Abstract
Glutamine synthetase is a ubiquitous central enzyme in nitrogen metabolism that is controlled by up to four regulatory mechanisms, including adenylylation of some or all of the twelve subunits by adenylyl transferase. It is considered a potential therapeutic target for the treatment of tuberculosis, being essential for the growth of Mycobacterium tuberculosis, and is found extracellularly only in the pathogenic Mycobacterium strains. Human glutamine synthetase is not regulated by the adenylylation mechanism, so the adenylylated form of bacterial glutamine synthetase is of particular interest. Previously published reports show that, when M. tuberculosis glutamine synthetase is expressed in Escherichia coli, the E. coli adenylyl transferase does not optimally adenylylate the M. tuberculosis glutamine synthetase. Here, we demonstrate the production of soluble adenylylated M. tuberulosis glutamine synthetase in E. coli by the co-expression of M. tuberculosis glutamine synthetase and M. tuberculosis adenylyl transferase. The differential inhibition of adenylylated M. tuberulosis glutamine synthetase and deadenylylated M. tuberulosis glutamine synthetase by ATP based scaffold inhibitors are reported. Compounds selected on the basis of their enzyme inhibition were also shown to inhibit M. tuberculosis in the BACTEC 460TB™ assay as well as the intracellular inhibition of M. tuberculosis in a mouse bone-marrow derived macrophage assay.
Introduction
Tuberculosis (TB) is a worldwide pandemic, caused by infection with the bacterium Mycobacterium tuberculosis. Although current treatment can be effective, existing drugs must be taken for at least six months to prevent relapse. Poor treatment compliance contributes directly to the emergence of multidrug- and extensively drug-resistant (MDR and XDR) strains of M. tuberculosis. New targets for drugs are therefore required. These new drugs should also simplify and shorten the treatment period, as well as reduce drug-drug interactions in patients co-infected with HIV. Drug discovery programmes for infectious diseases are normally focussed on pathogen proteins whose function is known to be essential to the bacterial cell, combined with a lack of mammalian homologues. One such potential drug target for TB is adenylylated glutamine synthetase (GS).
GS (EC 6.3.1.2) catalyzes the reversible conversion of l-glutamic acid, ATP and ammonia to l-glutamine, ADP and inorganic phosphate via a γ-glutamyl phosphate intermediate [1]. It is a central enzyme in nitrogen metabolism, and can be regulated by at least four different mechanisms: (a) adenylylation and deadenylylation of a conserved tyrosine residue, (b) conversion between a relaxed (inactive) and taut (active) state depending on the divalent metal cation present, (c) cumulative feedback inhibition by multiple end products of glutamine metabolism, and (d) repression and derepression of GS biosynthesis in response to nitrogen availability [1].
Three distinct forms of GS occur, with GS-I found only in bacteria (eubacteria) and archaea (archaebacteria) [2]. GS-II occurs only in eukaryotes, and soil-dwelling bacteria, while GS-III genes have been found only in a few bacterial species. Two significant prokaryotic GS-I sub-divisions exist: GS-Iα and GS-Iβ [3]. The GS-Iβ enzyme is regulated via the adenylylation/deadenylylation cascade, which does not occur in the GS-Iα or GS-II sub-divisions. M. tuberculosis and Escherichia coli GS are regulated in this manner, while the human homologue belongs to GS-II and is not subject to adenylylation, a difference that can be exploited by developing drugs that are only active against the adenylylated form of the enzyme.
The extent of adenylylation of the E. coli GS is regulated in response to the intracellular concentrations of 2-ketoglutarate and glutamine, via the reversible adenylylation of a tyrosine residue (Tyr397) in each subunit of GS [1, 4–8]. The presence of adenylylated GS predominates in a nitrogen-rich, carbon-limited media, while the deadenylylated form tends to predominate under conditions of nitrogen limitation [1, 4–15]. The regulation of the adenylylation state of GS is accomplished by three proteins: (1) uridylyltransferase/uridylyl-removing enzyme, (2) the signal transduction protein PII, and (3) adenylyltransferase or ATase. High intracellular concentrations of glutamine activate the uridylyl-removing enzyme, which causes the deuridylylation of PII. This interacts with the ATase, which then catalyses the adenylylation of the GS. A high intracellular 2-ketoglutarate concentration activates uridylyltransferase, which transfers UMP to each subunit of PII, forming PII-UMP. The PII-UMP interacts with the Atase, which in turn catalyses the removal of AMP from the GS. Research on the effect of glucose, ammonia and glutamic acid concentrations has shown that the adenylylation state of GS is a function of metabolic flux rather than absolute concentration only [10]. The activity of GS is therefore regulated by both the nature and the availability of the ammonia source [1,8]. The current view is that the level of GS activity is inversely related to the degree of adenylylation [reviewed in 1, 9, 10] and that adenylylated residues may be present on any number of subunits from zero to 12, depending on carbon and nitrogen availability [13, 16–21]. GS is therefore responsible for the assimilation of ammonia when the available ammonia in the environment is restricted, as well as for the formation of glutamine for the synthesis of protein and other nitrogen compounds. In ammonia-rich medium, the level of GS is low and GS functions primarily for the synthesis of glutamine.
A number of factors make GS a potential drug target in the fight against TB, including being considered essential for the survival of M. tuberculosis [22–25]. The GS inhibitor l-methionine-S,R-sulphoximine (MSO) inhibits growth of M. tuberculosis both in vitro and in vivo [22,23]. It is located extracellularly, a characteristic that is found only in the pathogenic mycobacteria such as M. tuberculosis and Mycobacterium bovis, and not with the non-pathogenic strains of Mycobacterium smegmatis and Mycobacterium phlei [21,22]. This location means a potential drug does not have to pass the M. tuberculosis cell wall barrier. It appears to play an important role in cell wall biosynthesis, in the form of a cell wall component found only in pathogenic mycobacteria: poly-l-glutamate/glutamine [26, 27].
M. tuberculosis GS (MtbGS) has previously been successfully expressed in heterologous systems including the non-pathogenic mycobacterial strain M. smegmatis and E. coli [28–30]. Mehta et al expressed M. tuberculosis GS in E. coli host strains that were deficient in either chromosomal GS, or both chromosomal GS and ATase [30]. They found that the E. coli ATase was inefficient in adenylylating the heterologous M. tuberculosis GS, with only ~25% of subunits being modified. A lack of E. coli ATase yielded completely deadenylylated M. tuberculosis GS. As a result no crystal structure exists for M. tuberculosis fully adenylylated GS [31]. A number of studies have been undertaken targeting MtbGS as a potential therapeutic target however, in none of these investigations was an attempt made to exploit the dichotomy between adenylylated and deadenylylated GS [32–39]. As outlined prokaryotic GS is regulated via a complex cascade that is based on the availability of NH4+ and glucose to the organism and the intracellular concentrations of 2-ketoglutarate and glutamine [1, 4–8]. This regulation results in the adenylylation or deadenylylation of the GS with a concomitant switch in the enzymes affinity from Mn2+ to Mg2+[1, 4–8, 30]. The adenylylation and the switch in metal ion specificity significantly impacts on the enzyme activity of GS and therefore probably impacts on the structure of the active site. It is proposed that this dichotomy may be exploitable in creating GS inhibitors that target only prokaryotic GS as only bacterial GS have regulation via the adenylylation/deadenylylation cascade.
Here, we describe the production of both the deadenylylated and adenylylated forms of MtbGS in E. coli. Deadenylylated MtbGS is produced by constitutive expression of MtbGS in an E. coli strain deficient in both E. coli GS and ATase activities, while adenylylated MtbGS is produced when co-expressed with an inducible M. tuberculosis ATase. Adenylylation was measured using the γ-glutamyl transferase assay, mass spectrometry and determination of phosphate content. IC50 values of the known GS inhibitors MSO and phosphinotricin (PhosT) were also determined. A battery of ATP scaffold compounds were identified and screened for their differential inhibitory effect on adenylylated MtbGS and deadeylylated MtbGS. Two of these compounds showed micromolar activity against MtbGS, acceptable activity against a cell-free and macrophage model of M. tuberculosis indicating their possible druggability. The two compounds identified here represent a good starting point for a hit-to-lead campaign to develop selective, druggable agents capable of selectively inhibiting the adenylated form of MtbGS in view of identifying novel agents against M. tuberculosis infection.
Experimental procedures
Plasmids and bacterial strains
E. coli JM109 (Promega Corporation) was used for cloning. Restriction endonucleases were purchased from Fermentas Life Sciences and Ex TaqTM DNA polymerase from TaKaRa Bio Inc.
M. tuberculosis glutamine synthetase glnA gene was PCR amplified from genomic DNA of M. tuberculosis H37Rv (ATCC 25618) using the oligonucleotide primers TB1: 5’-GATGGATCCACCCGATAACCAG-3’ and TB2: 5’-GATGGATCCTCGAAAAACCTCG-3’. The amplified DNA was then digested with BamHI (sites underlined in primer sequences) and ligated with similarly digested pBluescript SKII+, generating plasmid pTBSK, with the glnA gene under contol of the constitutive T3 promoter.
The M. tuberculosis H37Rv adenylyl transferase glnE gene was PCR amplified from genomic DNA using the oligonucleotide primers TBglnE-8: 5’-TAGCATATGGTCGTGACCAAAC-3’ and TBglnE-9: 5’-CAGGATCCTTAACTCCCGAACAC-3’. The amplified DNA was then co-digested with NdeI and BamHI (sites underlined in primer sequence) and ligated with NdeI and BglII digested pCDFDuet-1 (Novagen) to construct pTBGlnE, with glnE downstream of the IPTG-inducible T7lac promoter.
For MtbGS expression, the chromosomal adenylyl transferase glnE gene of the glutamine synthetase auxotroph YMC11 was deleted using the Quick and Easy E. coli Gene Deletion Kit (Gene Bridges, GmbH) to create the strain YMC11E [40]. The Novagen λDE3 Lysogenization Kit was used to create YMC11E(DE3).
Production of adenylylated and deadenylylated MtbGS
Deadenylylated MtbGS was expressed in YMC11E containing pTBSK. This strain was inoculated in 50 ml M9ZB medium (1% w/v N-Z-Amine A; 85 mM NaCl) containing M9 Salts (22 mM Na2HPO4; 22 mM KH2PO4; 18 mM NH4Cl; 8.5 mM NaCl) and supplemented with 100 μg.ml-1 ampicillin, 1mM MgSO4 and 4% (w/v) glucose. The inoculum was grown at 28°C for 16 hours with shaking at 220 rpm. Subsequently, 5 ml of the culture was transferred to 250 ml of the same medium, which was grown at 28°C for 16 hours. The cultures were harvested by centrifugation for 10 min at 16,300g (4°C) and the bacterial pellet used for MtbGS purification.
Adenylylated MtbGS was produced using YMC11E(DE3) containing both pTBSK and pTBglnE. This strain was inoculated into 50 ml of the same medium as above, including 50 μg.ml-1 streptomycin. The cultures were incubated at 33°C for 16 hours with shaking at 220 rpm. Thereafter, 5 ml was transferred to 250 ml of the same medium and grown at 33°C for a further 8 hours. M. tuberculosis adenylyl transferase expression was then induced by the addition 1 mM IPTG. After a further incubation at 33°C for 16 hours with shaking at 220 rpm, the cultures were harvested by centrifugation for 10 min at 16,300g (4°C) and the bacterial pellet used for MtbGS purification.
Purification of adenylylated and deadenylylated MtbGS
The purification of adenylylated and deadenylylated MtbGS from a cell free extract was carried out in a three stage process; a streptomycin sulphate precipitation to remove contaminating nucleic acid, an anion-exchange chromatography stage which removed the major fraction of contaminating proteins and an AMP-Sepharose affinity chromatography”polishing” stage which produced the purified enzyme (See Supporting Information for detail, S1 File).
Production and purification of adenylylated and deadenylylated E. coli GS
For comparison purposes, adenylylated and deadenylylated E. coli GS was also heterologously expressed and purified. Recombinant deadenylylated E. coli GS was produced in an E. coli strain lacking chromosomal glnA (GS) and glnE (ATase) genes. Adenylylated E. coli GS was produced in a strain lacking chromosomal glnA (GS) and glnD (uridilyl transferase) genes (See Supplementary Information for detail, S1 File) [41, 42]. Sodium dodecyl-sulphate polyacrylamide gel electrophoresis (SDS-PAGE) was used to analyse the molecular mass and purity of the isolated enzymes [43]. Protein concentrations were determined by using the Quant-ITTM Protein Assay Kit (Invitrogen, USA) used in conjunction with the QUBITTM fluorometer.
Determination of degree of adenylylation of purified GS
Mass spectrometry
A 40 μl aliquot of purified GS in 10 mM imidazole pH 7.0 was loaded on an OPTI-LYNX C4 trap cartridge at 100 μl/min using 2% ACN (Acetonitrile)/0.1% FA (Formic Acid). Samples were eluted using linear acetonitrile gradient (2–90% ACN/5% FA) in 5min and TOF-MS spectra, in the range 700–1700 mz, collected using QSTRA-Elite mass spectrometer with TurboIon source installed. The multiply charged series of TBGS was deconvoluted by the Bayesina Protein Recostruct tool of Bioanalyst QS 2.0 using a mass range of 40–70kD and signal to noise threshold of 3.
Hydrolysis of GS for determination of phosphate content
The relative phosphate content of the purified GS proteins was determined by hydrolysis to release the phosphate [44]. Purified GS (4 to 8 nmol in 1 ml) was digested by the addition of 4 M HCl (1 ml), and the mixture was evaporated to dryness. The residue was resuspended in 2 M HCl (1 ml) plus 20 μl of H2O2 (30% v/v), and the mixture was again evaporated to dryness. Five sequential additions of H2O2 (200 μl) with evaporation to dryness were used to complete the hydrolysis. The residue was resuspended in Milli-Q water (1 ml), and phosphate content was determined by using the BioVision Phosphate colorimetric assay kit as per supplier’s manual.
γ-Glutamyl transferase assay
The GS γ-glutamyl transferase enzyme activity was used to calculate the degree of adenylylation of the purified GS using the standard method as outlined by Shapiro and Stadtman [41].
Dose response assays: MtbGS inhibitor screening and specific activity determination
Standard GS inhibitors methionine sulphoximine (MSO), phosphinotricin (PhosT) and a battery of 213 compounds were tested for inhibition of the adenylylated and dedenylylated forms of MtbGS. Serial 4-fold dilutions of MSO and PhosT were prepared in DMSO. Two dilution series were set up, one starting at 1 mM and the other at 2 mM. Eight dilutions were performed in DMSO, to produce a concentration range of 1 mM– 61 nM or 2 mM—122 nM. Twenty μl of each dilution was distributed into duplicate wells of the reaction plate. Each well received 164 μl MtbGS in 50 mM HEPES, 4 mM NH4Cl, 1.8 mM MnCl2 or MgCl2. For adenylylated MtbGS, HEPES pH 6.95 and MnCl2 were used, while for deadenylylated MtbGS, HEPES pH 7.15 and MgCl2 were required. The plates were incubated at 37°C for 2 hours, after which the reactions were initiated by adding sodium glutamate and ATP to final concentrations of 4 mM and 0.8 mM respectively, with a final reaction volume of 200 μl. After further 2 hour incubation, the reactions were terminated by the addition of 1 μl 50% trichloro acetic acid to each well. Blank wells (no enzyme) were also prepared for each individual compound. In addition, each plate contained control wells (10% DMSO without inhibitor) and blank control wells (10% DMSO, no inhibitor, no enzyme). After termination of the reactions, ADP levels in each well were determined by HPLC.
HPLC based enzyme assay
The enzyme activity of GS was determined by measuring the ADP concentration by HPLC [45]. Samples were analysed on a Hewlett Packard series 1100 HPLC fitted with a Luna 5μm C18 column. Each sample was automatically injected (2 μl) and separated with a mobile phase containing 51 mM KH2PO4, PIC A Low UV Reagent, 25% (v/v) acetonitrile. An AMP, ADP and ATP standard was used to calibrate the HPLC and the concentration of ADP in each sample was determined by the area under the curve using Agilent Technologies ChemStation software. The ADP values in the blank wells were subtracted from enzyme-containing wells, and percentage enzyme activity in each well calculated relative to the average net ADP values of the control wells without inhibitor. The enzyme activity was expressed as a percentage of the maximum enzyme activity in the absence of any inhibitor.
HeLa cell cytotoxicity assay
HeLa cells (Human Negroid cervix epitheloid adenocarcinoma, ECACC) were routinely maintained as monolayer cell cultures in Eagle's minimal essential medium (EMEM) containing 5% fetal bovine serum, 2 mM L-glutamine and 50 μg/ml gentamicin at 37°C in a 5% CO2 incubator. To perform the cytotoxicity assay, the cells (3–19 passages) were used to inoculate 96-well microtiter plates at plating densities of 7000 cells/well. After incubating for 24 hours, the culture medium was replaced with medium containing serial dilutions of the experimental compounds. Each dilution series consisted of 8 x 3-fold serial dilutions, spanning the final concentration range 100–0.05 μM. Triplicate wells were used for each concentration point (n = 3). The dilutions were prepared from compound stocks of 10 mM in DMSO, thus the final DMSO content in the highest compound concentration wells was 1%. Control wells consisted of cells incubated in medium with 1% DMSO, while blank wells contained medium without cells. Emetine was used as a positive control drug standard. The plates were incubated for 48 hours after addition of the compounds. The cellular protein present after the incubation period was fixed to the bottom of each well with cold 50% TCA, washed in tap water and stained with 0.4% sulforhodamine B (SRB) in 1% acetic acid. Unbound dye was removed by washing with 1% acetic acid, after which protein-bound dye was solubilised with 10 mM Tris base and transferred to a duplicate 96-well plate. Optical density was measured at 540 nm using a Tecan Infinite F500 multiwell spectrophotometer. OD540 values of the blank wells were subtracted from the readings obtained for all the other wells, and percentage cell viability at each test compound concentration calculated relative to the untreated (DMSO alone) control wells. Percentage viability was plotted against Log (compound concentration) and the IC50 for each compound calculated from fitted non-linear regression dose-response curves using GraphPad Prism software.
Testing of compounds against M.tb strains in-vito and ex-vivo using the BACTEC 460TB™ assay
Bacterial strains
All M.tb strains used were from a strain bank kept in the Division Molecular Biology and Human Genetics of of Stellenbosch University. M.tb H37Rv reference strain (ATCC 25618) and a clinical isolate of M.tb (Beijing220) were used to evaluate compounds for anti-tuberculosis activity. The H37Rv strain was sensitive to the breakpoint concentrations (approximately 10x higher than their minimal inhibitory concentrations (MIC) of isoniazid (0.25ug/ml), ethambutol (9.4ug/ml), and rifampicin (2.0ug/ml) [46]. However, the Beijing220 clinical isolate was resistant to isoniazid and rifampicin [47, 48].
Test compounds
The compounds identified were used in the BACTEC 460TB™ assay [49]. The identified compounds were dissolved and diluted with 100% DMSO.
Bacterial selection
All mycobacterial colonies were cultured and selected from Lowenstein-Jensen slant [50] cultures followed by culture in Middlebrook 7H9 mycobacterial growth medium supplemented with OADC (0.005% v/v oleic acid (Merck), 0.5% w/v BSA Fraction V, 0.2% v/v glucose, 0.02% v/v catalase (Merck), w/v 0.085% NaCl). Cultures were stained by acid-fast staining (Ziehl-Neelsen) to control for contamination.
BACTEC 460TB™ system determination of mycobacterial growth
The BACTEC 460TB™ system has been devised to monitor mycobacterial growth of the slow growing species. The bacteria are grown on a radioactive substrate and the radioactive carbon dioxide produced is directly proportional to the mycobacterial growth rate. Read-out values are expressed as growth index (GI). M.tb reference strain H37Rv was cultured in 7H9 mycobacterial medium (Difco) enriched with ADC (0.5% w/v BSA Fraction V, 0.2% v/v glucose, 0.015% v/v catalase (Merck) with continuous stirring at 37°C. When cultures reached a density of approximately 0.16 at OD600 (one McFarland), 0.1 ml was inoculated into a BACTEC 12B medium vial. These primary cultures were incubated at 37°C until a growth index of 500 (± 50) was reached. These primary cultures were used for drug testing of known and unknown compounds. Compounds, resuspended in DMSO, were sterilized through a 13 mm organic solvent resistant syringe filter with 0.22 micron pore size (Millex-LG). Undiluted and sequentially diluted samples were tested for growth inhibitory activity. 0.1 ml of primary culture and 0.1 ml drug compound were added to a BACTEC 460TB™ vial, the vials incubated at 37°C, and the growth monitored every 24 hours. Controls included cultures with and without compound solvent. GI readings were continued until the controls reached the maximum GI value of or below 999. Control GI values between 50 and 800 are normally used to evaluate the efficacy of compounds with possible anti-tuberculosis activity.
Testing of screening hits against M.tb in a macrophage assay
M.tb cultures
M.tb clinical MDR strain (Beijing220) was cultured in 7H9 broth supplemented with 10% oleic acid-albumin-dextrose catalase (OADC, Difco, BD Biosciences, Mountain View, CA, USA) and 2% glycerol and 0.05% Tween 80 (Glickman et. al., 2000). Liquid cultures were grown for up to 3 weeks and stored at -80°C in 1ml aliquots with 15% glycerol. Clumps were eliminated by 30 passages through a needle (26-gauge 3/8; 0.45 x 10 for intradermal injection; BD Biosciences, USA). Before infection, viability of mycobacteria was evaluated by the propidium iodide exclusion method to ensure >90% viability. Contamination was checked by the Ziehl-Neelsen stain. The required amount of mycobacteria was spun down at 16 300xg for 5 min. The supernatant was removed and the bacteria resuspended in PBS (phosphate buffered saline) and passaged again 20 times and then used for macrophage infection.
Manipulation of mouse bone marrow-derived macrophages (MBMM)
This research study was approved by the Stellenbosch University Animal Ethics committee on Animal Care and Use and complies with the South African Animal Protection Act (Act no 71, 1962). Animal Ethics No. SU-ACUD14-00041. Mice were supplied by the Animal House of the University of Stellenbosch, South Africa. Mice were cared for in accordance with ethical laws on animal manipulation. Untreated mice were killed by cervical dislocation inside the Animal House 10 minutes after being received. Bone marrow cells were isolated from the femurs and tibias and differentiated into macrophages as previously described [51]. Bone marrow cells were obtained from femurs of 6–8 week-old C57BL/6 female mice and seeded into 24-well tissue culture plates. The culture medium was RPMI-1640 (Sigma, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 10% L-929 cell conditioned medium (a source of colony stimulating factor-1) with no antibiotics. Incubation was performed at 37°C, 5% CO2. At 5 days after seeding, adherent cells were washed twice with RPMI and re-fed with complete medium. Medium was then renewed every second day.
Macrophage infection and harvesting of TB
Frozen aliquots of M.tb (Beijing220) were removed from the -80°C freezer where they were kept for long-term storage and thawed and processed as indicated. Infection of the 7-Day old MBMM in 24-well plates was performed in triplicates with 100 μl of bacterial suspension relative to a multiplicity of infection (MOI) of 2, and incubated for 4 hrs at 37°C, 5% CO2. After the 4 hr incubation period, non-ingested M.tb (Beijing220) was removed by washing 4 times with ice-cold PBS. Fresh RPMI medium (1 ml) was added and again on day 2. At day 4 post infection the test compounds were added at the desired concentrations between 10 μM and 100 μM. Included were uninfected, infected (no drug) and an isoniazid drug controls. On day 2 post drug intervention the macrophages were washed 3 times with cold PBS (4°C) and then the bacteria harvested by adding 1 ml 0.025% SDS for 5 min to lyse the adherent macrophages. The contents of each well were placed into separate Eppendorf tubes and the bacteria were pelleted at 13000 rpm for 5 min. The bacteria was resuspended in 100 μl 7H9 Middlebrook medium and then inoculated into BACTEC 460TB™ vials. Growth was monitored over time until the BACTEC 460TB™ readings of the infected (no drug) control reached about 500. The percentage growth inhibition for each drug compound was determined by relating the linear growth unit readings to the infected (no drug) control.
Results
Production and purification of adenylylated and deadenylylated MtbGS
Expression strain E. coli YMC11E was constructed by deleting the chromosomal glnE gene in the GS auxotroph YMC11. For induction of M. tuberculosis ATase expression from pTBGlnE, the strain was lysogenised with λDE3 to create YMC11E(DE3).
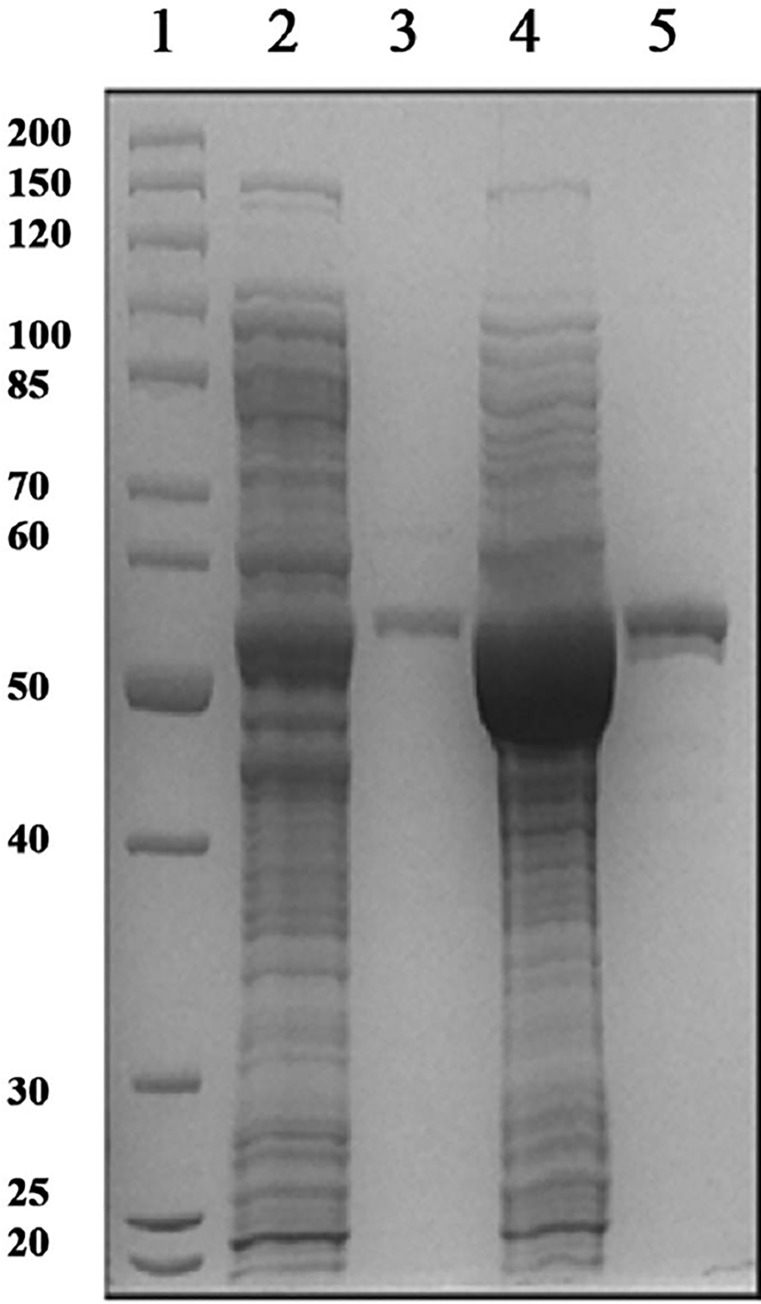
Deadenylylated MtbGS was expressed from pTBSK in E. coli YMC11E. Adenylylated MtbGS was produced in YMC11E(DE3) via co-expression of MtbGS in pTBSK and M. tuberculosis ATase in pTBGlnE. MtbGS was expressed constitutively, while ATase production was induced by the addition of IPTG. The two plasmids used contain compatible origins of replication and can thus be maintained stably in the same strain. Fig 1 illustrates a SDS-PAGE of the cell-free E. coli soluble extracts, as well as the purified adenylylated and deadenylylated forms of MtbGS. The average protein concentrations from three purifications of adenylylated and deadenylylated MtbGS are 120 ± 28 and 145 ± 49 μg.ml-1, respectively. The estimated size is in agreement with the theoretical ~53.4 kDa per subunit of the MtbGS dodecamer.
Fig 1. 12% SDS-PAGE gel of cell-free E. coli extracts, as well as purified adenylylated and deadenylylated MtbGS.
Lane 1: molecular weight marker with size in kDa indicated on left. Lane 2: Adenylylated MtbGS cell-free extract. Lane 3: Adenylylated MtbGS purified protein. Lane 4: Deadenylylated MtbGS cell-free extract. Lane 5: Deadenylylated MtbGS purified protein.
Determination of the adenylylation state of adenylylated and deadenylylated MtbGS
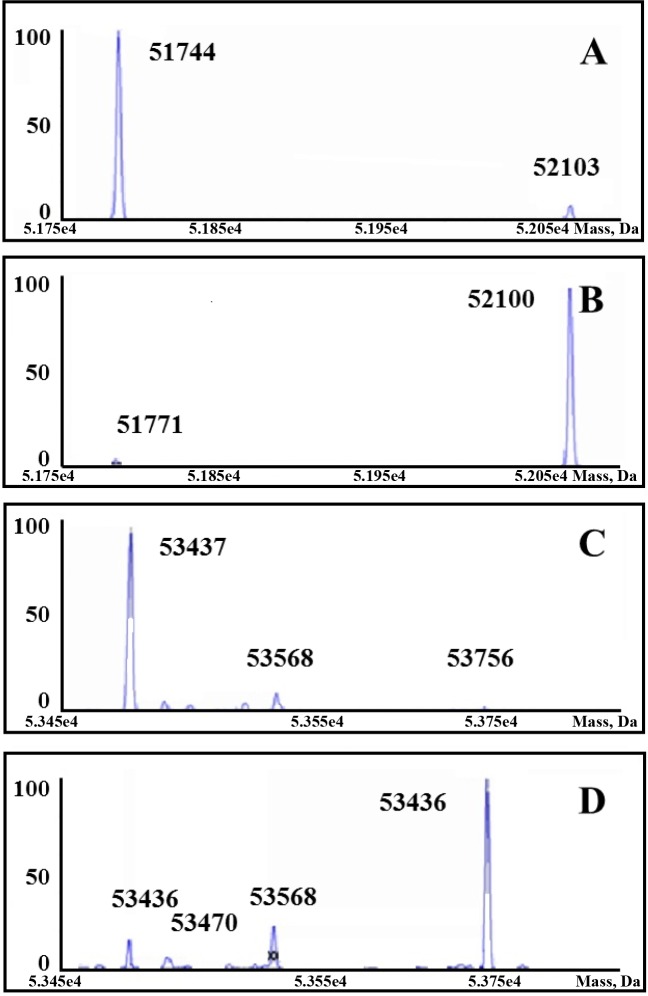
MS analysis of deadenylylated MtbGS, yielded a single peak with a calculated mass of 53,438.00 Da (Fig 2), in agreement with the theoretical mass of 53.4 kDa. Deadenylylated E. coli GS had a calculated mass of 51,773.00 Da (theoretical mass = 51.7 kDa). Adenylylated E. coli GS was calculated to be 52,102.00 Da (theoretical mass = 52.1 kDa), with a single peak indicating 100% adenylylation. The 330 Da adenyl moiety accounts for the mass difference between the adenylylated and deadenylylated forms of each enzyme. Adenylylated MtbGS would not ionise properly, therefore the spectra obtained (Fig 2) that shows 85% adenylylation, could therefore be potentially higher than 85%. In order to confirm adenylylation of MtbGS, other methods were also pursued.
Fig 2. Mass spectra of M. tuberculosis and E. coli GS purified proteins.
The calculated mass and presence of the major peak is shown within each spectrum (A) deadenylylated E. coli GS, (B) adenylylated E. coli GS, (C) deadenylylated M. tuberculosis GS, (D) adenylylated M. tuberculosis GS.
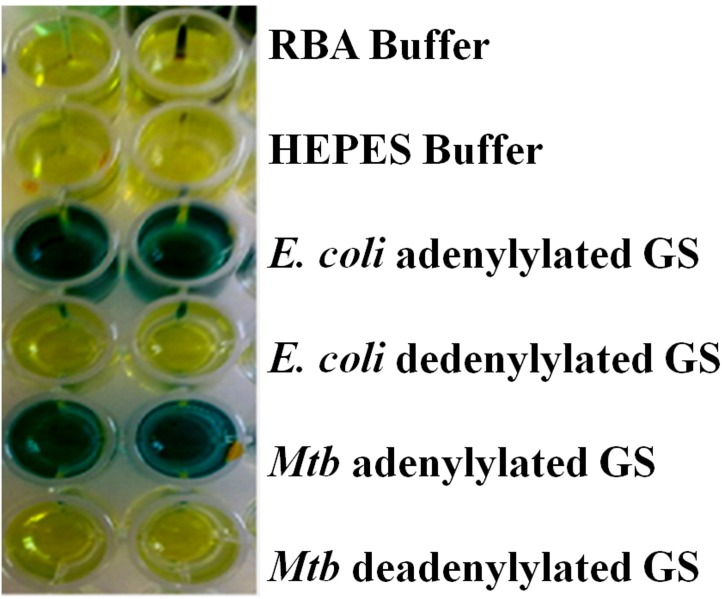
The hydrolysis of GS for determination of phosphate content was carried out on the purified enzymes to confirm the degree of adenylylation (Fig 3). Each adenyl moiety contains 1 phosphate, and 1 μM of GS (containing 12 subunits) should contain 12 μM of phosphate, if each subunit is adenylylated. The result obtained for the adenylylated MtbGS enzyme was the formation of 0.93 μM phosphate produced per μM GS active site, i.e. 93% adenylylated. In the case of the deadenylylated enzyme there was no formation of phosphate. The results obtained for E. coli GS were 0.95 μM and 0 μM for adenylylated and deadenylylated GS, respectively.
Fig 3. Hydrolysis of M. tuberculosis and E. coli GS purified protein (adenylylated and deadenylylated) for phosphate concentration determination.
A blue colour indicates the presence of free phosphate.
The γ-glutamyl transferase enzyme assay was carried out with purified GS to: i) confirm the functionality of the purified MtbGS enzymes, and ii) assess the degree of adenylylation of the purified enzyme. At a specific pH, the total enzyme activity of GS (both adenylylated and deadenylylated) occurs in the presence of Mn2+. At the same pH in the presence of Mn2+ and an excess Mg2+, only the deadenylylated component of the enzyme activity is measured. The resultant ratio is then used to calculate the degree of adenylylation of the enzyme. After three purifications, the typical degree of adenylylation for deadenylylated MtbGS is 3 ± 2%, while the adenylylated form is 68 ± 4% adenylylated. For comparison purposes, adenylylated and deadenylylated E. coli GS yielded results of 0% and 87 ± 3% adenylylation, respectively.
A summary of the adenylylation state of the M. tuberculosis GS and E. coli GS as determined by the different analysis techniques is out lined in Table 1.
Table 1. Summary of the adenylylation states of M. tuberculosis and E. coli GS determined by different methods.
(1) mass spectroscopy, (2) hydrolysis and phosphate concentration determination and (3) γ-glutamyl transferase assay.
| M. tuberculosis | E. coli | |||
|---|---|---|---|---|
| Deadenylylated | Adenylylated | Deadenylylated | Adenylylated | |
| MS | 0% | 85% | 7% | 100% |
| Phosphate Concentration | 0 μM | 0.93 μM | 0 μM | 0.95 μM |
| γ-glutamyl transferase | 3 ± 2% | 68 ± 4% | 0 | 87 ± 3% |
Specific activity, inhibitor studies, dose-response assays and M.tb BACTEC 460TB™ assays
The specific activity of the adenylylated and deadenylylated MtbGS were determined. The conversion of ATP to ADP was measured by HPLC and the typical specific activity of adenylylated and deadenylylated forms of M. tuberculosis GS are 0.010 and 0.015 μmol ADP/min/mg protein, respectively.
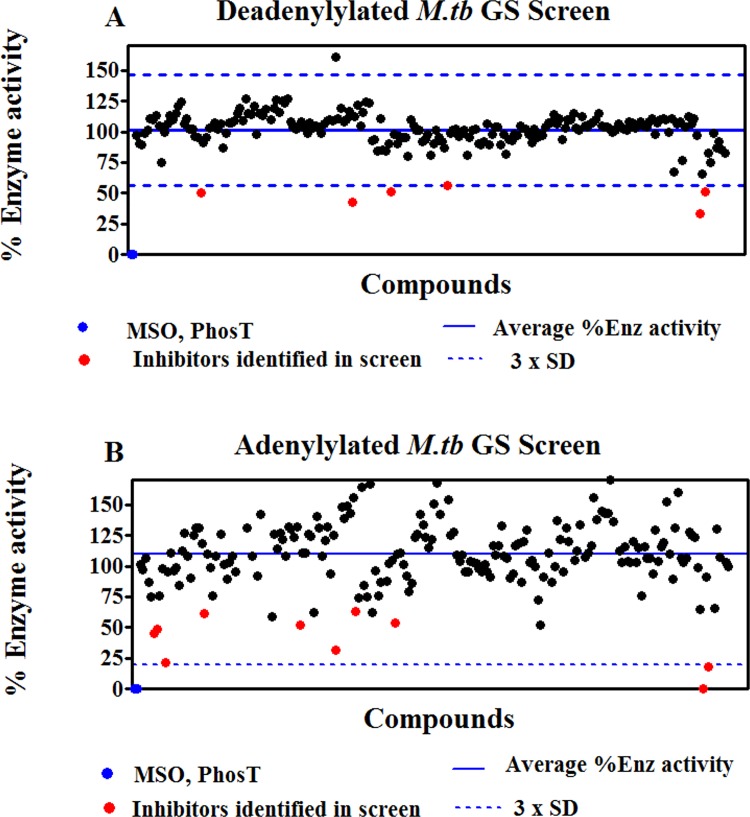
Inhibitor studies with a library of 213 ATP scaffold based inhibitors were carried out to reflect the differential inhibition of adenylated and deadenylated MtbGS. These assays were also performed using the pre-incubation protocol, where the inhibitors were allowed to interact with either adenylated or deadenylated MtbGS before the addition of substrates sodium glutamate and ATP. Based on the results illustrated with the deadenylylated enzyme in Fig 4A, six candidate inhibitors (indicated by the red data points) showed promising inhibitory activities. With the adenylylated enzyme, the % enzyme activities in the presence of the individual compounds varied widely, producing a pronounced scatter of values around the mean and consequently a very wide confidence interval (Fig 4B) nevertheless ten inhibitors (indicated by the red data points) showed promising inhibitory activities with one inhibitor yielding an inhibition of 99%. The scatter associated with the data when using the adenylylated GS is probably associated with inter subunit allosteric regulation of adenylylated GS [42]. Some compounds therefore allosterically activate the enzyme.
Fig 4. M. tuberculosis GS enzyme screen using the pre-incubation protocol.
The residual percentage adenylated (B) and deadenylated (A) GS enzyme activities after incubation with 10μM of the individual compounds are shown, along with horizontal lines depicting the average % enzyme activity obtained with all compounds, as well as an indication of the confidence interval of the results, expressed as the average activity ± 3 x standard deviation (SD). Percentage activity obtained with methionine sulfoximine (MSO) and phosphinothricin (PhosT) is shown in blue. Candidate inhibitors previously identified in the adenylated GS screen are shown as red datapoints. Additional compounds selected for further evaluation are shown as orange datapoints.
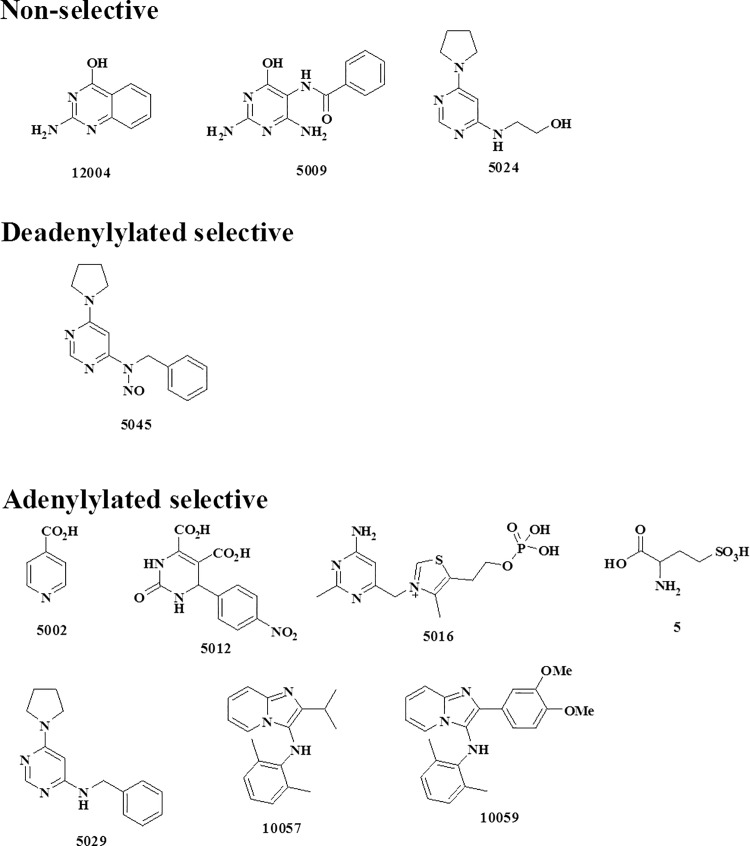
Fixed concentration evaluation of eleven candidate inhibitors were tested on both adenylated and deadenylated MtbGS. Based on the results indicated in Table 2 there is a clear indication of differential inhibition between the two forms of MtbGS. One compound (5045) displayed significant selectivity for the deadenylylated MtbGS, while seven compounds suggested specificity for the adenylylated MtbGS and three appeared relatively non-selective. The chemical structures of these candidate inhibitors are indicated in Fig 5.
Table 2. Fixed concentration evaluation of 11 candidate inhibitors in M.tuberculosis GS enzyme assays using the pre-incubation protocol.
The percentage enzyme inhibition obtained with the compounds (10μM final concentration) in 4 separate screens is shown, along with the average inhibition for each compound over the 4 screens and the corresponding standard deviation (SD). The colour coding of the highlighted table cells is clarified in the bottom legend. Compounds were defined as selective for the adenylylated or deadenylylated form of the enzyme based on at least a 30% difference in inhibition.
| Adenylated Mtb GS (% inhibition) | Deadenylated Mtb GS (% inhibition) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound (10μM) | Screen 1 | Screen 2 | Screen 3 | Screen 4 | AVG | SD | Screen 1 | Screen 2 | Screen 3 | Screen 4 | AVG | SD |
| PhosT | 100 | 95 | 97 | 100 | 98 | 2 | 100 | 98 | 100 | 100 | 100 | 1 |
| MSO | 100 | 100 | 96 | 100 | 99 | 2 | 100 | 100 | 99 | 100 | 100 | 1 |
| 5 | 48 | 33 | 40 | 29 | 38 | 8 | 0 | 0 | 6 | 0 | 2 | 3 |
| 5002 | 68 | 40 | 38 | 35 | 45 | 15 | 0 | 10 | 8 | 14 | 8 | 6 |
| 5009 | 37 | 50 | 50 | 22 | 40 | 13 | 57 | 41 | 44 | 44 | 47 | 7 |
| 5012 | 55 | 31 | 32 | 42 | 40 | 11 | 14 | 16 | 4 | 24 | 15 | 8 |
| 5016 | 51 | 43 | 46 | 30 | 43 | 9 | 7 | 19 | 3 | 13 | 11 | 7 |
| 5024 | 46 | 48 | 43 | 36 | 43 | 5 | 49 | 44 | 52 | 44 | 47 | 4 |
| 5029 | 79 | 46 | 56 | 60 | 60 | 14 | 5 | 8 | 7 | 9 | 7 | 2 |
| 5045 | 0 | 0 | 0 | 0 | 0 | 0 | 44 | 45 | 54 | 48 | 48 | 5 |
| 10057 | 100 | 99 | 98 | 100 | 99 | 1 | 67 | 62 | 47 | 58 | 59 | 9 |
| 10059 | 82 | 99 | 82 | 100 | 91 | 10 | 49 | 48 | 36 | 43 | 44 | 6 |
| 12004 | 39 | 32 | 33 | 30 | 34 | 4 | 50 | 43 | 38 | 38 | 42 | 6 |
| Non-selective | ||||||||||||
| Adenylylated | ||||||||||||
| Deadenylylated | ||||||||||||
Fig 5. Chemical structures of the 11 synthesized ATP scaffold based inhibitors which shown promising inhibition of M. tuberculosis GS enzymes.
They are divided into three distinct groups (1) non-selective (2) Deadenylylated selective and (3) Adenylylated selective.
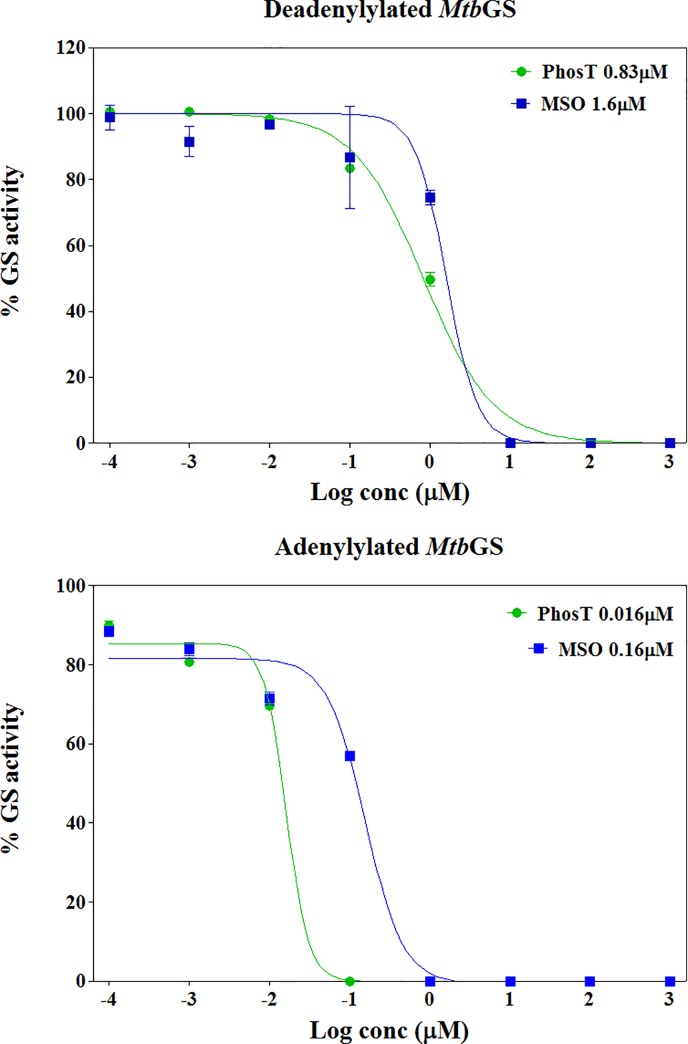
Dose-response assays were carried out to obtain a more accurate reflection of the activity of adenylated and deadenylated MtbGS. These assays were performed on the 2 forms of the enzyme to derive compound IC50 (50% inhibitory concentration) values, using the standard GS inhibitors MSO and PhosT. Dose-response assays were performed using a pre-incubation protocol, allowing the inhibitor to interact with the enzyme before addition of substrates sodium glutamate and ATP. Based on the results illustrated in Fig 6, IC50 values derived from the dose-response curves suggested, firstly, that adenylylated MtbGS is more sensitive to inhibition by the standard inhibitors than the deadenylylated form of the enzyme and, secondly, that PhosT is a more potent inhibitor of the Mtb enzymes than MSO. Compounds 10057 and 10059 both gave IC50 concentrations less than 100 μM. In duplicate assays run on different days in triplicate the IC50 concentrations obtained were 3.51 and 15.6 μM and 13.2 and 17.1 μM, for the adenylylated enzyme and deadenylylated enzyme, respectively.
Fig 6. Dose-response assays for MSO and PhosT standard inhibitors.
Percentage GS activity was plotted against log (compound concentration) and sigmoidal dose-response curves fitted to the data points using non-linear regression analysis. The curves were used to derive the IC50 values for the inhibitors (displayed in the top right legend).
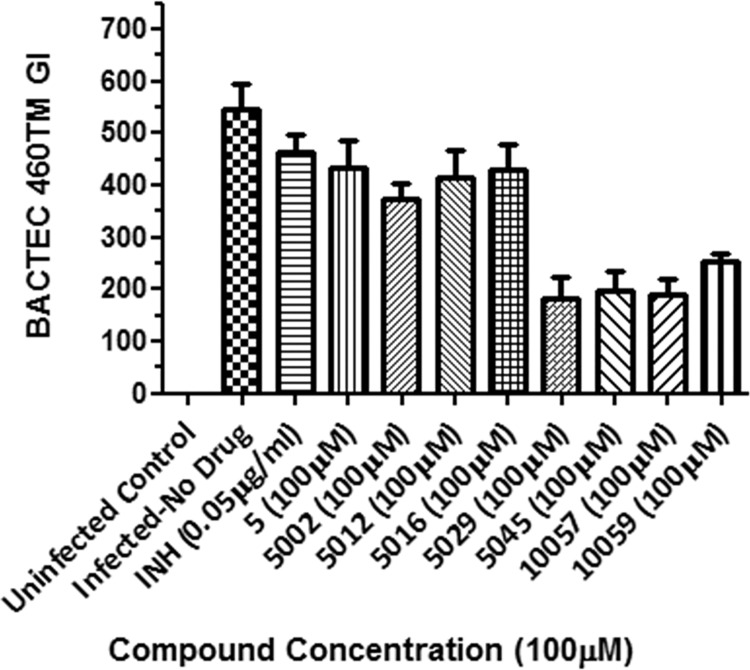
M.tb BACTEC 460TB™ assays were carried out on compounds identified following the primary screening for enzymatic activity against adenylylated and deadenylylated M.tb GS. Compounds identified (see Fig 5) were tested for activity against M.tb H37Rv reference strain for antibacterial activity. Growth inhibition was calculated as a percentage of the growth index (GI) at day 6 of incubation where the GI of 1.0% of the control culture equalled 50 GI units. Of the compounds directed against adenylylated GS (Fig 5), compound 10057 showed 53% (+/- 12.7) growth inhibition at 100 μM and also compound 5029 gave 66% (+/- 2.8) inhibition at 100uM against H37Rv on day 6 of incubation relative to the control cultures. However, the growth rate (ΔGI = GI at day+1 minus GI at day) in both cases continuously increased over the period of incubation and past day 6 of incubation indicating that bacterial doubling is taking place although at a slower rate. Most the compounds (5024, 12004, 5009, 5012, 5002, and 5016) showed no activity against M.tb H37Rv at 100 μM final concentration. Compound 5045, designed to be directed against the de-adenylylated GS (Fig 5) showed an inhibition of 98% (+/- 8.8) growth of the H37Rv mycobacterial cultures.
The 8 selective compounds identified in the enzyme assays namely compounds that were de-adenylylated selective (5, 5002, 5012, 5016, 5029, 10057, 10059) and deadenylylated selective (5045), were tested in mouse bone-marrow derived macrophages infected with a clinical multidrug-resistant strain actively spreading in the Western Cape region of South Africa (Johnson et. al., 2010; Leisching et. al., 2016). Only compounds 5029, 5045, 10057 and 10059 showed activity at a concentration of 100 μM (see Fig 7). The percentage killing exhibited by compounds 5029, 5045, 10057 and 10059 were 67%, 64%, 65% and 54% respectively at 100 μM.
Fig 7. Intracellular survival of M.tb (Beijing220) in mouse bone-marrow derived macrophages after 4-day post-infection period followed by intervention on D4 PI with different compounds, MOI 2:1, 4 day incubation at 37°C, 5% CO2.
Cells were sacrificed on Day 2 Post Drug Intervention. Only Drugs 5029, 5045, 10057 and 10059 showed moderate activity. Where not shown, error bars fall within symbols.
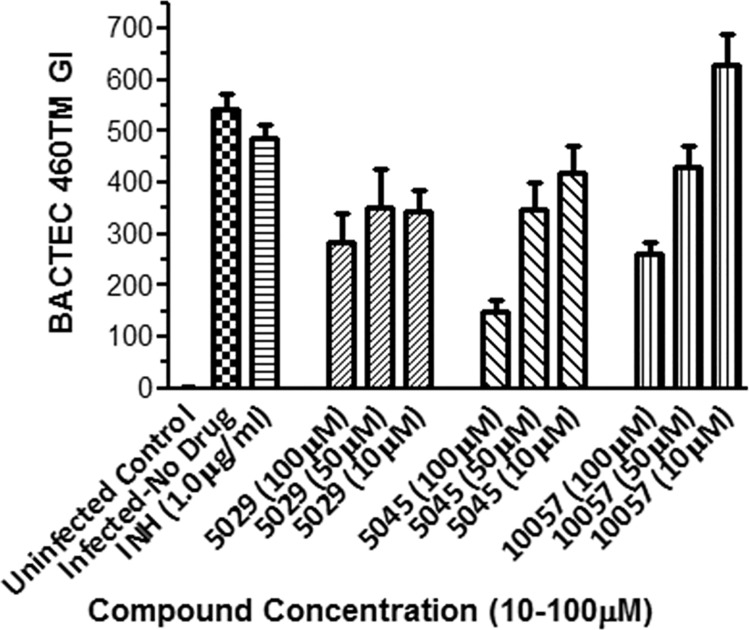
The most active compounds 5029, 5045 and 10057 were tested further in macrophages at concentrations 10 μM, 50 μM and 100 μM for 2 days (Fig 8). In this experiment compound 5045 showed the best activity (73% killing) at 100 μM. The growth inhibitory effect of compound 5045 correlated well with the high inhibition observed in the extracellular BACTEC assay against M.tb H37Rv (98% inhibition at 100μM).
Fig 8. Intracellular survival of M.tb (Beijing220) in mouse bone-marrow derived macrophages after 4-day post-infection period followed by intervention on D4 PI with different compounds, MOI 2:1, 4 day incubation at 37°C, 5% CO2.
Cells were sacrificed on Day 2 Post Drug Intervention. Drug 5045 showed the best activity (73% killing) at 100 μM. Where not shown, error bars fall within symbols.
Discussion
M. tuberculosis GS is a potentially valuable therapeutic target for TB drug intervention. Its regulation via adenylylation of a tyrosine residue on each subunit makes it distinct from the human form of the enzyme. Prokaryotic GS is regulated via a complex cascade that is based on the availability of NH4+ and glucose to the organism and the intracellular concentrations of 2-ketoglutarate and glutamine [1, 4–8]. This regulation results in the adenylylation or deadenylylation of the GS with a concomitant switch in the enzymes affinity from Mn2+ to Mg2+[1, 4–8, 30]. The coordination chemistry of Mn2+ to Mg2+ are significantly different enough to change the shape of the active site of the GS as is the effect of adenylylation. No crystal structure exits for the fully adenylylated form of the enzyme [31]. A number of studies have been undertaken targeting MtbGS as a potential therapeutic target however, in none of these investigations was an attempt made to exploit this dichotomy that exists between adenylylated and deadenylylated GS [32–38]. Previous reports of heterologous expression of MtbGS in E. coli have shown that the endogenous ATase activity of E. coli does not adenylylate MtbGS sufficiently, with only 25% of the MtbGS subunits produced displaying adenylylation [39]. The use of this expression system was therefore not considered optimal for the expression of adenylylated MtbGS for further study.
Here, we have described an E. coli production system, lacking endogenous GS and ATase activity, which utilises the co-expression of the M. tuberculosis ATase with MtbGS. Each gene was provided on a separate plasmid, the glnA gene on a pBluescript SKII+ plasmid with the ColE1 origin of replication, and the glnE gene on a pCDFDuet-1 plasmid containing a CloDF13 replicon. These replicons are compatible, and the two plasmids can be stably co-maintained, provided the relevant antibiotic selective pressure is exerted: ampicillin for pBluescript SKII+ and streptomycin for pCDFDuet-1[40]. In this way, we have produced MtbGS that has a better adenylylation state than any previously reported. Three methods were used to assess the adenylylation of MtbGS, and E. coli adenylylated and deadenylylated GS. The E. coli enzymes were produced recombinantly from pBluescript SKII+ in E. coli production strains lacking endogenous GS (for deadenylylated enzyme) or both GS and uridylyl transferase (adenylylated enzyme). Table 1 gives a summary of the results obtained. MS spectra showed distinct peaks for adenylylated and deadenylylated enzymes, with calculated masses agreeing with the theoretical values. Based on this data, it can be concluded that the adenylylation state of adenylylated MtbGS expressed in this novel system is at least 85% based on the MS data, but possibly up to 93% based on phosphate hydrolysis. Based on this data this investigation was carried out to demonstrate the proof-of-concept that compounds could be obtained that selectively inhibit either adenylylated or deadenylylated MtbGS an that these compounds would inhibit Mtb in macrophages.
The fixed concentration inhibition assays of the 213 ATP based scaffold inhibitors produced 11 potential inhibitors. The scatter associated with the data when using the adenylylated GS is probably associated with inter subunit allosteric regulation of adenylylated GS [42]. Some compounds therefore allosterically activate the enzyme. This would be achieved as a result of allosteric regulation occurring between two GS subunits as a result of the adenylylation. It is therefore conceivable that inhibitors binding one GS subunit may either activate or inhibit the binding of another inhibitor or ATP to the associated allosteric site. There was also differential inhibition of the adenylylated and deadenylylated MtbGS. Seven of the potential inhibitors were directed against adenylylated MtbGS and one was directed against deadenylylated MtbGS. Two of the inhibitors namely 10057 and 10059 showed promising anti-MtbGS activity against adenylylated MtbGS with 99% and 91% inhibition respectively. The IC50 concentrations obtained for compounds 10057 and 10059 were approximately 9.6 μM and 15 μM, for the adenylylated enzyme and deadenylylated enzyme, respectively. Taking into account the large variation in ATP and glutamine concentrations used in the screening assays similar low μM IC50 concentrations were obtained in the enzyme screens [32–38]. Imidazopyridine amides have been found to inhibit Mtb in other studies indicating an alternative mechanism via the cytochrome cytochrome bc1 complex impacting on the homeostasis of ATP synthesis [39]. The inhibition of glutamine synthetase may also impact the ATP homeostasis as the resultant accumulation of α-ketoglutarate, as a result of inhibiting glutamine synthetase, may lead to the slowing down of metabolic flux via the TCA cycle.
Compounds 10057 and 10059 are structurally very similar. Also in the macrophage assays compounds 10057, 10059 and 5029 which were GS-adenylylated specific, showed comparable activities in the BACTEC assays. This indicates that these compounds can inhibit M.tb as effectively in a macrophage environment. However, compound 5045, specific to the de-adenylylated form of GS, inhibited M.tb killing in both the in vitro and ex vivo macrophage models effectively at 98% and 73% respectively at 100μM. The GS adenylylation/deadenylylation status of the M.tb strains used in the BACTEC assay and the macrophage assays were not known which may explain the high killing effect of both the adenylylation- and de-adenylylation specific compounds. In further development, attention may be paid to improving the selectivity of the compounds vis-à-vis mammalian forms of GS, to reduce the possibility of side-effects.
Supporting information
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information file.
Funding Statement
This work forms part of a project (T80051) funded by The Innovation Fund, which is managed by the National Research Foundation (NRF) in South Africa.
References
- 1.Shapiro BM, Stadman ER. The regulation of glutamine synthesis in microorganisms. Ann Rev Microbiol. 1970;24: 501–524. [DOI] [PubMed] [Google Scholar]
- 2.Kumada Y, Benson DR, Hillemann TJ, Hosted DA, Rochefort CJ, Thompson W, et al. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci USA. 1993;90: 3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown JR, Masuchi Y, Robb FT, Doolittle WF. Evolutionary relationships of bacterial and archaeal glutamine synthetase genes. J Mol Evol. 1994;38: 566–576. [DOI] [PubMed] [Google Scholar]
- 4.Tyler B. Regulation of the assimilation of nitrogen compounds. Ann Rev Biochem. 1978;47: 1127–1162. doi: 10.1146/annurev.bi.47.070178.005403 [DOI] [PubMed] [Google Scholar]
- 5.Gaillardin CM, Magasanik B. Involvement of the product of the glnF gene in the autogenous regulation of glutamine synthetase formation in Klebsiella aerogenes. J Bacteriol. 1978;133: 1329–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foor F, Janssen KA, Magasanik B. Regulation of synthesis of glutamine synthetase by adenylylated glutamine synthetase. Proc Natl Acad Sci USA. 1975;72: 4844–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Janssen KA, Magasanik B. Glutamine synthetase of Klebsiella aerogenes: genetic and physiological properties of mutants in the adenylylation system. J Bacteriol. 1977;129: 993–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Senior PJ. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J Bacteriol. 1975;123: 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ginsberg A, Stadtman ER. Regulation of glutamine synthetase in Escherichia coli In: Prusiner SR, Stadman ER, editors. Enzymes of Glutamine Metabolism, Academic Press, New York, 1973; p. 9–44. [Google Scholar]
- 10.Wolhueter RM, Schutt H, Holzer H. Regulation of glutamine synthetase in vivo in E. coli In: Prusiner SR, Stadman ER, editors. Enzymes of Glutamine Metabolism, Academic Press, New York, 1973; p. 45–61. [Google Scholar]
- 11.Bender RA, Janssen KA Resnick AD, Blumenberg M, Foor F, Magasanik B. Biochemical parameters of glutamine synthetase from Klebsiella aerogenes. J Bacteriol. 1977;129: 1001–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom FR, Streicher SL, Tyler B. Regulation of enzyme synthesis by glutamine synthetase of Salmonella typhimurium: a factor in addition to glutamine synthetase is required for activation of enzyme formation. J Bacteriol. 1977;130: 983–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holzer H, Schutt H, Mašek Z, Mecke D. Regulation of two forms of glutamine synthetase in Escherichia coli by the ammonium content of the growth medium. Proc Natl Acad Sci USA. 1968;60: 721–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woolfolk CA, Shapiro B, Stadtman ER. Regulation of glutamine synthetase I. Purification and properties of glutamine synthetase from Escherichia coli. Arch Biochem Biophys. 1966;116: 177–192. [DOI] [PubMed] [Google Scholar]
- 15.Kustu SG, McKereghan K. Mutations affecting glutamine synthetase activity in Salmonella typhimurium. J Bacteriol. 1975;122: 1006–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shapiro BM, Kingdon HS, Stadtman ER. Regulation of glutamine synthetase. VII. Adenylyl glutamine synthetase: a new form of the enzyme with altered regulatory and kinetic properties. Proc Natl Acad Sci USA. 1967;58: 642–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapiro BM, Stadtman ER. 5'-adenylyl-O-tyrosine. The novel phosphodiester residue of adenylylated glutamine synthetase from Escherichia coli. J Bio. Chem. 1968;243: 3769–3771. [PubMed] [Google Scholar]
- 18.Kingdon HS, Shapiro BM, Stadtman ER. Regulation of glutamine synthetase. VIII. ATP: glutamine synthetase adenylyltransferase, an enzyme that catalyzes alterations in the regulatory properties of glutamine synthetase. Proc Natl Acad Sci USA. 1967;58: 1703–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mecke D, Wulff K, Liess K, Holzer H. Characterization of a glutamine synthetase inactivating enzyme from Escherichia coli. Biochem Biophys Res Commun. 1966;24: 452–458. [DOI] [PubMed] [Google Scholar]
- 20.Reitzer LJ, Magasanik B. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine In: Neidart FC, Ingarhou JLL, Low KB, Magasanik B, Schaechter M Nunberger HE, editors. Escherichia coli and Salmonella typhimurium. Cellular and molecular biology, American Society for Microbiology, Washington DC, 1987; p. 302–320. [Google Scholar]
- 21.Shapiro BM, Ginsburg A. Effects of specific divalent cations on some physical and chemical properties of glutamine synthetase from Escherichia coli. Taut and relaxed enzyme forms. Biochemistry. 1968;7: 2153–2167. [DOI] [PubMed] [Google Scholar]
- 22.Reynaud C, Etienne G, Payron P, Lenelle MA, Daffe M. Extracellular enzyme activities potentially involved in the pathogeniticty of Mycobacterium tuberculosis. Microbiol. 1998;144: 577–587. [DOI] [PubMed] [Google Scholar]
- 23.Harth G, Horwitz MA. An inhibitor of exported Mycobacterium tuberculosis glutamine synthetase selectively blocks the growth of pathogenic mycobacteria in axenic culture and in human monocytes: extracellular proteins as potential drug targets. J Exp Med. 1999;189: 1425–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harth G, Horwitz MA. Inhibition of Mycobacterium tuberculosis glutamine synthetase as a novel antiobiotic strategy against tuberculosis: demonstration of efficacy in vivo. Infect Immune. 2003;71: 456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tullius MV, Harth G, Horwitz MA. Glutamine synthetase GlnA1 is essential for growth of Mycobacterium tuberculosis in human THP-1 macrophages and guinea pigs. Infect Immun. 2003;71: 3927–3936. doi: 10.1128/IAI.71.7.3927-3936.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wietzerbin J, Lederer F, Petit JF. Structural study of the poly-l-glutamic acid of the cell wall of Mycobacterium tuberculosis var hominis, strain Brevannes. Biochem Biophys Res Commun. 1975;62: 246–252. [DOI] [PubMed] [Google Scholar]
- 27.Hirschfield GR, McNeil M, Brennan PJ. Peptidoglycan-associated polypeptides of Mycobacterium tuberculosis. J Bacteriol. 1990;172: 1005–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harth G, Horwitz MA. Expression and efficient transport of enzymatically active Mycobacterium tuberculosis glutamine synthetase in Mycobacterium smegmatis and evidence that information for export is contained within the protein. J Biol Chem. 1997;272: 22728–22735. [DOI] [PubMed] [Google Scholar]
- 29.Singh J, Joshi C, Bhatnagar R. Cloning and expression of mycobacterial glutamine synthetase gene in Escherichia coli. Biochem Biophys Res Comun. 2004;317:634–638. [DOI] [PubMed] [Google Scholar]
- 30.Mehta R, Pearson JT, Mahajan S, Nath A, Hickey MJ, Sherman DR, et al. Adenylylation and catalytic properties of Mycobacterium tuberculosis glutamine synthetase expressed in Escherichia coli versus Mycobacteria. J Biol Chem. 2004:279: 22477–22482. doi: 10.1074/jbc.M401652200 [DOI] [PubMed] [Google Scholar]
- 31.(2017.07.24) http://www.rcsb.org/pdb/home/home.do
- 32.Lagerlund O, Odell LR, Mowbray SL, Nilsson MT, Krajewski WW, Nordqvist A, et al. Microwave-enhanced alpha-acrylation of a protected glycine in water: evaluationof 3-phenylglycine derivatives as inhibitors of the tuberculosis enzyme, glutamine synthetase. Comb Chem High Throughput Screen. 2007;10: 783–789. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson MT, Krajewski W, Yellagunda S, Prabhumurthy S, Chamarahally GN, Siddanadappa C, et al. Structural basis for the inhibition of Mycobacterium tuberculosis glutamine synthetase by novel ATP-competitive inhibitors. J Mol Biol. 2009;393: 503–513. [DOI] [PubMed] [Google Scholar]
- 34.Odell LR, Nilsson MT, Gising J, Lagerhund O, Muthas D, Nordqvist A, et al. Functionalized 3-amino-imidazole[1,2a]pyridines: A novel class of drug-like Mycobacterium tuberculosis glutamine synthetase inhibitors. Bioorg Med Chem Lett. 2009;19: 4790–4793. doi: 10.1016/j.bmcl.2009.06.045 [DOI] [PubMed] [Google Scholar]
- 35.Gising J, Nilsson MT, Odell LR, Yahiaoui S, Lindh M, Iyer H, et al. Trisubstituted imidazoles as Mycobacterium tuberculosis glutamine synthetase inhibitors. J Med Chem. 2012;55: 2894–2898. doi: 10.1021/jm201212h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Couturier C, Silve S, Morales R, Pesseque B, Liopart S, Nair A, et al. Nanomolar inhibitors of Mycobacterium tuberculosis glutamine synthetase 1: Synthesis, biological evaluation and X-ray crystallographic studies. Bioorg Med Chem Lett. 2015;25: 1455–1459. doi: 10.1016/j.bmcl.2015.02.035 [DOI] [PubMed] [Google Scholar]
- 37.Mowbray SL, Kathiravan MK, Pandey AA, Odell LR. Inhibition of glutamine synthetase: A potential drug target in Mycobacterium tuberculosis. Molecules. 2014;19: 13161–13176. doi: 10.3390/molecules190913161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosikowska P, Bochno M, Maceginiak K, Forlani G, Kafarski P, Berlicki L. Bisphosphonic acids as effective inhibitors of Mycobacterium tuberculosis glutamine synthetase. J Enzyme Inhib Med Chem. 2016;31: 931–938. doi: 10.3109/14756366.2015.1070846 [DOI] [PubMed] [Google Scholar]
- 39.Pethe K, Bifani P, Jang J, Kang S, Park S, Ahn S, et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nature Medicine. 2013;19: 1157–1162. doi: 10.1038/nm.3262 [DOI] [PubMed] [Google Scholar]
- 40.Held D, Yaeger K, Novy R. New coexpression vectors for expanded compatibilities in E. coli. Innovations. 2003;18: 406. [Google Scholar]
- 41.Backman K Chen Y-M, Magasanik B. Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome. Proc Natl Acad Sci USA. 1981;78: 3743–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shapiro BM, Stadtman ER. Glutamine Synthetase (Escherichia coli) In: Hirs CHW, Timasheff SN, editors. Methods in Enzymology 130, Elsevier, 1970; p. 910–922. [Google Scholar]
- 43.Laemlli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227: 680–685. [DOI] [PubMed] [Google Scholar]
- 44.Ludden PW, Burris RH. Purification and properties of nitrogenase from Rhodospirillum rubrum, and evidence for phosphate, ribose and an adenine-like unit covalently bound to the iron protein. Biochem J. 1978;175: 251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kenyon CP, Steyn A, Roth RL, Steenkamp PA, Nkosi TC, Oldfield LC. The role of the C8 proton of ATP in the regulation of phosphoryl transfer within kinases and synthetases. BMC Biochem. 2011;12: 36 doi: 10.1186/1471-2091-12-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Collins L, Franzblau SG. Microplate alamar blue assay versus BACTEC 460 system for high-throughput screening of compounds against Mycobacterium tuberculosis and Mycobacterium avium. Antimicrob Agents Chemother. 1997;41(5):1004–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Johnson R, Warren RM, van der Spuy GD, Gey van Pittius NC, Theron D, Streicher EM, et al. Drug-resistant tuberculosis epidemic in the Western Cape driven by a virulent Beijing genotype strain. Int J Tuberc Lung Dis. 2010; 14(1):119–121. [PubMed] [Google Scholar]
- 48.Leisching G, Pietersen RD, Mpongoshe V, van Heerden C, van Helden P, Wiid I, et al. The host response to a clinical MDR mycobacterial strain cultured in a detergent-free environment: A global transcriptomics approach. PLoS One. 2016;11(4):e0153079 doi: 10.1371/journal.pone.0153079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siddiqi SH, Heifets LB, Cynamon MH, Hooper NM, Laszlo A, Libonati JP, et al. Rapid broth macrodilution method for determination of MICs for Mycobacterium avium isolates. J Clin Microbiol. 1993;31(9):2332–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.George G, Mony P, Kenneth J. Comparison of the efficacies of loop-mediated isothermal amplification, fluorescence smear microscopy and culture for the diagnosis of tuberculosis. PLoS One. 2011:6(6):e21007 doi: 10.1371/journal.pone.0021007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.De Chastellier C, Lang T, Thilo L. Phagocytic processing of the macrophage endoparasite, Mycobacterium avium, in comparison to phagosomes which contain Bacillus subtilis or latex beads. Eur J Cell Biol 1995;68:167–182. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information file.