ABSTRACT
During long-term lung infection in cystic fibrosis (CF) patients, Burkholderia cenocepacia faces multiple selective pressures in this highly stressful and fluctuating environment. As a consequence, the initial infecting strain undergoes genetic changes that result in the diversification of genotypes and phenotypes. Whether this clonal expansion influences the pathogenic potential is unclear. The virulence potential of 39 sequential B. cenocepacia (recA lineage IIIA) isolates, corresponding to 3 different clones retrieved from 3 chronically infected CF patients was compared in this study using the non-mammalian infection hosts Galleria mellonella and Caenorhabditis elegans. The isolates used in this retrospective study were picked randomly from selective agar plates as part of a CF Center routine, from the onset of infection until patients' death after 3.5 and 7.5 y or the more recent isolation date after 12.5 y of chronic infection. The infection models proved useful to assess virulence potential diversification, but for some isolates the relative values diverged in C. elegans and G. mellonella. Results also reinforce the concept of the occurrence of clonal diversification and co-existence of multiple phenotypes within the CF lungs, also with respect to pathogenicity. No clear trend of decrease (or increase) of the virulence potential throughout long-term infection was found but there is an apparent tendency for a clone/patient-dependent decrease of virulence when the G. mellonella model was used. The sole avirulent variant in both infection hosts was found to lack the small third replicon previously associated to virulence. Although possible, the in vivo loss of this nonessential megaplasmid was found to be a rare event (1 among a total of 64 isolates examined).
KEYWORDS: Burkholderia cenocepacia, Caenorhabditis elegans, cystic fibrosis chronic infection, Galleria mellonella, phenotypic diversification, virulence potential
Introduction
Cystic fibrosis (CF) patients are highly susceptible to chronic lung infections, which is the dominant cause of their premature death. Infections by Burkholderia cepacia complex (Bcc) bacteria are particularly feared since they generally lead to a more rapid decline in lung function and early death.1-3 During long-term respiratory infections, CF opportunistic pathogens face a highly stressful and fluctuating environment within the patient's airways, in particular due to the host immune system, antimicrobial therapy, reduced availability of oxygen and other nutrients.4-8 As a consequence, the initial infecting strain undergoes genetic changes resulting in parallel evolution within the lung with diversification of genotypes and phenotypes.7,9-13 The understanding of the mechanisms underlying microbial evolution within the CF host and its association with pathogenicity and persistence is crucial to deal with these chronic infections. To elucidate these mechanisms, our group has systematically compared the genome-wide expression patterns of 3 of the 11 Burkholderia cenocepacia sequential isolates examined in the present work, which were previously found to exhibit variation in relevant phenotypes in the context of bacterial pathogenesis.4-6,12 These isolates were retrieved from one CF patient (patient J) chronically infected during 3.5 y until death with cepacia syndrome.12 Late isolates (IST4113 and IST4134) were found to have a higher virulence potential compared to the first isolate (IST439) based on their higher ability to invade epithelial cells and to compromise epithelial monolayer integrity,5 suggesting an increase of B. cenocepacia virulence throughout the course of long-term lung infection. This result contrasts with the idea that Pseudomonas aeruginosa niche-specific selection reduces its ability to cause acute infections across a broad range of non-mammalian and mammalian infection hosts.14,15 However, even in P. aeruginosa this concept is debatable.16
In this work, we have used 2 non-mammalian infection models, the larva of the wax moth Galleria mellonella and the nematode Caenorhabditis elegans, to compare the virulence potential of a relatively large number (39) of B. cenocepacia variants from 3 different clones retrieved through the course of chronic lung infection of 3 different CF patients. Both infection hosts are attractive alternatives to traditional animal models in the study of bacterial virulence due to their low cost, simplicity of use and lack of ethical concerns.17 G. mellonella grows at 37°C (essential for the expression of several bacterial virulence factors) and has a relatively complex innate immune system,17 enabling a good correlation between bacterial pathogenicity in G. mellonella and mammalian infection models.18,19 On the other hand, C. elegans grows at 25°C and is suitable for rapid high-throughput screening in vivo.20 During the last decade, G. mellonella and C. elegans were established as infection models to assess both the virulence of different strains of Burkholderia cepacia complex (Bcc) species and the importance of specific bacterial genes in virulence, showing a good correlation with the pathogenicity observed in mice.21-24 The isolates compared in the present study concerning the virulence potential, as well as a number of phenotypes that have been related with virulence, were the 11 sequential B. cenocepacia (recA lineage IIIA) clonal isolates retrieved from the onset of infection until patient´s J death with the cepacia syndrome, including the 3 isolates examined before using human bronchial epithelial cells.4-6,12 Given that this patient died after 3.5 y of chronic infection and thus the original strain could not have evolved toward a very long-term chronic infection, we have extended this study to 2 additional CF patients infected during 12.5 and 7.5 y with different strains also belonging to the species B. cenocepacia (recA lineage IIIA). In this retrospective study more than one isolate per time point was analyzed, when available.
Results
Variation of the pathogenic potential of B. cenocepacia clonal isolates retrieved from 3 cystic fibrosis patients during chronic infection
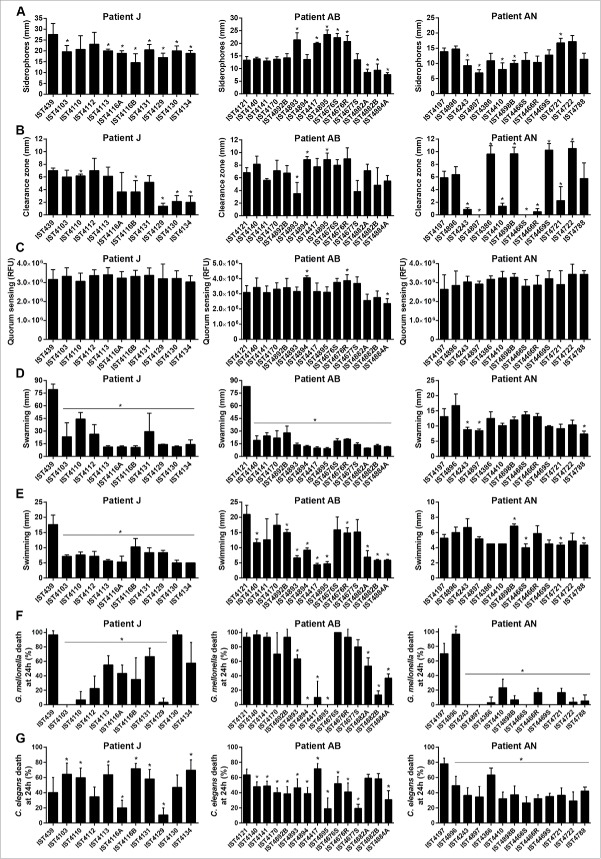
Galleria mellonella and Caenorhabditis elegans infection models were used to compare under standard conditions (25°C for C. elegans and 37°C for G. mellonella) the virulence potential of 11 B. cenocepacia recA lineage IIIA sequential isolates recovered from patient J, since the onset of the infection until death with cepacia syndrome, after 3.5 y of chronic lung infection.12 No consistent pattern of increase or decrease of the virulence potential was observed between early and late isolates in both infection models (Fig. 1A). Remarkably, in G. mellonella, the first isolate recovered from the patient (IST439) was found to be significantly more pathogenic than the late clonal isolates, IST4113 and IST4134, while the opposite was observed in C. elegans, as reported before using epithelial cells (Fig. 1A).12 Five of the clonal variants examined exhibited similar acute virulence levels in both infection models while 3 other isolates were more pathogenic for C. elegans than for G. mellonella and the remaining 3 isolates exhibited the opposite behavior (Fig. 1A). For example, IST4103 that effectively killed C. elegans, was poorly virulent in the G. mellonella model. Another remarkable result concerns isolate IST4129 that was recovered 3 months before the patient's death and found to be avirulent in both infection models. Also, 2 colony morphotypes obtained in the same isolation procedure, IST4116A and IST4116B, were found to exhibit different virulence potential in the C. elegans infection model. These two clonal variants also exhibited significant differences concerning most of the phenotypes tested before,12 or during the present study, as detailed below.
Figure 1.
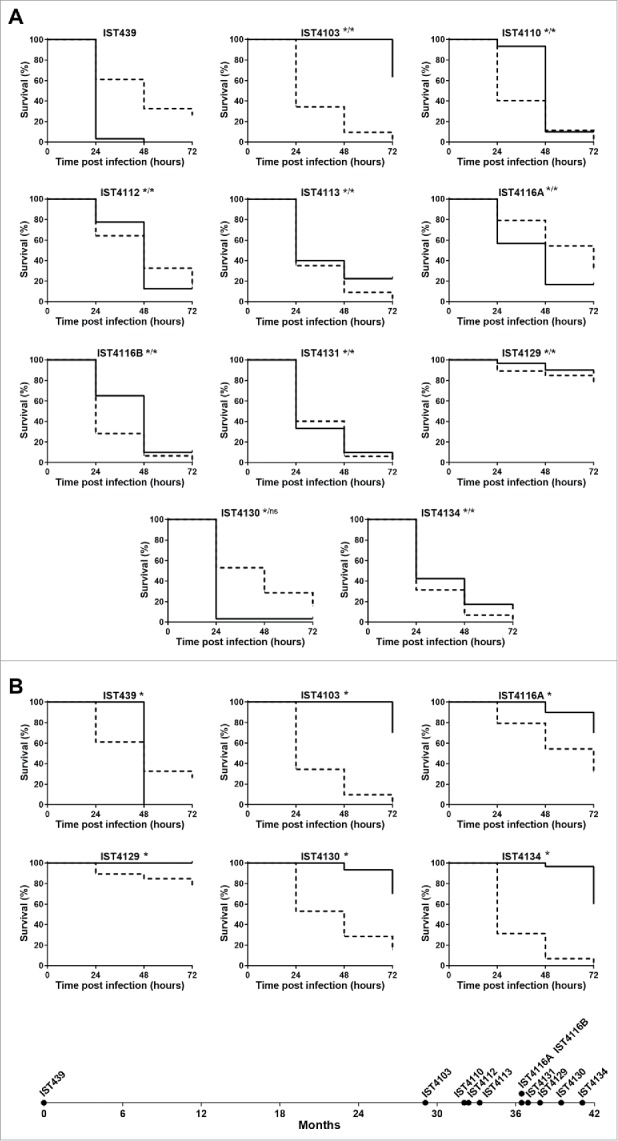
Kaplan-Meier graphs for Galleria mellonella (full lines) and Caenorhabditis elegans (dashed lines) survival after infection with B. urkholderia cenocepacia sequential isolates retrieved from patient J during 3.5 y of chronic infection until patient's death with the cepacia syndrome. (A) Results from G. mellonella assay performed at 37°C and C. elegans assay performed at 25°C, and (B) results from G. mellonella and C. elegans assays performed at 25°C. Statistical analysis was performed to compare the virulence potential between the first isolate, IST439, and each of the other isolates for C. elegans (left side of the slash) and G. mellonella (right side of the slash) (* p < 0.05, ns not significant, Mantel-Cox test).
Given that distinct standard temperatures were used to compare the 2 infection models (37°C for G. mellonella and 25°C for C. elegans) and this fact may affect the relative pathogenic potential of the different tested isolates because changes in temperature are known to alter the expression of B. cenocepacia genes, in particular virulence genes,23,25 6 isolates, representing different virulence profiles, were also tested in G. mellonella at 25°C (Fig. 1B). Compared with the virulence profiles obtained at standard temperatures (Fig. 1A), the most remarkable difference is the virulence attenuation for all the B. cenocepacia isolates in G. mellonella at 25°C compared with 37°C. Remarkably, IST439 was the only isolate that killed all larvae after 48 hours of infection at 25°C, even though its pathogenic potential in G. mellonella assays carried out at 25°C, versus 37°C, decreased significantly. Also, the results obtained at 25°C are consistent with the conclusions put forward at 37°C and the first isolate, IST439, is clearly the most virulent isolate tested at the lower temperature in G. mellonella.
The comparison of the pathogenic potential of isolates retrieved from patient J was extended to isolates also belonging to B. cenocepacia (recA lineage IIIA) retrieved from 2 other CF patients with longer infection periods: 12.5 y for patient AB and 7.5 y for patient AN (Figs. 2 and 3, respectively). Based on Random Amplified Polymorphic DNA (RAPD) analysis, the 15 isolates from patient AB and the 13 isolates from patient AN were confirmed as clonal isolates from 2 different original strains also different from patient J clone (Supplementary Fig. 1). The virulence potential of these 2 new sets of isolates was tested in C. elegans and G. mellonella infection models. Results showed that, again, no clear trend of alteration in virulence potential was observed between early and late isolates even though, as already suggested by the data obtained for patient J, there is an apparent tendency for virulence attenuation when the G. mellonella model was used, especially for patient´s AN clone (Figs. 2 and 3). In order to confirm the concept that multiple virulence phenotypes co-exist at each isolation time, whenever available, more than one isolate retrieved during the same isolation procedure from patients AB and AN were included in the analysis (Figs. 2 and 3). For a few isolation dates, no significant differences could be detected in both infection models between those isolates, even when they exhibited distinct morphotypes (e.g. IST4676S and IST4676R). However, several isolate sets retrieved during the same isolation procedure showed very distinct virulence potential, at least in one of the infection hosts. This is for example the case of early isolates from patient AN, IST4197 and IST4896, which exhibited distinct virulence potential in C. elegans and G. mellonella infection hosts. Also, IST4882A was found to be considerably more virulent than isolate IST4884A in C. elegans, but did not show a distinct virulence profile from IST4882B, while IST4882A and IST4882B have different virulence potential in G. mellonella at 24 hours.
Figure 2.

Kaplan-Meier graphs for Galleria mellonella (full lines) and Caenorhabditis elegans (dashed lines) survival after infection with Burkholderia cenocepacia clonal isolates retrieved from patient AB during 12.5 y of chronic infection. Results for G. mellonella and C. elegans were obtained as described in materials and methods and as used for patient J isolates in Figure 1. Statistical analysis was performed to compare the virulence potential between the first isolate retrieved from the patient, IST4121, and each of the other isolates for C. elegans (left side of the slash) and G. mellonella (right side of the slash) (* p < 0.05, ns not significant, Mantel-Cox test).
Figure 3.
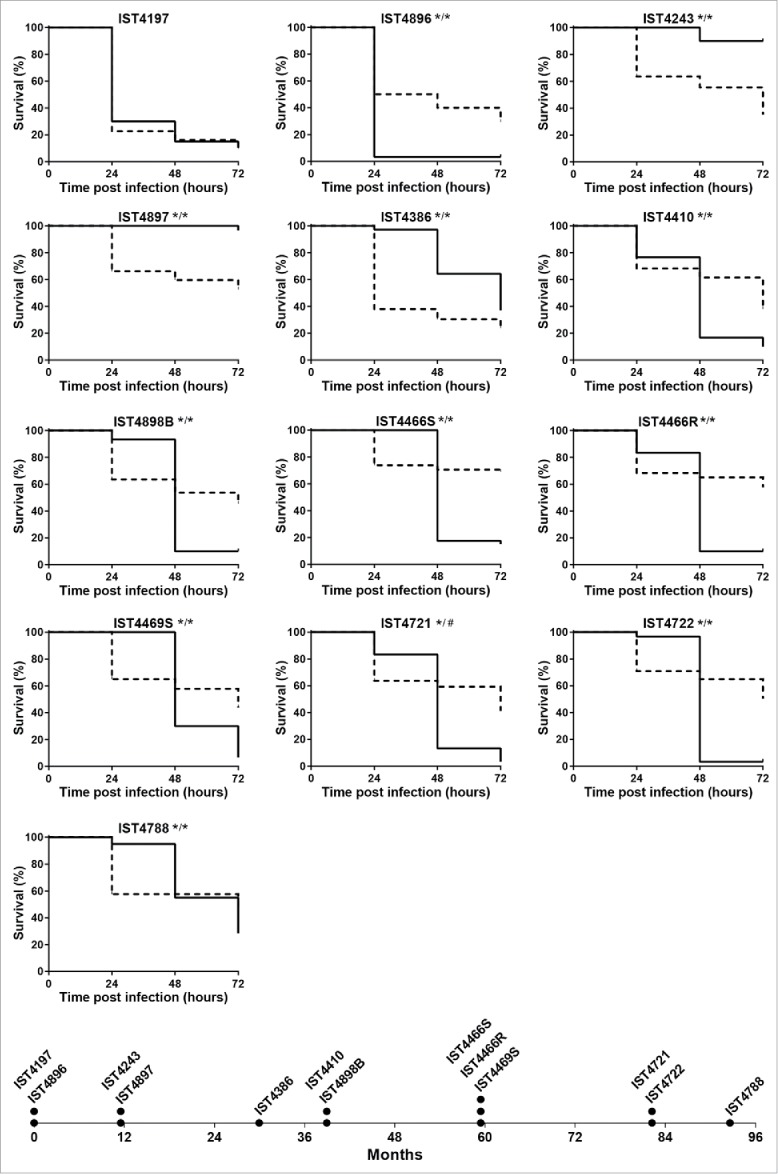
Kaplan-Meier graphs for Galleria mellonella (full lines) and Caenorhabditis elegans (dashed lines) survival after infection with Burkholderia cenocepacia clonal isolates retrieved from patient AN during 7.5 y of chronic infection. Results for G. mellonella and C. elegans were obtained as described in materials and methods and as used for patient J isolates in Figure 1. Statistical analysis was performed to compare the virulence potential between the first isolate, IST4197, and each of the other isolates for C. elegans (left side of the slash) and G. mellonella (right side of the slash) (* p < 0.05, Mantel-Cox test). The asterisk in both the left and right sides of the slash represents also the comparison of the virulence potential of IST4896, retrieved in the first isolation date together with IST4197, with remaining isolates in C. elegans and G. mellonella, respectively. The symbol # represents the only case were the statistical test gave distinct results: although no statistically significant difference was observed for IST4721 and IST4197 results in G. mellonella model, p < 0.05 between IST4721 and IST4896 in this infection host.
The virulence third replicon is missing in the highly attenuated virulence variant IST4129
Given that isolate IST4129 exhibited a highly attenuated virulence in both infection models and that this isolate was the only from the 11 isolates retrieved from patient J for which no aidA transcripts could be detected (unpublished data), it was considered of interest to examine the eventual absence of the third replicon in this clonal variant as well as in all the other clonal variants from patient J. The hypothesis put forward was that B. cenocepacia third replicon, previously described as encoding virulence factors, besides secondary metabolism and other accessory functions,26,27 could be missing in IST4129, or could at least be missing a portion of this replicon that includes the aidA gene. To test this hypothesis, we used an established strategy to confirm the presence of the third replicon based on a PCR screen with primer pairs used sequentially and designed to anneal to the highly conserved region concerning its replication origin.27 With the exception of IST4129, all the other isolates gave a positive PCR result for the presence of this megaplasmid with the first set of primers (repAFor and repARev) (Supplementary Fig. 2A). The negative result for IST4129 was confirmed with other 2 primer pairs (Supplementary Fig. 2B). Separation of undigested genomic DNA from the 11 sequential B. cenocepacia isolates retrieved from patient J by pulsed-field gel electrophoresis (PFGE) showed that IST4129 was the sole isolate lacking the smaller fragment with approximately 1 Mb (Fig. 4A), likely corresponding to the third replicon.26 An additional PFGE analysis of genomic macrofragments generated by the action of the endonuclease SpeI was performed and a 700 Kb band, of approximately the size of B. cenocepacia J2315 third replicon,28 was confirmed to be missing (Fig. 4B). However, isolate IST4103 that was found to exhibit an attenuated virulence in G. mellonella, but not in C. elegans, showed the same undigested and digested DNA PFGE profiles as all the other 9 isolates (Fig. 4).
Figure 4.
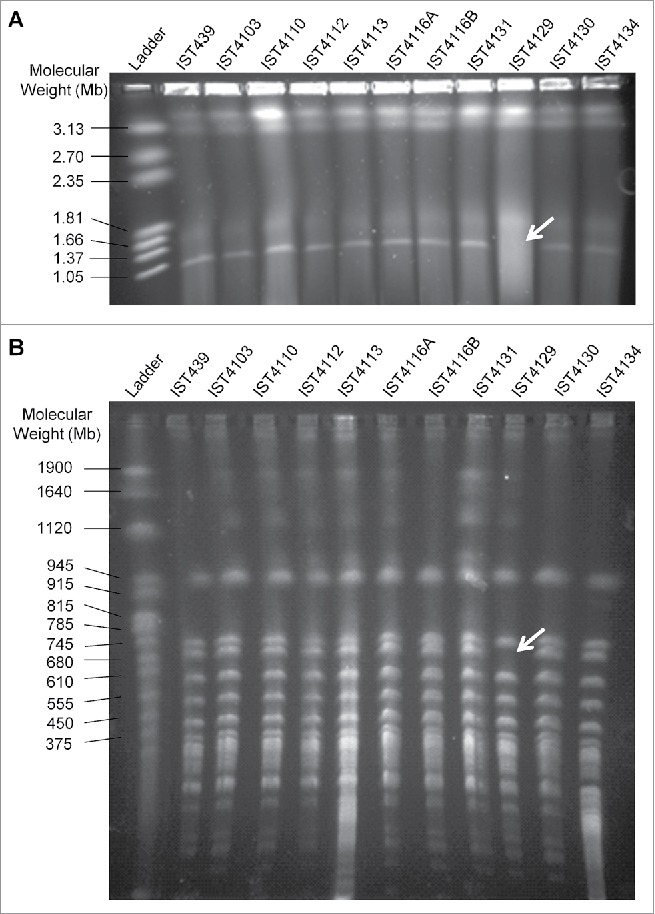
Visualization of genomic DNA from the 11 B. cenocepacia sequential isolates retrieved from CF patient J (A) undigested and (B) digested with SpeI. Genomic DNA was separated by pulsed-field gel electrophoresis. CHEF DNA Size Marker (Hansenula wingei) and Yeast Chromosome PFG Marker were used as ladder in (A) and (B), respectively. Arrows indicate the missing band in both gels.
Remarkably, none of the isolates from patients AB and AN exhibited the dramatic attenuated virulence registered in both infection models for isolate IST4129 from patient J (Figs. 2 and 3) and a PCR screening confirmed the presence of the third replicon in all the isolates from these patients that were tested (Supplementary Fig. 2A). This screening for the presence of the megaplasmid included not only the 28 isolates tested for virulence in C. elegans and G. mellonella but 25 additional isolates from patients AB and AN that were not included in the virulence screening.
Virulence-associated phenotypes of B. cenocepacia sequential clonal isolates retrieved from 3 CF patients
A number of relevant phenotypes previously shown to be involved in B. cenocepacia pathogenesis21,29,30,31 were systematically compared for all the 39 B. cenocepacia isolates retrieved from patients J, AB and AN tested for the virulence potential, namely, production of siderophores, extracellular polysaccharides (EPS), proteases, homoserine lactone (AHL) molecules, and swarming and swimming motilities. No clear correlation could be established between the various phenotypes characterized for the 3 B. cenocepacia clonal isolates with the respective virulence potential for the 2 infection models tested (Fig. 5). Concerning EPS production, all the B. cenocepacia clonal isolates from patient J were found to be producers, in agreement with what was previously reported.12 However, no EPS production could be detected for the late B. cenocepacia isolates IST4677S, IST4882A, IST4882B and IST4884A from patient AB and for the 9 B. cenocepacia isolates from patient AN retrieved after IST4897, reinforcing the idea that EPS production could be reduced or eliminated as the result of adaptation to long-term chronic infection.32 Remarkably, the first B. cenocepacia isolate retrieved from patient AB, IST4121, showed higher swarming values than the remaining isolates retrieved from this patient (Fig. 5D), similarly to what was reported before for the B. cenocepacia isolates from patient J12 and confirmed in this work. However, no clear decrease in the swarming motility was observed for the B. cenocepacia isolates retrieved from patient AN (Fig. 5D), as well as for the swimming motility of the B. cenocepacia isolates retrieved from patients AB and AN (Fig. 5E). The B. cenocepacia isolate that lost the third replicon, IST4129, showed values for all the phenotypes tested similar to those exhibited by the several isolates retrieved from patient J, suggesting little or no influence of the loss of the third replicon in all these phenotypes.
Figure 5.
Phenotypic characterization of Burkholderia cenocepacia isolates retrieved from patients J, AB and AN. (A) Siderophore production, (B) protease activity, (C) homoserine lactone signaling molecules detection, (D) swarming and (E) swimming motility, (F) percentage of Galleria mellonella death at 24 h after infection and (G) percentage of Caenorhabditis elegans death at 24 h after infection. Values in (F) and (G) were taken from those presented in Figures 1, 2 and 3. Error bars represent standard deviations.
Discussion
The cystic fibrosis (CF) airways represent a complex and diverse ecosystem, where the highly plastic genome of Burkholderia cepacia complex (Bcc) bacteria enables a marked genetic adaptation to multiple selective pressures during chronic infection.8,33,34 However, the influence that the mechanisms and dynamics of Bcc evolution during the course of CF chronic infection has on the virulence potential of these bacteria remains poorly understood. To get insights into the outcome of the alterations taking place in this group of opportunistic bacteria in the CF lung during chronic infection, the acute pathogenicity of a total of 39 sequential Burkholderia cenocepacia (recA lineage IIIA) isolates, corresponding to 3 different clones retrieved from 3 chronically infected CF patients (J, AN and AB) from the onset of the infection for a period of 3.5, 7.5 and 12.5 years, was assessed by the determination of mortality of the 2 established non-mammalian infection models Galleria mellonella and Caenorhabditis elegans. Both models were successfully used to differentiate the pathogenic potential of the clonal variants sequentially isolated from the 3 patients. The large intraclonal variation in virulence potential registered for the B. cenocepacia (recA lineage IIIA) isolates tested was already observed for Pseudomonas aeruginosa using mammalian and non-mammalian infection hosts, including mice models with different genetic backgrounds.14,16 However, in our study, no consistent progressive pattern of decrease (or increase) of the acute virulence of all the B. cenocepacia clonal isolates retrieved from the 3 patients tested along the course of infection and lung deterioration was observed in both infection models. These results are in line with recent comparative genomic studies involving clinical isolates of P. aeruginosa and Bcc bacteria retrieved from CF infected patients, suggesting that the bacterial infecting population at any given time point possesses significant diversity, where mutations are not fixed and instead coexist in the form of different sub-lineages along the course of the infection.9-11,35 Depending on the varying selective pressures acting on allele diversity that occur concomitantly with disease progression and management, the predominance of these different lineages can vary over time.9-11,35
In P. aeruginosa, it is accepted that the virulence potential is reduced throughout chronic infection, based on the comparative assessment of the virulence of late isolates with the respective early isolates in both mammalian and non-mammalian infection models.14,16 The systematic comparison carried out in our study of the virulence potential of such a relatively large number of variants (39), corresponding to 3 different clones obtained from 3 patients, was feasible because C. elegans and G. mellonella were used as infection models. Given that results obtained from other authors who explored non-mammalian and mammalian infection models were found to be consistent when the variation of P. aeruginosa virulence potential during long-term CF infection was examined,14 it is not unlikely that the results from our analysis may reflect the relative virulence levels assessed in mammalian infection hosts. However, for several of the B. cenocepacia isolates tested, divergent results were obtained concerning the virulence potential assessed using the 2 infection models, such as the remarkable case of patient's J isolate IST4103 that effectively killed C. elegans, being poorly virulent in the G. mellonella model. Differences in the temperature at which most of the virulence assays are performed (37°C for G. mellonella vs. 25°C for C. elegans) could not explain the differences observed in the virulence potential registered between both infection models for several isolates, even though growth temperature is recognized to alter gene expression, in particular the expression of virulence genes, as it is the case of the increased cepI expression in B. cenocepacia J2315 cells grown at 37°C compared with 20°C.23,25 However, the differences registered in the relative values determined for a number of the isolates tested when C. elegans or G. mellonella were used as infection models recommends caution in drawing definitive conclusions. Remarkably, B. cenocepacia isolates from patient AN who died after 7.5 y of infection showed an apparent decrease in the virulence potential along chronic infection when tested in G. mellonella whereas such decrease is not clear for isolates from patient AB who was still alive after 12.5 y of chronic infection. These results are questioning the generalized idea that the establishment of a persistent infection requires the loss of virulence factors and, consequently, the decrease of the virulence potential.14,15,36
Results obtained in this study support the idea that a more complete evaluation of B. cenocepacia pathogenic potential can only be achieved using multiple infection models since each host has specific features and limitations. In fact, the divergence in the pathogenic potential of similar Burkholderia strains in the same infection model or of the same strain in different non-mammalian infection models was previously observed and proposed to be dependent on specific virulence factors.23,24 Also, it was found that only a few B. cenocepacia virulence factors are required for full pathogenicity in multiple hosts, these including genes involved in quorum sensing signaling, siderophore production, lipopolysaccharide (LPS) biosynthesis, and purine, pyrimidine and shikimate biosynthesis.23,24 Moreover, it was found that most of the B. cenocepacia mutants for virulence factors that were already tested in several infection models showed attenuation in only one infection host, similarly to what was described for P. aeruginosa.37,38 However, only single mutants were tested for both species, which does not reflect the complex scenario that occurs in infected CF patients. For P. aeruginosa, it was recognized that genes required for pathogenicity in one strain might not be required or be predictive of virulence in other strains.39 This was clearly demonstrated when some of the virulence factors identified in P. aeruginosa PA14 were screened in other clinical isolates, and no correlation could be established between virulence and the presence or absence of these specific genes.39 For this reason, P. aeruginosa virulence is considered multifactorial and combinatorial, resulting from a pool of virulence-related genes that interact in several combinations in different genetic backgrounds. Remarkably, although a number of relevant phenotypes related to virulence in bacteria23,29-31 were systematically examined for all the tested isolates for the virulence potential and found to vary for the different clonal variants, no correlation could be established between a specific phenotype and the relative virulence potential or with the progress of the chronic infection. The variation registered for a number of relevant phenotypes tested is consistent with the occurrence of clonal expansion during long-term infection, with no consistent time-dependent alteration pattern, as reported before.12 Results of this systematic study clearly indicate that chronic pathogenesis of Burkholderia infections is multifactorial. This is further highlighted by a comparative genomics analysis carried out for patient J isolates against a complete draft genome of the first isolate, under an adaptive evolutionary perspective (SC dos Santos, CP Coutinho, M Dillon, AS Moreira, VS Cooper, and I Sá-Correia, manuscript in preparation), where, among a relatively small number of accumulated mutations, no mutational events that can be clearly linked to variations in bacterial virulence could be detected. The exception is the loss of the third replicon in isolate IST4129, shown in this work to be an infrequent event (as discussed below).
Although the virulence phenotype for each isolate results from the combined expression of several virulence factors, some correlations might be established between the observed variation in the pathogenic potential in each model with specific phenotypic and molecular features. Among the intriguing results obtained in this work is the contradictory observation regarding the comparison of the virulence potential exhibited by the first isolate IST439 obtained from patient J compared with the late isolates IST4113 and IST4134. Isolate IST439 exhibited lower virulence potential compared to IST4113 and IST4134 when assessed in this study using C. elegans or in a previous study using epithelial cells,5 but it was more virulent than the late isolates when G. mellonella was used as infection model. The sole known feature of isolate IST439 compared with all the others that might be considered of interest, at this moment, to try to explain such remarkable divergence on the virulence potential when assessed in the nematode infection model or in epithelial cells compared with the insect infection model, is related with the presence of the O-antigen subunit in the IST439 lipopolysaccharide (LPS), which is absent in all the other subsequent B. cenocepacia isolates from patient J (Maldonado et al., unpublished data). LPS composition was already suggested to affect B. cenocepacia virulence potential in both non-mammalian models,21-23,40,41 and it is possible that differences in B. cenocepacia virulence potential between infection models might be related with the differential recognition and response to LPS composition by the different immune systems. In fact, there is evidence for an important role played by the O-antigen in Gram-negative bacteria virulence. For example, it was shown that a Salmonella typhimurium mutant missing the entire O-antigen is avirulent in G. mellonella host model, and the shortening of the O-antigen chain length in a mutant lacking the enzymes involved in the synthesis of long and very long O-antigen reduced its pathogenic potential by one-half compared to wild-type.42 Another noteworthy observation of this work is the highly attenuated virulence of IST4129 in both infection models. Except for this isolate, mRNA levels from the aidA gene were detected in all isolates retrieved from patient J (unpublished data). This information led us to hypothesize and subsequently confirm the lack in isolate IST4129 of the third replicon, where the aidA gene is located. This replicon was reported to be associated with virulence in this species.26 This result strongly suggests, for the first time, the in vivo loss of the nonessential virulence third replicon in B. cenocepacia. Based on previous studies26,27 and in the screening performed in this work comprising the 39 isolates examined throughout this study plus 20 5 additional isolates from patients AB and AN, all of them found to have the third replicon, it is possible to conclude that the in vivo loss of this megaplasmid, although possible, is a rare event.
Concluding remarks
This study supports the concept of the occurrence during cystic fibrosis long-term lung infection of Burkholderia cenocepacia clonal expansion and diversification of the pathogenic potential, assessed using Galleria mellonella and Caenorhabditis elegans. However, no strain/patient-independent pattern of reduction (or increase) of the virulence potential was registered during long-term infection. Moreover, no correlation could be established between several relevant phenotypes related with bacterial virulence and the virulence potential. Studies such as this one on the adaptation of a group of bacteria with inherent genomic plasticity to a highly stressful environment are ideal to gain insights into the dynamics of phenotype alteration during chronic infections. This study also shows that the 2 established non-mammalian infection models C. elegans and G. mellonella are able to differentiate the pathogenic potential of B. cenocepacia clonal variants. However, divergent results were obtained when the 2 infection hosts were used for some of the isolates tested, as would be expected considering the multifactorial and combinatorial nature of B. cenocepacia virulence and the dependence of the virulence potential on specific virulence factors in different infection models. This study also provides the first evidence for the in vivo infrequent loss of the nonessential third replicon in B. cenocepacia and its relation with the marked attenuation of the pathogenic potential in different infection models.
Materials and methods
Bacterial isolates and growth conditions
The 39 Burkholderia cenocepacia clinical isolates examined in this study were recovered from the sputum of 3 cystic fibrosis (CF) patients under surveillance at the major Portuguese CF Center in Hospital de Santa Maria (HSM) from Centro Hospitalar Lisboa Norte EPE, Lisbon. According to the hospital's routine, sputum samples are obtained from CF patients every 2 to 3 months, during periodic consultations to monitor their clinical status, or more often for patients showing clinical deterioration. This retrospective study used isolates that were selected at random among the isolated colonies obtained in selective Burkholderia cepacia Selectatab medium. For several isolation dates only one isolate was available for this study but for other dates 2 or 3 different isolates corresponding to different colonies obtained in the isolation plate were tested. The eleven isolates retrieved from patient J and examined during this study were previously identified as belonging to B. cenocepacia recA lineage IIIA and confirmed to be clonal variants.12,43 The other isolates included in this study were retrieved from patients AB and AN, also infected with B. cenocepacia recA lineage IIIA (43,44 and results of the present work), were randomly selected from the onset of the infection until patient's death after 7.5 y (patient AN) or until the last isolation date after 12.5 y (patient AB). Bacterial growth was carried out in Lysogeny Broth, Lennox (LB; Conda, Pronadisa) liquid medium, at 37°C and 250 rpm, or in LB agar plates prepared by supplementation of LB medium with 2% of agar (Iberagar, Portugal).
Molecular characterization of B. cenocepacia isolates
Species identification and RAPD typing of Bcc isolates
Genomic DNA was prepared from fresh overnight cultures using the Puregene DNA isolation kit (cell and tissue kit) (Gentra Systems), followed by quantification of DNA concentration using a ND-1000 spectrophotometer (NanoDrop Technologies). Species identification was carried out based on the amplification of fragments of recA gene with species specific primers.45 Strain assignment was performed by Random Amplified Polymorphic DNA (RAPD).46 Genotyping of all the isolates used in this study was performed using primer 270 (5′-TGCGCGCGGG-3′), followed by confirmation of the strain types established by this primer with primers 208 (5′-ACGGCCGACC-3′) and 272 (5′-AGCGGGCCAA-3′). RAPD profiles were visually compared and analyzed by GelJ software.47 Similarity coefficients were calculated using the RAPD profiles obtained with each primer for the complete set of isolates examined using Pearson coefficient and Unweighted Pair Group Method with Arithmetic Mean (UPGMA) linkage method.
Analysis of B. cenocepacia clonal variants genomic DNA by pulsed field gel electrophoresis (PFGE)
Genomic DNA was prepared in situ in agarose blocks based on the method described by Schoonmaker et al. (1992).48 Briefly, cells grown in LB broth until mid-exponential phase were washed twice, and resuspended in Pett IV solution (1 M NaCl and 10 mM Tris-HCl, pH 7.6). Equal volumes of these cell suspensions and 1.5% (w/v) low-melting-point agarose (FMC Bioproducts, Rockland, Me., USA) were mixed, and 100 μl aliquots were dispensed into insert molds to solidify. Gel plugs were treated with a lysis solution (6 mM Tris-HCl, 1 M NaCl, 0.01 M EDTA, 0.5% Brij-58, 0.2% deoxycholate and 0.5% N-lauroylsarcosine, pH 7.6) overnight at 37°C. The purification of DNA in gel plugs was performed by the standard proteinase K method (0.5 M EDTA, 1% (w/v) N-lauroylsarcosine, 1 mg/ml proteinase K) at 50°C for 24 h, followed by an additional period of 24 h in the same fresh solution without proteinase K at 50°C. Gel plugs were finally washed several times with TE buffer (10 mM Tris-HCl and 1 mM EDTA, pH 7.6) at 37°C, and then stored at 4°C in TE buffer. These gel plugs were used directly for chromosome separation by PFGE or after digestion with SpeI to separate the generated macrofragments by PFGE.
For restriction endonuclease digestion, slices (2 mm) were cut from the agarose plug and equilibrated with 150 µl of restriction endonuclease buffer (supplied with the enzyme by the manufacturer) at 4°C for 1 h. The buffer was replaced with fresh buffer, and DNA was digested overnight with 20 U of SpeI. After complete digestion, the buffer was removed, and slices were equilibrated for 1 h in 500 µl of TE buffer at 4°C and then loaded into 1% (w/v) Pulse-Field grade agarose (Nzytech, Lda - genes and enzymes, Lisbon, Portugal) gels that were prepared in 0.5× TBE running buffer (0.45 M Tris-Base, 0.45 M boric acid and 0.01 M EDTA (pH 8.0)).
PFGE was carried out using a Gene Navigator apparatus (Pharmacia-LKB, Uppsala, Sweden). Intact chromosomes or the corresponding SpeI restriction fragments were separated using an initial pulse time of 400 s that was increased to 700 s over 48 h at 100 V at 8°C or an initial pulse time of 5 s that was increased to 80 s over 24 h at 200 V at 8°C, respectively. CHEF DNA Size Marker (from Hansenula wingei, Biorad) or Yeast Chromosome PFG Marker (New England Biolabs) were used as size standards for complete chromosome separation or macrofragment separation, respectively. Gels were stained with GelRed (GelRed™, Biotium) and photographed under UV light illumination.
PCR screening for the presence of the third replicon
The screening for the detection of the third replicon in all the isolates examined in this study was performed using sequentially 3 sets of primers designed to anneal to the highly conserved origin of replication in the third replicon, as previously described.27
Virulence potential assessment
Nematode slow killing assays
Aliquots of 50 µl of B. cenocepacia cultures were plated onto the surface of 35 mm diameter Petri dishes containing 4 ml of nematode growth medium II (NGMII; 0.3% NaCl, 0.35% bacto peptone, 5 μg/ml cholesterol, 1 mM CaCl2, 1 mM MgSO4, 25 mM potassium phosphate buffer pH 6.0, 2 µg/ml uracil, 0.0025% nystatin, 1.7% bacto agar)49 and incubated for 24 h at 37°C. Nematode slow-killing assays were performed using L4-stage synchronized larvae of Caenorhabditis elegans strain DH26,50 based on methods described previously.51 This C. elegans strain is sterile at 25°C, thus allowing the score of worms without the interference of progeny. Results are the mean values of at least 2 independent experiments with at least 7 plates per experiment using a nematode population of 178 ± 39 worms. Escherichia coli OP50 was used as a negative control.
Galleria mellonella killing assays
B. cenocepacia isolates were grown in LB broth until mid-exponential phase. After dilution to a standardized OD640 nm of 0.2 ± 0.02 in 0.9% NaCl (w/v), 100 μl of these cell suspensions were plated onto LB plates and incubated for 24 h at 37°C. Cells were washed with 1.5 ml of 0.9% NaCl (w/v), centrifuged at 8000 rpm and 4°C for 5 minutes, and resuspended in 10 mM MgSO4 with 1.2 mg/ml ampicillin (inclusion of antibiotic in the inoculum was previously proposed21 and used to prevent contamination by bacteria naturally present on the caterpillar or in their surface). As described before,52 a micrometer was adapted to control the injection volume of a microsyringe used to inject 3.5 µl of a bacterial suspension containing 1 × 106 CFU/larvae into the haemocoel of the caterpillars via the hindmost left proleg, which had been surface-sterilized with 70% (v/v) ethanol. Larvae were placed in Petri dishes and stored in the dark at 37°C, the standard temperature for the killing assays,21 or at 25°C to examine the temperature effect in comparative virulence assays. Results are the mean values of at least 3 independent experiments that gave similar results using 10 larvae per treatment for each isolate, recording their survival at 24 h intervals until 72 h. Larvae were considered dead when no movement was observed in response to touch with a pipette tip. As negative control, at least 5 larvae were injected with 3.5 µl of 10 mM MgSO4 plus 1.2 mg/ml ampicillin in each assay to monitor any problem related to the injection process.
Phenotypic characterization of B. cenocepacia isolates
Swarming and swimming assays
B. cenocepacia isolates were grown until the beginning of the stationary phase of growth and 2 µl of these cell cultures were used to inoculate swim agar plates containing 25 ml of 1% (wt/vol) tryptone, 0.5% (wt/vol) NaCl and 0.3% (wt/vol) agar or swarm plates containing 25 ml of 0.8% (wt/vol) nutrient broth, 0.5% (wt/vol) glucose and 0.5% (wt/vol) agar. These swim and swarm plates were incubated for 24 h at 37°C in stacks with 4 plates, and the diameter of the growth was measured. Values represent the means for at least 3 independent experiments.
Determination of siderophore production, protease activity and EPS production
For the detection of siderophore production, CAS plates were inoculated with 5 µl of B. cenocepacia cultures grown until the beginning of the stationary phase, and incubated for 48 h at 37°C. Halo diameter was measured to determine siderophore production, and values represent the mean of at least 3 independent experiments.
For the determination of protease activity, 5 µl of B. cenocepacia cultures grown until the beginning of the stationary phase were used to inoculate skimmed milk agar plates (2% skimmed milk, LB broth, 1% agar). Plates were incubated during 48 h at 37°C, and the clearance zone was measured. Results are the mean values of at least 3 independent experiments.
Extracellular polysaccharides (EPS) production was visualized using mannitol agar plates supplemented with Congo red, as previously described,24 after 24 h of incubation at 37°C and using 5 µl of B. cenocepacia cultures grown until the beginning of the stationary phase.
AHL quantification
Cell cultures of B. cenocepacia isolates grown until the beginning of the stationary phase were centrifuged at 6,500 rpm for 5 minutes with subsequent filter sterilization of supernatants. One hundred microliters of each supernatant was mixed with an equal volume of a culture of Pseudomonas putida F117(pAS-C8-Gmr),53 kindly provided by Professor Leo Eberl, in exponential growth phase. After 6 h of incubation at 30°C in BRAND cellGrade™ plates, fluorescence was measured with a FilterMax F5 Multimode Microplate Reader (Molecular Devices), with an excitation wavelength of 485 nm and an emission detection of 535 nm, and connected to the SoftMax® Pro Microplate reader. The relative fluorescence units (RFU) were determined for at least 3 independent supernatants of each isolate.
Statistical analysis
Statistical analysis was performed using Prism GraphPad software 6.05 (GraphPad Software, San Diego, CA). The statistical significance of the variation in virulence potential between the first isolate and each of the other isolates was determined using the Mantel-Cox test, while the statistical differences regarding the variation in phenotypes was determined using a 2-tailed Student's t-test. In both cases, P ≤ 0.05 was considered statistically significant.
Ethics
Research studies involving the clinical Bcc isolates examined in this study that were obtained as part of the hospital routine were approved by the hospital ethics committee and the patients' anonymity is preserved.
Supplementary Material
Abbreviations
- Bcc
Burkholderia cepacia complex
- CF
cystic fibrosis
- HSM
Hospital de Santa Maria, Centro Hospitalar Lisboa Norte
- LPS
lipopolysaccharide
- PFGE
Pulsed Field Gel Electrophoresis
- RAPD
Random Amplified Polymorphic DNA
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors acknowledge the supply of C. elegans DH26 by Caenorhabditis Genetics Center (CGC, University of Minnesota), and Professor Leo Eberl for P. putida F117(pAS-C8-Gmr). The contribution of Luís Lito, José Melo-Cristino, Celeste Barreto, Pilar Azevedo and Luísa Pereira, over the last 21 years of epidemiological surveillance of Bcc respiratory infections in CF patients receiving care at Hospital de Santa Maria, Centro Hospitalar Lisboa Norte, in Lisbon, is gratefully acknowledged.
Funding
Funding provided by iBB - Institute for Bioengineering and Biosciences from Programa Operacional Regional de Lisboa 2020 (Project N. 007317) and from the Portuguese Foundation for Science and Technology (FCT) (UID/BIO/04565/2013) is acknowledged. FCT also supported PhD and postdoctoral fellowships to A.S.M. (SFRH/BD/82162/2011), C.P.C. (SFRH/BPD/81220/2011), S.C.S. (SFRH/BPD/75483/2010), D.M.H. (SFRH/BPD/91831/2012), S.A.S. (SFRH/BPD/102006/2014) and C.G.R. (SFRH/BPD/75718/2011).
Author contributions
ASM performed Bcc species identification and RAPD typing of selected isolates, as well as the molecular biology analysis to search for the presence of the third replicon with SCS collaboration. CPC performed RFLP-PFGE analysis. ASM, SAS, CPC, and AO prepared the cell cultures used in virulence assays. SAS, CGR and JHL performed the C. elegans assays, DMH and AMF the G. mellonella assays, and ASM and CPC the phenotypic assays. ASM prepared the figures and contributed to the writing of the manuscript under the scientific supervision of ISC, who conceived and coordinated the study. All authors read and approved the final manuscript.
ORCID
Dalila Mil-Homens http://orcid.org/0000-0002-9727-3579
Sílvia A. Sousa http://orcid.org/0000-0001-9291-9169
Arsénio M. Fialho http://orcid.org/0000-0002-8066-5787
Jorge H. Leitão http://orcid.org/0000-0001-8850-274X
Isabel Sá-Correia http://orcid.org/0000-0003-2208-5183
References
- [1].Drevinek P, Mahenthiralingam E. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin Microbiol Infect 2010; 16:821-30; PMID:20880411; http://dx.doi.org/ 10.1111/j.1469-0691.2010.03237.x [DOI] [PubMed] [Google Scholar]
- [2].Lipuma JJ. The changing microbial epidemiology in cystic fibrosis. Clin Microbiol Rev 2010; 23:299-323; PMID:20375354; http://dx.doi.org/ 10.1128/CMR.00068-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mahenthiralingam E, Baldwin A, Vandamme P. Burkholderia cepacia complex infection in patients with cystic fibrosis. J Med Microbiol 2002; 51:533-8; PMID:12132768; http://dx.doi.org/ 10.1099/0022-1317-51-7-533 [DOI] [PubMed] [Google Scholar]
- [4].Madeira A, Santos PM, Coutinho CP, Pinto-de-Oliveira A, Sá-Correia I. Quantitative proteomics (2D DIGE) reveals molecular strategies employed by Burkholderia cenocepacia to adapt to the airways of cystic fibrosis patients under antimicrobial therapy. Proteomics 2011; 11:1313-28; PMID:21337515; http://dx.doi.org/ 10.1002/pmic.201000457 [DOI] [PubMed] [Google Scholar]
- [5].Madeira A, dos Santos SC, Santos PM, Coutinho CP, Tyrrell J, McClean S, Callaghan M, Sá-Correia I. Proteomic profiling of Burkholderia cenocepacia clonal isolates with different virulence potential retrieved from a cystic fibrosis patient during chronic lung infection. PLoS One 2013; 8:e83065; PMID:24349432; http://dx.doi.org/ 10.1371/journal.pone.0083065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Mira NP, Madeira A, Moreira AS, Coutinho CP, Sá-Correia I. Genomic expression analysis reveals strategies of Burkholderia cenocepacia to adapt to cystic fibrosis patients' airways and antimicrobial therapy. PLoS One 2011; 6:e28831; PMID:22216120; http://dx.doi.org/ 10.1371/journal.pone.0028831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lieberman TD, Michel JB, Aingaran M, Potter-Bynoe G, Roux D, Davis MR Jr, Skurnik D, Leiby N, LiPuma JJ, Goldberg JB, et al.. Parallel bacterial evolution within multiple patients identifies candidate pathogenicity genes. Nat Genet 2011; 43:1275-80; PMID:22081229; http://dx.doi.org/ 10.1038/ng.997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Döring G, Parameswaran IG, Murphy TF. Differential adaptation of microbial pathogens to airways of patients with cystic fibrosis and chronic obstructive pulmonary disease. FEMS Microbiol Rev 2011; 35:124-46; PMID:20584083; http://dx.doi.org/ 10.1111/j.1574-6976.2010.00237.x [DOI] [PubMed] [Google Scholar]
- [9].Lieberman TD, Flett KB, Yelin I, Martin TR, McAdam AJ, Priebe GP, Kishony R. Genetic variation of a bacterial pathogen within individuals with cystic fibrosis provides a record of selective pressures. Nat Genet 2014; 46:82-7; PMID:24316980; http://dx.doi.org/ 10.1038/ng.2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cramer N, Klockgether J, Wrasman K, Schmidt M, Davenport CF, Tummler B. Microevolution of the major common Pseudomonas aeruginosa clones C and PA14 in cystic fibrosis lungs. Environ Microbiol 2011; 13:1690-704; PMID:21492363; http://dx.doi.org/ 10.1111/j.1462-2920.2011.02483.x [DOI] [PubMed] [Google Scholar]
- [11].Huse HK, Kwon T, Zlosnik JE, Speert DP, Marcotte EM, Whiteley M. Parallel evolution in Pseudomonas aeruginosa over 39,000 generations in vivo. MBio 2010; 1:e00199-10; PMID:20856824; http://dx.doi.org/ 10.1128/mBio.00199-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Coutinho CP, de Carvalho CCCR, Madeira A, Pinto-de-Oliveira A, Sá-Correia I. Burkholderia cenocepacia phenotypic clonal variation during three and a half years of residence in the lungs of a cystic fibrosis patient. Infect Immun 2011; 79:2950-60; PMID:21536796; http://dx.doi.org/ 10.1128/IAI.01366-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moreira AS, Coutinho CP, Azevedo P, Lito L, Melo-Cristino J, Sá-Correia I. Burkholderia dolosa phenotypic variation during the decline in lung function of a cystic fibrosis patient during 5.5 years of chronic colonization. J Med Microbiol 2014; 63:594-601; PMID:24469681; http://dx.doi.org/ 10.1099/jmm.0.069849-0 [DOI] [PubMed] [Google Scholar]
- [14].Lore NI, Cigana C, De Fino I, Riva C, Juhas M, Schwager S, Eberl L, Bragonzi A. Cystic fibrosis-niche adaptation of Pseudomonas aeruginosa reduces virulence in multiple infection hosts. PLoS One 2012; 7:e35648; PMID:22558188; http://dx.doi.org/ 10.1371/journal.pone.0035648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Friman VP, Ghoul M, Molin S, Johansen HK, Buckling A. Pseudomonas aeruginosa adaptation to lungs of cystic fibrosis patients leads to lowered resistance to phage and protist enemies. PLoS One 2013; 8:e75380; PMID:24069407; http://dx.doi.org/ 10.1371/journal.pone.0075380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bragonzi A, Paroni M, Nonis A, Cramer N, Montanari S, Rejman J, Di Serio C, Döring G, Tümmler B. Pseudomonas aeruginosa microevolution during cystic fibrosis lung infection establishes clones with adapted virulence. Am J Respir Crit Care Med 2009; 180:138-45; PMID:19423715; http://dx.doi.org/ 10.1164/rccm.200812-1943OC [DOI] [PubMed] [Google Scholar]
- [17].Lopez Hernandez Y, Yero D, Pinos-Rodriguez JM, Gibert I. Animals devoid of pulmonary system as infection models in the study of lung bacterial pathogens. Frontiers Microbiol 2015; 6:38; PMID:25699030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hoffmann JA. Innate immunity of insects. Curr Opin Immunol 1995; 7:4-10; PMID:7772280; http://dx.doi.org/ 10.1016/0952-7915(95)80022-0 [DOI] [PubMed] [Google Scholar]
- [19].Kavanagh K, Reeves EP. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol Rev 2004; 28:101-12; PMID:14975532; http://dx.doi.org/ 10.1016/j.femsre.2003.09.002 [DOI] [PubMed] [Google Scholar]
- [20].Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol 2010; 10:47-58; PMID:20029447; http://dx.doi.org/ 10.1038/nri2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Seed KD, Dennis JJ. Development of Galleria mellonella as an alternative infection model for the Burkholderia cepacia complex. Infect Immun 2008; 76:1267-75; PMID:18195031; http://dx.doi.org/ 10.1128/IAI.01249-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Cardona ST, Wopperer J, Eberl L, Valvano MA. Diverse pathogenicity of Burkholderia cepacia complex strains in the Caenorhabditis elegans host model. FEMS Microbiol Lett 2005; 250:97-104; PMID:16043310; http://dx.doi.org/ 10.1016/j.femsle.2005.06.050 [DOI] [PubMed] [Google Scholar]
- [23].Uehlinger S, Schwager S, Bernier SP, Riedel K, Nguyen DT, Sokol PA, Eberl L. Identification of specific and universal virulence factors in Burkholderia cenocepacia strains by using multiple infection hosts. Infect Immun 2009; 77:4102-10; PMID:19528212; http://dx.doi.org/ 10.1128/IAI.00398-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schwager S, Agnoli K, Kothe M, Feldmann F, Givskov M, Carlier A, Eberl L. Identification of Burkholderia cenocepacia strain H111 virulence factors using nonmammalian infection hosts. Infect Immun 2013; 81:143-53; PMID:23090963; http://dx.doi.org/ 10.1128/IAI.00768-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sass AM, Schmerk C, Agnoli K, Norville PJ, Eberl L, Valvano MA, Mahenthiralingam E. The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. ISME J 2013; 7:1568-81; PMID:23486248; http://dx.doi.org/ 10.1038/ismej.2013.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Agnoli K, Schwager S, Uehlinger S, Vergunst A, Viteri DF, Nguyen DT, Sokol PA, Carlier A, Eberl L. Exposing the third chromosome of Burkholderia cepacia complex strains as a virulence plasmid. Mol Microbiol 2012; 83:362-78; PMID:22171913; http://dx.doi.org/ 10.1111/j.1365-2958.2011.07937.x [DOI] [PubMed] [Google Scholar]
- [27].Agnoli K, Frauenknecht C, Freitag R, Schwager S, Jenul C, Vergunst A, Carlier A, Eberl L. The third replicon of members of the Burkholderia cepacia complex, plasmid pC3, plays a role in stress tolerance. Appl Environ Microbiol 2014; 80:1340-8; PMID:24334662; http://dx.doi.org/ 10.1128/AEM.03330-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Holden MT, Seth-Smith HM, Crossman LC, Sebaihia M, Bentley SD, Cerdeno-Tarraga AM, Thomson NR, Bason N, Quail MA, Sharp S, et al.. The genome of Burkholderia cenocepacia J2315, an epidemic pathogen of cystic fibrosis patients. J Bacteriol 2009; 191:261-77; PMID:18931103; http://dx.doi.org/ 10.1128/JB.01230-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Silva IN, Tavares AC, Ferreira AS, Moreira LM. Stress conditions triggering mucoid morphotype variation in Burkholderia species and effect on virulence in Galleria mellonella and biofilm formation in vitro. PLoS One 2013; 8:e82522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tomich M, Herfst CA, Golden JW, Mohr CD. Role of flagella in host cell invasion by Burkholderia cepacia. Infect Immun 2002; 70:1799-806; PMID:11895941; http://dx.doi.org/ 10.1128/IAI.70.4.1799-1806.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Urban TA, Griffith A, Torok AM, Smolkin ME, Burns JL, Goldberg JB. Contribution of Burkholderia cenocepacia flagella to infectivity and inflammation. Infect Immun 2004; 72:5126-34; PMID:15322006; http://dx.doi.org/ 10.1128/IAI.72.9.5126-5134.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zlosnik JE, Hird TJ, Fraenkel MC, Moreira LM, Henry DA, Speert DP. Differential mucoid exopolysaccharide production by members of the Burkholderia cepacia complex. J Clin Microbiol 2008; 46:1470-3; PMID:18256220; http://dx.doi.org/ 10.1128/JCM.02273-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Sharma P, Gupta SK, Rolain JM. Whole genome sequencing of bacteria in cystic fibrosis as a model for bacterial genome adaptation and evolution. Expert Rev Anti Infect Ther 2014; 12:343-55; PMID:24502835; http://dx.doi.org/ 10.1586/14787210.2014.887441 [DOI] [PubMed] [Google Scholar]
- [34].Folkesson A, Jelsbak L, Yang L, Johansen HK, Ciofu O, Hoiby N, Molin S. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol 2012; 10:841-51; PMID:23147702; http://dx.doi.org/ 10.1038/nrmicro2907 [DOI] [PubMed] [Google Scholar]
- [35].Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc Natl Acad Sci USA 2013; 110:E250-9; PMID:23271804; http://dx.doi.org/ 10.1073/pnas.1207025110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Smith EE, Buckley DG, Wu ZN, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, et al.. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci USA 2006; 103:8487-92; PMID:16687478; http://dx.doi.org/ 10.1073/pnas.0602138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Dubern JF, Cigana C, De Simone M, Lazenby J, Juhas M, Schwager S, Bianconi I, Döring G, Eberl L, Williams P, et al.. Integrated whole-genome screening for Pseudomonas aeruginosa virulence genes using multiple disease models reveals that pathogenicity is host specific. Environ Microbiol 2015; 17(11):4379-93; PMID:25845292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hilker R, Munder A, Klockgether J, Losada PM, Chouvarine P, Cramer N, Davenport CF, Dethlefsen S, Fischer S, Peng H, et al.. Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ Microbiol 2015; 17:29-46; PMID:25156090; http://dx.doi.org/ 10.1111/1462-2920.12606 [DOI] [PubMed] [Google Scholar]
- [39].Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, et al.. Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol 2006; 7:R90; PMID:17038190; http://dx.doi.org/ 10.1186/gb-2006-7-10-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Loutet SA, Flannagan RS, Kooi C, Sokol PA, Valvano MA. A complete lipopolysaccharide inner core oligosaccharide is required for resistance of Burkholderia cenocepacia to antimicrobial peptides and bacterial survival in vivo. J Bacteriol 2006; 188:2073-80; PMID:16513737; http://dx.doi.org/ 10.1128/JB.188.6.2073-2080.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ortega X, Hunt TA, Loutet S, Vinion-Dubiel AD, Datta A, Choudhury B, Goldberg JB, Carlson R, Valvano MA. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J Bacteriol 2005; 187:1324-33; PMID:15687196; http://dx.doi.org/ 10.1128/JB.187.4.1324-1333.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Bender JK, Wille T, Blank K, Lange A, Gerlach RG. LPS structure and PhoQ activity are important for Salmonella typhimurium virulence in the Galleria mellonella infection model. PLoS One 2013; 8:e73287; PMID:23951347; http://dx.doi.org/ 10.1371/journal.pone.0073287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cunha MV, Leitão JH, Mahenthiralingam E, Vandamme P, Lito L, Barreto C, Salgado MJ, Sá-Correia I. Molecular analysis of Burkholderia cepacia complex isolates from a Portuguese cystic fibrosis center: a 7-year study. J Clin Microbiol 2003; 41:4113-20; PMID:12958234; http://dx.doi.org/ 10.1128/JCM.41.9.4113-4120.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cunha MV, Pinto-de-Oliveira A, Meirinhos-Soares L, Salgado MJ, Melo-Cristino J, Correia S, Barreto C, Sá-Correia I. Exceptionally high representation of Burkholderia cepacia among B. cepacia complex isolates recovered from the major portuguese cystic fibrosis center. J Clin Microbiol 2007; 45:1628-33; PMID:17360834; http://dx.doi.org/ 10.1128/JCM.00234-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P. DNA-Based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J Clin Microbiol 2000; 38:3165-73; PMID:10970351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mahenthiralingam E, Campbell ME, Henry DA, Speert DP. Epidemiology of Burkholderia cepacia infection in patients with cystic fibrosis: analysis by randomly amplified polymorphic DNA fingerprinting. J Clin Microbiol 1996; 34:2914-20; PMID:8940422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Heras J, Dominguez C, Mata E, Pascual V, Lozano C, Torres C, Zarazaga M. GelJ - a tool for analyzing DNA fingerprint gel images. BMC Bioinformatics 2015; 16:270; PMID:26307353; http://dx.doi.org/ 10.1186/s12859-015-0703-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Schoonmaker D, Heimberger T, Birkhead G. Comparison of ribotyping and restriction enzyme analysis using pulsed-field gel electrophoresis for distinguishing Legionella pneumophila isolates obtained during a nosocomial outbreak. J Clin Microbiol 1992; 30:1491-8; PMID:1320629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tan MW, Mahajan-Miklos S, Ausubel FM. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 1999; 96:715-20; PMID:9892699; http://dx.doi.org/ 10.1073/pnas.96.2.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roberts TM, Ward S. Membrane flow during nematode spermiogenesis. J Cell Biol 1982; 92:113-20; PMID:7056795; http://dx.doi.org/ 10.1083/jcb.92.1.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sousa SA, Ramos CG, Almeida F, Meirinhos-Soares L, Wopperer J, Schwager S, Eberl L, Leitão JH. Burkholderia cenocepacia J2315 acyl carrier protein: a potential target for antimicrobials' development? Microbial Pathogenesis 2008; 45:331-6; PMID:18771721; http://dx.doi.org/ 10.1016/j.micpath.2008.08.002 [DOI] [PubMed] [Google Scholar]
- [52].Mil-Homens D, Rocha EP, Fialho AM. Genome-wide analysis of DNA repeats in Burkholderia cenocepacia J2315 identifies a novel adhesin-like gene unique to epidemic-associated strains of the ET-12 lineage. Microbiology 2010; 156:1084-96; PMID:20019083; http://dx.doi.org/ 10.1099/mic.0.032623-0 [DOI] [PubMed] [Google Scholar]
- [53].Steidle A, Sigl K, Schuhegger R, Ihring A, Schmid M, Gantner S, Stoffels M, Riedel K, Givskov M, Hartmann A, et al.. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl Environ Microbiol 2001; 67:5761-70; PMID:11722933; http://dx.doi.org/ 10.1128/AEM.67.12.5761-5770.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.