Abstract
Diffuse large B-cell lymphoma (DLBCL) is an aggressive malignancy and the most common subtype of non-Hodgkin lymphoma in China. However, many cases still remain biologically and clinically heterogeneous, indicating that the DLBCL mechanism remains unclear. MicroRNAs (miRNAs) are critically responsible for lymphomagenesis. We found that plasma miR-21 level was significantly higher in B-cell lymphoma. However, the exact contribution of miR-21 in DLBCL remains unknown.
To determine the function and mechanism of miR-21 in DLBCL, miR-21 and phosphatase and tensin homolog (PTEN) expressions were examined through real-time PCR and immunohistochemical methods. Moreover, the effects of antisense oligonucleotide (ASO) targeting miR-21 (ASO-21) were observed in DLCBL cell line.
MiR-21 expressions in cell line and tissues of patients were significantly higher than those in normal controls, which were inversely correlated with PTEN expression. MiR-21 expression was significantly higher in stage III/IV patients than in stage I/II patients. PTEN protein was expressed positively in only 6 patients with DLBCL (6/26). MiR-21 expression level in the PTEN-negative group was 11.73 (2.13–64.29), which was significantly higher than that in the PTEN-positive group (1.04, 0.67–15.15; P = .038). After down-regulating the miR-21 expression, apoptosis of DLBCL cells increased and PTEN protein was up-regulated in ASO-21-treated cells compared with SCO-21-treated cells by western blot.
These results suggested that miR-21 affects apoptosis of lymphoma cells by regulating the expression of PTEN in DLBCL, which may be associated with increased poor prognosis for DLBCL patients and represents a useful approach for DLBCL treatment.
Keywords: diffuse large B-cell lymphoma, microRNA, miR-21, phosphatase and tensin homologtarget gene
1. Introduction
Diffuse large B-cell lymphoma (DLBCL) is a malignancy of large transformed B lymphocytes. This disease is the most common subtype of non-Hodgkin lymphoma, accounting for approximately 35% to 40% of non-Hodgkin lymphoma.[1] Possible factors leading to high incidence of lymphoma include virus infection, genomic translocations or genetic alterations, environmental factors, chemical factors, and immune system disorders. Although DLBCL is a curable lymphoma in advanced stages, up to 40% of patients eventually relapse or fail to achieve remission, indicating that the mechanism of DLBCL remains unclear.
MicroRNAs (miRs) are short noncoding RNAs (20–24 nucleotides) that control gene expression by targeting specific genes in a post-transcriptional way.[2,3] miRs are involved in the pathogenesis of tumors acting as oncogenes or tumor suppressor genes.[4–7] Thus, miRs are excellent tools for advance molecular diagnostics and targeted molecular therapy in cancer. Among cancer-related miRs, miR-21 has been invariably and consistently found to be overexpressed in almost every diverse type of malignant tumors,[8–11] and has been reported to be mediated in cancer-related processes. We have showed that plasma miR-21 level in patients with B-cell lymphoma is higher than that in normal controls.[12] MiR-21 is closely associated with DLBCL as an onco-miRNA; however, only few studies have analyzed its mechanism. Phosphatase and tensin homolog (PTEN) has recently become a candidate target of miR-21 in tumorigenesis, such as in cancers of the breast, colon, lung, pancreas, prostate, and stomach.[13] PTEN is the candidate target of miR-21 in tumorigenesis.[14] Previous studies have indicated that PTEN is down-regulated in DLBCL as a tumor suppressor gene.[15] However, no investigation has been carried out to identify the association between deficiency of expression and miR-21. In the present study, we investigate the mechanism and clinical significance of miR-21 in DLBCL.
2. Methods
2.1. Patients and tissue samples
A total of 36 tissue samples were collected from 26 patients with DLBCL and 10 normal controls. The patients were diagnosed at the Hematology Department, Tianjin Medical University General Hospital. Clinical and pathological data of the patients are displayed in Table 1. All cases were divided into germinal center-type DLBCL (GCB-DLBCL) and nongerminal center-type by b-cell lymphoma 6, CD10, and multiple myeloma oncogene 1. Tissue samples were immediately fixed in 10% formalin and embedded using paraffin. Normal lymphocytes in the peripheral blood were also collected from 8 healthy donors (3 females and 5 males). Both tumor and nontumor samples were confirmed by 2 pathologists as per the 2008 WHO classification of morphological criteria for the diagnosis of lymphoma pathology. The pathological stage, grade, and nodal status were appraised by an experienced pathologist. The study was approved by the Ethics Committee of Tianjin Medical University. Informed written consent was obtained from all patients or their parents in accordance with the Declaration of Helsinki.
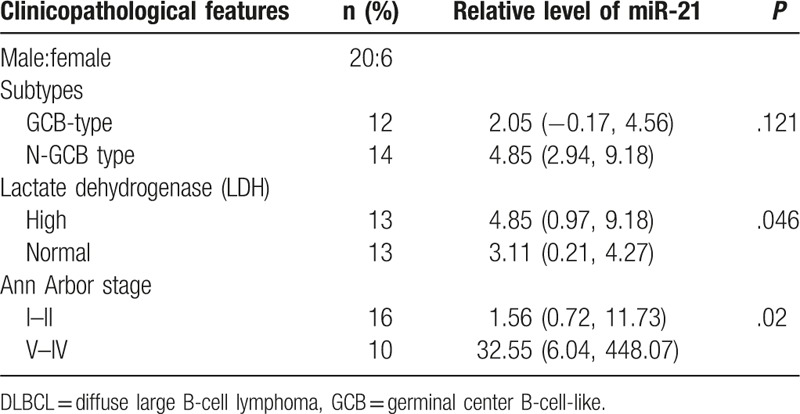
Table 1.
Summary of clinical details of the DLBCL patients (n = 26).
2.2. Cell culture, gene knockdown, and transduction
The DLBCL cell line CRL-2630 was thawed and cultured in RPMI1640 containing 10% fetal calf serum in a 5% CO2 humidified incubator at 37°C. For transfection, cells were plated at a density of 106 cells/well in 24-well dishes with 500 μL of medium, and then transfected with oligonucleotides using an electroporation system at a final working concentration of 200 nM. The antisense oligonucleotide (ASO-21) and the scrambled control oligonucleotide (SCO-21) were 5-TCAACATCAGTCTGATAAGCTA-3 and 5-TAACGTCACTTCGACTGAACTGCT-3, respectively. After 2 days, the cells were collected, resuspended in fresh medium, and used in subsequent experiments.
2.3. Real-time quantitative PCR
Recover All Total Nucleic Acid Isolation kit (Ambion) was used to extract total RNA from the primary specimens embedded in paraffin according to the instructions of the manufacturer. RNA was extracted from cells using Trizol reagent (Invitrogen, Paisley, UK) as described by the manufacturer. Quantitative stem-loop reverse transcription (RT) was then performed with a Taqman miRNA RT kit (Applied Biosystems), using 1 μg of total RNA per reaction (Applied Biosystems). All reactions were performed in triplicate. MiR-21 expression was detected using the ABI 7500 quantitative PCR system with the SYBGREEN MIX (Applied Biosystems) according to the product manual, and U6 was used as a control. Mean cycle threshold (Ct) values for miRNAs were quantified by sequence detection system software (SDS, version 2.1; Applied Biosystems). The miRNA expression was normalized to U6 mRNA expression, △△Ct = (CtmiR-21-CtU6) tumor − (CtmiR-21-CtU6) normal, 2-△△Ct value representative of the level of miR-21 expression, and P values were calculated using Mann–Whitney U test.
2.4. Immunohistochemical staining
A total of 26 DLBCL cases were classified as either GCB or non-GCB. The selected tumor tissues were used to construct tissue microarray slides. Paraffin sections were cut and mounted on glass slides, and 5-μm sections from formalin-fixed and paraffin-embedded specimens were deparaffinized using xylene and rehydrated in graded ethanol. Samples were then preincubated with 3% H2O2 to eliminate endogenous peroxidase activity. Antigen retrieval was achieved by heating the sections for 2 minutes at 100°C in citric acid buffer (0.01 mol/L, pH 6.0). Immunohistochemistry was performed using the 2-step method with primary antibody, including heat-induced antigen-retrieval procedures. Sections were incubated overnight at 37°C with primary antibody PTEN (1:100 dilutin; Sigma, St. Louis, MO). After the primary antibody was washed off, the components of the EnVision Detection System were applied with an antimouse polymer (Envision 1/HRP/Mo, Dako, Glostrup, Denmark). PBS buffer, instead of primary antibody, was used as a negative control. Immunostaining was classified based on staining intensity and percentage of positive tumor cells. Staining intensity was determined as 0 (absent), 1 (weak), 2 (moderate), or 3 (strong). To assess the positive degree of antigen expression, the expression levels of the antigens were semiquantified using an immunohistochemistry score (H-score method) according to the reference,[16] which was calculated by multiplying the staining intensity with the percentage of positive tumor cells. The H-score ranged from 0 to 300, where H-score = (1 × %1+) + (2 × %2+) + (3 × %3+). Patients with an immunohistochemistry score ≤140 were considered to have negative or weak immunoreactivity, whereas those with a score of >140 were classified to be moderately to strongly immunoreactive, which will be used for subsequent analysis.
2.5. Apoptosis analysis
An annexin V-PI apoptosis detection kit (BD Biosciences, Franklin Lakes, NJ) was used to assess apoptosis. The cells were collected and resuspended in fresh medium. The cells were then exposed to 100 μmol/L etoposide for 4 hours, after which, the assays were obtained. The cells were stained with Annexin V and propidiom iodide (PI) at room temperature for 15 minutes in the dark. Cell apoptosis was analyzed using FACSCalibur flow cytometry.
2.6. Western blot analysis
Total proteins of CRL-2630 cells were extracted using RIPA (Biotech, Beijing, China), and the protein concentration was determined using the commercial Bradford reagent assay (Bio-Rad, Hercules, CA). Approximately 50 μg of total protein was used for each treatment to detect PTEN/GAPDH. Solubilized proteins were separated using 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred onto polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford) using a Trans-Blot Cell system (Bio-Rad, Hercules, CA) in transfer buffer (25 mM Tris, 190 mM glycine,10% methanol) at 80 V for 1 hour at room temperature. The membranes were blocked with 5% skim milk in PBS for 1 hour, and then incubated with antibodies (PTEN, 1:1000, Santa Cruz, CA) overnight at 4°C. After washing, the membrane was incubated with appropriate HRP-conjugated secondary antibody (1:5000) for 1 hour at room temperature. The blots were revealed with an enhanced chemiluminescence system (Gene). The GAPDH antibody (Santa Cruz, CA) as a control was diluted (1:1000) and developed using the secondary antibody and chemiluminescence system previously described.
2.7. Statistical analyses
The results were analyzed using SPSS 18.0. Measurement data were presented as median and interquartile range. MiR-21 expression levels from the 2 groups were compared and analyzed using Mann–Whitney U test. P < .05 was considered significant.
3. Results
3.1. MiR-21 overexpression in DLBCL patients
We extracted the total RNA from the paraffin-embedded tissues and normal lymph nodes to examine miR-21 expression in 26 DLBCL patients. The level of miR-21 was 2.7 (0.13–5.25) in DLBCL tissues, which was significantly higher than that in normal lymph nodes (−0.34, −1.51 to 1.39; P = .008) (Fig. 1A). We also extracted total RNA from CRL-2630 cells and normal lymphocytes; the results showed that the miR-21 level was up-regulated in CRL-2630 cells (Fig. 1B).
Figure 1.
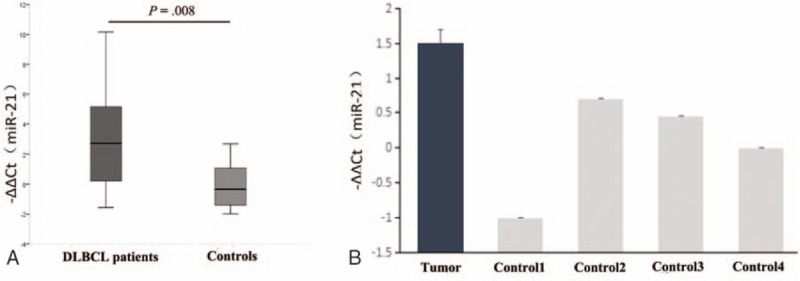
Quantitative RT-PCR analysis of miR-21 in specimens of primary DLBCL and DLBCL cell line. (A) The level of miR-21 was significantly higher in DLBCL patients (n = 26) than in healthy controls (n = 10; P = .008). (B) Quantitative PCR analysis of mature miR-21 in normal cells and DLBCL cell line. Results showed that the level of miR-21 was significantly higher in DLBCL line than in healthy controls. P values were calculated using Mann–Whitney U test (P < .05). DLBCL = diffuse large B-cell lymphoma.
Moreover, we found that miR-21 expression from DLBCL patients positively correlated with the level of serum lactate dehydrogenase (LDH) (P = .046). MiR-21 expression from stage III/IV patients were significantly higher than that from stage I/II patients (P = .02) (Table 1).
3.2. PTEN protein expressed lowly in DLBCL
We found that miR-21 was overexpressed in CRL-2630 cell lines and paraffin-embedded tissues of DLBCL patients. We then conducted immunohistochemical staining with paraffin-embedded tissues of DLBCL patients. Results showed that miR-21 expression level was negatively correlated with PTEN protein level. PTEN protein was expressed positively in 7 normal controls (7/10), whereas only in 6 patients with DLBCL (6/26). PTEN was expressed prominently in the nucleus, and certain amounts were observed in the cell membrane and cytoplasm of malignant cells, with positive rates of 23% (Fig. 2). MiR-21 expression levels in the PTEN-negative group were 11.73 (2.13–64.29), which was significantly higher than that in the PTEN-positive group (1.04, 0.67–15.15; P = .038).
Figure 2.
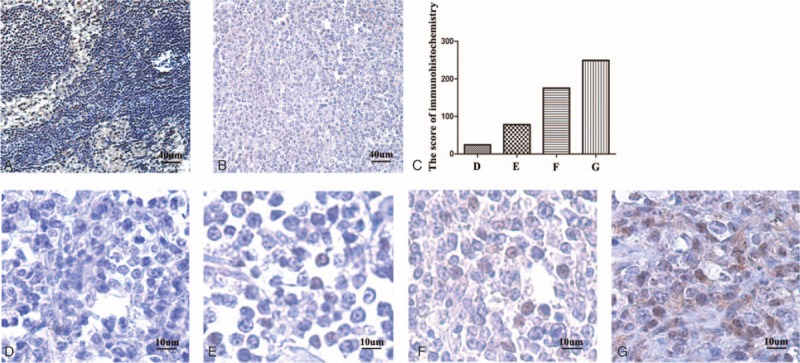
The expression of PTEN protein in DLBCL patients. The expression of PTEN protein was observed by microscope in normal control (A), DLBCL patients (B) in 10 times, and DLBCL patients in different positive rates in 40 times (C–G). Immunoreactivity was classified as follows: 0 (D), 1+ (E), 2+ (F), and 3+ (G). DLBCL = diffuse large B-cell lymphoma, PTEN = phosphatase and tensin homolog.
3.3. MiR-21 induced down-regulation of PTEN in DLBCL
After the transfection of ASO-21, the mRNA expression level of miRNA-21 was down-regulated by 30% compared with the internal reference U6 snRNA. Apoptosis analysis was performed to investigate whether DLBCL cell apoptosis could be influenced after miR-21 expression inhibition and examine the effects of ASOs targeted to miR-21 (ASO-21) compared with cells treated with SCO-21 in tumor cells. We found that ASO-21 increased the apoptosis of CRL-2630 cells after exposure to 100 μmol/L etoposide for 4 hours (Table 2, Fig. 3A and B).[17] We detected significant differences in the cell apoptosis between SCO-21-treated and ASO-21-treated cells. We then conducted Western blot analyses with SCO-21- and ASO-21-treated cells (Fig. 3C). Western blot analysis revealed that PTEN was up-regulated in ASO-21-treated cells compared with SCO-21-treated cells. These results reflect that miR-21 contributes to lymphomagenesis by reducing apoptotic activity, with PTEN as its probable target gene.
Table 2.
Apoptosis ratio of CRL-2630 cells after transfection of ASO-21.
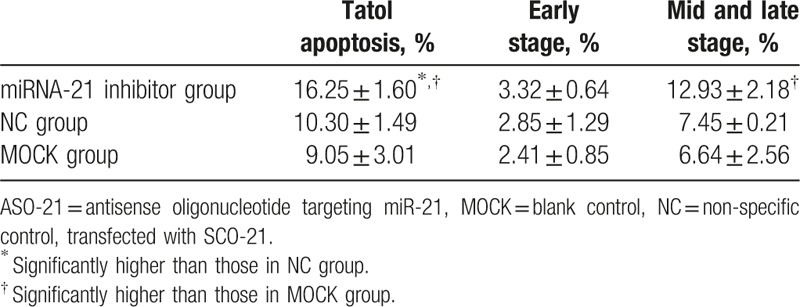
Figure 3.
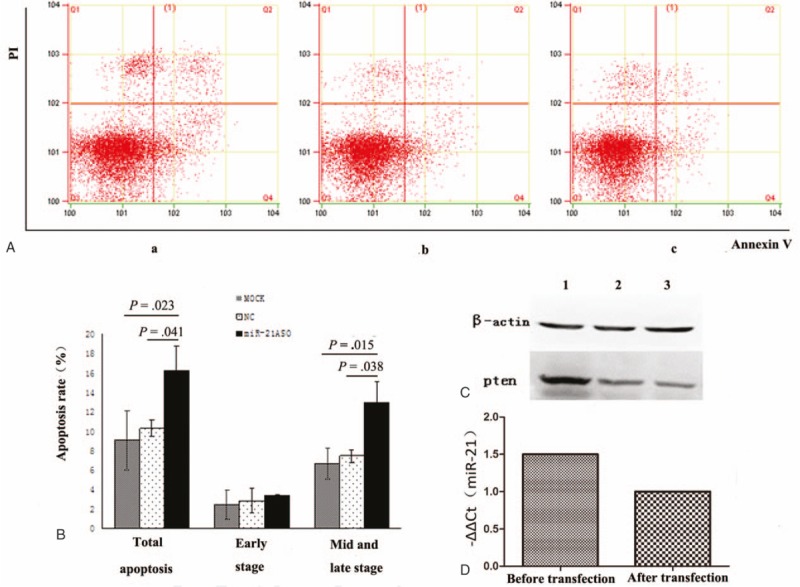
The function of miR-21 in DLBCL. miR-21 induced apoptosis of tumor cells in DLBCL. ASO-21 and SCO-21-treated cells were exposed to 100 μmol/L etoposide for 4 hours. (A) Flow cytometric analysis of ASO-21-treated and SCO-21-treated DLBCL cells. Apoptotic cells are in Q2 and Q4. Apoptotic rate (Q2 + Q4) of ASO-21-treated cells (a) showed significantly higher percentage than those of SCO-21-treated cells (b) and normal cells (c). (B) Percentages of apoptotic cells (Q2 + Q4). Symbols and bars indicate means and SDs of triplicate samples. Total apoptosis rate of miR-21ASO group significantly increased compared with NC group (P = .041) and MOCK group (P = .023); and mid and late stage of apoptosis of miR-21 ASO group significantly increased compared with NC group (P = .038) and MOCK group (P = .015). (C) PTEN protein was detected using Western blot 48 hours after transfection. 1 = ASO-21 group, 2 = normal control group, 3 = MOCK group. ASO-21 = antisense oligonucleotide targeting miR-21, DLBCL = diffuse large B-cell lymphoma, MOCK = blank control, NC = non-specific control, transfected with SCO-21, PTEN = phosphatase and tensin homolog, SCO = scrambled control oligonucleotide.
4. Discussion
Diffuse large B-cell lymphoma is characterized by marked clinical and pathological heterogeneity reflected at the molecular level. Gene expression and immunohistochemical studies have revealed the presence of at least 2 distinct molecular subtypes of DLBCL that represent the postulated cell of origin, the germinal center B-cell-like (GCB), and the activated B-cell-like (ABC).[18] MiR-21 up-regulation commonly occurs in tumor cells or in body fluids of patients with DLBCL.[19] We have showed that plasma miR-21 level in patients with B-cell lymphoma is significantly higher than in normal controls.[20] miR-21 mRNA was extracted from tumor tissues and was detected using quantitative RT-PCR, which showed higher miR-21 mRNA expression in lymphoma. The current study provides further evidence showing that circulating miRNAs detected in the serum differed between patients with cancer and healthy individuals. The results also suggest that circulating miRNAs may be from tumor cells. In terms of miR-21 sources, previous studies have shown that high miR-21 expression levels originate not only from tumor cells but also from circulating B cells in AIDS-related non-Hodgkin lymphoma.[21] Therefore, other sources of high miR-21 expression exist. Our data showed that DLBCL cell line CRL-2630 and specimens of primary lymphoma overexpressed miR-21. Moreover, high miR-21 expression was associated with LDH (P < .05). The elevated LDH suggested high tumor burden and poor prognosis, which can also mean that higher levels of miR-21 may indirectly suggest poor prognosis. These corollaries are consistent with our previous findings in patients with B-cell lymphoma, showing high miR-21 expression associated with poor prognosis. We found no significant correlation between miR-21 expression and the 2 subtypes in these cases. Lawrie et al[22] suggested that higher miR-21 expression in DLBCL can be associated with improved relapse-free survival times, but not overall survival, and that expression level was higher in DLBCL cases with an ABC-type immunophenotype than with GCB-type.[23] However, they did not describe the differences between the 2 subtypes in the subsequent study on plasma miR-21 in DLBCL patients. Therefore, further study is needed to confirm this claim. The 5-year survival rates between GCB-type and ABC-type were, respectively, 74% and 36% after chemotherapy, which showed that the study of the miR-21 difference between different types is meaningful. However, in the current study, no significant difference is shown because the sample size may be limited; thus, a study involving larger sample size is necessary to further clarify the miR-21 mechanism. To explore the miR-21-intervening biological behavior of tumor cells in DLBCL, we transfected miR-21 ASOs into DLBCL cell lines through electroporation. The incidence of cell apoptosis increased after miR-21 transfection. Medina et al[24] showed that miR-21 overexpression results in a pre-B malignant lymphoid-like phenotype. When miR-21 was inactivated, the tumors completely regressed after a few days. This result provides a theoretical basis for the applied miR-21 inhibitor to targeted therapy, and also suggests the possibility of treating cancer by targeting the apoptotic pathway. MiRNA molecules regulate gene expression at the post-transcriptional level. The pathogenesis of DLBCL involves a variety of regulatory factors acting in multiple signaling pathways. PTEN is a candidate target of miR-21 in tumorigenesis, such as in breast cancer,[25] lung cancer,[26] and colon cancer.[27] Previous studies have indicated that PTEN is down-regulated in DLBCL as a tumor suppressor gene if the loss of expression is associated with miR-21 levels. In the present study, results of immunohistochemical staining with paraffin-embedded DLBCL tissue showed that the miR-21 expression level was negatively correlated with PTEN protein level. In the Western blot analyses of SCO-21 and ASO-21-treated cells, we found that PTEN was up-regulated in ASO-21-treated cells compared with SCO-21-treated cells. Therefore, miR-21 down-regulates the tumor suppressor (PTEN) in DLBCL. Gu et al also found that miR-21 may target Pdcd4 and PTEN in DLBCL cell lines OCI-LY3 and OCI-LY10, suggesting the relevance of miR-21 in tumor signaling pathway. Studies have indicated that miR-21 may be targeted by a multiple of genes, and it appears to be cell type-specific. Thus, the functions of miR-21 in the pathogenesis of these diseases remain largely unknown.
5. Conclusions
In conclusion, we showed that miR-21 up-regulation contributed to the pathogenesis of DLBCL by inducing apoptosis of tumor cells, and a higher level of specific miR-21 was associated with diagnosis and prognosis of patients with DLBCL. miR-21 may affect apoptosis by regulating the expression of PTEN. These findings provide new insights into the pathogenesis of DLBCL and suggest that targeting miR-21 may represent a useful approach in the treatment of DLBCL.
Footnotes
Abbreviations: ABC = activated B-cell-like, ASO = antisense oligonucleotide, DLBCL = diffuse large B-cell lymphoma, GCB = germinal center B-cell-like, LDH = lactate dehydrogenase, miRNAs = microRNAs, PI = propidiom iodide, PTEN = phosphatase and tensin homolog, PVDF = polyvinylidene difluoride, SCO = scrambled control oligonucleotide, SDS-PAGE = sodium dodecyl sulfate–polyacrylamide gel electrophoresis.
J.S., Q.S., and C.L. contributed equally to this work.
Funding: This work was supported by the National Natural Science Foundation of China (Grant nos. 81570106, 81770110, 81600093, 81600088); the anticancer major special project of Tianjin (Grant nos. 12ZCDZSY18000), the Tianjin Municipal Natural Science Foundation (Grant nos. 14JCYBJC25400, 15JCYBJC24300), Tianjin Key Projects of Health and Family Planning Commision(Grant no. 15KG150).
The authors report no conflicts of interest.
References
- [1].Coiffier B. Diffuse large cell lymphoma. Curr Opin Oncol 2001;13:325–34. [DOI] [PubMed] [Google Scholar]
- [2].Baek D, Villen J, Shin C, et al. The impact of microRNAs on protein output. Nature 2008;455:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Sellbach M, Schwanhausser B, Thierfelder N, et al. Widespread changes in protein synthesis induced by microRNAs. Nature 2008;455:58–63. [DOI] [PubMed] [Google Scholar]
- [4].Ni Y, Meng L, Wang L, et al. MicroRNA-143 functions as a tumor suppressor in human esophageal squamous cell carcinoma. Gene 2013;517:197–204. [DOI] [PubMed] [Google Scholar]
- [5].Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nature Rev Cancer 2006;6:259–69. [DOI] [PubMed] [Google Scholar]
- [6].Yang M, Liu R, Sheng J, et al. Differential expression profiles of microRNAs as potential biomarkers for the early diagnosis of esophageal squamous cell carcinoma. Oncol Rep 2013;29:169–76. [DOI] [PubMed] [Google Scholar]
- [7].Matsushima K, Isomoto H, Yamaguchi N, et al. MiRNA-205 modulates cellular invasion and migration via regulating zinc finger E-box binding homeobox 2 expression in esophageal squamous cell carcinoma cells. J Translat Med 2011;9:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 2005;65:6029–33. [DOI] [PubMed] [Google Scholar]
- [9].Xu LF, Wu ZP, Chen Y, et al. MicroRNA-21(miR-21) regulates cellular proliferation, invasion, migration, and apoptosis by targeting RTEN, RECK and Bcl-s in lung squamous carcinoma, Gejiu City, China. PLoS One 2014;9: e103698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lv L, Huang F, Mao H, et al. MicroRNA-21 is overexpressed in renal cell carcinoma. Int J Biol Markers 2013;28:201–7. [DOI] [PubMed] [Google Scholar]
- [11].Hiyoshi Y, Kamohara H, Karashima R, et al. MiroRNA-21 regulates the proliferation and invasion in esophageal squamous cell carcinoma. Clin Cancer Res 2009;15:1915–22. [DOI] [PubMed] [Google Scholar]
- [12].Ge TT, Liang Y, Fu R, et al. [Expressions of miR-21, miR-155 and miR-210 in plasma of patients with lymphoma and its clinical significance]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2012;20:305–9. [PubMed] [Google Scholar]
- [13].Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006;103:2257–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Roy S, Yu Y, Padhye SB, et al. Difluorinated-curcumin (CDF) restores PTEN expression in colon cancer cells by down-regulating miR-21. PLoS One 2013;8:e68543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Pfeifer M, Grau M, Lenze D, et al. PTEN loss defines a PI3K/AKT pathway-dependent germinal center subtype of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A 2013;110:12420–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Parris TZ, Aziz L, Kovacs A, et al. Clinical relevance of breast cancer-related genes as potential biomarkers for oral squamous cell carcinoma. BMC Cancer 2014;14:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yamanaka Y, Tagawa H, Takahashi N, et al. Aberrant overexpression of microRNAs activate AKT signaling via down-regulation of tumor suppressors in natural killer-cell lymphoma/leukemia. Blood 2009;114:3265–75. [DOI] [PubMed] [Google Scholar]
- [18].Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004;103:275–82. [DOI] [PubMed] [Google Scholar]
- [19].Baraniskin A, Kuhnhenn J, Schlegel U, et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 2011;117:3140–6. [DOI] [PubMed] [Google Scholar]
- [20].Ge TT, Liang Y, Fu R, et al. [Expressions of miR-21, miR-155 and miR-210 in plasma of patients with lymphoma and its clinical significance]. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2012;20:305–9. [PubMed] [Google Scholar]
- [21].Thapa DR, Bhatia K, Bream JH, et al. B-cell activation induced microRNA-21 is elevated in circulating B cells preceding the diagnosis of AIDS-related non-Hodgkin lymphomas. AIDS 2012;26:1177–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 2008;141:672–5. [DOI] [PubMed] [Google Scholar]
- [23].Lawrie CH, Soneji S, Marafioti T, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer 2007;121:1156–61. [DOI] [PubMed] [Google Scholar]
- [24].Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010;467:86–90. [DOI] [PubMed] [Google Scholar]
- [25].Han M, Liu M, Wang Y, et al. Antagonism of miR-21 reverses epithelial-mesenchymal transition and cancer stem cell phenotype through AKT/ERK1/2 inactivation by targeting PTEN. PLoS One 2012;7:e39520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhang W, Bai W, Zhang W. MiR-21 suppresses the anticancer activities of curcumin by targeting PTEN gene in human non-small cell lung cancer A549 cells. Clin Transl Oncol 2014;16:708–13. [DOI] [PubMed] [Google Scholar]
- [27].Deng J, Lei W, Fu JC, et al. Targeting miR-21 enhances the sensitivity of human colon cancer HT-29 cells to chemoradiotherapy in vitro. Biochem Biophys Res Commun 2014;443:189–95. [DOI] [PubMed] [Google Scholar]


