To the Editor
Vibrio mimicus strain SCCF01 was isolated from the yellow catfish (Pelteobagrus fulvidraco) during a mass mortality event at an aquaculture facility. We sequenced the genome of strain SCCF01, then predicted possible effective targets for subunit vaccine and identified possible virulence genes for live attenuated vaccine. The genome of SCCF01 consists of 2 circular chromosomes of 3,213,040 and 1,272,975 bp with G+C content of 46.61% and 45.88%, respectively. Subcellular localization, number of transmembrane helices and adhesion probability were predicted by Vaxign programs. Several candidate virulence genes were identified using the Virulence Factors Database. The genomic information presented herein will be useful for future studies investigating the pathogenesis of V. mimicus and with vaccine development.
Vibrio mimicus is an enteric pathogen that is widely distributed in aquatic environments that can infect and cause disease in fish.1,2 Additionally, the pathogen is zoonotic and can cause bacterial gastroenteritis and food poisoning in humans if infected fish are consumed.2 Hence, understanding the pathogenesis of V. mimicus and developing a vaccine to treat infected fish hosts is an important research direction. In this study, we revived V. mimicus strain SCCF01 from the farmed yellow catfish (Pelteobagrus fulvidraco) in Sichuan Province, China, during a mass mortality event.3 Our objectives were to: (1) characterize the SCCF01 genome, (2) predict possible effective targets for subunit vaccine, (3) identify possible virulence genes that could lead to development of attenuated V. mimicus for vaccine development, and (4) identify the ability of colonization and adhesion of V. mimicus infection in yellow catfish.
The whole-genome sequence of SCCF01 strain was determined with single molecule real-time (SMRT) sequencing platform PacBio RS II. Single SMRT cells produced 35,089 reads for 306,431,068 bases (N50 size 12,135 bp and mean read length 8,732 bp). Subsequently, the sequence reads were assembled using the PacBio SMRT Analysis (ver. 2.3.0) with default options. Genome annotation was performed using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). The rRNA/tRNA genes were identified by Barrnap version 0.6 (www.vicbioinformatics.com/software.barrnap.shtml) and tRNAscan-SE,4 respectively. The annotated genome was performed with the software Vaxign to further analyze subcellular localization, number of transmembrane helices and adhesion probability.5 In addition, possible virulence genes in SCCF01 were identified by Virulence Factors Database and software BLASTp.6,7 Bacterial levels of yellow catfish challenged with SCCF01 via water bath immersion were measured using qPCR.8 The entire experimental procedure was approved by the Committee of the Ethics on Animal Care and Experiments at Sichuan Agricultural University and was carried out in accordance with the approved guidelines.
Based on our analysis, the genome of V. mimicus strain SCCF01 is 4.49 Mb in size and consists of 2 circular chromosomes of 3,213,040 and 1,272,975 bp with G+C content of 46.61% and 45.88%, respectively (Table 1). The following rules for bacterial subunit vaccine development, (1) it is often preferred to identify outer membrane(especially adhesins) or secreted proteins as vaccine targets.5 (2) more than one transmembrance helix is hard to be isolated from a recombinant Escherichi. coli strain.9 (3) virulence factors are better targets.10 The results of vaccine targets prediction (34 potential vaccine candidates are marked in red) and virulence genes annotation are summarized in Table S1 and Table S2, respectively. These results are useful to predict possible effective targets and helpful for development of DNA and subunit vaccines.
Table 1.
General genome features of Vibrio mimicus strain SCCF01.
V. mimicus strain SCCF01 colonized the mucosal surfaces of the skin, gills, and intestines of yellow catfish within 24 hours post-exposure (Fig. 1). Systemic infection in multiple organs occurred within 36 hours (Fig. 1). These results showed that V. mimicus is able to infect yellow catfish quickly. The results also showed that SCCF01 can efficiently adhere to mucosal surfaces, which is promising for attenuated vaccine development.
Figure 1.
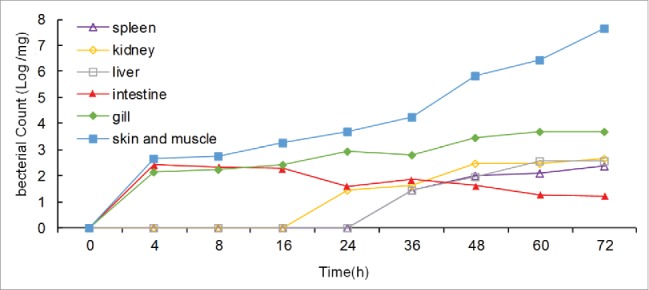
Infection kinetics of V. mimicus strain SCCF01 in yellow catfish. Following immersion infection with strain SCCF01 at a dose of 1.0 × 106 cfu/ml for 30 minutes, the fish were sampled in triplicate at 0, 4, 8, 16, 24, 36, 48, 60, and 72 hours. The organs (spleen, kidney, liver, intestine gill skin and muscle) from 3 fish were mixed and homogenized. Bacterial count was measured by using qPCR.8
Herein, we report the complete genome sequence of V. mimicus strain SCCF01 associated with yellow catfish infection. Multiple possible virulence genes were identified, which could be targeted for future work on creating an attenuated pathogen. In particular, HlyA (VMH), thermolabile hemolysin (TLH), outer membrane protein (Omp), accessory colonization factor (ACF) and toxin-coregulated pilus (TCP) are known to be associated with virulence in other pathogens.11 The virulence genes can be identified from the genome sequence of strain SCCF01 (Table S2). Given the adhesion properties of SCCF01 demonstrated by qPCR in the challenge experiment, creating an attenuated pathogen for use in therapeutic treatment may be possible. Live attenuated enteric pathogens (e.g., Vibrio cholera, Edwardsiella tarda) have been used previously as vaccines,12,13 because they can enter the host, replicate on mucosal surfaces, and stimulate an immune response.14 Our results also suggested that oral administration of an attenuated SCCF01 may be possible. Degradation of antigens in the highly tolerogenic gut environment is a main reason for limited efficacy of oral vaccines in fish.14,15
In conclusion, the results of the genome sequence analysis reported here for V. mimicus provides genetic basis for the development of subunit and live attenuated vaccine. Moreover, immersion challenge test indicated that strain SCCF01 has the ability of colonization and adhesion to the mucosal surfaces, which is promising for attenuated vaccine development.
Nucleotide sequence accession numbers
The complete genome sequence of Vibrio mimicus strain SCCF01 has been deposited at GenBank under the accession no. CP016383 (chromosome I) and CP016384 (chromosome II). This strain has been deposited at College of Veterinary Medicine, Sichuan Agricultural University under the accession number CVM2013034.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed
Acknowledgments
We thank Tianjin Biochip Corporation (TianJin, China) for PacBio sequencing and de novo assembly of the genome. We also thank Dr. Matthew Gray (University of Tennessee) for providing editorial comments on an earlier draft of our manuscript.
Funding
This work was supported by the Sichuan Technology Support Planning (No 2014NZ0003).
References
- [1].Chowdhury MA, Yamanaka H, Miyoshi S, Aziz KM, Shinoda S. Ecology of Vibrio mimicus in aquatic environments. Appl Environ Microbiol 1989; 55:2073-78; PMID:2782878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Chitov T, Kirikaew P, Yungyune P, Ruengprapan N, Sontikun K. An incidence of large foodborne outbreak associated with Vibrio mimicus. Eur J Clin Microbiol Infect Dis 2009; 28:421-24; http://dx.doi.org/ 10.1007/s10096-008-0639-7 [DOI] [PubMed] [Google Scholar]
- [3].Geng Y, Liu D, Han S, Zhou Y, Wang KY, Huang XL, Chen DF, Peng X, Lai WM. Outbreaks of vibriosis associated with Vibrio mimicus in freshwater catfish in China. Aquaculture 2014; 433:82-84; http://dx.doi.org/ 10.1016/j.aquaculture.2014.05.053 [DOI] [Google Scholar]
- [4].Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 1997; 25:955-64; PMID:9023104; http://dx.doi.org/ 10.1093/nar/25.5.0955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].He Y, Xiang Z, Mobley HLT. Vaxign: The First Web-Based Vaccine Design Program for Reverse Vaccinology and Applications for Vaccine Development. Biomed Res Int 2010; 2010:297505-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 2005; 33:325-28; PMID:15653633; http://dx.doi.org/ 10.1093/nar/gki008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chen L, Xiong Z, Sun L, Yang J, Jin Q. VFDB 2012 update: toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res 2012; 40:641-45; http://dx.doi.org/ 10.1093/nar/gkr989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wang HB. Establishment of TaqMan real-time PCR for important pathogenic Vibrios. Shandong University Master's Thesis, 2009. [Google Scholar]
- [9].Pizza M, Scarlato V, Masignani V, Giuliani MM, Aric∫ B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 2000; 287:1816-20; PMID:10710308; http://dx.doi.org/ 10.1126/science.287.5459.1816 [DOI] [PubMed] [Google Scholar]
- [10].Soares SC, Trost E, Ramos RTJ, Carneiro AR, Santos AR, Pinto AC, Barbosa E, Aburjaile F, Ali A, Diniz CAA. Genome sequence of Corynebacterium pseudotuberculosis biovar equi strain 258 and prediction of antigenic targets to improve biotechnological vaccine production. J Biotechnol 2013; 167:135-41; PMID:23201561; http://dx.doi.org/ 10.1016/j.jbiotec.2012.11.003 [DOI] [PubMed] [Google Scholar]
- [11].Rosenberg E, Delong EF, Lory S, Stackebrandt E, Thompson F. The prokaryotes: Gammaproteobacteria In: Gomez-Gil B, Thompson CC, Matsumura Y, Sawabe T, Iida T, Christen R, Thompson F, Sawabe T editors. The famlily vibrionaceae. Berlin Heidelberg (Germany: ): Springer; 2013. p. 660-747. [Google Scholar]
- [12].Wang Q, Zhao Y, Qin L, Zhang Y. Surface-exposed expression of Edwardsiella tarda EseB in live attenuated Vibrio anguillarum based on novel surface display systems. Aquaculture Res 2009; 40:1459-67; http://dx.doi.org/ 10.1111/j.1365-2109.2009.02176.x [DOI] [Google Scholar]
- [13].Wang Y, Yang W, Wang Q, Qu J, Zhang Y. Presenting a foreign antigen on live attenuated Edwardsiella tarda using twin-arginine translocation signal peptide as a multivalent vaccine. J Biotechnol 2013; 168:710-17; PMID:23994481; http://dx.doi.org/ 10.1016/j.jbiotec.2013.08.018 [DOI] [PubMed] [Google Scholar]
- [14].Embregts CWE, Forlenza M. Oral vaccination of fish: lessons from humans and veterinary species. Dev Comp Immunol 2016; 64:118-37; PMID:27018298; http://dx.doi.org/ 10.1016/j.dci.2016.03.024 [DOI] [PubMed] [Google Scholar]
- [15].Mutoloki S, Munang'andu HM, Evensen Ø. Oral vaccination of fish - antigen preparations, uptake, and immune induction. Front Immunol 2014; 6:519. [DOI] [PMC free article] [PubMed] [Google Scholar]


