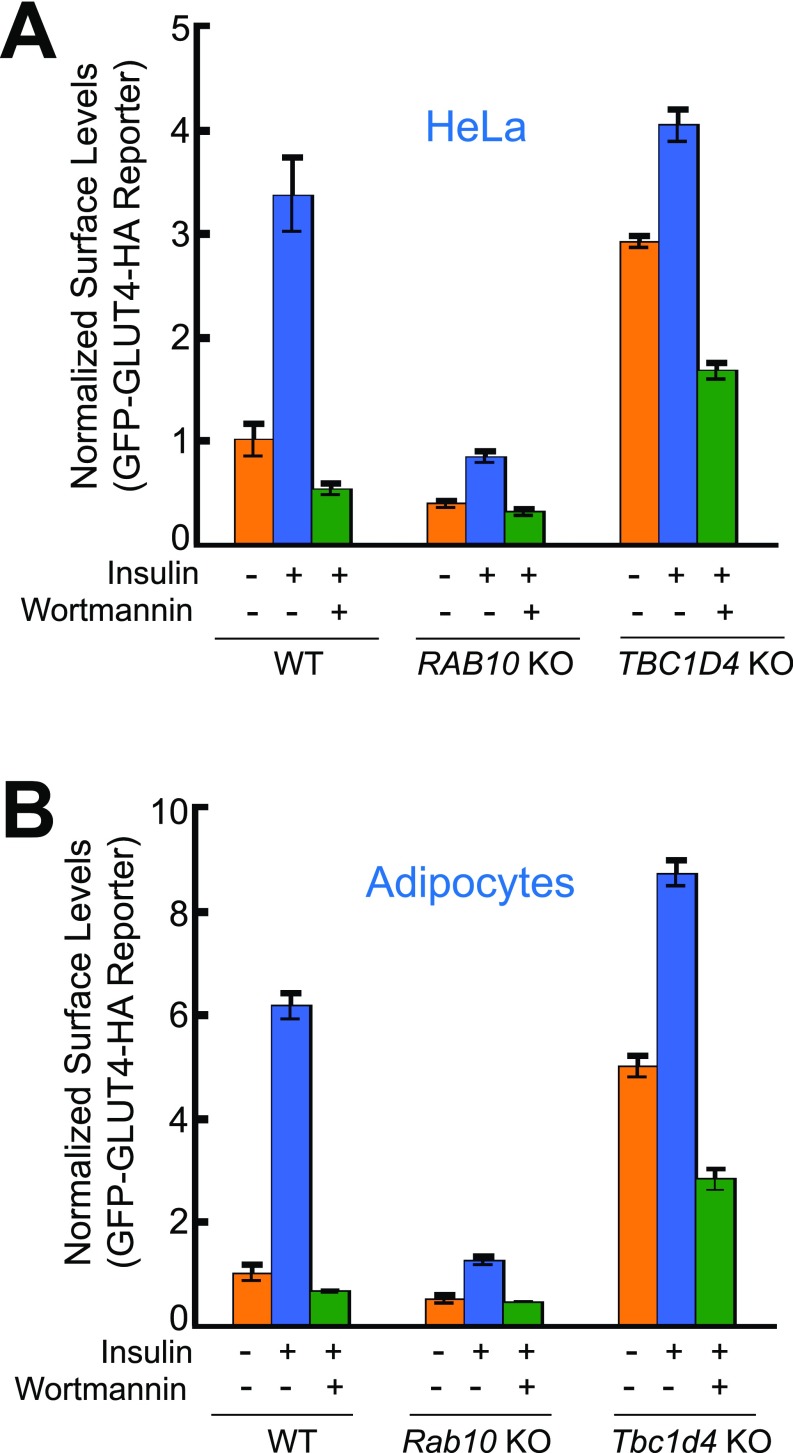
To identify exocytic regulators using CRISPR screens, we needed to first develop a facile screening platform that allows the selection of live cells using FACS. Since insulin-regulated trafficking of glucose transporters is also observed in cancer cells, including HeLa cells (30–32), we established a HeLa cell line stably expressing a dual-tag GLUT4 reporter (Fig. 1A). As expected, we observed that insulin treatment markedly increased the surface levels of the GLUT4 reporter in this cell line (Fig. S1A) (32). Insulin-stimulated GLUT4 translocation was abolished by wortmannin, a phosphoinositide 3-kinase inhibitor that blocks insulin signaling (Fig. S1A). Knockout of RAB10, which encodes a known positive regulator (33), abrogated insulin-stimulated GLUT4 translocation in HeLa cells (Fig. S1A). Knockout of TBC1D4, which encodes a known negative regulator (34), resulted in constitutive surface translocation of GLUT4 in HeLa cells (Fig. S1A). These data correlate well with the observations in adipocytes (33, 34) (Fig. S1B), suggesting that the regulatory components of the GLUT4 exocytic pathway in adipocytes are conserved in HeLa cells. Therefore we chose to perform genome-wide CRISPR screens in the readily expandable HeLa cells and then extended the findings to physiologically relevant insulin-responsive cell types.
Fig. 1.
Dissection of insulin-stimulated GLUT4 exocytosis using CRISPR genetic screens. (A) Diagram of the GFP-GLUT4-HA reporter used to monitor insulin-dependent GLUT4 trafficking. After the translocation of the reporter, the HA epitope is exposed to the cell exterior (60). (B) Illustration of the genome-wide genetic screen of insulin-stimulated GLUT4 exocytosis. (C) Ranking of genes in the CRISPR screen based on the P value. Each dot represents a gene. Genes above the horizontal line were tested in the pooled secondary screen. A gene is shown as a large dot if it was validated in the secondary screen. Other genes are shown as small gray dots. Selected known or validated regulators of GLUT4 exocytosis are labeled. (D) Selected genes from the screen were individually mutated in mouse adipocytes using CRISPR-Cas9. Effects of the mutations on insulin-stimulated GLUT4 exocytosis were measured by flow cytometry. Error bars indicate SD. ***P < 0.001.
Fig. S1.
Comparative analyses of insulin-stimulated GLUT4 exocytosis in adipocytes and HeLa cells. HeLa cells (A) and mouse adipocytes (B) expressing the GFP-GLUT4-HA reporter were either untreated or treated with 100 nM insulin for 30 min before the surface reporters were stained with anti-HA antibodies and APC-conjugated secondary antibodies. To inhibit insulin signaling, the cells were incubated with 100 nM wortmannin for 10 min before insulin stimulation. The ratio of APC and GFP fluorescence was measured by flow cytometry, and the mean fluorescence was normalized to that of untreated WT cells. Error bars indicate SD. RAB10- and TBC1D4-KO cell lines were generated using CRISPR-Cas9 genome editing.
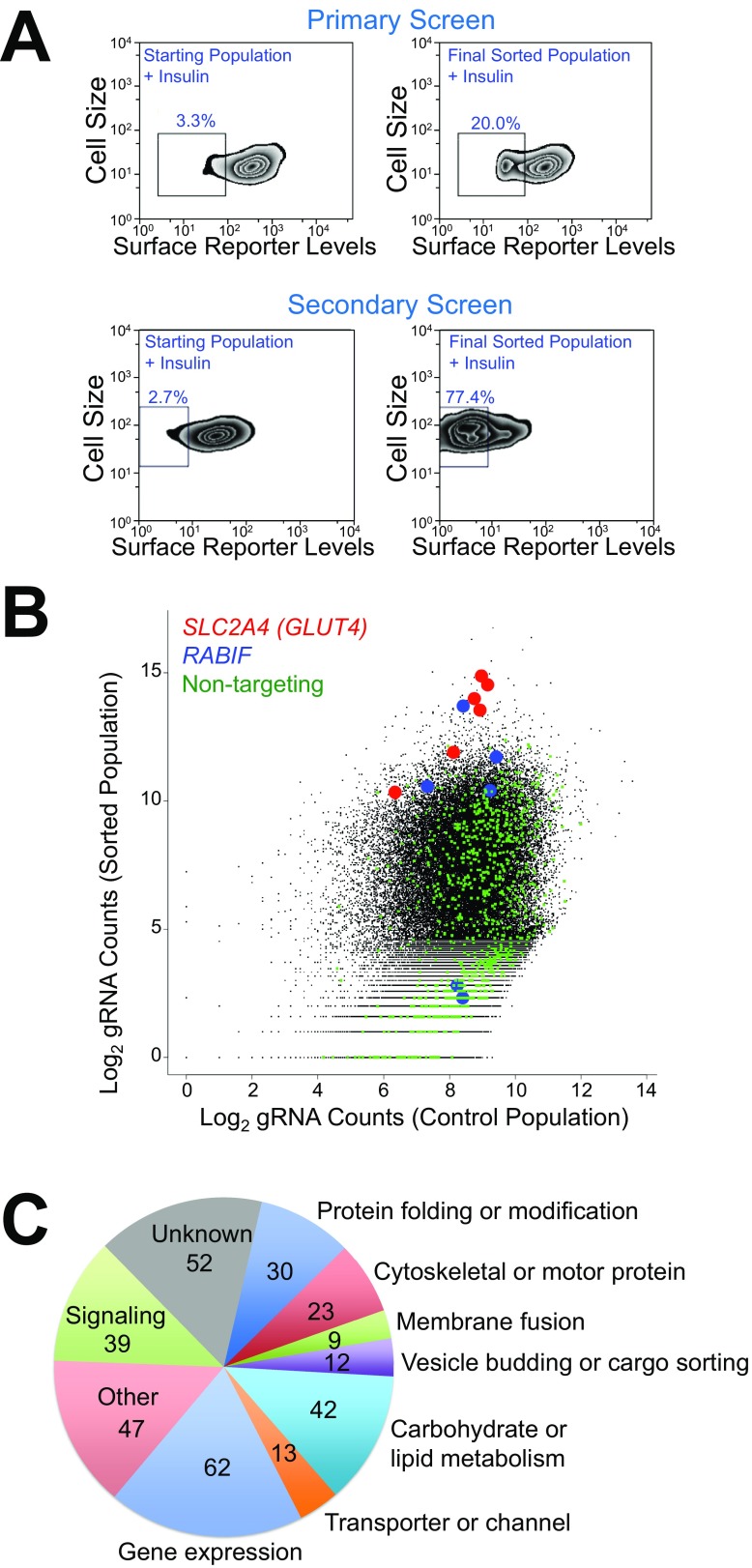
HeLa cells expressing the GLUT4 reporter were mutagenized by a pooled lentiviral CRISPR library containing 123,411 independent single-guide RNAs (sgRNAs) targeting 19,050 protein-coding genes and 1,864 miRNAs (35). FACS was used to sort mutant cells exhibiting reduced surface levels of the GLUT4 reporter after insulin stimulation (Fig. 1B and Fig. S2A). To effectively enrich true positives, the mutagenized cells were successively sorted for three rounds before sgRNAs were recovered and analyzed by deep sequencing. The abundance of many sgRNAs in the sorted populations was substantially increased compared with unsorted control populations grown under the same condition (Dataset S1). For example, sgRNAs targeting SLC2A4 (encoding GLUT4) and RABIF were significantly enriched (Fig. S2B). By contrast, nontargeting control sgRNAs overall exhibited no enrichment (Fig. S2B). Genes were ranked based on the enrichment of their targeting sgRNAs using the MAGeCK (model-based analysis of genome-wide CRISPR-Cas9 knockout) algorithm (Datasets S1 and S2) (36).
Fig. S2.
CRISPR-Cas9 genetic screens of insulin-stimulated GLUT4 exocytosis. (A) Flow cytometry analysis of the starting mutant population and the final sorted population in the genome-wide primary screen (Top) and the secondary screen (Bottom). HeLa cells were treated with 100 nM insulin for 30 min before the cells were stained and analyzed by flow cytometry. (B) Abundance of sgRNAs in the genome-wide screen and the passage control. (C) Summary of genes validated in the secondary screen.
To distinguish true regulators from false positives, we performed deeper secondary screens by building a pooled CRISPR library targeting the top 598 candidate genes from the genome-wide screen with 10 new sgRNAs for each gene (Dataset S3). HeLa cells expressing the GLUT4 reporter were mutagenized by the secondary CRISPR library and sorted by FACS (Fig. S1A). Enrichment of sgRNAs in the sorted populations was then determined (Dataset S4). A gene is considered significant only if its corresponding sgRNAs were enriched in both the primary and secondary screens (Dataset S4). The screens identified known regulators of insulin-dependent GLUT4 translocation, including Rab10, exocyst subunits, and insulin-signaling molecules (Fig. 1C). Recovery of these known regulators indicates that the screens were sensitive and specific. The great majority of the identified genes, however, were not previously linked to the GLUT4 exocytic pathway. These genes encode known or predicted regulators involved in vesicle budding or fusion, cargo sorting, signal transduction, and cell motility (Fig. S2C). Other genes encode enzymes catalyzing lipid or carbohydrate metabolism, mediators of gene expression, as well as factors lacking annotated functions (Fig. S2C).
To further validate the screen results, we designed individual sgRNAs targeting selected genes from the screens including RAB10, VPS35, and RABIF. Using flow cytometry-based assays, we observed that individual knockout of these genes abrogated insulin-stimulated GLUT4 translocation in adipocytes (Fig. 1D), in agreement with the findings of the screens.
RABIF Plays a Critical Role in Insulin-Stimulated GLUT4 Exocytosis.
Next we focused on the molecular mechanism of a regulator identified in the CRISPR screens, the 14-kDa soluble protein RABIF/MSS4 (“RABIF” is the preferred name, because “MSS4” also refers to 1-phosphatidylinositol-4-phosphate 5-kinase). Expressed in most mammalian tissues (29, 37), RABIF exhibits no sequence similarity with other mammalian proteins. In vitro, RABIF and its yeast homolog DSS4 can accelerate GDP release from Rab GTPases (28, 29), but their biological function was still unclear.
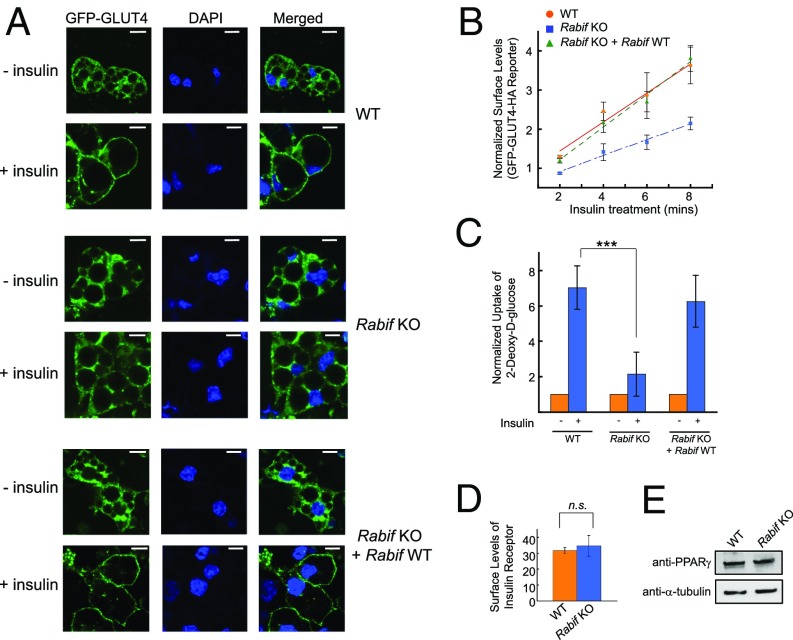
Confocal imaging showed that insulin-stimulated GLUT4 translocation was diminished in RABIF-KO adipocytes (Fig. 2A), confirming the flow cytometry data (Fig. 1D). Importantly, the translocation defects were fully rescued by the expression of the WT RABIF gene (Fig. 2A), excluding off-target effects. Further analyses revealed that the translocation defects were caused by a substantial decrease in the rate of GLUT4 exocytosis in RABIF-KO cells (Fig. 2B). We next measured insulin-stimulated glucose uptake in reporter-free adipocytes. Insulin stimulated the uptake of 2-deoxy-d-glucose into WT adipocytes, but the uptake was strongly reduced in RABIF-KO cells (Fig. 2C), consistent with the GLUT4 exocytosis defects observed in the knockout cells (Figs. 1D and 2A). By contrast, the surface levels of insulin receptor remained intact in RABIF-KO adipocytes (Fig. 2D), suggesting that insulin receptor translocates to the plasma membrane through a distinct route. The differentiation of adipocytes was not affected in RABIF-KO cells, as indicated by the normal expression of peroxisome proliferator-activated receptor gamma (PPARγ), an adipocyte marker (Fig. 2E). These data established that RABIF plays a critical role in insulin-stimulated GLUT4 exocytosis.
Fig. 2.
RABIF plays a critical role in insulin-stimulated GLUT4 exocytosis. (A) WT or Rabif-KO adipocytes were either untreated or treated with 100 nM insulin for 30 min before the localization of the GLUT4 reporter was visualized by confocal microscopy. (Scale bars: 10 µm.) (B) Normalized surface levels of the GLUT4 reporter in WT or mutant adipocytes. After serum starvation, the cells were treated with 200 µM dynasore for 5 min at 37 °C before 100 nM insulin was added. The cells were harvested for analysis at the indicated time points. Error bars indicate SD. (C) Normalized 2-deoxy-d-glucose uptake into WT or mutant adipocytes. ***P < 0.001. Error bars indicate SEM from four independent experiments. (D) Normalized surface levels of insulin receptor in WT or mutant adipocytes. n.s., not significant. (E) Immunoblots showing the expression levels of PPARγ and α-tubulin in WT or mutant adipocytes.
RABIF Recognizes the Exocytic Rab GTPase Rab10.
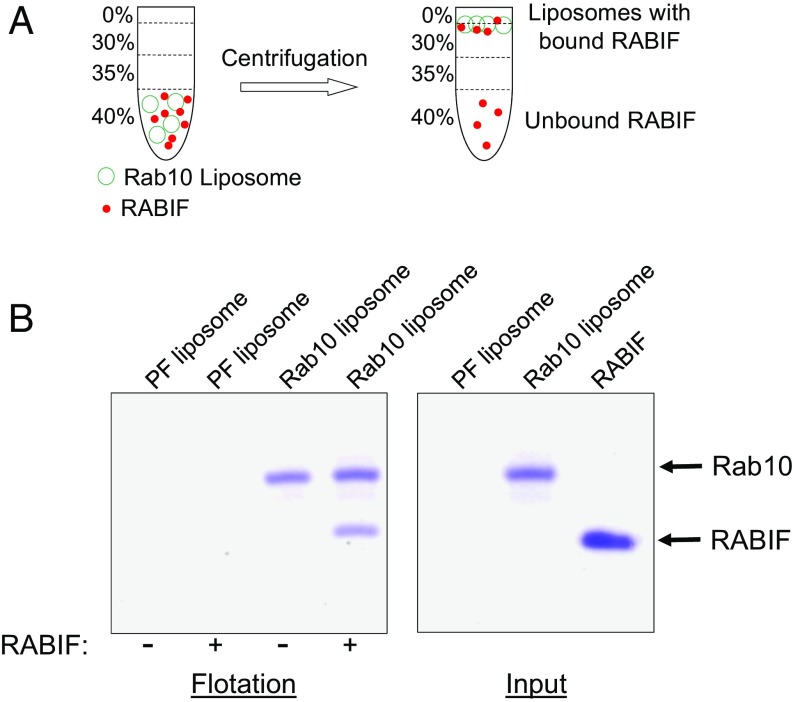
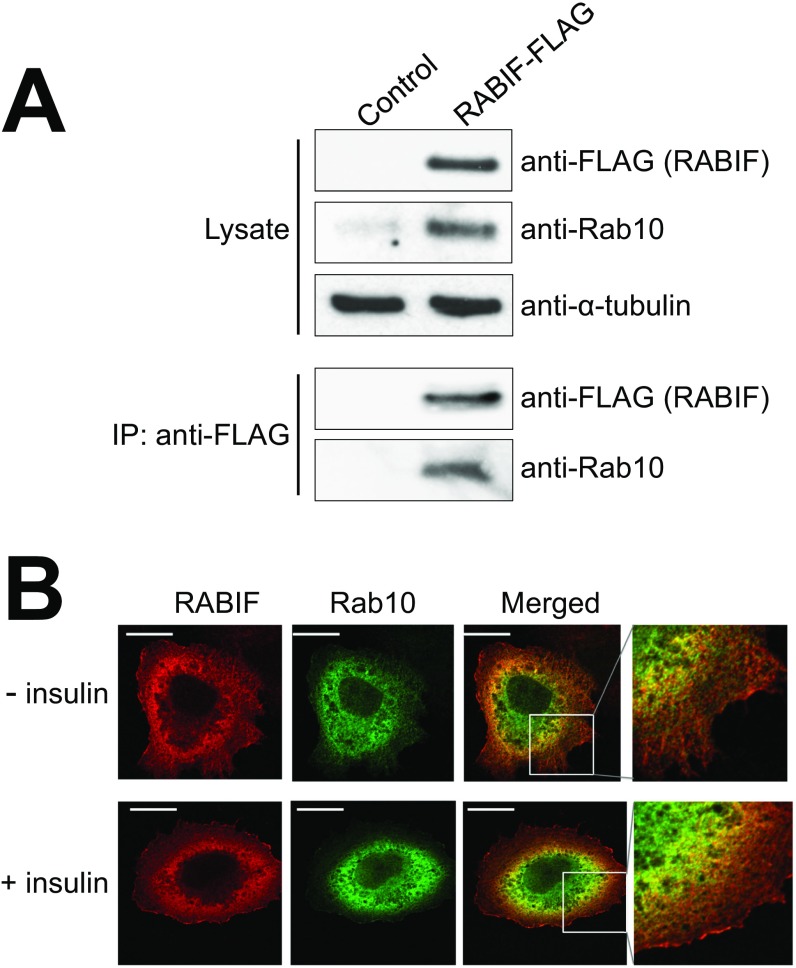
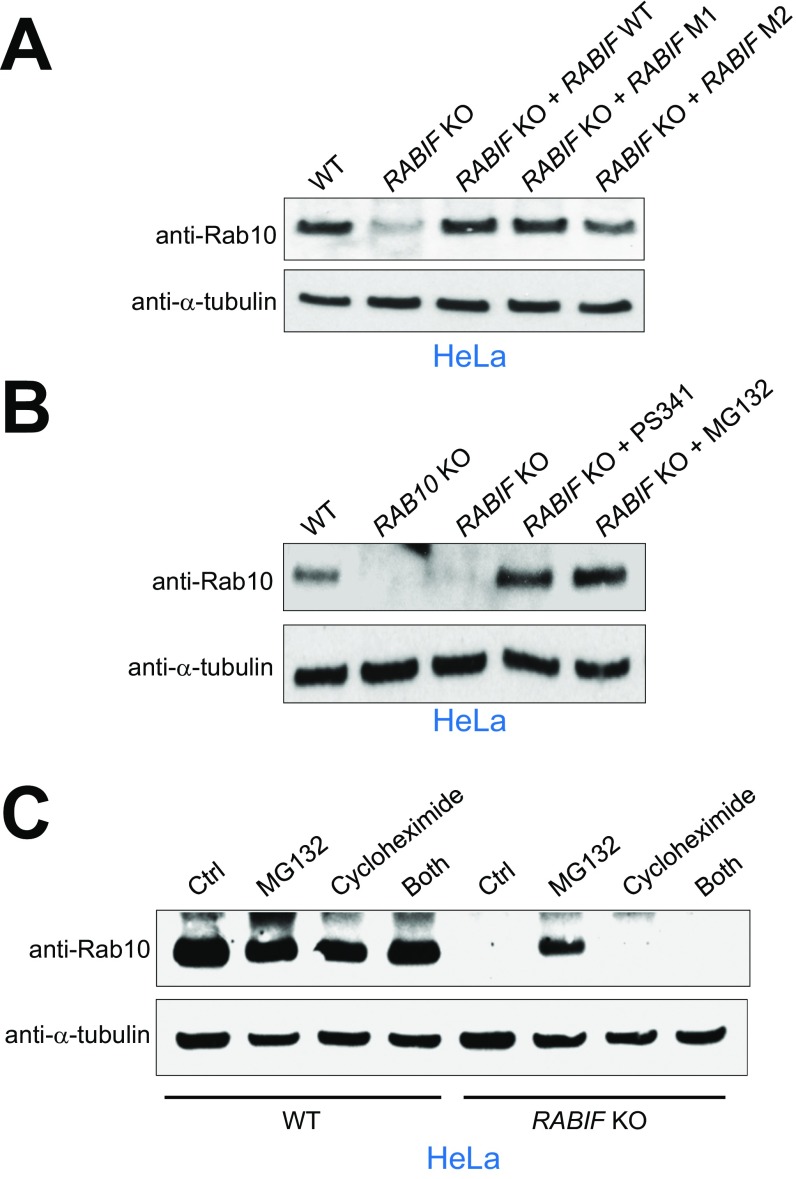
Our CRISPR screens isolated Rab10, a known regulator of insulin-stimulated GLUT4 exocytosis (Fig. 1C and Fig. S1) (33). RABIF stimulates GDP release from Rab10 in vitro (38), raising the possibility that RABIF promotes GLUT4 exocytosis by regulating the activity of Rab10. Using a liposome coflotation assay (Fig. 3A), we observed that recombinant RABIF bound to membrane-anchored recombinant Rab10 to form a complex with an estimated stoichiometry of 1:1 (Fig. 3B). In a coimmunoprecipitation assay using detergent-solubilized cell lysates, FLAG-tagged RABIF bound to endogenous Rab10 (Fig. S3A). Confocal imaging showed that RABIF and Rab10 exhibited partial colocalization in the cell (Fig. S3B), in agreement with the ability of RABIF and Rab10 to form complexes. These data suggest that RABIF may directly regulate Rab10 in insulin-stimulated GLUT4 exocytosis.
Fig. 3.
RABIF directly interacts with Rab10. (A) Diagram of the liposome coflotation assay. (B) Coomassie blue-stained denaturing gels showing the interaction between recombinant RABIF and liposome-anchored Rab10 proteins in the liposome coflotation assay. PF, protein free.
Fig. S3.
Rab10 and RABIF interact with each other and partially colocalize in the cell. (A) RABIF-FLAG was transiently expressed in RABIF-KO HeLa cells. RABIF-FLAG was immunoprecipitated using anti-FLAG antibodies, and the presence of Rab10 in the precipitates was detected by immunoblotting. (B) Representative confocal images showing the localization of Rab10-Myc and RABIF-FLAG in HeLa cells with or without 30 min of insulin treatment (100 nM). (Scale bars: 10 µm.) (Magnification: B, Right, 40×.)
RABIF Does Not Function as a GEF in GLUT4 Exocytosis.
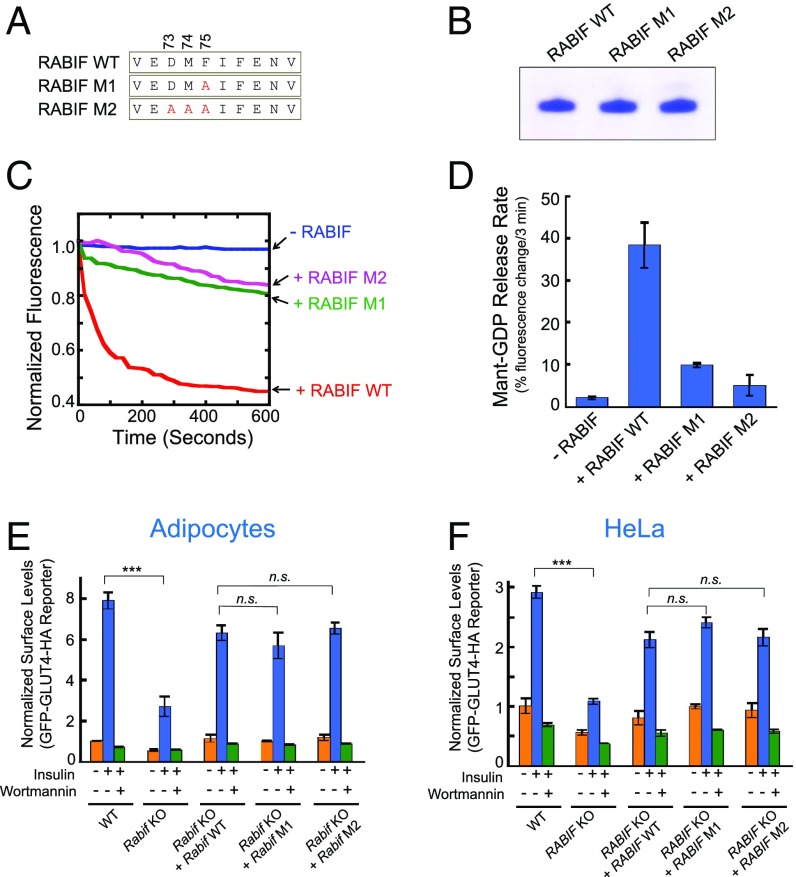
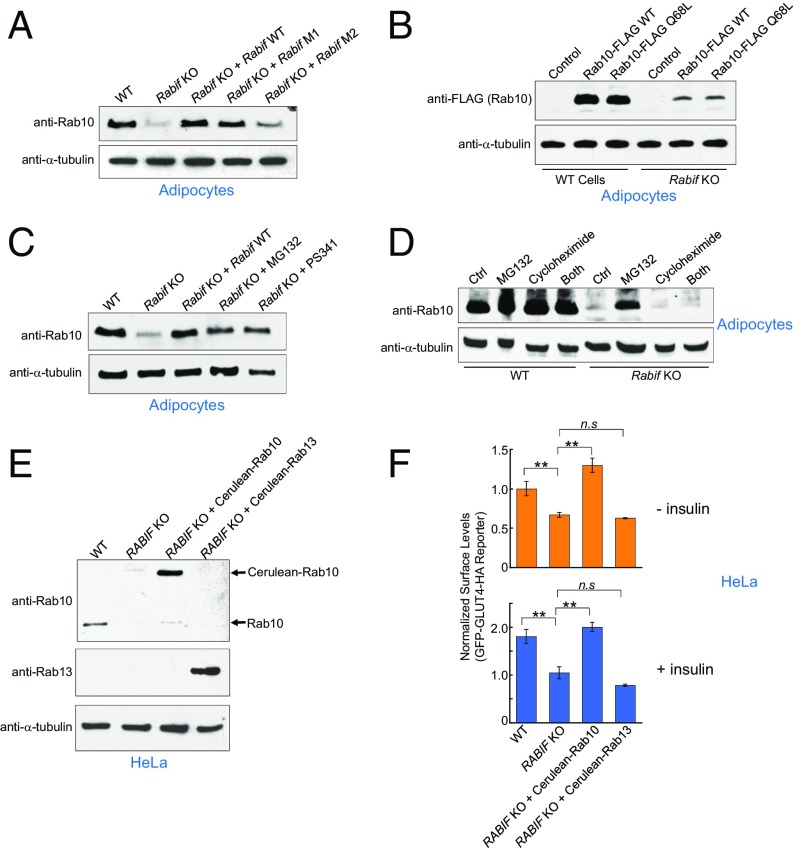
To regulate vesicle transport, Rab GTPases must be converted from their GDP-bound inactive state to the GTP-bound active conformation, a process stimulated by GEFs (39–43). RABIF is a putative GEF because it exhibits GDP-releasing catalytic activities in vitro (28, 29, 44, 45). Thus, we first examined whether RABIF functions as a GEF for Rab10 in GLUT4 exocytosis. Structural and biochemical studies suggested that the conserved residues D73, M74, and F75 of RABIF are located at the binding interface with Rab8 (Fig. 4A) (45, 46). Given the sequence similarity of Rab8 and Rab10, these residues are likely involved in Rab10 binding as well. In an in vitro assay, RABIF efficiently stimulated GDP release from prenylated Rab10 (Fig. 4 B–D). The GDP-releasing activity of RABIF was strongly reduced by the single F75A substitution (M1) and was largely abolished by the triple D73A/M74A/F75A mutations (M2) (Fig. 4 B–D).
Fig. 4.
RABIF does not function as a GEF in GLUT4 exocytosis. (A) Diagram showing the RABIF point mutations predicted to impair its interaction with Rab10. (B) Coomassie blue-stained gel showing purified WT and mutant RABIF proteins. (C) Kinetics of fluorescence changes resulting from RABIF-catalyzed mant-GDP release. The reactions were carried out in the presence of WT or mutant RABIF, using prenylated Rab10 as the substrate. (D) Initial rates of the reactions in C. Data are shown as percentage of fluorescence change within the first 3 min of the reactions. Error bars indicate SD. (E) Normalized surface levels of the GLUT4 reporter in the indicated adipocytes. n.s., not significant. ***P < 0.001. Error bars indicate SD. (F) Normalized surface levels of the GLUT4 reporter in the indicated HeLa cells. ***P < 0.001. Error bars indicate SD.
To examine the effects of these mutations on RABIF function in GLUT4 exocytosis, genes encoding WT or mutant RABIF proteins were introduced into RABIF-KO adipocytes. Surprisingly, we observed the RABIF mutants rescued GLUT4 exocytosis to similar levels as WT RABIF in adipocytes (Fig. 4E). Similar results were obtained in GLUT4 translocation assays carried out in HeLa cells (Fig. 4F). These results suggest that the GDP-releasing activity of RABIF is not required for its stimulatory function in GLUT4 exocytosis.
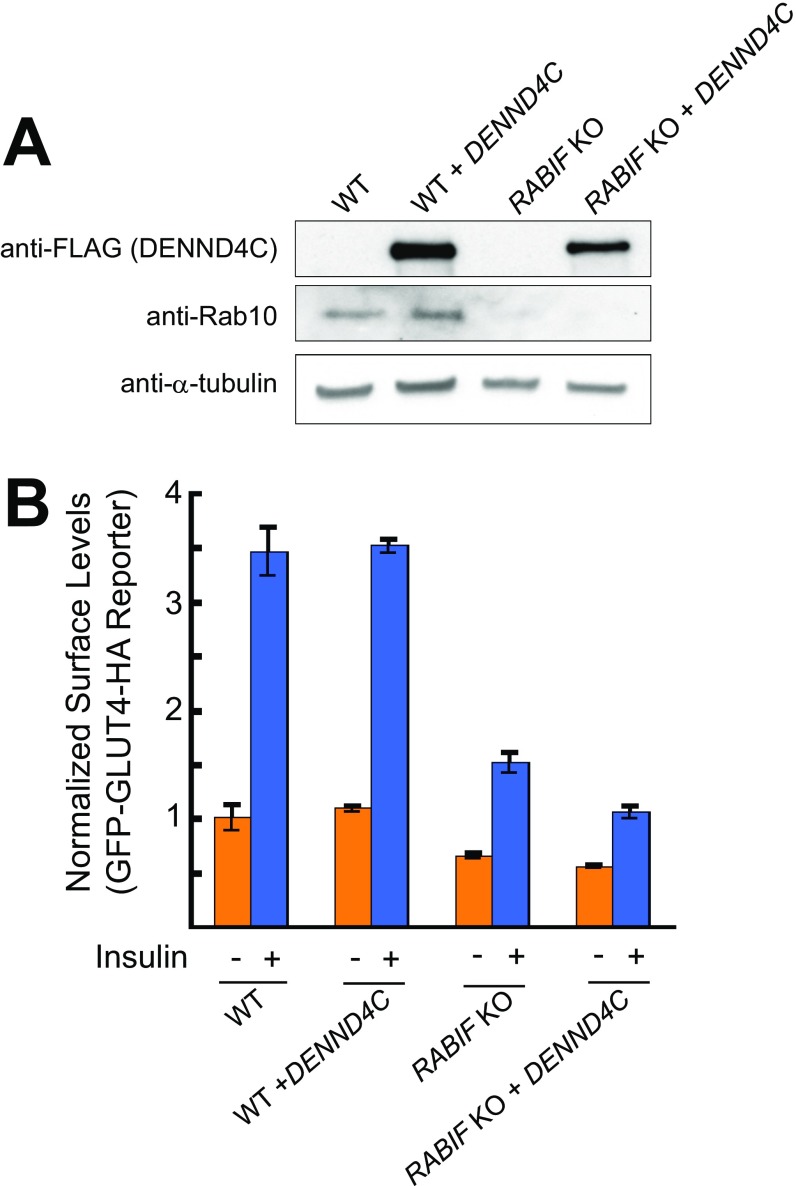
Given the robust GDP-releasing activity exhibited by RABIF in vitro, it is surprising that this activity is dispensable for RABIF function in GLUT4 exocytosis. However, this result is in line with the previous finding that the DENN (“differentially expressed in neoplastic versus normal cells”) domain protein DENND4C serves as the cognate GEF for Rab10 in GLUT4 exocytosis (47, 48). We next overexpressed DENND4C in RABIF-KO cells to examine whether it is functionally equivalent to RABIF. However, DENND4C overexpression failed to rescue GLUT4 exocytosis in RABIF-KO cells (Fig. S4). Thus, the GLUT4 exocytosis defects in RABIF-KO cells were not caused by a lack of GEF for Rab10. Together, these results indicate that RABIF does not act as a GEF in GLUT4 exocytosis.
Fig. S4.
Overexpression of DENND4C does not rescue GLUT4 translocation defects in RABIF-KO cells. (A) Immunoblots showing the expression of DENND4C, Rab10, and α-tubulin in WT or RABIF-KO HeLa cells. (B) Effects of DENND4C overexpression on GLUT4 reporter translocation were measured by flow cytometry. Error bars indicate SD.
RABIF Regulates the Stability of Rab10 in GLUT4 Exocytosis.
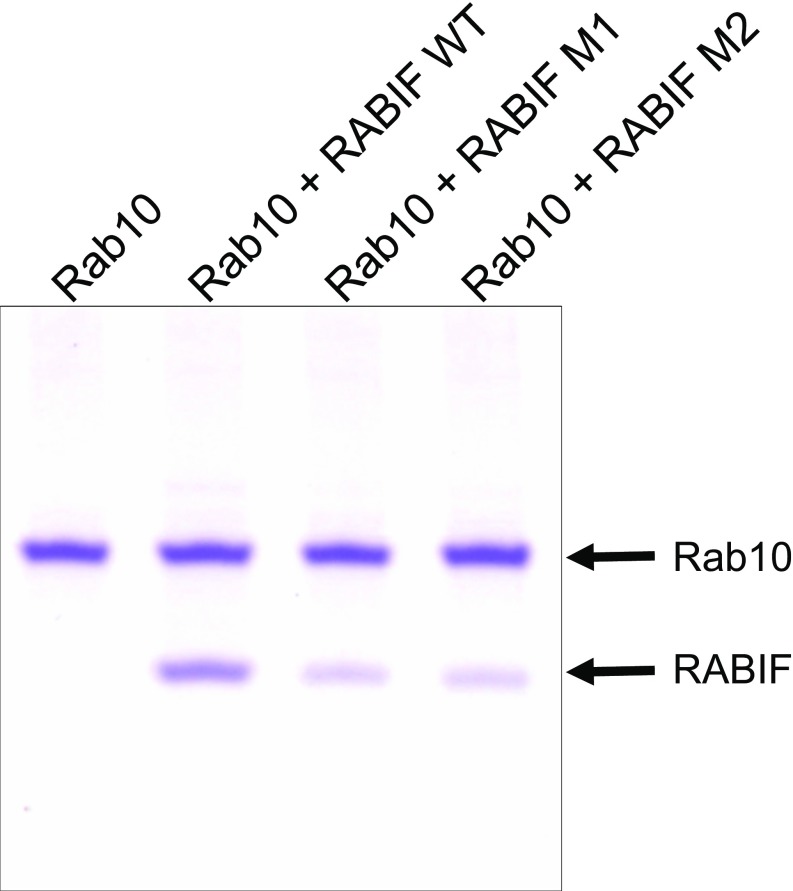
If RABIF does not function as a GEF, how does it regulate Rab10 in GLUT4 exocytosis? When analyzing RABIF-KO cells, we unexpectedly observed that RAB10 expression levels were dramatically reduced in RABIF-KO adipocytes and HeLa cells (Fig. 5A and Fig. S5A). Rab10 expression was rescued by the introduction of WT RABIF gene (Fig. 5A and Fig. S5A). Importantly, Rab10 expression was also restored by the genes encoding the RABIF mutants defective in GDP-releasing activities (Fig. 5A and Fig. S5A). Thus, RABIF is required for the expression of Rab10, and this function does not require its GEF catalytic activity. By contrast, overexpression of DENND4C failed to rescue Rab10 expression in RABIF-KO cells (Fig. S4A), as is consistent with its inability to restore GLUT4 exocytosis in RABIF-KO cells (Fig. S4B). Thus, even though DENND4C physically interacts with Rab10, it does not stabilize Rab10 as does RABIF. Interestingly, the RABIF mutations only partially reduced the binding of RABIF to Rab10 (Fig. S6). The remaining binding likely accounts for the activity of RABIF in stabilizing Rab10. Collectively, these results raise the possibility that RABIF promotes GLUT4 exocytosis by stabilizing Rab10 protein rather than by regulating its binding to guanine nucleotides.
Fig. 5.
RABIF regulates the stability of Rab10. (A) Immunoblots showing the expression levels of endogenous Rab10 and α-tubulin in the indicated adipocytes. (B) Lentiviral expression of FLAG-tagged Rab10 in WT or Rabif-KO adipocytes. The levels of tagged Rab10 and endogenous α-tubulin were probed by immunoblotting. (C) Rabif-KO adipocytes were either untreated or treated with 10 µM MG132 or 100 nM PS341 for 24 h. The expression levels of endogenous Rab10 and α-tubulin were probed by immunoblotting. (D) WT and Rabif-KO adipocytes were treated with 10 µM MG132 or 100 nM PS341 for 24 h in the absence or presence of 100 ng/mL cycloheximide. The expression levels of endogenous Rab10 and α-tubulin were probed by immunoblotting. (E) WT or RABIF-KO HeLa cells were either untransfected or transiently transfected with cerulean-tagged Rab10 or Rab13. The expression levels of Rab10, Rab13, and α-tubulin were probed by immunoblotting. (F) Flow cytometry measurements of the surface levels of the GLUT4 reporter using cells from E. **P < 0.01. n.s., not significant. Error bars indicate SD.
Fig. S5.
RABIF regulates the stability of Rab10 in HeLa cells. (A) Immunoblots showing the expression levels of endogenous Rab10 and α-tubulin in the indicated HeLa cells. (B) RABIF-KO HeLa cells were either untreated or treated with the indicated proteasome inhibitors as described in Fig. 5C. The expression levels of endogenous Rab10 and α-tubulin were probed by immunoblotting. (C) WT or RABIF-KO HeLa cells were treated with the indicated inhibitors as described in Fig. 5D. The expression levels of endogenous Rab10 and α-tubulin were probed by immunoblotting. Ctrl, DMSO treatment.
Fig. S6.
The GEF mutations of RABIF reduce but do not abolish the RABIF–Rab10 interaction. Coomassie blue-stained gel shows the interaction of Rab10 with WT RABIF or RABIF mutants in a liposome coflotation assay.
Next we sought to further explore how RABIF regulates the expression levels of Rab10. A lentiviral expression system was used to express FLAG-tagged Rab10 in adipocytes. While FLAG-tagged Rab10 was readily expressed in WT cells, little expression was observed in RABIF-KO cells (Fig. 5B). Since the lentiviral expression system used a viral promoter at a chromosomal location distinct from the endogenous RAB10 gene, these results suggested that the loss of Rab10 expression in RABIF-KO cells was not due to transcriptional or epigenetic silencing. Rab10 expression was rescued by the proteasome inhibitor MG132 or PS341, indicating that Rab10 was subject to proteasomal degradation in RABIF-KO cells (Fig. 5 C and D and Fig. S5 B and C). The restoration of Rab10 expression by proteasome inhibitors was abrogated by cotreatment with cycloheximide, a protein synthesis inhibitor (Fig. 5D and Fig. S5C), suggesting that the Rab10 proteins that accumulated during proteasome inhibitor treatments were from a newly synthesized pool. Thus, Rab10 proteins can be efficiently synthesized in the absence of RABIF, but they are rapidly degraded by the proteasome.
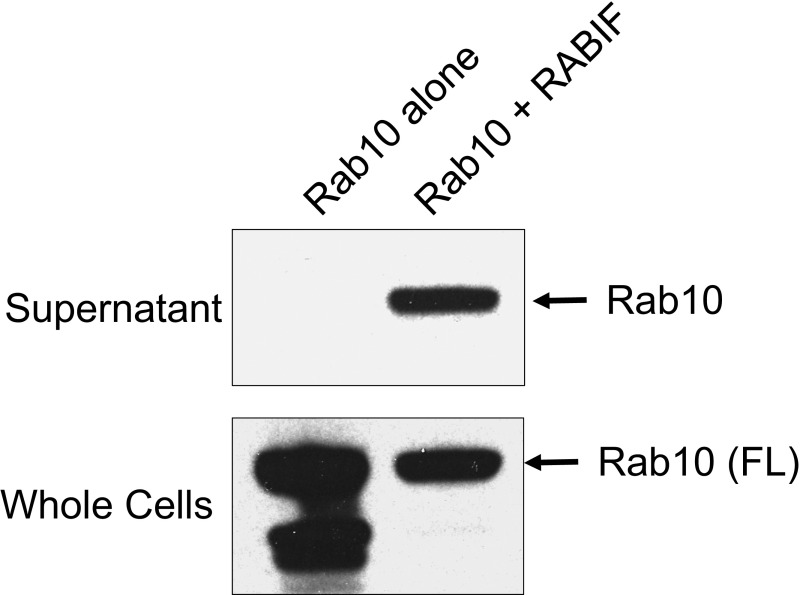
RABIF may stabilize an incompletely folded form of Rab10 protein that is unstable in the absence of RABIF. Alternatively, RABIF may protect fully folded Rab10 from proteosomal degradation (e.g., by masking degrons). To distinguish between these two possibilities, we reconstituted RABIF and Rab10 activities in Escherichia coli cells, which lack a proteasomal degradation system. We observed that recombinant Rab10 protein was expressed in E. coli cells with or without RABIF coexpression (Fig. S7). However, soluble Rab10 protein was obtained only from E. coli extracts coexpressing RABIF, whereas Rab10 protein was mainly found in the insoluble fraction in the absence of RABIF (Fig. S7). These results suggest that, during its folding and maturation process, Rab10 protein exists in an intrinsically unstable, aggregation-prone intermediate stage that must be stabilized by RABIF.
Fig. S7.
RABIF stabilizes Rab10 protein in E. coli. The RABIF gene was cloned into the pET28a-based SUMO vector (1), while Rab10 was cloned into the pGEX-4T3 vector. GST-Rab10 was expressed in BL21 E. coli cells either alone or together with RABIF. After the addition of 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), the cells were incubated at 37 °C for 3.5 h. After cell lysis and centrifugation, Rab10 proteins in the supernatant were precipitated using glutathione beads. Rab10 proteins from the supernatant and whole cells were probed by immunoblotting using anti-Rab10 antibodies.
We next examined whether the RABIF-KO phenotype can be rescued by forced expression of Rab10. Since lentiviral expression in adipocytes failed to express the Rab10 protein (Fig. 5B), we took advantage of a transient expression system in HeLa cells that permits high levels of exogenous protein expression. Our comparative analysis had clearly established that the molecular mechanism of RABIF is conserved in adipocytes and HeLa cells. Therefore, the RABIF functions revealed in HeLa cells would be expected to be conserved in adipocytes. Rab10 was fused to the cerulean fluorescent protein, and the fusion protein was transiently expressed in HeLa cells. While untagged Rab10 failed to be expressed, cerulean-tagged Rab10 was expressed at the expected size in RABIF-KO cells (Fig. 5E), as is consistent with the notion that fluorescent protein tags enhance the stability of target proteins to which they are fused (49). Strikingly, expression of cerulean-Rab10 fully rescued the surface levels of the GLUT4 reporter in RABIF-KO HeLa cells (Fig. 5F), indicating that restoration of Rab10 expression is sufficient to rescue exocytosis defects caused by RABIF deletion. By contrast, expression of cerulean-Rab13 failed to restore surface levels of GLUT4 (Fig. 5 E and F). Together, these data strongly suggest that RABIF promotes GLUT4 exocytosis by stabilizing Rab10 protein but is not required for the downstream functions of Rab10 in vesicle transport. Thus, RABIF functions as a Rab10 chaperone in GLUT4 exocytosis. Unlike the classic Hsp70 and Hsp90 chaperones (50), RABIF does not harbor an ATPase domain and thus cannot undergo ATP-dependent cycles to bind and release from its substrates. Instead, our data suggest that RABIF acts as an ATP-independent holdase chaperone that binds and stabilizes Rab10 protein in GLUT4 exocytosis.
RABIF Regulates a Group of Exocytic Rab GTPases.
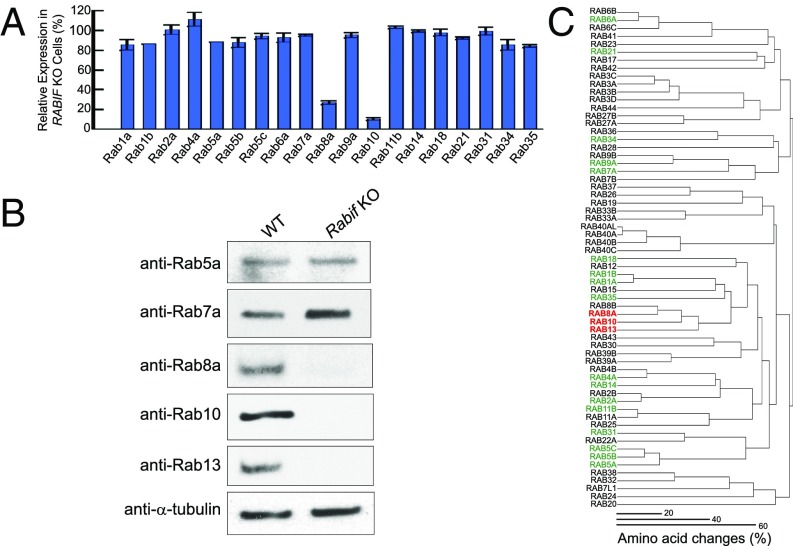
Finally, we sought to determine whether the holdase chaperone function of RABIF is restricted to Rab10 in a specific vesicle-transport pathway. To this end, we used MS-based proteomics to systematically analyze the protein-expression profiles of WT and RABIF-KO cells. Among the 19 Rabs detected by MS, the Rab10 level was reduced by more than 10-fold in RABIF-null cells (Fig. 6A and Dataset S5), in agreement with the immunoblotting results (Figs. 5D and 6B). In addition to Rab10, Rab8a expression was also diminished in RABIF-KO cells, whereas other Rabs detected by MS were not significantly affected (Fig. 6A). The reduction of Rab8a expression was confirmed by immunoblotting using anti-Rab8a antibodies (Fig. 6B). Immunoblotting using anti-Rab5a or anti-Rab7a antibodies showed that these Rab GTPases were not significantly affected by RABIF knockout (Fig. 6B), as is consistent with the MS data (Fig. 6A).
Fig. 6.
RABIF regulates a subset of related Rab GTPases. (A) Proteomic analysis of Rab expression in RABIF-KO HeLa cells. Data are presented as percentage of expression levels in WT cells. Average values of two technical replicates are shown (Rab1b levels were identical in the replicates, while Rab5a was quantified in only one replicate). (B) Immunoblots showing the expression levels of selected Rabs in the WT or Rabif-KO adipocytes. (C) Dendrogram showing the relatedness of human Rab proteins to each other. Proteins influenced by RABIF are highlighted in red. Rabs that were not influenced by RABIF are labeled in green. Rabs not detected by MS are shown in black.
Interestingly, Rab10 and Rab8a are related Rab GTPases according to a hierarchical cluster plot we compiled based on the sequence similarity of all the known Rab family proteins encoded by the human genome (Fig. 6C). The hierarchical cluster plot showed that Rab13 is also related to Rab8 and Rab10. While not recovered in our proteomic analysis (Fig. 6A), Rab13 was detected by immunoblotting using anti-Rab13 antibodies (Fig. 6B). As predicted by the hierarchical cluster plot, Rab13 expression was abolished in RABIF-KO cells (Fig. 6B). The next group of Rab GTPases exhibiting significant sequence similarity to Rab10 includes Rab1a, Rab1b, and Rab18. However, the expression levels of these Rab GTPases were not significantly affected in RABIF-KO cells (Fig. 6A). These results demonstrated that RABIF selectively regulates the stability of a subset of Rab GTPases. Despite their sequence relatedness, the RABIF-controlled Rab GTPases play distinct roles in exocytosis and cannot functionally replace each other (51–53). Thus, the requirement of a Rab-stabilizing holdase chaperone is not limited to a specific vesicle-transport pathway.
Discussion
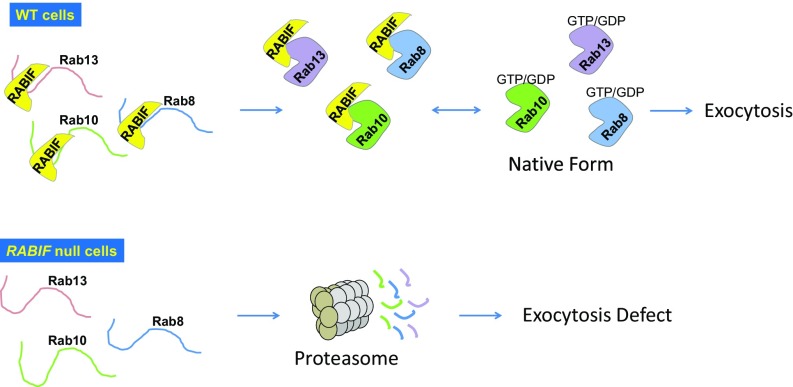
RABIF and its yeast homolog DSS4 were originally isolated as the first putative Rab GEFs (28, 29), but insights into their molecular mechanisms were impeded by the lack of a loss-of-function phenotype. In this work, we identified a critical role of RABIF in GLUT4 exocytosis, which subsequently enabled us to uncover a highly unusual mechanism by which RABIF regulates vesicle transport. Instead of functioning as a Rab GEF as previously postulated, RABIF is a holdase chaperone that is crucial for the expression of its cognate Rab GTPases (Fig. 7). The holdase chaperone model is supported by five lines of evidence. First, mutations that abrogate the GDP-releasing activity of RABIF do not affect its stimulatory function in exocytosis. Second, Rab10 protein can be efficiently synthesized in the absence of RABIF but is rapidly eliminated by proteasomal degradation. Third, overexpression of a cognate GEF of Rab10 (DENND4C) fails to restore Rab10 expression or exocytosis defects in RABIF-KO cells, suggesting that the RABIF-mutant cells do not lack a GEF. This observation also suggests that the Rab-recognizing mode of RABIF is fundamentally distinct from that of GEFs. Fourth, restoration of Rab10 expression rescues exocytosis defects, bypassing the requirement for RABIF. Thus, RABIF regulates the folding and maturation of Rabs without being involved in their downstream functions in vesicle transport. Finally, RABIF stabilizes Rab10 protein when reconstituted in bacterial cells.
Fig. 7.
Model illustrating the Rab-stabilizing holdase chaperone function of RABIF.
While our mechanistic studies were carried out primarily in adipocytes, the key findings were verified in an unrelated cell type, i.e., HeLa cells. Thus, the holdase chaperone model proposed here likely represents a conserved feature of RABIF. Like RABIF, DSS4 stimulates GDP release from the yeast exocytic Rab Sec4p. However, Sec2p is the known cognate GEF for Sec4p in yeast (54, 55), suggesting that DSS4 does not act as a GEF for Sec4p in vivo. This scenario is remarkably similar to the role of DENND4C as the Rab10 GEF in GLUT4 exocytosis, suggesting that DSS4 may also act as a holdase chaperone for Sec4p. The holdase chaperone model readily explains the previous observation that overexpression of DSS4 suppressed detrimental mutations in Sec4p (28).
Since RABIF possesses the unique property of binding Rab mutants, including those unable to accommodate guanine nucleotides, it was speculated to act as a chaperone for nucleotide-free Rabs (56, 57). This model, however, was not tested due to a lack of a clear biological function for RABIF or DSS4. Small GTPases constantly switch between different conformational states and can exist as nucleotide-free forms in their life cycles (41). Since nucleotide-free small GTPases are often unstable (58), it is possible that the nucleotide-free state represents the form of Rabs stabilized by RABIF. In support of this notion, RABIF-bound Rab8 is in its nucleotide-free state according the crystal structure of the complex (45). Interestingly, RABIF recognizes the first 58 residues of Rab8 (45), suggesting that it may also engage in the earlier stages of the Rab-folding pathway even before the complete synthesis of the Rab protein. No matter the conformational state of its Rab targets, RABIF is clearly essential to the stabilization of an incompletely folded form of Rab protein but is not involved in the downstream functions of Rabs in vesicle transport. In mammalian cells lacking RABIF, the unstable Rab intermediate is efficiently degraded by the proteasomal system, likely through the recognition of exposed hydrophobic motifs on the Rab protein. In E. coli cells, the Rab protein forms insoluble aggregates without the assistance of RABIF. In this regard, the Rab-stabilizing function of RABIF is analogous to the roles of histone chaperones in assisting nucleosome assembly (59).
By nature, a holdase chaperone stabilizes its substrate at a nonaggregated state but dissociates before the substrate progresses to a more stable conformation. RABIF-bound Rabs associate with GTP inefficiently due to the disruption of the nucleotide-binding pocket (45). Thus, RABIF dissociation is a prerequisite for Rab10 to enter the Rab cycle to regulate vesicle transport. The dissociation of RABIF is likely facilitated by other Rab-binding proteins such as GEFs in conjunction with guanine nucleotide association.
Our findings raised important questions regarding the role of RABIF as a Rab-stabilizing holdase chaperone. First, can RABIF act as a bona fide GEF in a vesicle-transport pathway? RABIF clearly does not function as a GEF for Rab10 in GLUT4 exocytosis, in which the GDP-releasing activity of RABIF is likely a by-product of its chaperone function and thus plays no physiological role. However, we cannot rule out the possibility that RABIF serves as a GEF for a Rab GTPase via a binding mode fundamentally distinct from its association with Rab10. It is even possible that RABIF serves as both a holdase chaperone and a GEF for a Rab GTPase. Further studies will be needed to examine these possibilities. Another question raised by this work is whether other Rab GTPases also involve cognate holdase chaperones in their life cycles. The Rab10-stabilizing function of RABIF demonstrates that a Rab GTPase requires a holdase chaperone during its normal maturation process. We further showed that RABIF controls the stability of a group of related but functionally distinct Rabs, suggesting that the holdase chaperone function is not limited to a specific vesicle transport pathway. Rab GTPases appear to adopt similar tertiary structures and undergo nucleotide-controlled regulatory steps that are conserved across pathways and species (41). Thus, we expect that requirement of a holdase chaperone is a general feature of Rab GTPases. A large number of Rab-associated proteins have been identified, including those lacking functional annotations. Further studies will be needed to determine whether these molecules function as holdase chaperones for their cognate Rab GTPases.
Experimental Procedures
Generation of GLUT4 Reporter Cell Lines.
HeLa cells, 293T cells, and mouse preadipocytes (derived from inguinal white adipocyte tissues, a gift from Shingo Kajimura, University of California, San Francisco) were maintained in DMEM supplemented with 10% FBS and penicillin/streptomycin. To differentiate into adipocytes, preadipocytes were cultured to ∼95% confluence before a differentiation mixture was added at the following concentrations: 5 µg/mL insulin (no. I0516; Sigma), 1 nM Triiodo-l-thyronine (T3; no. T2877; Sigma), 125 mM indomethacin (no. I-7378; Sigma), 5 µM dexamethasone (no. D1756; Sigma), and 0.5 mM isobutylmethylxanthine (IBMX; no. I5879; Sigma). After 2 d, the cells were switched to DMEM supplemented with 10% FBS, 5 µg/mL insulin, and 1 nM T3. After another 2 d, fresh medium of the same composition was supplied. Differentiated adipocytes were usually analyzed 6 d after addition of the differentiation mixture.
To generate cell lines expressing the GFP-GLUT4-HA reporter, lentiviruses were produced by transfecting 293T cells with a mixture of plasmids including GFP-GLUT4-HA (60), pAdVAntage (no. E1711; Promega), pCMV-VSVG, and psPax2. Lentiviral particles were collected 40 h after transfection and every 24 h thereafter for a total of four collections. Lentiviruses were pooled and concentrated by centrifugation in a Beckman SW28 rotor at 25,000 rpm for 1.5 h. The viral pellets were resuspended in PBS and used to transduce HeLa cells and preadipocytes. HeLa cells expressing the reporter were enriched for the strongest responses to insulin using FACS on a MoFlo cell sorter (Beckman Coulter). A clonal cell line with the strongest insulin response was used in the genetic screens. Mouse preadipocytes expressing the reporter were similarly generated except that pooled cell populations were used. The cell lines used in this study were authenticated and routinely tested for mycoplasma contamination.
Flow Cytometry Analysis of Insulin-Stimulated GLUT4 Exocytosis.
HeLa cells or adipocytes were washed three times with the KRH buffer [121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 0.33 mM CaCl2, and 12 mM Hepes (pH 7.0)]. After incubation in the KRH buffer for 2 h, the cells were treated with 100 nM insulin for 30 min. When applicable, 100 nM wortmannin (no. W1628; Sigma) was added 10 min before insulin treatment. After insulin stimulation, the cells were rapidly chilled in an ice bath, and their surface reporters were stained using anti-HA antibodies (no. 901501; BioLegend) and allophycocyanin (APC)-conjugated secondary antibodies (no. 17-4014; eBioscience). The cells were dissociated from the plates using Accutase (no. AT 104; Innovative Cell Technologies), and their APC and GFP fluorescence were measured on a CyAN ADP analyzer (Beckman Coulter, Inc.). To calculate normalized surface levels of the GLUT4 reporter, the mean surface GLUT4 reporter fluorescence (HA-APC) was divided by the mean total GLUT4 reporter fluorescence (GFP). The obtained values were then normalized to those of WT untreated samples. To measure the surface levels of insulin receptor, anti-insulin receptor antibodies (no. ab69508; Abcam) were used in the place of anti-HA antibodies. Data from populations of ∼5,000 cells were analyzed using the FlowJo software. Statistical significance was calculated based on experiments run in biological triplicate.
Genome-Wide CRISPR Mutagenesis of HeLa Reporter Cells.
HeLa cells expressing the GFP-GLUT4-HA reporter were mutagenized using the GeCKO V2 CRISPR Knockout Pooled Library (no. 1000000048; Addgene), following previously described procedures with minor modifications (12, 16–18, 35, 61). When delivered into targeted cells, Cas9 and sgRNAs encoded by the library introduced loss-of-function indel mutations through nonhomologous end joining (7, 9, 10, 15, 17, 62, 63). Lentiviruses were produced by transfecting the GeCKO V2 library plasmids (Parts A and B) into 293T cells using procedures similar to the generation of GFP-GLUT4-HA reporter cell lines. Starting at 48 h after transfection, media containing lentiviruses were collected every 24 h for a total of four collections. Lentiviral particles were pelleted in a Beckman SW28 ultracentrifuge rotor at 25,000 rpm for 1.5 h. The lentiviral pellets were resuspended in PBS and stored at −70 °C.
To test viral titers, 1 million HeLa cells were seeded into each well of a 12-well plate. The cells were spin-infected by lentiviruses produced from the Part A or B of the CRISPR library at 860 × g for 2 h. The plate was subsequently transferred to a 37 °C incubator. On the following day, the cells were dissociated and seeded in replicate wells of a 24-well plate. Fresh medium was supplied on the following day, and 1 µg/mL puromycin (no. P8833; Sigma) was added to half of the duplicate wells. After 24 h, attached cells were washed once with PBS and were counted using CountBright beads (no. C36950; Thermo Fisher) on a CyAN ADP analyzer. Numbers of cells in puromycin-treated wells were divided by those in the parallel untreated wells to calculate multiplicity of infection (MOI). Viral concentrations that yielded an MOI of ∼0.4 were chosen for large-scale preparations.
In large-scale preparations, 45 million HeLa reporter cells were seeded for each part of the library. Small-scale replicates of these mutagenized populations were separately treated with puromycin and counted to verify the MOI. After puromycin treatment, the large-scale mutant populations were combined at a 1:1 ratio and were frozen on the sixth day after viral transduction.
Genome-Wide CRISPR Screens.
Forty million mutagenized HeLa reporter cells were seeded at 1.2 million cells per 10-cm dish. On the following day, the cells were incubated in the KRH buffer for 2 h before treatment with insulin for 30 min. The dishes were subsequently chilled in an ice bath, and the cells were stained with anti-HA antibodies and APC-conjugated secondary antibodies. After dissociation from the plates by Accutase, the cells were concentrated by centrifugation and sorted by FACS. The bottom 3% of the cells were collected and expanded. Two additional rounds of selection were performed using the same fluorescence gating.
Illumina Deep Sequencing.
Genomic DNA was isolated using a genomic DNA isolation kit (no. K0721; Thermo Fisher). The unsorted control population contained 50 million cells, whereas the sorted populations contained 5 million cells. The isolated genomic DNA was used as template to amplify guide sequences. In the first round of PCR, each reaction was performed in a total volume of 100 µL containing 10 µg genomic DNA and the primers forward: AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG and reverse: AATGGACTATCATATGCTTACCGTAACTTGAAAGTATTTCG.
The second round of PCR reactions was performed in a total volume of 50 µL, using 5 µL of the PCR products from the first round as template. Of the 12 forward barcoded primers (F01–F12), six were used for the sorted populations, and six were used for the unsorted control population. The barcoded forward primers and the reverse primer (R01) are listed below with barcodes highlighted in bold. Stagger sequences are shown at the 5′ end of the barcode in lowercase letters, while the priming sites are shown to the 3′ end of the barcode in lowercase letters.
| F01 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTtAAGTAGAGtcttgtggaaaggacgaaacaccg |
| F02 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTatACACGATCtcttgtggaaaggacgaaacaccg |
| F03 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTgatCGCGCGGTtcttgtggaaaggacgaaacaccg |
| F04 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTcgatCATGATCGtcttgtggaaaggacgaaacaccg |
| F05 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTtcgatCGTTACCAtcttgtggaaaggacgaaacaccg |
| F06 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTatcgatTCCTTGGTtcttgtggaaaggacgaaacaccg |
| F07 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTgatcgatAACGCATTtcttgtggaaaggacgaaacaccg |
| F08 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTcgatcgatACAGGTATtcttgtggaaaggacgaaacaccg |
| F09 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTacgatcgatAGGTAAGGtcttgtggaaaggacgaaacaccg |
| F10 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTtAACAATGGtcttgtggaaaggacgaaacaccg |
| F11 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTatACTGTATCtcttgtggaaaggacgaaacaccg |
| F12 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTgatAGGTCGCAtcttgtggaaaggacgaaacaccg |
| R01 | CAAGCAGAAGACGGCATACGAGATAAGTAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTtTCTACTATTCTTTCCCCTGCACTGT |
PCR products were pooled, purified using a gel purification kit (no. 740609; Clontech), and sequenced on an Illumina HiSeq 2000 sequencing system using 1 × 125 v4 Chemistry. Sequencing reads were demultiplexed and processed to contain only the 20-bp unique guide sequences using the FASTX-Toolkit (hannonlab.cshl.edu/fastx_toolkit/). Readcount tables and gene enrichment analysis were performed using the MAGeCK algorithm (https://sourceforge.net/projects/mageck/).
Pooled Secondary CRISPR Screens.
We constructed a pooled secondary CRISPR library based on the guide sequences of an activity-optimized library (19). Ten sgRNAs were selected for the top 598 genes from the genome-wide screen. The secondary library also contained 1,000 nontargeting control sgRNAs and guides targeting 572 unrelated genes. Oligonucleotides containing the guide sequences were synthesized by CustomArray and amplified by PCR using the primers ArrayF: TAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCG and ArrayR: ACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC.
PCR products were ligated into the pLenti-CRISPR vector (no. 49535; Addgene) using a Gibson assembly kit (no. GA1200; Synthetic Genomics). The pLenti-CRISPR vector was digested using BsmBI followed by alkaline phosphatase treatment and gel purification. Each Gibson assembly reaction contained 100 ng vector and 40 ng PCR products in triplicates. The reactions were subsequently dialyzed against deionized water and transformed into electrocompetent E. coli cells (no. 60242; Lucigen). E. coli colonies were counted to ensure >20× coverage of the library.
HeLa cells expressing the GLUT4 reporter were mutagenized by the secondary CRISPR library and sorted as described in the primary screens.
CRISPR-Cas9 Genome Editing of Candidate Genes.
To edit a candidate gene, two independent guide sequences were selected within the early constitutive exons of the gene. Oligonucleotides containing one guide sequence were cloned into the pLenti-CRISPR-V2 vector (no. 52961; Addgene) as we previously described (64). Oligonucleotides containing the other guide sequence were cloned in a modified version of the CRISPR vector in which the puromycin selection marker was replaced with a hygromycin selection marker (pLenti-CRISPR-Hygro). Lentiviruses produced from the CRISPR plasmids were used to infect target cells. The infected cells were consecutively selected using 1 µg/mL puromycin and 500 µg/mL hygromycin B (no. 10687010; Thermo). The list of sgRNAs used to edit individual genes is given below.
| Species | Gene | Target 1 | Target 2 |
| Mouse | Rabif | GAACGAGCTCGTGTCAGCCG | CATGAGAAAGAAGCCAGATC |
| Mouse | Rab10 | CCACTCCCGAGTCCCCGATC | GTTCTCAAAGCTTTTACCGT |
| Mouse | Vps35 | AAAGTTTTTCCTGCTCATCC | TTACCAGGCATCTTTTCATC |
| Mouse | Tbc1d4 | AGCCGGAAGCGCTTGTCGCC | ATCTGTGACTCGGGGTCGTC |
| Human | RABIF | GCACCCGGGAGCCGCAACGC | TCCTGGAGGAGATCGCCGTC |
| Human | RAB10 | CCTGATCGGGGATTCCGGAG | ATCAAAACAGTTGAATTACA |
| Human | TBC1D4 | AAGTCAGCCAGGTCCTCTCC | CTGGGTCATCCTCCCCAGAC |
Immunoblotting and Immunoprecipitation.
Cells grown in 24-well plates were lysed in 1× SDS protein sample buffer, and the cell lysates were resolved on 8% Bis-Tris SDS/PAGE. Proteins were detected using primary antibodies and horseradish peroxidase-conjugated secondary antibodies. Primary antibodies used in immunoblotting were anti-Rab10 (no. 8127; Cell Signaling Technology and no. MABN730; Millipore), anti-Rab5a (no. 3547; Cell Signaling Technology), anti-Rab7a (no. 9367; Cell Signaling Technology), anti-Rab8a (no. 6975; Cell Signaling Technology), anti-Rab13 (no. MAB8305; R&D Systems), anti–PPAR-γ (no. A304-460; Bethyl Laboratories), anti–α-tubulin (no. 14-4502-82; eBioscience), and anti-FLAG (no. F1804; Sigma) antibodies.
In immunoprecipitation experiments, cells were lysed in a buffer containing 25 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, 5% glycerol, and a protease inhibitor mixture. Proteins were precipitated by using anti-FLAG magnetic beads (no. M8823; Sigma). Proteins in the precipitates were resolved on SDS/PAGE and were detected by immunoblotting.
Immunostaining and Imaging.
HeLa cells were seeded on coverslips coated with fibronectin (no. F1144; Sigma). The cells were fixed using 2% paraformaldehyde and were permeabilized in PBS supplemented with 5% FBS and 0.2% saponin. Antigens were stained using the following primary antibodies: anti-Myc (clone 9E10; Santa Cruz Biotechnology) and anti-FLAG (no. F7425; Sigma) antibodies. The cells were subsequently incubated with Alexa Fluor 488- or Alexa Fluor 568-conjugated secondary antibodies. After mounting on glass slides using the ProLong Antifade mountant with DAPI (no. P36931; Thermo), the cells were visualized on a Carl Zeiss LSM780 confocal microscope. Cell images were captured and processed using the Carl Zeiss Zen 2 and Adobe Photoshop software. To visualize the GFP-GLUT4-HA reporter in adipocytes, the cells were fixed and permeabilized similarly as HeLa cells.
MS.
Quantitative proteomic analysis of protein levels was performed using stable isotope labeling with amino acids in cell culture (SILAC) and MS. Cells were grown in SILAC medium (no. 88423; Thermo) supplemented with 10% dialyzed FB Essence (no. 3100; Seradigm). WT HeLa cells were grown in the presence of light lysine and arginine (no. L1262 and A5131; Sigma), whereas RABIF-null cells were grown in the presence of heavy lysine and arginine (no. CNLM-291 and CNLM-539; Cambridge Isotope Laboratories). After 5 d, the cells were harvested at ∼60% confluence in a lysis buffer [4% SDS and 50 mM Tris⋅HCl (pH 6.8)]. The cell lysates were processed for MS analysis following the filter-aided sample preparation (FASP) protocol (65, 66). Briefly, after the addition of 20 mM DTT, equal amounts of whole-cell lysates were mixed and loaded onto a spin filter with a cutoff of 30 kDa. The sample was then washed with the UA solution [8 M urea and 0.1 M Tris⋅HCl (pH 7.9)] and was alkylated using 0.1 M iodoacetamide. The sample was further washed with the UA solution and equilibrated with 0.1 M ammonium bicarbonate and 0.01% deoxycholic acid. The sample was then digested using 1% (wt/wt) trypsin at 37 °C for 16 h. The resulting tryptic peptides were eluded by centrifugation and were acidified using formic acid. Deoxycholic acid was removed using phase transfer with ethyl acetate. The tryptic peptides were fractionated by a Pierce high-pH reversed-phase spin column using 18-step gradients (4% acetonitrile for the first fraction, 1% increment for each fraction to the 17th fraction, and 50% acetonitrile for the 18th fraction). The fractions were dried using vacuum centrifugation.
One third of each fraction (5 µL) from the high-pH fractionation was analyzed by ultra performance LC (UPLC)-MS/MS. The tryptic peptides were loaded onto a Waters nanoACQUITY UPLC BEH C18 column (130 Å, 1.7 µm × 75 µm × 250 mm) equilibrated with 0.1% formic acid/3% acetonitrile/water. Mobile phase A was 0.1% formic acid/water, while phase B was 0.1% formic acid/acetonitrile. The peptides were eluted at 0.3 mL/min using a gradient of 3–8% B (0–5 min) and 8–35% B (5–123 min).
Precursor ions between 300–1,800 m/z (1 × 106 ions, 60,000 resolution) were scanned on a LTQ Orbitrap Velos mass spectrometer. The 10 most intense ions for MS/MS were selected with 180-s dynamic exclusion, 10 ppm exclusion width, with a repeat count = 1, and a 30-s repeat duration. Ions with unassigned charge state and MH+1 were excluded from MS/MS. Maximal ion injection times were 500 ms for FT (one microscan) and 250 ms for LTQ, and the automatic gain control (AGC) setting was 1 × 104. The normalized collision energy was 35% with activation Q 0.25 for 10 ms.
Raw data files from MS were searched against the UniProt human proteome database (Consortium, 2015) (total 88,479 entries), using the MaxQuant/Andromeda search engine (version 1.5.2.8) (67). Searches allowed trypsin specificity with two missed cleavages and included fixed Cys carbamidomethylation and variable acetylation (protein N terminus) and methionine oxidation. Mass tolerances were set to 20 ppm (first search) and 4.5 ppm (main search) for precursor ions and 0.5 Da for ion trap mass spectrometry (ITMS) MS/MS ions. MaxQuant/Andromeda used the top eight MS/MS peaks per 100 Da and seven amino acid minimum peptide length, with a 0.01 false discovery rate for both protein and peptide identification. For SILAC ratio measurements, a minimum of two independent peptide ratios were used to calculate a protein ratio.
Recombinant Protein Expression and Purification.
Recombinant Rab10 proteins were produced in Sf9 insect cells using baculovirus infection. Full-length mouse Rab10 gene was cloned into the baculovirus transfer vector pFastBac to generate a construct encoding a His6-tagged Rab10 protein with a tobacco etch virus (TEV) protease cleavage site. His6-Rab10 was expressed in Sf9 cells as we previously described for other proteins (68, 69). The cells were harvested in a lysis buffer [25 mM Hepes (pH 7.4), 400 mM KCl, 10% glycerol, 20 mM imidazole, 5 mM MgCl2, 1% Triton X-100, 2 mM β-mercaptoethanol, and a protease inhibitor mixture]. Rab10 proteins were purified by nickel-affinity chromatography, and the His6 tag was removed by TEV protease digestion.
Recombinant RABIF proteins were expressed and purified from E. coli as we previously described for other soluble proteins (70–72). The human RABIF gene was cloned into a pET28a-based SUMO (small ubiquitin-related modifier) vector. Purified His6-SUMO-RABIF fusion proteins were digested by SUMO proteases to obtain untagged RABIF proteins. RABIF mutants were generated by site-directed mutagenesis and were expressed using the same procedure used for the WT protein.
Liposome Coflotation Assay.
Protein-free or Rab10 liposomes were prepared using POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), following a liposome reconstitution protocol previously established in our group (70, 73, 74). Soluble RABIF proteins were incubated with liposomes at 4 °C with gentle agitation. After 1 h, an equal volume of 80% nycodenz (wt/vol) in reconstitution buffer was added, and the solution was transferred to 5 × 41 mm centrifuge tubes. The liposomes were overlaid with 200 μL each of 35% and 30% Nycodenz and then with 20 μL reconstitution buffer on the top. The gradients were centrifuged for 4 h at 52,000 rpm in a Beckman SW55 rotor. Samples were collected from the 0/30% Nycodenz interface (2 × 20 μL) and analyzed by SDS/PAGE.
Guanine Nucleotide Release Assay.
Recombinant Rab10 proteins were incubated with the fluorescent GDP analog 2'-(or-3′)-O-(N-Methylanthraniloyl) Guanosine 5′-Diphosphate (mant-GDP; no. M12414; Molecular Probes) at room temperature for 1 h in a loading buffer [20 mM Hepes (pH 7.4), 50 mM NaCl, 5 mM EDTA, and a 25-fold molar excess of mant-GDP]. The loading reaction was terminated by the addition of 10 mM MgCl2. Free mant-GDP was removed using desalting columns (no. 17-0853-02; GE Healthcare). Mant-GDP–bound RAB10 was diluted to 0.2 µM using the exchange buffer [20 mM Hepes (pH 7.4), 50 mM NaCl, and 5 mM MgCl2] in the absence or presence of 10 μM RABIF and 100 µM unlabeled GDP. Fluorescence changes associated with mant-GDP release were measured on a SpectraMax M5 microplate reader (Molecular Devices) at the excitation wavelength of 365 nm and emission wavelength of 440 nm. Initial rates of the mant-GDP release were calculated based on the percentage of fluorescence change within the first 3 min of the reactions.
Glucose Uptake Assay.
Preadipocytes were differentiated in 12-well plates as described above. On day 6, adipocytes were washed three times with the KRH buffer [121 mM NaCl, 4.9 mM KCl, 1.2 mM MgSO4, 0.33 mM CaCl2, and 12 mM Hepes (pH 7.0)]. After incubation in KRH buffer for 2 h, the cells were treated with 100 nM insulin for 30 min. Uptake was initiated by the addition of 100 µL of 0.67 mM 2-deoxy-[3H]-glucose (7.5 µCi/mM). The uptake was terminated after 1 min by washing the cells three times with ice-cold PBS. The cells were lysed in 10% SDS, and the radiolabeled glucose was quantified using a Beckman LS6500 liquid scintillation counter. Glucose uptake was measured in duplicate in all experiments, and the mean of the measurements was used for analysis. Nonspecific glucose uptake was measured in the presence of 50 µM cytochalasin B (no. 228090010; Acros Organics) and was subtracted from the measurements. Glucose uptake rates of the insulin-treated samples were normalized to those of the corresponding untreated controls.
Dendrogram Generation.
Amino acid sequences for 63 human Rab proteins were retrieved from the National Center for Biotechnology Information database. A distance matrix for percent difference was generated using BLAST for pairwise comparisons, and the first comparison available was used when multiple comparisons were present. Hierarchical clustering was performed in R using the command ‘hclust’ within the package ‘stats’ using the complete method. These clusters were then used to create the dendrogram using the package ggdendro in R (https://www.r-project.org/).
Supplementary Material
Acknowledgments
We thank Drs. Shingo Kajimura, Frances Brodsky, Gus Lienhard, Joaquin Espinosa, Gia Voeltz, and Cynthia Mastick for providing reagents; Drs. David James, David Sabatini, Tim Wang, Juan Bonifacino, Soyeon Park, and David Gershlick for insightful advice; Tom Blumenthal, Ding Xue, Ben Weaver, and Dwight Klemm for helpful discussions; Yuming Han, Thomas Lee, Katrina Diener, Jeff Reece, and Molishree Joshi for technical assistance; and members of the J.S. group for discussions and assistance. This work was supported by NIH Grants DK095367 (to J.S.) and GM102217 (to J.S.), a Pew Scholar Award (to J.S.), a University of Colorado Seed Grant (to J.S.), and an American Heart Association Predoctoral Fellowship (to D.R.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712176114/-/DCSupplemental.
References
- 1.Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- 2.Wickner W, Schekman R. Membrane fusion. Nat Struct Mol Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schekman R, Novick P. 23 genes, 23 years later. Cell. 2004;116(2 Suppl):S13–S15, and 11 pp following S19. doi: 10.1016/s0092-8674(03)00972-3. [DOI] [PubMed] [Google Scholar]
- 4.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wandinger-Ness A, Zerial M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb Perspect Biol. 2014;6:a022616. doi: 10.1101/cshperspect.a022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bryant NJ, Govers R, James DE. Regulated transport of the glucose transporter GLUT4. Nat Rev Mol Cell Biol. 2002;3:267–277. doi: 10.1038/nrm782. [DOI] [PubMed] [Google Scholar]
- 7.Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 8.Gilbert LA, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014;159:647–661. doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koike-Yusa H, Li Y, Tan EP, Velasco-Herrera MdelC, Yusa K. Genome-wide recessive genetic screening in mammalian cells with a lentiviral CRISPR-guide RNA library. Nat Biotechnol. 2014;32:267–273. doi: 10.1038/nbt.2800. [DOI] [PubMed] [Google Scholar]
- 14.Zhou Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature. 2014;509:487–491. doi: 10.1038/nature13166. [DOI] [PubMed] [Google Scholar]
- 15.Hart T, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163:1515–1526. doi: 10.1016/j.cell.2015.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Sidik SM, et al. A genome-wide CRISPR screen in toxoplasma identifies essential apicomplexan genes. Cell. 2016;166:1423–1435.e1412. doi: 10.1016/j.cell.2016.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marceau CD, et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature. 2016;535:159–163. doi: 10.1038/nature18631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang R, et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature. 2016;535:164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang S, Czech MP. The GLUT4 glucose transporter. Cell Metab. 2007;5:237–252. doi: 10.1016/j.cmet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Antonescu CN, McGraw TE, Klip A. Reciprocal regulation of endocytosis and metabolism. Cold Spring Harb Perspect Biol. 2014;6:a016964. doi: 10.1101/cshperspect.a016964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aslamy A, Thurmond DC. Exocytosis proteins as novel targets for diabetes prevention and/or remediation? Am J Physiol Regul Integr Comp Physiol. 2017;312:R739–R752. doi: 10.1152/ajpregu.00002.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 24.Jewell JL, Oh E, Thurmond DC. Exocytosis mechanisms underlying insulin release and glucose uptake: Conserved roles for Munc18c and syntaxin 4. Am J Physiol Regul Integr Comp Physiol. 2010;298:R517–R531. doi: 10.1152/ajpregu.00597.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ezcurra M, Reimann F, Gribble FM, Emery E. Molecular mechanisms of incretin hormone secretion. Curr Opin Pharmacol. 2013;13:922–927. doi: 10.1016/j.coph.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaisano HY, Macdonald PE, Vranic M. Glucagon secretion and signaling in the development of diabetes. Front Physiol. 2012;3:349. doi: 10.3389/fphys.2012.00349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lieberman J. The ABCs of granule-mediated cytotoxicity: New weapons in the arsenal. Nat Rev Immunol. 2003;3:361–370. doi: 10.1038/nri1083. [DOI] [PubMed] [Google Scholar]
- 28.Moya M, Roberts D, Novick P. DSS4-1 is a dominant suppressor of sec4-8 that encodes a nucleotide exchange protein that aids Sec4p function. Nature. 1993;361:460–463. doi: 10.1038/361460a0. [DOI] [PubMed] [Google Scholar]
- 29.Burton J, Roberts D, Montaldi M, Novick P, De Camilli P. A mammalian guanine-nucleotide-releasing protein enhances function of yeast secretory protein Sec4. Nature. 1993;361:464–467. doi: 10.1038/361464a0. [DOI] [PubMed] [Google Scholar]
- 30.Simpson IA, et al. The facilitative glucose transporter GLUT3: 20 years of distinction. Am J Physiol Endocrinol Metab. 2008;295:E242–E253. doi: 10.1152/ajpendo.90388.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adekola K, Rosen ST, Shanmugam M. Glucose transporters in cancer metabolism. Curr Opin Oncol. 2012;24:650–654. doi: 10.1097/CCO.0b013e328356da72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trefely S, et al. Kinome screen identifies PFKFB3 and glucose metabolism as important regulators of the insulin/insulin-like growth factor (IGF)-1 signaling pathway. J Biol Chem. 2015;290:25834–25846. doi: 10.1074/jbc.M115.658815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sano H, et al. Rab10, a target of the AS160 Rab GAP, is required for insulin-stimulated translocation of GLUT4 to the adipocyte plasma membrane. Cell Metab. 2007;5:293–303. doi: 10.1016/j.cmet.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 34.Eguez L, et al. Full intracellular retention of GLUT4 requires AS160 Rab GTPase activating protein. Cell Metab. 2005;2:263–272. doi: 10.1016/j.cmet.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li W, et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 2014;15:554. doi: 10.1186/s13059-014-0554-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 38.Wixler V, et al. Identification and characterisation of novel Mss4-binding Rab GTPases. Biol Chem. 2011;392:239–248. doi: 10.1515/BC.2011.022. [DOI] [PubMed] [Google Scholar]
- 39.Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25:414–419. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langemeyer L, et al. Diversity and plasticity in Rab GTPase nucleotide release mechanism has consequences for Rab activation and inactivation. Elife. 2014;3:e01623. doi: 10.7554/eLife.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–659. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segev N. Ypt and Rab GTPases: Insight into functions through novel interactions. Curr Opin Cell Biol. 2001;13:500–511. doi: 10.1016/s0955-0674(00)00242-8. [DOI] [PubMed] [Google Scholar]
- 43.Barr FA. Review series: Rab GTPases and membrane identity: Causal or inconsequential? J Cell Biol. 2013;202:191–199. doi: 10.1083/jcb.201306010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhu Z, Delprato A, Merithew E, Lambright DG. Determinants of the broad recognition of exocytic Rab GTPases by Mss4. Biochemistry. 2001;40:15699–15706. doi: 10.1021/bi0116792. [DOI] [PubMed] [Google Scholar]
- 45.Itzen A, Pylypenko O, Goody RS, Alexandrov K, Rak A. Nucleotide exchange via local protein unfolding–Structure of Rab8 in complex with MSS4. EMBO J. 2006;25:1445–1455. doi: 10.1038/sj.emboj.7601044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu Z, Dumas JJ, Lietzke SE, Lambright DG. A helical turn motif in Mss4 is a critical determinant of Rab binding and nucleotide release. Biochemistry. 2001;40:3027–3036. doi: 10.1021/bi002680o. [DOI] [PubMed] [Google Scholar]
- 47.Sano H, Peck GR, Kettenbach AN, Gerber SA, Lienhard GE. Insulin-stimulated GLUT4 protein translocation in adipocytes requires the Rab10 guanine nucleotide exchange factor Dennd4C. J Biol Chem. 2011;286:16541–16545. doi: 10.1074/jbc.C111.228908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marat AL, Dokainish H, McPherson PS. DENN domain proteins: Regulators of Rab GTPases. J Biol Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pédelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 50.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 51.Ioannou MS, et al. DENND2B activates Rab13 at the leading edge of migrating cells and promotes metastatic behavior. J Cell Biol. 2015;208:629–648. doi: 10.1083/jcb.201407068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y, Bilan PJ, Liu Z, Klip A. Rab8A and Rab13 are activated by insulin and regulate GLUT4 translocation in muscle cells. Proc Natl Acad Sci USA. 2010;107:19909–19914. doi: 10.1073/pnas.1009523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bravo-Cordero JJ, et al. MT1-MMP proinvasive activity is regulated by a novel Rab8-dependent exocytic pathway. EMBO J. 2007;26:1499–1510. doi: 10.1038/sj.emboj.7601606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walch-Solimena C, Collins RN, Novick PJ. Sec2p mediates nucleotide exchange on Sec4p and is involved in polarized delivery of post-Golgi vesicles. J Cell Biol. 1997;137:1495–1509. doi: 10.1083/jcb.137.7.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong G, Medkova M, Novick P, Reinisch KM. A catalytic coiled coil: Structural insights into the activation of the Rab GTPase Sec4p by Sec2p. Mol Cell. 2007;25:455–462. doi: 10.1016/j.molcel.2007.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nuoffer C, Wu SK, Dascher C, Balch WE. Mss4 does not function as an exchange factor for Rab in endoplasmic reticulum to Golgi transport. Mol Biol Cell. 1997;8:1305–1316. doi: 10.1091/mbc.8.7.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Collins RN, Brennwald P, Garrett M, Lauring A, Novick P. Interactions of nucleotide release factor Dss4p with Sec4p in the post-Golgi secretory pathway of yeast. J Biol Chem. 1997;272:18281–18289. doi: 10.1074/jbc.272.29.18281. [DOI] [PubMed] [Google Scholar]
- 58.Smith SJ, Rittinger K. Preparation of GTPases for structural and biophysical analysis. Methods Mol Biol. 2002;189:13–24. doi: 10.1385/1-59259-281-3:013. [DOI] [PubMed] [Google Scholar]
- 59.Burgess RJ, Zhang Z. Histone chaperones in nucleosome assembly and human disease. Nat Struct Mol Biol. 2013;20:14–22. doi: 10.1038/nsmb.2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muretta JM, Romenskaia I, Mastick CC. Insulin releases Glut4 from static storage compartments into cycling endosomes and increases the rate constant for Glut4 exocytosis. J Biol Chem. 2008;283:311–323. doi: 10.1074/jbc.M705756200. [DOI] [PubMed] [Google Scholar]
- 61.Parnas O, et al. A genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162:675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xiong X, Chen M, Lim WA, Zhao D, Qi LS. CRISPR/Cas9 for human genome engineering and disease research. Annu Rev Genomics Hum Genet. 2016;17:131–154. doi: 10.1146/annurev-genom-083115-022258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Davis EM, et al. Comparative haploid genetic screens reveal divergent pathways in the biogenesis and trafficking of glycophosphatidylinositol-anchored proteins. Cell Rep. 2015;11:1727–1736. doi: 10.1016/j.celrep.2015.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wiśniewski JR, Zougman A, Nagaraj N, Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 66.Erde J, Loo RR, Loo JA. Enhanced FASP (eFASP) to increase proteome coverage and sample recovery for quantitative proteomic experiments. J Proteome Res. 2014;13:1885–1895. doi: 10.1021/pr4010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cox J, et al. Andromeda: A peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 68.Yu H, et al. Extended synaptotagmins are Ca2+-dependent lipid transfer proteins at membrane contact sites. Proc Natl Acad Sci USA. 2016;113:4362–4367. doi: 10.1073/pnas.1517259113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu H, et al. Comparative studies of Munc18c and Munc18-1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc Natl Acad Sci USA. 2013;110:E3271–E3280. doi: 10.1073/pnas.1311232110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yu H, Rathore SS, Shen J. Synip arrests soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-dependent membrane fusion as a selective target membrane SNARE-binding inhibitor. J Biol Chem. 2013;288:18885–18893. doi: 10.1074/jbc.M113.465450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yu H, Rathore SS, Davis EM, Ouyang Y, Shen J. Doc2b promotes GLUT4 exocytosis by activating the SNARE-mediated fusion reaction in a calcium- and membrane bending-dependent manner. Mol Biol Cell. 2013;24:1176–1184. doi: 10.1091/mbc.E12-11-0810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rathore SS, et al. Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc Natl Acad Sci USA. 2010;107:22399–22406. doi: 10.1073/pnas.1012997108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rathore SS, Ghosh N, Ouyang Y, Shen J. Topological arrangement of the intracellular membrane fusion machinery. Mol Biol Cell. 2011;22:2612–2619. doi: 10.1091/mbc.E11-03-0190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen J, Rathore SS, Khandan L, Rothman JE. SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18-1 activation of membrane fusion. J Cell Biol. 2010;190:55–63. doi: 10.1083/jcb.201003148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.