Salmonella is found in a large variety of environments including soil and water, as well as in the gut flora of warm-blooded animals. One of the requirements to survive in those diverse milieus is its capacity to adapt its metabolism. Salmonella displays phenotypic heterogeneity, a built-in mechanism devoted to generate variability within isogenic populations at the level of the individual, to cope with an ever changing environment. The ability to achieve such heterogeneity can further be considered as dynamic source to drive diversity that benefits bacterial populations inasmuch as variant subpopulations could adapt more efficiently to environmental perturbations and exploit new niches.1,2 Combining biochemistry, proteomics, microbial genetics, competitive infections, and computational approaches has allowed the study of phenotypic heterogeneity of bacterial pathogens revealing the versatile adaptation of Salmonella to a complex nutritional landscape in infected host cells. It was found that Salmonella degrades major nutrients as glycerol, fatty acids, N-acetylglucosamine, glucose, lactate, and arginine to nourish itself during the successive stages of infection.3 Nevertheless, such studies did not yield a resolution at the single host cell or single bacterium level, which would be needed to draw direct conclusions on bacterial heterogeneity. They only allowed the analysis of averaged information of the measured population.
Methods that overcome these hurdles are presented in this issue of Virulence by Diacovich et al. who developed a set of fluorescence probes to explore metabolic pathways used by individual Salmonella during macrophage infection focusing on sugar and fatty acid catabolism (Fig. 1A). They fused promoters of key catabolic enzymes and regulators upstream to a mutant of the green fluorescent protein (gfpmut3) that was original developed by Raphael Valdivia for studies of intracellular bacterial pathogens.4 Such reporters can be analyzed by FACS or microscopy read-outs with the possibility to peer at single infected cells or even at single bacteria. A powerful feature of the presented tools is the possibility to combine fluorescence sensitivity with reporters for distinct metabolic pathways of the sugar and lipid metabolism. Another strength of the fluorescence reporter approach is its adaptability for in vivo experiments and the possibility to scale them via multiplexing for high-throughput screenings followed by high-content analysis. Such usage requires rigorous testing of the read-out robustness to enable automatic computer-assisted analysis. Apart from the novel reporters presented by Diacovich, fluorescent assays have been broadly explored in the context of Salmonella infections to decipher its intracellular trafficking within infected host cells and its communication with distinct host compartments. For example, on the host side it revealed the implication of autophagy during Salmonella invasion, and the precise intracellular localization of the internalized pathogen.5 On the pathogen side, fluorescent approaches have been used to monitor the growth rates of Salmonella, the expression of the bacterial type 3 secretion system or its capacity to adapt to hostile host environments, such as low pH.6,7 The reporters are mostly based on genetically-encoded proteins that can readily be introduced into Salmonella and do not require the addition of other compounds that would render the assays more complicated.
Figure 1.
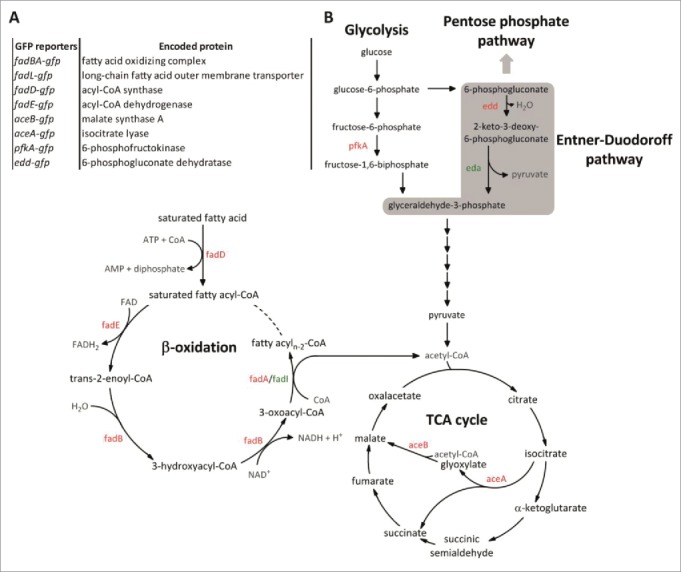
(A) List of fluorescence transcriptional reporters used by Diacovich et al. They were made by cloning the promoter region of selected genes upstream of the fluorescent reporter gene gfpmut3A. The reporters are functional for FACS and microscopy read-outs. (B) Schematic representation of the catabolic part of the central carbon metabolism in S. enterica and elements that were studied by Diacovich et al. The enzymes that correspond to the studied transcriptional fusions are indicated in red and other relevant metabolic enzymes are highlighted in green (eda: 2-keto-3-deoxygluconate-6-phosphate aldolase and fadI: acetyl-CoA acyltransferase). The scheme was generated using the BioCyc Database Collection.11
Using their newly developed metabolic promoter-reporter fusions, the authors described the existence of different subpopulations of Salmonella with differential catabolic behavior either favoring sugars or lipids as carbon source, and they also found that the distribution among these populations changed in response to different substrates.8 Surprisingly, alterations in the glucose and fatty acid availability in the cell culture medium had a strong effect on bacteria within the Salmonella containing vacuole (SCV) of macrophages suggesting a connection between this compartment and the extracellular environment.
The main finding by Diacovich and colleagues was the identification of Entner-Doudoroff pathway (ED) as the most relevant pathway used by intracellular Salmonella to break down glucose (Fig. 1B). At first glance, the prevalence of ED pathway could look disadvantageous as it generates only one ATP per glucose instead of the 2 produced by glycolysis. However, the ED pathway requires 3.5 times less enzymatic protein to achieve the same flux as the glycolytic pathway. Considering this, Flamholz et al. proposed that the choice between glycolytic and ED pathway may reflect a tradeoff between ATP yield and protein costs.9 Taking into account that the SCV contains a limited amount of building blocks to produce large amounts of biologic macromolecules, it seems worthwhile to save resources by choosing “affordable” pathways in terms of protein synthesis. Potentially, this notion is also supported by the increased amount of NADPH through the ED pathway that favors anabolic pathways.10
With the novel reporters the relevance of different, namely sugar versus fatty acid, catabolic pathways during Salmonella infection can be studied in detail in different cell types as well as in vivo. For example, it will be interesting to monitor Salmonella metabolism in different host cells types, including epithelial and dendritic cells. The bacterial reporters can also be combined with reporters for different intracellular localization, host function as well as the expression of virulence genes. The combination of cutting-edge microscopy and surgery procedures allow the tracing of the infection within different organs under well-controlled conditions. This could help to reveal the perplexing data on fadD. Another open question is how the bacteria within the SCV sense the nutrition availability in the extracellular space. How are these 2 spaces connected and which pathways are involved here. Beyond the question of nutrition, this is also relevant for the sensibility of the pathogen toward different antibiotics. It turns out that the understanding of “the individual” during bacterial infection does not only matter for the host (cell type specificities, organ specificities), but is also relevant for the pathogen. Together, the development of novel reporters for the real-time tracing of fundamental physiologic parameters of the pathogen is very welcome to decipher how Salmonella makes itself at home in distinct environments during the course of infection.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded by the ANR (LabEx IBEID, project StopBugEntry and project AutoHostPath) and the European Commission (Consolidator Grant EndoSubvert).
References
- [1].Avery SV. Microbial cell individuality and the underlying sources of heterogeneity. Nat Rev Microbiol 2006; 4:577-87; PMID:16845428; http://dx.doi.org/ 10.1038/nrmicro1460 [DOI] [PubMed] [Google Scholar]
- [2].Lidstrom ME, Konopka MC. The role of physiological heterogeneity in microbial population behavior. Nat Chem Biol 2010; 6:705-12; PMID:20852608; http://dx.doi.org/ 10.1038/nchembio.436 [DOI] [PubMed] [Google Scholar]
- [3].Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, Farhan H, Mazé A, Bumann D. Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS Pathog 2013; 9:e1003301; PMID:23633950; http://dx.doi.org/ 10.1371/journal.ppat.1003301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Cormack BP, Valdivia RH, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP). Gene 1996; 173:33-8; PMID:8707053; http://dx.doi.org/ 10.1016/0378-1119(95)00685-0 [DOI] [PubMed] [Google Scholar]
- [5].Kreibich S, Emmenlauer M, Fredlund J, Ramo P, Munz C, Dehio C, Enninga J, Hardt WD. Autophagy proteins promote repair of endosomal membranes damaged by the salmonella type three secretion system 1. Cell Host Microb 2015; 18:527-37; http://dx.doi.org/ 10.1016/j.chom.2015.10.015 [DOI] [PubMed] [Google Scholar]
- [6].Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. Dynamics of intracellular bacterial replication at the single cell level. Proc Natl Acad Sci U S A 2010; 107:3746-51; PMID:20133586; http://dx.doi.org/ 10.1073/pnas.1000041107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Viala JP, Meresse S, Pocachard B, Guilhon AA, Aussel L, Barras F. Sensing and adaptation to low pH mediated by inducible amino acid decarboxylases in Salmonella. PloS One 2011; 6:e22397; PMID:21799843; http://dx.doi.org/ 10.1371/journal.pone.0022397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Diacovich L, Lorenzi L, Tomassetti M, Meresse S, Gramajo H. The infectious intracellular lifestyle of Salmonella enterica relies on the adaptation to nutritional conditions within the Salmonella-containing vacuole. Virulence 2016:0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Flamholz A, Noor E, Bar-Even A, Liebermeister W, Milo R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc Natl Acad Sci U S A 2013; 110:10039-44; PMID:23630264; http://dx.doi.org/ 10.1073/pnas.1215283110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. New York: Garland Science, 2002. [Google Scholar]
- [11].Caspi R, Billington R, Ferrer L, Foerster H, Fulcher CA, Keseler IM, Kothari A, Krummenacker M, Latendresse M, Mueller LA, et al.. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucl Acids Res 2016; 44:D471-80; PMID:26527732; http://dx.doi.org/ 10.1093/nar/gkv1164 [DOI] [PMC free article] [PubMed] [Google Scholar]


