Nobs et al. show that PPARγ drives pathogenic type-2 effector responses in the lung in both T cells and DCs by controlling IL-33–driven Th2 effector function and lung DC migration and Th2 priming capacity.
Abstract
Type-2 immune responses are well-established drivers of chronic inflammatory diseases, such as asthma, and represent a large burden on public health systems. The transcription factor PPARγ is known to promote M2-macrophage and alveolar macrophage development. Here, we report that PPARγ plays a key role in both T cells and dendritic cells (DCs) for development of type-2 immune responses. It is predominantly expressed in mouse Th2 cells in vitro and in vivo as well as human Th2 cells from allergic patients. Using conditional knockouts, we show that PPARγ signaling in T cells, although largely dispensable for IL-4 induction, is critical for IL-33–driven Th2 effector function in type-2 allergic airway responses. Furthermore, we demonstrate that IL-4 and IL-33 promote up-regulation of PPARγ in lung-resident CD11b+ DCs, which enhances migration to draining lymph nodes and Th2 priming capacity. Thus, we uncover a surprising proinflammatory role for PPARγ and establish it as a novel, important mediator of DC–T cell interactions in type-2 immunity.
Introduction
Type-2 immune responses are thought to have evolved as protective mechanisms against parasitic infections, especially against helminths. More recently, they have also been associated with wound repair and reestablishing tissue homeostasis (Wynn, 2015). Because of increased hygiene and possibly other factors, diseases characterized by aberrant forms of type-2 immunity, including allergies, have become a major health burden in western societies. Asthma is a prime example of a widespread, chronic inflammatory disease affecting 300 million people worldwide. This disease of the respiratory tract is classically associated with reversible airway obstruction, airway hyperresponsiveness, infiltration of eosinophils, mucus production, and a Th2-type inflammation (Gregory and Lloyd, 2011). It is generally induced by allergens, such as house dust-mite fecal pellets (von Mutius, 2009). Allergens are inhaled and, upon reaching the airways, are recognized by epithelial cells through pattern-recognition receptors, leading to the secretion of inflammatory mediators, such as thymic stromal lymphopoietin and IL-33, which, in turn, activate group 2 innate lymphocytes (ILC2s) and DCs to initiate allergen-specific immune responses (Willart et al., 2012). DCs, as specialized APCs, are essential for the uptake, transport, and subsequent presentation of these innocuous antigens to T cells (van Rijt et al., 2005), which are the main drivers of allergy-associated inflammation in the lung once individuals are reexposed to the allergen (Kopf et al., 1993).
Peroxisome proliferator-activated receptor γ (PPARγ) is a lipid-activated transcription factor that has an important role in regulating genes associated with lipid metabolism as well as being essential for adipocyte development. In the immune system, PPARγ is thought to have an important role in polarization of macrophages toward an M2 or anti-inflammatory phenotypes (Bouhlel et al., 2007), and PPARγ, acting in CD4+ T cells, has been suggested to inhibit Th17 differentiation and thereby suppress autoimmunity in the central nervous system (Klotz et al., 2009). More recently, our laboratory showed that PPARγ is essential for the development of alveolar macrophages (AMs) in the lung and that, in its absence, animals develop pulmonary alveolar proteinosis (Schneider et al., 2014b). In the context of pulmonary, allergic inflammation, it has been shown that treatment with PPARγ agonists, such as rosiglitazone, dampens inflammation, and that has been linked to an inhibitory role in DCs and eosinophils (Woerly et al., 2003; Hammad et al., 2004). However, the underlying mechanism, and which cell types are actually targeted by these agents, is largely unclear. To more thoroughly address the role of PPARγ in type-2 immunity, we studied the cell-intrinsic role of PPARγ in two key immune cell types in this context, i.e., antigen-presenting DCs as initiators and Th2 cells as drivers of type-2 responses.
We find that PPARγ, in both T cells and DCs, controls development of type-2 immunity. In CD4+ T cells, PPARγ is highly expressed in both mouse and human Th2 cells and intrinsically controls Th2 differentiation and effector function. In addition, in lung CD11b+ DCs, PPARγ intrinsically controls priming of naive T cells toward Th2 polarization in vivo. Thus, we uncover a surprising and, thus far, unappreciated, proinflammatory role of PPARγ in type-2 immunity.
Results
PPARγ intrinsically controls Th2 effector function in vivo
We aimed to address the role of PPARγ comprehensively in the context of allergic inflammation and decided to focus first on T cells as key drivers (i.e., Th2 cells) and regulators (i.e., regulatory T cells [Treg cells]) of type-2 immune responses. For this purpose, we generated T cell–specific PPARγ KO animals by crossing Ppargfl/fl to mice expressing Cre under the Cd4 promoter. To assess the specificity and efficiency of CD4–Cre-mediated deletion, we crossed Cd4–Cre animals to the Rosa26-RFP-Cre reporter strain (Cd4-Cre Rosa26f/l+ RFP) and evaluated RFP expression in different cell types. 85–90% of CD4+ and CD8+ T cells were found to be RFP+, whereas other cell types were barely affected, with the exception of ILCs, where 10% of cells were found to express RFP (Fig. S1 A).
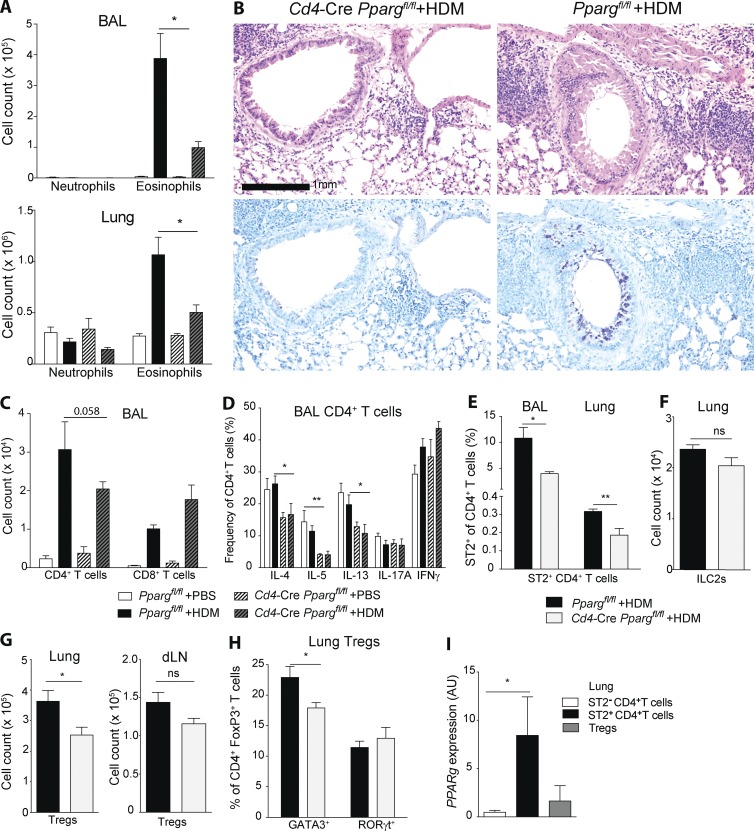
To evaluate the T cell intrinsic role of PPARγ in pulmonary allergic immunity, we sensitized Ppargfl/fl Cd4–Cre and Cre-negative controls with house dust mite extract (HDM) as a common model for type-2 allergic inflammation. To evaluate the impact of T cell–specific deletion of PPARγ on the development of asthmatic inflammation, we quantified pulmonary eosinophilia, one of the classical hallmarks of this disease, in animals subsequently challenged with PBS or HDM, as indicated in the scheme (Fig. S1 B). HDM challenge induced significant recruitment of eosinophils into the bronchoalveolar lavage (BAL) and lungs of control animals (Fig. 1 A). However, the number of eosinophils was strongly reduced in Ppargfl/fl Cd4–Cre mice, whereas the number of neutrophils was unchanged (Fig. 1 A). In addition, peribronchial inflammation, as well as the number of mucus cells, was strongly reduced in T cell–specific, PPARγ KO animals (Fig. 1 B), and a trend toward lower levels of IgE could be observed (Fig. S1 C). Furthermore, the number of CD4+ T cells was slightly reduced in the in the BAL of Ppargfl/fl Cd4–Cre animals (Fig. 1 C). To evaluate whether intrinsic deletion of PPARγ in CD4+ T cells might directly affect polarization and the T cell effector response, we measured their cytokine profile (i.e., Th1, Th2, Th17) and ST2 expression. The frequency of IL-4–, IL-5–, and IL-13–expressing cells was strikingly reduced, whereas IFN-γ– or IL-17–expressing cells were enhanced or unchanged, respectively, in Ppargfl/fl Cd4–Cre mice in the BAL (Fig. 1 D). Consistent with a reduction of Th2 cytokine-producing cells, the frequency of CD4+ T cells expressing ST2 (IL-33R), a surface marker of Th2 cells (Lambrecht et al., 2000) was reduced in the absence of PPARγ (Fig. 1 E), whereas pulmonary ILC2 cells remained unaffected (Fig. 1 F). Characterizing the Treg compartment of HDM-treated animals, we found that the number of Treg cells was significantly reduced in the lung, but not in lung-draining LNs, of Ppargfl/fl Cd4–Cre mice compared with controls (Fig. 1 G). Treg cells were recently described to consist of distinct subsets that can be distinguished by expression of the transcription factors RORγt and GATA3 (Wohlfert et al., 2011; Ohnmacht et al., 2015). We, therefore, examined the impact of T cell–specific deletion of PPARγ on the makeup of the Treg compartment. Indeed, we found that the frequency of GATA3-expressing CD4+FoxP3+ cells was reduced, whereas Treg cells expressing RORγt were unaffected by the loss of PPARγ (Fig. 1 H). Corroborating a role of PPARγ in Th2 and Treg development, we found that Pparg expression was highly expressed in pulmonary ST2+CD4+ T cells and, to a lesser extent, in Treg cells (i.e., Foxp3-GFP+CD4+ T cells isolated from DEREG mice) compared with ST2−CD4+ (non-Th2) cells at the peak of HDM-induced lung inflammation (Fig. 1 I).
Figure 1.
PPARγ in T cells mediates development of pulmonary allergic inflammation. (A–I) Ppargfl/fl and Cd4-CrePpargfl/fl mice were sensitized intratracheally with 10 µg HDM on d 0 and subsequently challenged intratracheally with 10 µg HDM or PBS on days 7–11. Animals were analyzed on day 14. Flow cytometry was used to characterize and quantitate BAL and lung cell populations, including eosinophils (A) and neutrophils (B). (B, top) Hematoxylin and eosin histology. (B, bottom) PAS and Alcian blue histology. CD4+ and CD8+ T cells (C) and BAL CD4+ T cells (D) were restimulated with PMA/ionomycin for 4 h. Shown is the frequency of CD4+ T cells that produced the indicated cytokines. The data presented are pooled from two independent experiments (n = 5–11/group). Shown are ST2+CD4+ Th2 cells (E), lin−CD90+CD127+CD25+ ILC2s (F), Foxp3+CD4+ Treg cells (G), and GATA3+ and RORγt+ cells (H) among CD4+FoxP3+ Treg (n = 4–6/group). (I) DEREG mice were treated with HDM as described in this legend. GFP+CD4+ Treg, ST2+CD4+ Th2, and ST2−CD4+ non-Th2 cells were sorted on day 14 by flow cytometry. Shown is the Pparg mRNA expression measured by quantitative PCR (n = 3/group). The data are representative of three experiments. Data are means ± SEM and the sample size (n). The Student’s t test (unpaired) was used. *, P < 0.05; **, P < 0.01.
To evaluate whether that deficiency in T cell–driven, type-2 immunity was specific to the HDM model, we tested Ppargfl/fl Cd4–Cre animals in the widely used OVA/Alum experimental asthma model (Fig. S1 D). Very similar to the HDM model, the number of eosinophils in the BAL was strongly reduced, whereas the number of neutrophils was somewhat increased (Fig. S1 E). Again, a trend toward fewer CD4+ T cells infiltrating in the BAL could be observed in the BAL of T cell–specific PPARγ KO animals (Fig. S1 F). Moreover, IL-4+ and IL-5+ CD4+ T cells, but not IFN-γ+ and IL-17+ CD4+ T cells, were significantly impaired if they intrinsically lacked PPARγ (Fig. S1 G). Collectively, these results suggest that PPARγ critically and specifically regulates Th2 effector function in vivo in different settings as well as having a minor role in regulating the phenotype of lung Treg cells.
PPARγ in T cells intrinsically controls Th2 effector program
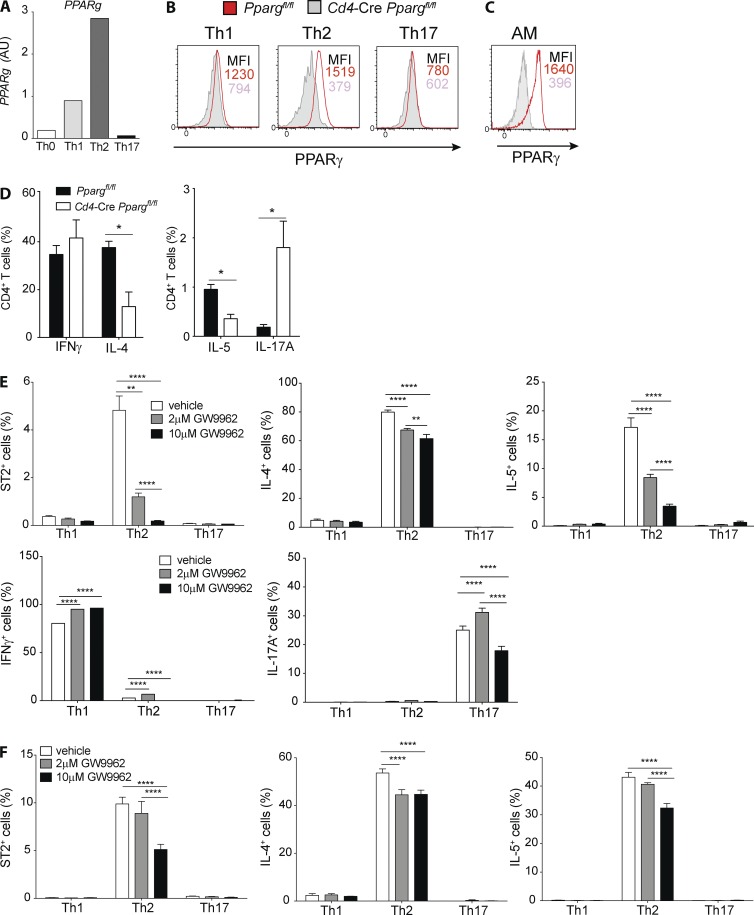
Having observed a profound deficiency in Th2 effector function in vivo, we wanted to more thoroughly address the underlying mechanism using an in vitro co-culture system. When naive CD4+ T cells were polarized into Th1, Th2, and Th17 subsets in vitro and PPARγ expression was evaluated, it became evident that expression of PPARγ mRNA and protein was highest in Th2 cells (Fig. 2, A and B), almost reaching expression levels of AMs (Fig. 2 C). Furthermore, under neutral conditions (i.e., peptide stimulation alone), T helper cell cytokine production was significantly changed in the absence of PPARγ. The frequency of cells expressing IL-4, and in particular IL-5, was strongly reduced (Fig. 2 D), Conversely, IFN-γ producers were comparable, and IL-17A producers were even enhanced in the absence of PPARγ (Fig. 2 D). To address whether PPARγ was required for differentiation of Th2 cells or for maintenance of effector function, naive Smarta-1 transgenic CD4+ T cells were co-cultured with splenic DCs and peptides in the presence of the PPARγ-antagonist GW9662. The antagonist was added either from the beginning or after 4 d of culturing. Examining the frequencies of ST2+, IL-4+, and IL-5+ cells, it became evident that PPARγ blockade during the entire differentiation period strikingly affected generation of ST2+ and IL-5+ cells in a dose-dependent manner, whereas IL-4+ cells were moderately reduced (Fig. 2 E). In contrast, IFN-γ remained unaffected and, paradoxically, IL-17 production was increased or decreased dependent on the amount of inhibitor (Fig. 2 E). Furthermore, PPARγ blockade in already-differentiated cells inhibited ST2, IL-4, and IL-5 production, although not as pronounced as with blockade throughout the entire co-culture period. Conversely, no effect was seen on IL-17A and IFN-γ production (Fig. S1, H and I). In conclusion, these results suggested that PPARγ promotes Th2 differentiation and effector function, in particular, for expression of IL-5 and ST2.
Figure 2.
PPARγ is highly expressed in mouse T cells and controls Th2 polarization in vitro. Naive splenic CD4+ T cells were sorted from Ppargfl/fl and Cd4-CrePpargfl/fl mice and were co-cultured for 4 d with splenic DCs and soluble αCD3 mAbs (2 µg/ml). Shown are the Pparg mRNA of Ppargfl/fl cells (WT; A) and the intracellular protein expression of PPARγ in Ppargfl/fl cells (B), with the corresponding Cd4-CrePpargfl/fl controls and PPARγ expression in AMs as a comparison (C). (D) Cytokine production profile after 4 h restimulation with PMA/ionomycin (n = 4/condition). (E and F) naive, splenic, Smarta-1, transgenic CD4+ T cells were co-cultured with splenic DCs and 10 nM gp61 peptide in polarizing conditions in the presence of PPARγ antagonist GW9662 at the indicated concentrations or in vehicle (DMSO) as a control. Shown are the frequencies of ST2+ and cytokine-producing cells of indicated cytokines after restimulation with PMA/ionomycin. (E) GW9662 was added from the beginning and cells were analyzed after 4 d of culture (n = 4/condition). (F) Cells were cultured for 4 d before addition of GW9662 and analyzed 24 h later (n = 4/condition). The data are representative of three independent experiments. Data are means ± SEM and the sample size (n). ANOVA (one-way) was used. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
PPARγ is dispensable for Treg function in vitro
Because we had observed a difference in the phenotype of Treg cells intrinsically lacking PPARγ in pulmonary models of type-2 immunity, we wanted to explore the possibility that PPARγ directly regulated Treg function. For that purpose, we conducted Treg suppression assays with naive CD4+CD25− T cells from WT and Treg cells isolated from Ppargfl/fl Cd4–Cre or Ppargfl/fl mice. PPARγ-deficient Treg cells, sorted from HDM-inflamed lungs (Fig. S1, J and K) or naive spleens (not depicted), suppressed proliferation and cytokine production comparably in this setting. Overall, these results suggest that the main functions of Treg cells were intact in the absence of PPARγ.
PPARγ suppresses a Th17/Th22 transcriptional program and is essential for IL-33–induced CD4+ T cell effector cytokine production
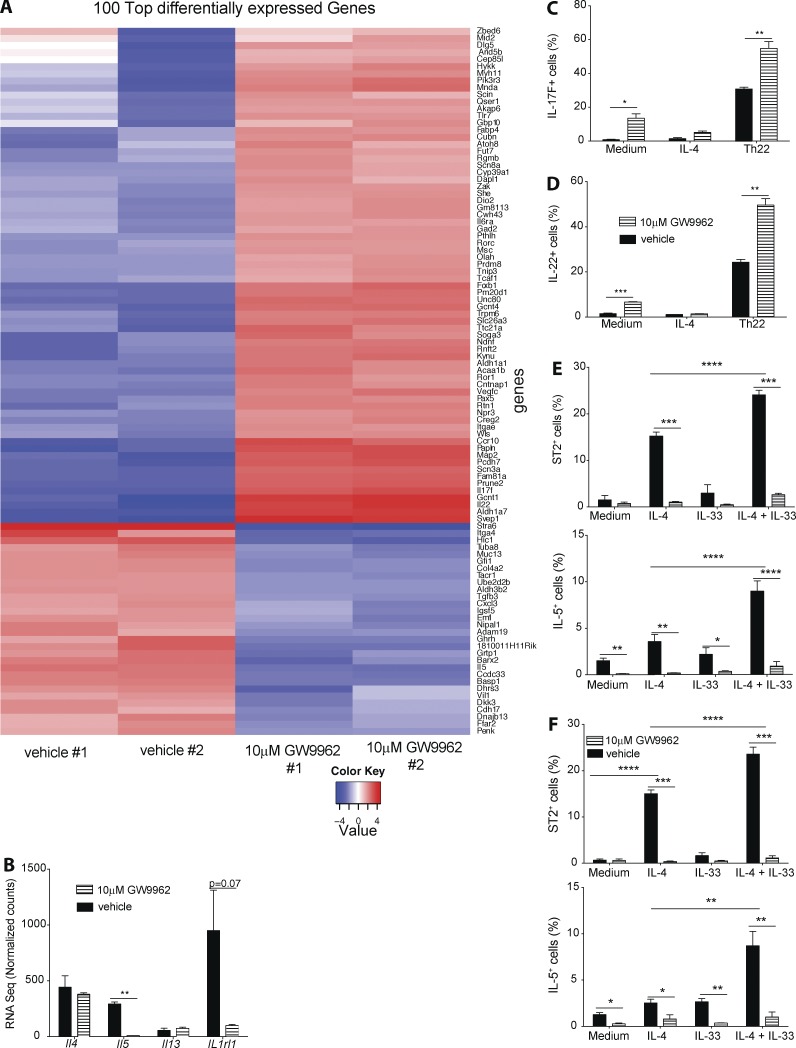
To better characterize the role of PPARγ in controlling Th2 effector function, we performed RNA sequencing of T cells co-cultured with DCs and PPARγ antagonist or vehicle, similar to the setup described in Fig. 2. Examining the most differentially regulated genes confirmed a strong reduction in expression of Il5 and Il1rl1 (IL-33 receptor; ST2) RNA (Fig. 3, A and B). In addition, multiple genes associated with Th17 effector function were significantly increased in expression, including Il17f, Il22, and Rorc. No significant differences could be detected in Il4 and Il13 RNA levels (Fig. 3 B). To confirm enhanced expression of Th17-associated cytokines IL-17F and IL-22, Smarta-1 splenic CD4+ T cells were co-cultured with DCs in the presence of polarizing cytokines. The frequency of both IL-17F– and IL-22–expressing CD4+ T cells was significantly higher in PPARγ antagonist-treated cells, in particular, under Th22-polarizing conditions (Fig. 3, C and D). Having established that PPARγ predominantly promotes IL-5 and ST2 expression in vitro, we hypothesized that the general impairment of Th2 immunity observed in vivo is mainly due to the lack of PPARγ-deficient Th2 cells to respond to IL-33 produced by lung epithelia but are absent in our in vitro culture protocol. To test that hypothesis, we co-cultured Smarta-1 splenic CD4+ T cells with WT or Il1rl1−/− splenic DCs in the presence of IL-4 and IL-33 and PPARγ antagonist. IL-4, alone, potently induced ST2 cell surface expression on CD4+ T cells and IL-5 production, and that was further enhanced when IL-33 was added to the culture (Fig. 3 E). Because IL-33 alone had no effect, IL-4 together with IL-33 seems to act in synergy. This significant increase in both ST2 and IL-5 was independent of ST2 (IL-33R) expression on splenic DCs because co-cultures using Il1rl1−/− splenic DCs revealed the same results (Fig. 3 F). Overall, these results suggested that PPARγ both suppressed a Th17-associated effector program and promoted Th2 responsiveness to IL-33, which in vivo strongly promotes Th2 effector function.
Figure 3.
PPARγ suppresses a Th17/Th22 transcriptional program and is essential for IL-33–induced CD4+ T cell effector cytokine production. Naive splenic Smarta-1 transgenic CD4+ T cells were co-cultured with splenic DCs and 10 nM gp33 peptide in Th2 polarizing conditions in the presence of PPARγ antagonist GW9662 or vehicle (DMSO) as a control. After 4 d of co-culture, RNA sequencing of CD4+ T cells was performed (n = 2). Heat map of the 100 most-differentially regulated genes (A) and expression levels of selected Th2 cytokines (B). (C) Naive splenic Smarta-1 transgenic CD4+ T cells were co-cultured with splenic DCs and 10 nM gp61 peptide in Th2 or Th22 polarizing conditions in the presence of PPARγ antagonist GW9662 or vehicle (DMSO) as a control. (C and D) Shown is the production of IL-17F (C) and IL-22 (D) after 4-h restimulation with PMA/ionomycin (n = 4/condition). (E and F) Naive splenic Smarta-1 transgenic CD4+ T cells were co-cultured with splenic DCs purified from WT or Il1rl1−/− mice and 1 nM gp61 peptide with indicated cytokines and in the presence of PPARγ antagonist GW9662 or vehicle (DMSO) as a control. Frequencies of IL-5+ (E) and ST2+ (F) of CD4+ cells after 4 h restimulation with PMA/ionomycin (n = 4/condition) for co-culture with) WT (E and Il1rl1−/− (F) splenic DCs (n = 4/condition). Data are means ± SEM and are representative of two independent experiments. The Student’s t test (unpaired) was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
PPARγ is highly expressed and strongly associated with CRTh2+ type-2 human memory T cells
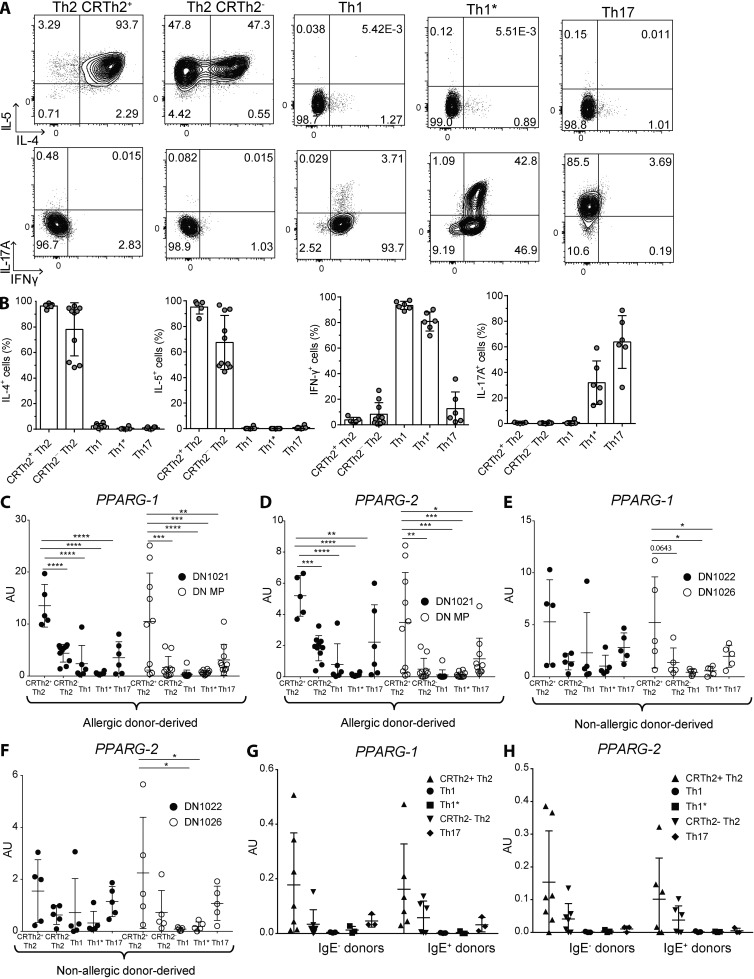
Having established an essential role of PPARγ in controlling Th2 effector function in mice, we wanted to investigate whether this transcription factor could also be used as a marker and potential therapeutic target in human-memory Th2 cells. For that purpose, T cell clones were generated from allergic and non-allergic donors by sorting different T helper cell subsets according to differential chemokine receptor expression and subsequent culture in vitro. That lead to very distinct cytokine expression profiles of individual clones, as recently described (Becattini et al., 2015). T cell clones from the CXCR3+CCR4−CCR6− subset (enriched in Th1 cells) were characterized by expressing high levels of IFN-γ, whereas T cells clones from the CXCR3+CCR4−CCR6+ subset (enriched in Th1*) in addition also expressed significant levels of IL-17A (Fig. 4, A and B). Conversely, T cell clones from the CRTh2+CCR4+ and, to a lesser extent, CRTh2−CCR4+ subset (enriched in Th2) expressed high levels of IL-4 and IL-5 (Fig. 4, A and B). Clones from the CXCR3−CCR4+CCR6+ subset (enriched in Th17), in contrast, almost exclusively expressed IL-17A (Fig. 4, A and B). Having validated the distinct cytokine expression patterns, we determined PPARG expression levels in all of these different clones under different stimulation conditions. Strikingly, after stimulation with PMA/ionomycin, both PPARG transcripts were expressed by far the highest in CRTh2+CCR4+ cells derived from allergic donors (Fig. 4, C and D), as opposed to all other types of clones. Similarly, stimulation with αCD3 also revealed a specific expression of PPARG in CRTh2+CCR4+ clones (Fig. S2, A and B). HDM-reactive CRTh2+CCR4+ Th2 cells from an HDM-allergic donor also expressed higher PPARγ mRNA compared with HDM-reactive CCR4− non-Th2 cells (not depicted). In clones derived from non-allergic donors, a similar expression pattern could be observed, although PPARG expression levels were generally lower than in cells derived from IgE− individuals (Fig. 4, E and F). To corroborate these results, memory CD4+ T cells were isolated directly ex vivo from multiple allergic and non-allergic donors, and PPARG expression was quantified. Similar to the pattern observed in the donor-derived clones, the highest expressing samples were found in the CRTh2+CCR4+ subset, although no differences could be observed between IgE+ and IgE− individuals (Fig. 4, G and H). We hypothesized that the lack of difference between allergic and non-allergic donors in the ex vivo samples could reflect the need for PPARG to be induced by an activating stimulus. To address that question, selected CRTh2+ Th2 and Th1 clones were stimulated with αCD3 for different times, and PPARG expression was measured. Indeed PPARG expression was strongly induced after activation in CRTh2+ Th2 clones but not in Th1 cells (Fig. S2, E and F). Collectively, these results suggested that PPARγ is a specific marker for allergic CRTh2+ Th2 cells in humans and that its expression in human T cells is dependent on an activating stimulus.
Figure 4.
PPARγ is highly expressed in human effector memory Th2 cells. Effector memory T cells were sorted from PBMCs of human allergic and non-allergic donors, according to chemokine receptor expression and subsequently cultured in vitro as described in the Materials and methods section. T cells were pregated as CD4+CD8–CD14–CD19–CD25–CD56–CD45RA– CCR7– cells, and subsequently, T cell subsets were identified as follows: CRTh2+CCR4+CXCR3–CCR6– (enriched in inflammatory Th2 cells); CCR4+CRTh2–CXCR3–CCR6– (enriched in Th2 cells); CXCR3+CCR4–CCR6– (enriched in Th1 cells); CXCR3+CCR6+CCR4– (enriched in Th1* cells); and CCR4+CCR6+CXCR3– (enriched in Th17 cells). (A and B) Representative dot plots of the cytokine production of the different T cell clone types analyzed from an allergic donor after restimulation with PMA/ionomycin (A) and a summary of all clones tested for that donor (B). (C–F) mRNA expression of PPARG-1 and PPARG-2 of two allergic (IgE+; C and D) and two non-allergic (IgE−; E and F) donor-derived clones after restimulation with PMA/ionomycin.(G and H) mRNA expression of PPARG-1 (G) and PPARG-2 (H) of memory T cell subsets sorted directly ex vivo from PBMCs. Data are means ± SEM, and the sample size is visible in each graph. ANOVA (one-way) was used. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
PPARγ in CD11c+ cells controls Th2 immunity in vivo
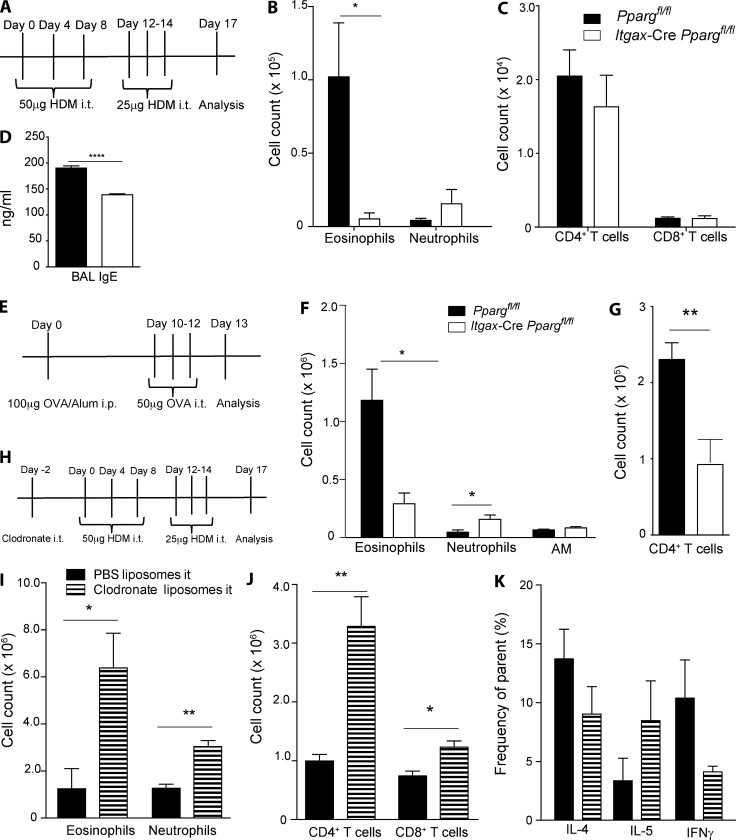
After having established a critical role for PPARγ in the regulation of Th2 immunity in T cells in vivo and having discovered that PPARγ regulates Th2 responsiveness to tissue-derived signaling molecule IL-33, we sought to further characterize the role of this transcription factor in other cells, which have an important role in allergic inflammation. Indeed, we were particularly interested in cells that are known to interact closely with T cells and the lung epithelium. We decided to focus on the mononuclear phagocyte compartment of the lung, which is essential for induction and regulation of pulmonary immunity (Kopf et al., 2015) and which has been shown to respond strongly to tissue-derived signals, including IL-33 (Kurowska-Stolarska et al., 2009; de Kleer et al., 2016). In the lung, this compartment is mainly composed of DCs and AMs, and both of those cell types express CD11c (Schneider et al., 2014b; Nobs et al., 2015). We, therefore, crossed our Ppargfl/fl animals to mice expressing Cre under the CD11c promoter (Ppargfl/fl Itgax–Cre) to specifically target these cells. To check specificity and efficacy of Itgax-Cre activity, we generated Itgax-Cre, Rosa26 flstopfl-RFP reporter mice. Both in naive (Fig. S3 A) and inflamed lungs (Fig. S3, B and C), conventional DC subsets and monocyte-derived DCs, as well as AMs, were efficiently targeted by CD11c-Cre–mediated deletion. To avoid misinterpretation of the results from potential targeting of other cells, key cell types in pulmonary allergic inflammation were also evaluated for their RFP expression. Indeed, only very few eosinophils, neutrophils, and epithelial cells exhibited CD11c-Cre activity (Fig. S3, B and C). We initially characterized Ppargfl/fl Itgax-Cre mice in the naive state. These animals have immature AMs (Fig. S3, D and E) because of the requirement of PPARγ in AM fetal development, as recently described (Schneider et al., 2014b). This developmentally arrested population expresses higher levels of CD11b, is more autofluorescent (Fig. S3 F), and is largely nonfunctional, leading to the development of pulmonary alveolar proteinosis in Ppargfl/fl Itgax-Cre mice (Schneider et al., 2014b). The DC compartment in these animals, in contrast, develops normally (Fig. S3, G and H). To study the role of PPARγ in CD11c+ mononuclear phagocytes, Ppargfl/fl Itgax-Cre animals were tested in an HDM model of pulmonary allergic inflammation (Fig. 5 A). Interestingly, infiltration of eosinophils was strongly reduced in the absence of PPARγ in CD11c+ cells (Fig. 5 B). Although the total number of CD4+ and CD8+ T cells was unaffected (Fig. 5 C), the level of BAL IgE was significantly reduced in Ppargfl/fl Itgax-Cre mice (Fig. 5 D). To test whether these surprising findings were specific to HDM as the allergen, we subjected CD11c-specific PPARγ KO mice to the classical OVA/Alum model (Fig. 5 E). The results observed using OVA, instead of HDM, were very similar because, again, a significant reduction in pulmonary eosinophilia could be observed (Fig. 5 F). In addition, a moderate decrease in the number of CD4+ T cells was also visible in this model (Fig. 5 G). Overall, these results indicated that PPARγ in a CD11c-expressing cell type is required for induction of asthmatic inflammation. This result immediately raised the question as to the identity of this cell type. Because it was already clear that AMs were nonfunctional in the absence of PPARγ, from earlier studies (Schneider et al., 2014b), we wanted to elucidate the general function of these cells in our setting. For this purpose, AMs were depleted with clodronate before subsequent sensitization and challenge with HDM (Fig. 5 H). Clodronate specifically eliminates AMs (Fig. S4 A), whereas lung-resident DCs are not affected (Fig. S4, B–D). AM depletion before HDM treatment resulted in a much more-pronounced inflammation, with increased infiltration of eosinophils and neutrophils (Fig. 5 I). Moreover, the total number of lung CD8+ and CD4+ T cells (Fig. 5 J) were also increased in the absence of AMs, whereas the T helper cell subset differentiation at the single-cell level was not significantly affected (Fig. 5 K). These data confirm previous studies showing that AMs interfere with T cell activation and, therefore, suppress lung inflammation (Thepen et al., 1989; Holt et al., 1993; Soroosh et al., 2013). Because AM depletion and PPARγ deficiency in AMs and DCs had the opposite effects on type-2 inflammation, we hypothesized that PPARγ deficiency in DCs, rather than AMs, was responsible for impaired type-2 inflammation in the lung.
Figure 5.
PPARγ in CD11c+ cells intrinsically mediates pulmonary allergic inflammation. (A–D) Ppargfl/fl and Cd11c-CrePpargfl/fl mice were sensitized and challenged intratracheally (i.t.), as indicated in the scheme with HDM extract. Total cell numbers in the BAL of eosinophils, neutrophils and AMs (B), CD4+ and CD8+ T cells (C), and the total amount of IgE in the BAL (D; n = 5/group). (E–G) WT and Cd11c-CrePpargfl/fl mice sensitized with OVA/alum i.p. and challenged i.t. A scheme of the immunization protocol (E); total cell numbers in the BAL of eosinophils, neutrophils, and AMs (F); and the total number of CD4+ T cells in the BAL (G; n = 5/group). (H–K) AMs were depleted with clodronate before HDM sensitization and challenge, as depicted in the scheme (H). The number of lung eosinophils and neutrophils (I) and CD4+ and CD8+ T cells (J). (K) Depicted is the frequency of cytokine-producing cells of CD4+ T cells after restimulation with PMA/ionomycin (n = 4/group). The data are representative of two experiments each and are means ± SEM, with the sample size (n). The Student’s t test (unpaired) was used. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
PPARγ in DCs controls type-2 immunity in vivo
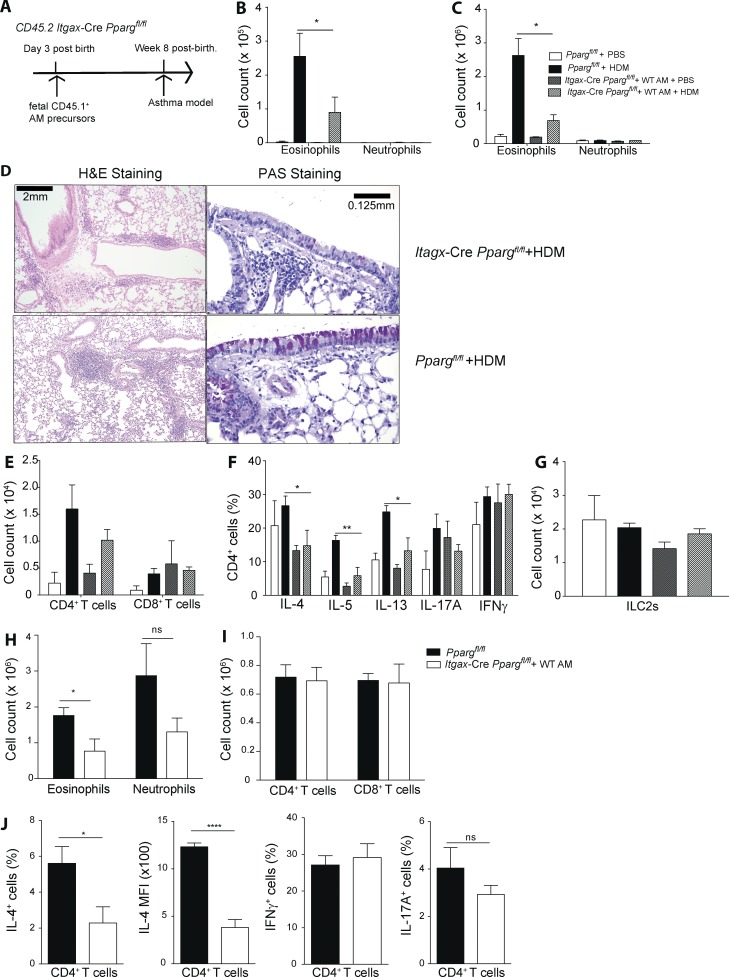
To address the role of PPARγ specifically in pulmonary DCs, neonatal Ppargfl/fl Itgax-Cre animals (CD45.2) were treated intratracheally with CD45.1+ cells harvested from the lungs of WT E17.5 embryos, which contain immediate AM precursors (Fig. 6 A). 8 wk later, AM-reconstituted, adult Ppargfl/fl Itgax-Cre mice were then subjected to HDM-induced experimental asthma. Lung analysis of Ppargfl/fl Itgax-Cre revealed that mice were completely reconstituted with WT AMs (Fig. S4 E) because the WT precursors outcompeted the PPARγ KO cells (Schneider et al., 2014b), leading to a functional AM compartment (Schneider et al., 2014a). HDM immunization and subsequent challenge with HDM or PBS revealed that HDM-treated animals, which lacked PPARγ only in DCs, exhibited a pronounced reduction in pulmonary eosinophilia in both BAL (Fig. 6 B) and lung (Fig. 6 C), similar to the phenotype observed in Ppargfl/fl Itgax-Cre mice, which had not been reconstituted (Fig. 5 B). Furthermore, histological analysis showed that they had a lower frequency of mucus-producing cells and reduced areas of peribronchial lymphoid follicles (Fig. 6 D). In addition, a trend toward fewer CD4+ T cells in the BAL was also observed (Fig. 6 E). Because DCs are potent inducers of T cell–mediated immunity, we carefully analyzed the CD4+ T cell response in AM-reconstituted Ppargfl/fl Itgax-Cre animals. The frequency of IL-4+, IL-5+, and IL-13+ cells was significantly reduced in animals lacking PPARγ specifically in DCs (Fig. 6 F). No significant differences could be seen in the number of ILC2s (Fig. 6 G). To corroborate those results, DC-specific PPARγ KO animals were also tested in the OVA/Alum experimental asthma model. Similar to the results obtained with HDM-induced allergic inflammation, the number of eosinophils in the lung was significantly reduced in AM-reconstituted Ppargfl/fl Itgax-Cre mice (Fig. 6 H). Although no differences could be observed in the number of CD4+ or CD8+ T cells (Fig. 6 I), the frequency of IL-4+CD4+ T cells (Fig. 6 J) as well as the amount of IL-4 (mean fluorescence intensity [MFI]) produced at the single-cell level was significantly reduced if the lung DC compartment was devoid of PPARγ (Fig. 6 J), whereas the frequencies of IFN-γ– and IL-17A–producing cells were not different from Ppargfl/fl controls (Fig. 6 J), Overall, these results indicated that PPARγ intrinsically in lung-resident DCs promotes induction of Th2 immunity in vivo.
Figure 6.
PPARγ in DCs modulates Th2 polarization in vivo. (A) The generation of Cd11c-CrePpargfl/fl mice containing WT AMs is shown schematically. Fetal (E17.5) lung AM precursors from WT CD45.1+ C57BL/6 embryos were transferred intranasally to neonatal (day 3 after birth) Ppargfl/fl and Cd11c-CrePpargfl/fl mice. After 8 wk, animals showed a reconstitution of Cd11c-CrePpargfl/fl mice with mature WT AMs, and they were subsequently used in HDM or OVA/alum asthma protocols, as described in Fig. 1 and Fig. S1. (B) Animals were sensitized with HDM and challenged with HDM or PBS with total cell numbers in the BAL (B) and lung (C) of eosinophils and neutrophils (n = 4–8/group). (D, left) Hematoxylin and eosin (H&E) histology. (D, right) PAS and Alcian blue histology. (E) Representative sections of total cell numbers of CD4+ and CD8+ T cells in the BAL. (F) BAL CD4+ T cells were restimulated with PMA/ionomycin for 4 h, and intracellular cytokine production was quantified, with the frequency of indicated cytokines (n = 4–8/group). (G) Total number of lung ILC2s are shown. (H–J) WT and Cd11c-CrePpargfl/fl mice were sensitized with OVA/alum i.p. and challenged with OVA intratracheally (H). Shown are total cell numbers in the lung of eosinophils and neutrophils (I) and CD4+ and CD8+ T cells (J). (J) BAL CD4+ T cells were restimulated with PMA/ionomycin for 4 h, and intracellular cytokine production was quantified. Frequency of IFN-γ+, IL-17A+, and IL-4+ cells as well as the MFI for IL-4 (n = 5/group). The data are representative of two experiments for each panel and are means ± SEM, with the sample size (n). The Student’s t test (unpaired) was used. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
PPARγ is dispensable for antigen uptake but modulates migration of lung CD11b+ DCs to the lung draining LNs (dLNs)
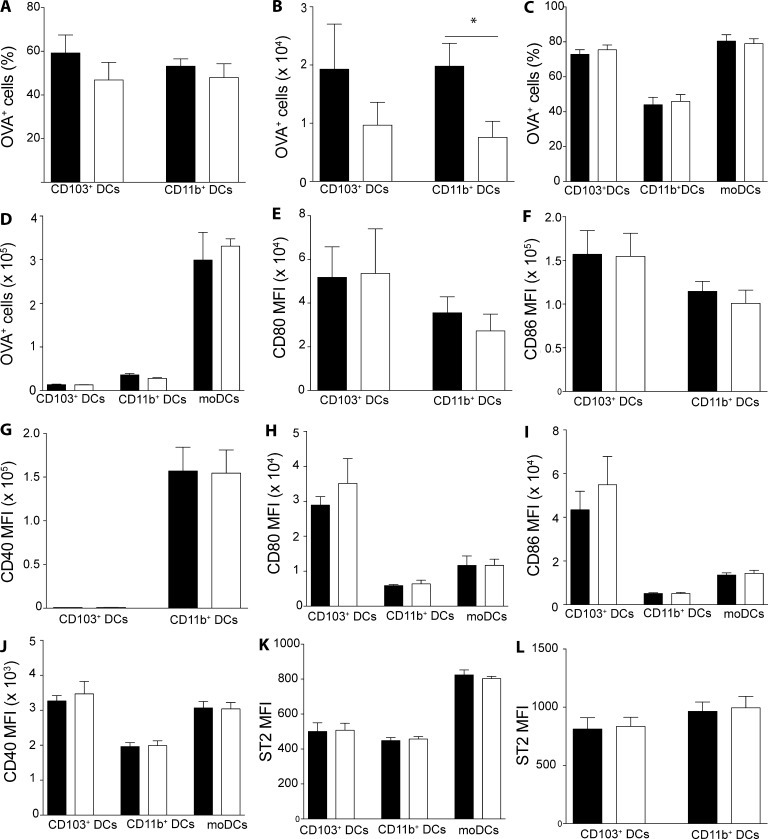
Having established that PPARγ in lung DCs mediates induction of type-2 immunity, we wanted to further elucidate the underlying mechanisms. One of the central aspects of DC biology is the capacity to take up antigen and transport it to the dLNs to enable priming of naive T cells. To assess a potential role for PPARγ in this process, we administered AF-488 labeled OVA (OVA-AF488) with HDM to AM-reconstituted Ppargfl/fl Itgax-Cre mice and evaluated the uptake and transport of OVA by lung DCs. Although the frequency of OVA+ cells among CD103+ and CD11b+DCs in the lung dLN was similar between DC-specific PPARγ KO animals and controls (Fig. 7 A), the total number of OVA-carrying CD11b+ DCs was significantly reduced in AM-reconstituted Ppargfl/fl Itgax-Cre mice (Fig. 7 B). In the lung, no differences could be observed in the frequency of OVA+ cells within each DC subset (Fig. 7 C) or in the total number of OVA+ DCs (Fig. 7 D). To elucidate whether DC activation was somehow impaired in the absence of PPARγ, we analyzed the surface expression of a variety of activation markers on DCs in the lung dLN as well as the lung. No significant differences in CD80, CD86, or CD40 could be observed in migratory LN DCs or lung-resident DCs (Fig. 7 J). Similarly, no difference could be observed in the expression of the IL-33 receptor ST2 between WT and PPARγ-deficient cells for dLNs (Fig. 7 K) and lung (Fig. 7 L). Overall, these results suggested that PPARγ is dispensable for antigen uptake and activation of lung DCs but allows better migration of antigen-loaded lung CD11b+ DCs to the lung dLN.
Figure 7.
PPARγ is largely dispensable for lung DC activation and antigen uptake but mediates antigen transport by CD11b+ DCs to the dLN. WT and AM-reconstituted Cd11c-CrePpargfl/fl were injected intratracheally with 100 µg OVA-AF488 and 100 µg HDM and sacrificed 24 h later for analysis of lung and dLNs by flow cytometry. DC subsets were identified as CD45+ Siglec-F−CD11c+MHCIIhigh for the lung and CD45+autofluorescent−CD11c+MHCIIhigh for the lung and dLNs, respectively. Frequency of OVA-AF488+ among each DC subset in the lung dLN (A) and the total number of OVA-AF488+ DCs (B). Frequency of OVA-AF488+ among each DC subset in the lung (C) and the total the total number of OVA-AF488+ DCs (D). MFIs of indicated DC activation markers for lung dLN (E–G) and lung (H–J) DC subsets as a summary of FACS data. (K and L) MFI of ST2-expressing cells among DCs in the lung (K) and the lung dLN (L; n = 4–6/group). The data are representative of two experiments and are means ± SEM. The Student’s t test (unpaired) was used. *, P < 0.05.
PPARγ intrinsically modulates Th2 polarization capacity in lung DCs
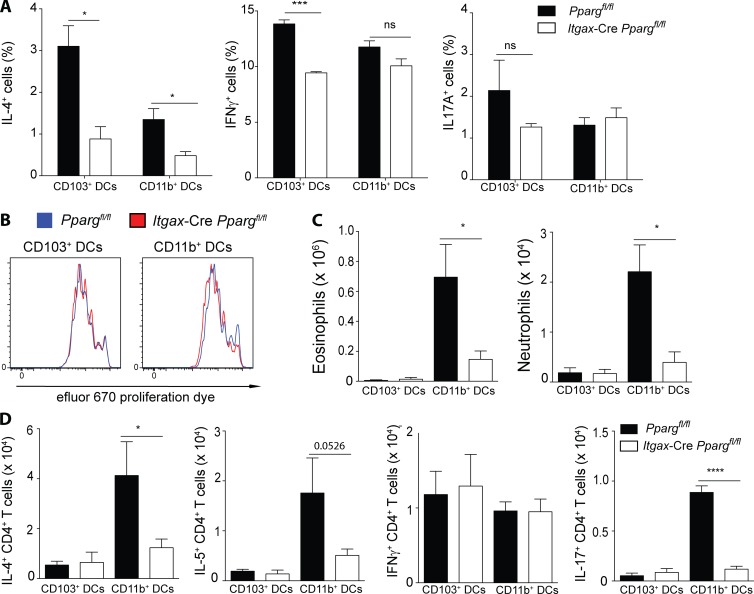
To address a direct DC-intrinsic role of PPARγ in priming naive CD4+ T cells, we sorted naive migratory lung DCs from AM-reconstituted Ppargfl/fl Itgax-Cre mice and co-cultured them with OTII-transgenic, naive CD4+ T cells in vitro. Evaluating the CD4+ T cell cytokine production profile revealed that the frequency of IL-4–producing cells was significantly reduced if DCs lacked PPARγ in both CD103+ and CD11b+ conventional DC subsets analyzed (Fig. 8 A). In contrast, Th1 and Th17 polarization was unaffected in cultures containing PPARγ-deficient CD11b+ DCs and was slightly reduced in cultures containing PPARγ-deficient CD103+ DCs (Fig. 8 A). Induction of CD4+ T cell proliferation appeared to be also completely intact, despite the DC-intrinsic deficiency of PPARγ (Fig. 8 B). Having established a modulating role of PPARγ for lung DC-induced Th2 differentiation in vitro and CD11b+ DC-mediated antigen transport, we aimed to elucidate the importance of PPARγ for each of the two major lung DC subsets, CD103+ and CD11b+ DCs, in an in vivo setting. For that purpose, Ppargfl/fl Itgax-Cre mice reconstituted with WT AMs were injected with HDM, and lung DCs were sorted and transferred to naive WT recipients after 24 h. These animals were then subsequently challenged with HDM and the type-2 immune response was evaluated. Strikingly, transfer of either WT or Pparg-deficient CD103+ DCs poorly induced eosinophil and neutrophil recruitment (Fig. 8 C). In contrast, transfer of HDM-pulsed CD11b+ DCs resulted in a potent eosinophilia and weak neutrophilia in the lung (Fig. 8 C), which was strongly dependent on the presence of PPARγ in the transferred DCs. Examining the CD4+ T cell response, it was evident that transfer of HDM-pulsed CD103+ DCs mainly led to the generation of IFN-γ–producing CD4+ T cells, whereas CD11b+ DCs induced predominant Th2 and weaker Th1 and Th17 responses (Fig. 8 D). Both Th2 and Th17 CD4+ T cell polarization was strongly dependent on the DC-intrinsic presence of PPARγ because, in its absence, IL-4–, IL-5–, and IL-17A–producing cells were significantly reduced, whereas the number of IFN-γ+ cells was unaffected (Fig. 8 D). Collectively, these results suggested that, although PPARγ regulates DC-induced T helper cell polarization in both lung-resident, conventional DC subsets, in vivo it is mainly the CD11b+ subset that is strongly dependent on PPARγ for induction of potent type-2 immune responses.
Figure 8.
PPARγ in DCs intrinsically controls Th2 polarization. Lung DCs from Ppargfl/fl and Cd11c-CrePpargfl/fl mice, which had been reconstituted with WT AMs were sorted and cultured in vitro for 4 d with OTII cells and 10 nM OVA323-339 peptide. Frequency of IL-4+, IFNγ+ and IL-17A+ CD4+ T cells (A) and the proliferation of efluor-670 labeled OTII cells (B; n = 3/condition) are shown. (C and D) Ppargfl/fl and Cd11c-CrePpargfl/fl mice, which had been reconstituted with WT AMs were injected with 100 µg HDM. 24 h after infection, the lung conventional DC subsets of each genotype were sorted and transferred intratracheally to naive WT recipients. After 10 d, those animals were challenged for a consecutive 5 d with 10 µg HDM and were subsequently analyzed on day 17. Total number of eosinophils and neutrophils (C) and the total numbers of cytokine producing CD4+ T cells in the BAL (D; n = 5/group). Data are means ± SEM and the sample size (n) and are representative of two independent experiments. ANOVA (one-way) was used. *, P < 0.05; ***, P < 0.001; ****, P < 0.0001.
IL-33 receptor and IL-4 receptor signaling controls PPARγ expression and Th2 polarization
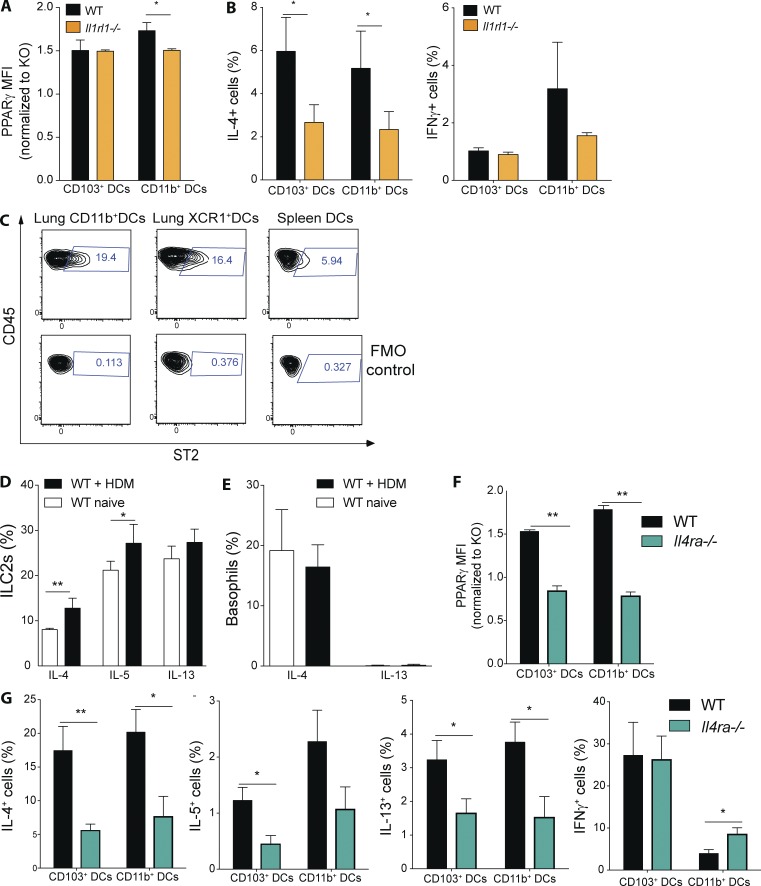
Having observed a major connection between IL-33 and PPARγ in regulating Th2 function, we were interested in examining whether IL-33 might also have a role in regulating PPARγ in lung DCs. For that purpose, we measured PPARγ expression in lung DC subsets in naive WT and Il1rl1−/− animals. Interestingly, we found that PPARγ expression (i.e., MFI) was slightly reduced in lung CD11b+ DCs but not in CD103+ DCs from Il1rl1−/− mice (Fig. 9 A). To address the impact of blocked IL-33 receptor signaling on lung DC-mediated priming of naive T cells generally, lung DCs from naive WT and Il1rl1−/− animals were sorted and co-cultured with Smarta-1 transgenic T cells. Indeed, the frequency of IL-4+ CD4+ T cells was reduced if the CD11b+ and CD103+ DC lacked IL-33 receptor signaling, whereas no significant difference could be observed for IFN-γ (Fig. 9 B). As ST2 was found to be dispensable for splenic, DC-mediated T cell priming in this assay (Fig. 3 F), we wanted to evaluate whether differences in ST2 expression between lung and splenic DCs could explain that discrepancy. Indeed, we found only a small fraction of splenic DCs express the IL-33 receptor (Fig. 9 C). Conversely, lung DCs exhibited a significantly higher proportion of ST2+ cells (Fig. 9 C).
Figure 9.
IL-4 receptor and IL-33 receptor signaling controls PPARγ expression and DC-mediated Th2 polarization. (A) PPARγ expression in lung DCs from naive WT and Il1rl1−/− mice was measured using flow cytometry. Shown is the MFI of PPARγ normalized to the PPARγ-deficient control of the indicated DC subsets (n = 3/group). (B) Lung DCs from WT and Il1rl1−/− animals were isolated and co-cultured in vitro for 4 d with naive, splenic, Smarta-1, transgenic CD4+ T cells and 1 nM gp61 peptide. Frequency of IL-4+ and IFN-γ+ CD4+ T cells (n = 10/condition). (C) Surface expression of ST2 on WT lung and splenic DCs was evaluated using flow cytometry. Shown are representative FACS plots including fluorescence minus one (FMO) controls (n = 3/group). (D and E) WT animals received PBS or 100 µg HDM intratracheally and lungs were analyzed for cytokine-producing cells 18 h after infection. Shown is the frequency of cytokine-producing ILC2s (D) and basophils (E) of indicated cytokines (n = 4/group). (F) PPARγ expression in lung DCs from naive WT and Il4ra−/− mice was measured using flow cytometry. Shown is the MFI of PPARγ normalized to the PPARγ-deficient control of the indicated DC subsets (n = 3/group). (G) Lung DCs from WT and Il4ra−/− animals were isolated and co-cultured in vitro for 4 d with naive, splenic, Smarta-1, transgenic CD4+ T cells and 1 nM gp61 peptide. Shown are the frequencies of IL-4+, IL-5+, IL-13+, and IFN-γ+ CD4+ T cells (n = 8/condition) Data are means ± SEM and the sample size (n). ANOVA (one way) was used. *, P < 0.05; **, P < 0.01.
IL-4 receptor signaling has been shown to induce PPARγ in macrophages (Szanto et al., 2010); therefore, we hypothesized that, in the context of allergic inflammation, that might also be the case in lung DCs. To address that question we initially analyzed the cytokine production of lung-resident cells in naive and HDM-injected WT animals. Indeed, we found that already in the steady-state a significant fraction of ILC2s produces IL-4, IL-5, and IL-13 and that, for IL-4 in particular, a significant increase in the frequency of IL-4+ cells can be observed after HDM administration (Fig. 9 D). Similarly, basophils were also found to express significant levels of IL-4 but not IL-13 in the steady state, but that was not further enhanced after HDM injection (Fig. 9 E). Having established the presence of IL-4– and/or IL-13–producing cells in both naive and HDM-treated lungs, we were interested to examine the lung DC compartment in animals devoid of IL-4 receptor signaling. For that purpose, we analyzed PPARγ expression in Il4r−/− animals, which cannot respond to IL-4 or IL-13 (Noben-Trauth et al., 1997). Indeed, we found that both CD11b+ and CD103+ DCs from Il4r-deficient animals exhibited a strongly reduced level of PPARγ expression in the steady state (Fig. 9 F). Furthermore, when these cells were co-cultured with transgenic CD4+T cells, we observed a significant reduction in type-2 cytokine, but not IFN-γ–producing cells, for both lung DC subsets (Fig. 9 G).
Overall, these results suggested that both IL-33 and IL-4 receptor signaling has a role in regulating PPARγ and DC capacity to promote Th2 priming.
Discussion
Dysregulated type-2 immune responses are responsible for some of the most-common, chronic inflammatory diseases, and to date, no therapies with high specificity and efficacy are available for treatment of conditions, such as asthma (Tan et al., 2016). This is also because the underlying molecular mechanisms remain incompletely understood. Here, we report that the transcription factor PPARγ regulates pulmonary type-2 immunity at multiple levels, including the initiation and the exacerbation stage. We show that PPARγ is highly and specifically expressed in both mouse and human Th2 cells, as opposed to other Th subsets. Indeed, we demonstrate that PPARγ, although having only a minor direct role in regulating Th2 differentiation, controls Th2 sensitivity to IL-33 and, thus, has a major impact on Th2 effector function in vivo. In this context, mice lacking PPARγ exclusively in T cells develop reduced type-2 inflammatory responses in different models of pulmonary allergic inflammation. Both pulmonary eosinophilia and mucus production were found to be strongly reduced in T cell–specific PPARγ KO animals. More specifically, we found PPARγ directly controlled IL-5 production and, more importantly, ST2 expression in vitro and in vivo. IL-33 receptor signaling has been shown to strongly promote Th2 effector function (Schmitz et al., 2005), and indeed, we found that IL-33, in conjunction with IL-4, strongly promoted IL-5 and ST2 expression, compared with IL-4 alone, suggesting the existence of a positive feedback loop between the IL-33 receptor stimulation and ST2 expression levels. Likely the lack of ST2 induction in vivo in PPARγ-deficient T cells is responsible for the general impairment of Th2 effector function observed in allergic inflammation because PPARγ appears to have a significant, but only minor, direct role in regulating IL-4 expression. In addition, the observation that pharmacologic blockade of PPARγ in differentiated Th2 cells significantly inhibited effector cytokine production suggests that PPARγ is more important in maintenance of effector function than in Th2 differentiation by itself. Indeed, this observation was corroborated with human data, in which PPARG expression was found to be particularly high in CRTh2+ Th2 cells, which also express high levels of IL-5, and only marginally higher levels of IL-4 compared with CRTh2− Th2 cells. This suggests that, in humans, PPARγ has mainly a role in these “pathogenic” Th2 cells, as opposed to all Th2 cells. As pharmacologic inhibition of PPARγ in differentiated Th2 cells was able to significantly attenuate their capacity to secrete type-2 cytokines, which suggests a potential therapeutic target for human therapy. This is in line with recent evidence that some polymorphisms in PPARG in humans are associated with increased risk of developing asthma (Li et al., 2015).
In addition to controlling Th2 effector responses in vivo intrinsically in T cells, we found PPARγ also regulates the ability of lung-resident CD11b+ DCs to induce Th2 immunity in vitro and in vivo. How DCs induce distinct T helper cell effector profiles and how different subsets contribute to the generation of the large diversity of different T cell responses in vivo has been intensely investigated in recent years (Kopf et al., 2015), and our results identify PPARγ as a new lung DC-intrinsic regulator of type-2 immunity. We found PPARγ to modulate the ability of CD11b+ DCs for antigen transport to the lung dLN in the context of HDM-induced inflammation, as well as in prime, naive CD4+ T cells toward Th2 polarization. Indeed, the requirement for PPARγ in CD11b+ DCs to induce potent Th2 effector responses in vivo, likely represents a combined effect of these two deficiencies. Indeed, these results also confirm CD11b+ DCs as the main drivers of type-2 inflammation in the HDM model (Plantinga et al., 2013). Interestingly, we found that, similar to T cells, PPARγ links CD11b+ DC-mediated Th2 immunity to the epithelium-derived cytokine IL-33. Recent evidence shows that IL-33 promotes DC-mediated Th2 priming in HDM-induced inflammation, and it appears that it directly controls PPARγ expression in DCs in vivo (de Kleer et al., 2016). Indeed, we could confirm the notion that the lack of IL-33 receptor signaling impairs the capacity of lung CD11b+ DCs to prime Th2 cytokine production. Interestingly, this was not the case for splenic DCs. Likely this can be explained by the much lower frequency of ST2+ DCs in the spleen compared with the lung. These results, therefore, highlight the specific importance of IL-33 in lung immunity. In addition to the role of IL-33 in regulating PPARγ in lung CD11b+ DCs, we found it to strongly depend on IL-4 receptor signaling. IL-4 receptor–deficient lung CD11b+ DCs were found to have impaired capacity to prime naive CD4+ T cells toward Th2, which is line with the notion that IL-4 can promote lung DC-mediated Th2 priming (Webb et al., 2007). Thus, it appears that PPARγ integrates multiple tissue– and immune cell–derived, external cues to promote induction of Th2 immunity by lung-resident CD11b+ DCs. The transcriptional program in lung CD11b+ DCs controlled by PPARγ to promote Th2 polarization likely involves regulation of lipid metabolism, and indeed, this link has been suggested to explain some of the effects observed in in vitro–derived mouse and human DCs using PPARγ-activating ligands (Klotz et al., 2007; Szatmari et al., 2007). Collectively, the results presented in this article establish PPARγ as a potent proinflammatory factor in type-2 immunity by acting as a sensor for multiple, external signals in regulating the interaction between T cells and DCs during the induction and promotion of allergic lung inflammation. Indeed, they strongly contrast with the current view of this transcription factor as being more anti-inflammatory (Croasdell et al., 2015). These earlier conclusions were largely based on studies administering synthetic PPARγ agonists (i.e., thiazolidinediones) in different experimental models of asthma (Trifilieff et al., 2003; Woerly et al., 2003; Zhao et al., 2014). Aside from the possibility that the effects observed in earlier studies could be explained by off-target effects of PPARγ agonists (Nemenoff, 2007; Sauer, 2015), this discrepancy could also suggest that, in addition to a role in T cells and DCs, PPARγ may have opposing functions in other cell types. Indeed, PPARγ is expressed by lung epithelial cells (Honda et al., 2004) and also by other lung-resident, immune cell types, such as ILC2s (Robinette et al., 2015). The cell-type–specific roles of PPARγ in these cell types remain to be elucidated. In summary, our results establish PPARγ as an important driver of lung type-2 immune responses by regulating the interaction between DCs and CD4+ T cells and acting as an integrator of immune cell– and tissue-derived signals to control induction and promotion pulmonary allergic inflammation.
Materials and methods
Mice
Ppargfl/fl mice (Imai et al., 2004) were backcrossed for eight generations to C57BL/6 before crossing them to Tg(Itgax-cre)1-1Reiz (Cd11c-Cre) mice (Caton et al., 2007) or Cd4-cre mice (Wolfer et al., 2001) to generate mice with deficiency in PPARγ in CD11c+ cells (Cd11c-CrePpargfl/fl) and T cells (Cd4-CrePpargfl/fl), respectively. Gt(ROSA)26Sortm1Hjf (Rosa26fl/fl-RFP) mice (Luche et al., 2007) were crossed with Cd11c-Cre and Cd4-Cre mice. OT-II mice (JAX 004194; The Jackson Laboratory), Smarta-1 mice (Oxenius et al., 1998), and Il4r−/− mice (Barner et al., 1998). All mice, including C57BL/6J WT mice, were either bred and maintained in individually ventilated cages under specific pathogen-free conditions at ETH Phenomics Center, except the Il1rl1−/− mice (Townsend et al., 2000), which were provided by D. Pinschewer (University of Basel, Basel, Switzerland). For all strains expressing Cre, littermate controls were used, whereas for other experiments, C57BL/6J mice were used as controls. Mice used for experiments were between 6 and 14 wk old, unless otherwise stated. All animal experiments were approved by the local animal ethics committee (Kantonales Veterinaersamt Zurich) and were performed according to local rules [Swiss Animal Protection Ordinance (TschV), Zurich, Switzerland] and Swiss animal protection law (TschG).
Cell suspension preparations
Mice were sacrificed by an overdose of sodium pentobarbital by i.p. injection. Organs were removed and then processed according to the following procedure: spleens and LNs were minced and then digested with 2 mg/ml of type IV collagenase (Worthington) and 0.02 mg/ml DNaseI (Sigma) at 37°C for 45 min and subsequently passed through a 70-µm cell strainer. Lungs were digested with 1 mg/ml hyaluronidase (Sigma), 25 µg/ml collagenase XI (Sigma), 50 µg/ml Liberase TM (Roche), and 0.02 mg/ml DNaseI (Sigma) at 37°C for 45 min and subsequently passed through a 70-µm cell strainer. ACK buffer was used for erythrocyte lysis for all organs.
Flow cytometry
Multiparameter analysis was performed on a FACSCanto II or LSR Fortessa (BD) and analyzed with FlowJo software (Tree Star). Fluorochrome-conjugated or biotinylated mAbs specific to mouse CD11c (N418), CD11b (M1/70), SiglecF (E50−2440; BD Biosciences), CD103 (2E7), CD45 (30-F11), CD45.2 (104), CD4 (GK1.5), CD8α (53-6.7), MHC class II (M5/114.15.2; eBioscience), CD64 (X54-5/7.1), CD19 (6D5), CD3e (145-2C11), CD127 (A7R34), CD90.2 (30-H12), TCR-β (H57-597) NK1.1 (PK136, eBioscience), ST2 (DIH9), CD25(PC61), GITR (DTA-1) FcεRIα (MAR-1; eBioscience), XCR1 (ZET), Ly-6G (1A8-Ly6g), IL-4 (11B11), IL-5 (TRFK5), GATA3 (TWAJ; eBioscience), RORγt (AFKJS-9; e Bioscience), IL-17A (eBio17B7; eBioscience), IL-13 (eBio13A; eBioscience), IFN-γ (XMG1.2), IL-22 (IL22JOP; eBioscience), IL-17F (SHLR17; eBioscience) were purchased from BioLegend unless otherwise stated. For intracellular staining of transcription factors, cells were fixed and stained using the intracellular fixation and permeabilization kit (eBioscience) according to the manufacturer’s instructions. PPARγ was stained using monoclonal rabbit anti-PPARγ (81B8; Cell Signaling Technology), followed by staining with goat anti–rabbit IgG antibodies (Invitrogen). Dead cells were excluded using the live/dead marker eFluor780 (eBioscience). Before all staining, FcγIII/II receptors were blocked by incubation with homemade anti-CD16/32 (2.4G2). Annexin V on apoptotic cells was stained using the Annexin V detection kit (eBioscience) according to the manufacturer’s instructions. eFluor 670 proliferation dye (eBioscience) and CFSE (eBioscience) were used according to the manufacturer’s instructions.
Mouse models of airway inflammation
For the OVA model, animals were injected i.p. with 100 µg OVA (Invitrogen) resuspended in aluminum hydroxide solution (Serva). At the indicated times, animals were challenged with intratracheal injection of 20 µg of OVA in PBS. For the HDM model, animals received either 10 µg or 50 µg HDM (Greer) intratracheally for sensitization and were then challenged at the indicated times with either 10 µg or 20 µg HDM extract, depending on the set of experiments and on the batch of HDM used. The HDM batches were standardized for their activity in vivo to allow comparison of experiments with different batches, and the amounts used are indicated in each figure.
DC–T cell co-culture
DCs were sorted from processed organs as described in Cell suspension preparations. CD4+ T cells were obtained with CD4 MACS-bead (Miltenyi) pre-enrichment and subsequent to FACS-sorting from naive splenocytes. CD4+ T cells were sorted as CD4+CD11c− autofluorescent-negative cells. T cells and DCs were then cultured together for 4 d in complete IMDM medium (Life Technologies). For cultures using OTII cells varying concentrations of OVA323-339 peptide (Mimotopes Australia) was added. For cultures using Smarta-1 CD4+ T cells, varying concentrations of Gp61-80 (Mimotopes Australia) peptide was added. For cultures using Ppargfl/fl and Cd4-CrePpargfl/fl animals, plates were coated with 2 µg/ml αCD3. For in vitro polarizing conditions the following setup was used: Th0 was medium only; Th1 was 20 ng/ml IL-12; Th2 was 100 ng/ml IL-4 + 20 µg/ml αIFN-γ; Th17 was 1 ng/ml TGF-β1 + 50 ng/ml IL-6 + 20 µg/ml αIFN-γ; and Th22 was 20 ng/ml IL-6 + 20 ng/ml IL-23. For co-cultures with IL-33, all samples were cultured with 20 µg/ml αIFN-γ, 100 ng/ml IL-4, 100 ng/ml IL-33, or 100 ng/ml IL-4 + 100 ng/ml IL-33,respectively. Before flow cytometric analysis, cells were restimulated with PMA (Sigma)/ionomycin (Sigma) in the presence of monensin (Sigma). For the cultures using PPARγ antagonist GW9962 (Sigma), cells were either cultured in the presence of the inhibitor from the beginning, with analysis after 4 d, or the inhibitor was added after 4 d of culture for 24 h together with αCD3.
Treg suppression assay
Treg cells were sorted as CD4+CD25+GITR+ T cells after MACS-bead (Miltenyi) pre-enrichment from the indicated organs. Treg cells were then cultured at the indicated ratios to naive T cells in the presence of splenic DCs and αCD3. Proliferation of effector T cells was evaluated on day 3 and cytokine production on day 4 of co-culture.
Antibody ELISA
At the indicated times, BAL fluid was measured for HDM-specific IgE antibody isotype or cytokine levels. For HDM-specific IgE, 96-well plates (Maxisorp; Nunc) were coated with HDM in PBS overnight at 4°C. Plates were washed and incubated with PBS-1% BSA for 2 h at room temperature for blocking. Samples from BAL fluids were incubated at room temperature for 2 h. Plates were then washed five times. For IgE, plates were then incubated with alkaline–phosphate-labeled goat anti–mouse antibodies to IgE (SouthernBiotech Technologies, Inc.) at a 1:1,000 dilution in PBS-0.1% BSA at room temperature for 2 h. Subsequently, plates were washed five times, and substrate p-nitrophenyl phosphate (Sigma) was added. ODs were measured on an ELISA reader (Bucher Biotec) at 405 nm.
DC-mediated antigen uptake and transport
Animals received 100 µg OVA-AlexaFluor488 (Invitrogen) and 100 µg HDM (Greer) intratracheally. Lungs and dLNs were collected 24 h after infection and analyzed by flow cytometry.
Clodronate treatment
For the depletion of AMs, mice were treated with 100 µl clodronate liposomes intratracheally 2 d before sensitization with HDM. Clodronate was a gift from Roche. Control mice were treated with PBS liposomes.
Bone marrow chimeras
For BM chimeras, C57BL/6 CD45.2+ mice were lethally irradiated (9.5 gray, using a cesium source) and reconstituted with 5–10 × 106 BM cells of the background and with the ratio indicated for each experiment. Mice were analyzed 10 wk after reconstitution.
Transfer of AM precursors into neonates
Lungs from E17.5 embryos from CD45.1+ WT animals were harvested, minced, and digested with collagenase IV as described above in Cell suspension preparations. CD45+ cells were then purified using MACS beads (Miltenyi), and cells were then transferred intranasally into day 1–7 postbirth neonates to allow reconstitution of a WT AM compartment.
Quantitative real-time PCR
For analysis of mRNA expression, RNA was isolated from cells with TRIzol reagent (Invitrogen) and was reverse-transcribed with GoScript reverse transcription according to the manufacturer’s instructions (Promega). Quantitative real-time RT-PCR was performed with Kapa SYBR Fast. For human samples, the expression of PPARG-1 (forward primer: 5′-AAAGAAGCCGACACTAAACC-3′ and reverse primer: 5′-CTTCCATTACGGAGAGATCC-3′) and PPARG-2 (forward primer: 5′-AGGCGAGGGTCTTGACAG-3′ and reverse primer: 5′-GATGCGGATGGCCACCTCTTT-3′) were normalized to that of housekeeping gene TBP (forward primer: 5′-TTGACCTAAAGACCATTGCACTTC-3′ and reverse primer: 5′-TGTTCTTCACTCTTGGCTCC-3′). For mouse samples, Pparg (forward primer: 5′-GTGATGGAAGACCACTCGCATT-3′ and reverse primer: 5′-CCATGAGGGAGTTAGAAGGTTC-3′) expression was normalized to Tbp (forward primer: 5′-TTGACCTAAAGACCATTGCACTTC-3′ and reverse primer: 5′-TTCTCATGATGACTGCAGCAAA-3′).
Human blood samples
Blood samples were obtained from the Swiss Blood Donation Center and from allergic donors. All blood donors provided written, informed consent forms approved by the ethics committee of Canton of Ticino before inclusion in this study. Human primary cell protocols were approved by the Federal Office of Public Health (authorization. A000197/2 to F. Sallusto). IgE concentration in plasma was measured by ELISA.
Isolation of human T cell clones and HDM-reactive T cells
PBMCs were isolated with Ficoll-Paque Plus (GE Healthcare). CD4+ T cells were isolated by positive selection with CD4 magnetic micro-beads (Miltenyi Biotec). CD4+ T cells were then stained with the following mAbs: anti–CD4-PE Texas Red (S3.5), anti–CD45RA-QD655 (MEM-56; Invitrogen), anti–CCR7-Brilliant Violet 421 (G043H7), anti–CXCR3-Alexa Fluor 647 (G025H7; BioLegend), anti–CCR6-PE (11A9), anti–CCR4-PE-Cy7 (1G1), anti–CRTh2-FITC (BM16; BD), anti–CD8-PE-Cy5 (B9.11), anti–CD14-PE-Cy5 (RMO52), anti–CD19-PE-Cy5 (J3-119), anti–CD25-PE-Cy5 (B1.49.9), and anti-CD56-PE-Cy5 (N901; Beckman Coulter). CD4+ effector memory T cell subsets were FACS-sorted (FACSAria III; BD) as follows and after gating on CD4+CD8–CD14–CD19–CD25–CD56–CD45RA–CCR7– cells: CRTh2+CCR4+CXCR3–CCR6– (enriched in inflammatory Th2 cells); CCR4+CRTh2–CXCR3–CCR6– (enriched in Th2 cells); CXCR3+CCR4–CCR6– (enriched in Th1 cells); CXCR3+CCR6+CCR4– (enriched in Th1* cells); CCR4+CCR6+CXCR3– (enriched in Th17 cells; Messi et al., 2003; Acosta-Rodriguez et al., 2007). To obtain T cell clones, T cells from each subset were plated at 0.5 cells/well in 384-well plates in RPMI-1650 complete medium (2 mM glutamine, 1% nonessential aa, 1% sodium pyruvate, 50 µM 2-mercaptoethanol, 1% penicillin/streptomycin [all from Life Technologies] and 5% human serum [Swiss Red Cross]) in the presence of 1 µg/ml PHA (Remel), irradiated allogeneic PBMCs (2.5 × 104 per well), and 500 IU/ml IL-2. T cell clones were grown in IL-2–containing RPMI-1650 complete medium and used on day 18–20 after initial stimulation. For the analysis of HDM-reactive T cell subsets sorted from the blood of HDM-IgE+ donors were cultured at a ratio of 2:1 with irradiated autologous monocytes prepulsed for 5 h with 30 µg/ml HDM extract (Greer). On day 6, activated T cells were sorted as ICOS+CD25+ after staining with anti-ICOS–Pacific Blue (C398.4A) and anti-CD25–Brilliant Violet 785 (BC96; BioLegend) mAbs and analyzed immediately or after expansion in IL-2–containing complete medium.
Analysis of human T cells
T cell clones and HDM-reactive T cells were screened for their cytokine expression profile by intracellular cytokine staining. Cells were stimulated with 0.2 µM PMA (Sigma) and 1 µg/ml ionomycin (Sigma) for 5 h in RPMI-1640 complete medium. Brefeldin A (Sigma) was added at 10 µg/ml after 2.5 h of stimulation. T cells were stained with Live/Dead Fixable Aqua Dead Cell Stain kit (Life Technologies, Molecular Probes). T cells were then fixed and permeabilized with Cytofix/Cytoperm (BD), according to the manufacturer’s instructions, and stained with IL-4-Alexa Fluor 488 (8D4-8; eBioscience), IL-5-APC (TRFK5; BD), IL-17-Brilliant Violet 605 (BL168), and IFN-γ-Brilliant Violet 650 (4S.B3; BioLegend). PPAR-γ mRNA expression was measured by quantitative PCR on 5 × 105 T cells, resting or stimulated for 4 h with 0.2 µM PMA and 1 µg/ml ionomycin, or for 4 h and 16 h with immobilized anti-CD3 (Lanzavecchia and Scheidegger, 1987; TR66) and anti-CD28 (CD28.2) mAbs (BD), both at 1 µg/ml, in RPMI-1640 complete medium.
Histology
Mouse lungs from HDM-treated animals were removed and fixed in 4% formalin. Subsequently, they were processed and then stained with either hematoxylin and eosin or periodic acid–Schiff and Alcian blue.
RNA sequencing
Indicated cells were collected into TRIzol (Life Technologies), phase separation was achieved with the addition of chloroform (Sigma), and RNA was precipitated from the aqueous layer with isopropanol (Sigma) using glycogen (Roche) as a carrier. The TruSeq RNA Stranded sample kit (Illumina) was used to construct the sequencing libraries. In brief, total RNA samples (100 ng) were poly(A)-enriched and reverse-transcribed into double-stranded cDNA. TruSeq adapters were then ligated to double-stranded cDNA. Fragments containing TruSeq adapters on both ends were selectively enriched with PCR and were subsequently sequenced on the Illumina NextSeq 500 in single-end mode, 150 cycles, at the Functional Genomics Center Zurich (GEO accession no. GSE100588).
Statistical analysis
Means, SDs, SEMs, Student’s t test (unpaired), and ANOVAs (one-way) were calculated with Prism software (GraphPad Software). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Online supplemental material
Fig. S1 shows that PPARγ in T cells mediates development of pulmonary allergic inflammation in an OVA/alum model and is not required for Treg suppressive capacity in vitro. Fig. S2 shows that PPARγ is highly expressed in human effector memory Th2 cells. Fig. S3 shows that PPARγ is required for terminal differentiation of AMs. Fig. S4 shows that intratracheal administration of clodronate specifically depletes AMs.
Supplementary Material
Acknowledgments
We acknowledge the use of the ImmGen database as an informative tool for the design of this study. We thank Franziska Ampenberger for technical assistance.
We thank Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (grants 310030B and 141175) and Eidgenössische Technische Hochschule Zürich (grant 34/13-1) for supporting this study. This work was also partially supported by European Research Council (grant 323183, PREDICT, to F. Sallusto).
The authors declare no competing financial interests.
Author contributions: S.P. Nobs, S. Natali, and K. Okreglicka performed the experiments; S.P. Nobs, S. Natali, L. Pohlmeier, K. Okreglicka, and M. Kurrer analyzed data; S.P. Nobs, C. Schneider, F. Sallusto, and M. Kopf designed the experiments; and S.P. Nobs and M. Kopf wrote the manuscript.
Footnotes
Abbreviations used:
- AM
- alveolar macrophage
- BAL
- bronchoalveolar lavage
- dLN
- draining LN
- HDM
- house dust mite extract
- MFI
- mean fluorescence intensity
References
- Acosta-Rodriguez E.V., Rivino L., Geginat J., Jarrossay D., Gattorno M., Lanzavecchia A., Sallusto F., and Napolitani G.. 2007. Surface phenotype and antigenic specificity of human interleukin 17-producing T helper memory cells. Nat. Immunol. 8:639–646. 10.1038/ni1467 [DOI] [PubMed] [Google Scholar]
- Barner M., Mohrs M., Brombacher F., and Kopf M.. 1998. Differences between IL-4R α-deficient and IL-4-deficient mice reveal a role for IL-13 in the regulation of Th2 responses. Curr. Biol. 8:669–672. 10.1016/S0960-9822(98)70256-8 [DOI] [PubMed] [Google Scholar]
- Becattini S., Latorre D., Mele F., Foglierini M., De Gregorio C., Cassotta A., Fernandez B., Kelderman S., Schumacher T.N., Corti D., et al. 2015. T cell immunity. Functional heterogeneity of human memory CD4+ T cell clones primed by pathogens or vaccines. Science. 347:400–406. 10.1126/science.1260668 [DOI] [PubMed] [Google Scholar]
- Bouhlel M.A., Derudas B., Rigamonti E., Dièvart R., Brozek J., Haulon S., Zawadzki C., Jude B., Torpier G., Marx N., et al. 2007. PPARγ activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 6:137–143. 10.1016/j.cmet.2007.06.010 [DOI] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croasdell A., Duffney P.F., Kim N., Lacy S.H., Sime P.J., and Phipps R.P.. 2015. PPARγ and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015:549691 10.1155/2015/549691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kleer I.M., Kool M., de Bruijn M.J., Willart M., van Moorleghem J., Schuijs M.J., Plantinga M., Beyaert R., Hams E., Fallon P.G., et al. 2016. Perinatal activation of the interleukin-33 pathway promotes type 2 immunity in the developing lung. Immunity. 45:1285–1298. 10.1016/j.immuni.2016.10.031 [DOI] [PubMed] [Google Scholar]
- Gregory L.G., and Lloyd C.M.. 2011. Orchestrating house dust mite-associated allergy in the lung. Trends Immunol. 32:402–411. 10.1016/j.it.2011.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H., de Heer H.J., Soullié T., Angeli V., Trottein F., Hoogsteden H.C., and Lambrecht B.N.. 2004. Activation of peroxisome proliferator-activated receptor-γ in dendritic cells inhibits the development of eosinophilic airway inflammation in a mouse model of asthma. Am. J. Pathol. 164:263–271. 10.1016/S0002-9440(10)63116-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt P.G., Oliver J., Bilyk N., McMenamin C., McMenamin P.G., Kraal G., and Thepen T.. 1993. Downregulation of the antigen presenting cell function(s) of pulmonary dendritic cells in vivo by resident alveolar macrophages. J. Exp. Med. 177:397–407. 10.1084/jem.177.2.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Marquillies P., Capron M., and Dombrowicz D.. 2004. Peroxisome proliferator-activated receptor γ is expressed in airways and inhibits features of airway remodeling in a mouse asthma model. J. Allergy Clin. Immunol. 113:882–888. 10.1016/j.jaci.2004.02.036 [DOI] [PubMed] [Google Scholar]
- Imai T., Takakuwa R., Marchand S., Dentz E., Bornert J.M., Messaddeq N., Wendling O., Mark M., Desvergne B., Wahli W., et al. 2004. Peroxisome proliferator-activated receptor γ is required in mature white and brown adipocytes for their survival in the mouse. Proc. Natl. Acad. Sci. USA. 101:4543–4547. 10.1073/pnas.0400356101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz L., Dani I., Edenhofer F., Nolden L., Evert B., Paul B., Kolanus W., Klockgether T., Knolle P., and Diehl L.. 2007. Peroxisome proliferator-activated receptor γ control of dendritic cell function contributes to development of CD4+ T cell anergy. J. Immunol. 178:2122–2131. 10.4049/jimmunol.178.4.2122 [DOI] [PubMed] [Google Scholar]
- Klotz L., Burgdorf S., Dani I., Saijo K., Flossdorf J., Hucke S., Alferink J., Nowak N., Beyer M., Mayer G., et al. 2009. The nuclear receptor PPARγ selectively inhibits Th17 differentiation in a T cell-intrinsic fashion and suppresses CNS autoimmunity. J. Exp. Med. 206:2079–2089 (published erratum appears in J. Exp. Med. 2009. 206:3159). 10.1084/jem.20082771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopf M., Le Gros G., Bachmann M., Lamers M.C., Bluethmann H., and Köhler G.. 1993. Disruption of the murine IL-4 gene blocks Th2 cytokine responses. Nature. 362:245–248. 10.1038/362245a0 [DOI] [PubMed] [Google Scholar]
- Kopf M., Schneider C., and Nobs S.P.. 2015. The development and function of lung-resident macrophages and dendritic cells. Nat. Immunol. 16:36–44. 10.1038/ni.3052 [DOI] [PubMed] [Google Scholar]
- Kurowska-Stolarska M., Stolarski B., Kewin P., Murphy G., Corrigan C.J., Ying S., Pitman N., Mirchandani A., Rana B., van Rooijen N., et al. 2009. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J. Immunol. 183:6469–6477. 10.4049/jimmunol.0901575 [DOI] [PubMed] [Google Scholar]
- Lambrecht B.N., De Veerman M., Coyle A.J., Gutierrez-Ramos J.C., Thielemans K., and Pauwels R.A.. 2000. Myeloid dendritic cells induce Th2 responses to inhaled antigen, leading to eosinophilic airway inflammation. J. Clin. Invest. 106:551–559. 10.1172/JCI8107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanzavecchia A., and Scheidegger D.. 1987. The use of hybrid hybridomas to target human cytotoxic T lymphocytes. Eur. J. Immunol. 17:105–111. 10.1002/eji.1830170118 [DOI] [PubMed] [Google Scholar]
- Li W., Dai W., Sun J., Zhang W., Jiang Y., Ma C., Wang C., and He J.. 2015. Association of peroxisome proliferator-activated receptor-γ gene polymorphisms and gene-gene interaction with asthma risk in a Chinese adults population. Int. J. Clin. Exp. Med. 8:19346–19352. [PMC free article] [PubMed] [Google Scholar]
- Luche H., Weber O., Nageswara Rao T., Blum C., and Fehling H.J.. 2007. Faithful activation of an extra-bright red fluorescent protein in “knock-in” Cre-reporter mice ideally suited for lineage tracing studies. Eur. J. Immunol. 37:43–53. 10.1002/eji.200636745 [DOI] [PubMed] [Google Scholar]
- Messi M., Giacchetto I., Nagata K., Lanzavecchia A., Natoli G., and Sallusto F.. 2003. Memory and flexibility of cytokine gene expression as separable properties of human TH1 and TH2 lymphocytes. Nat. Immunol. 4:78–86. 10.1038/ni872 [DOI] [PubMed] [Google Scholar]
- Nemenoff R.A. 2007. Peroxisome proliferator-activated receptor-γ in lung cancer: Defining specific versus “off-target” effectors. J. Thorac. Oncol. 2:989–992. 10.1097/JTO.0b013e318158cf0a [DOI] [PubMed] [Google Scholar]
- Noben-Trauth N., Shultz L.D., Brombacher F., Urban J.F. Jr., Gu H., and Paul W.E.. 1997. An interleukin 4 (IL-4)-independent pathway for CD4+ T cell IL-4 production is revealed in IL-4 receptor-deficient mice. Proc. Natl. Acad. Sci. USA. 94:10838–10843. 10.1073/pnas.94.20.10838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobs S.P., Schneider C., Dietrich M.G., Brocker T., Rolink A., Hirsch E., and Kopf M.. 2015. PI3-Kinase-γ has a distinct and essential role in lung-specific dendritic cell development. Immunity. 43:674–689. 10.1016/j.immuni.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Ohnmacht C., Park J.H., Cording S., Wing J.B., Atarashi K., Obata Y., Gaboriau-Routhiau V., Marques R., Dulauroy S., Fedoseeva M., et al. 2015. Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science. 349:989–993. 10.1126/science.aac4263 [DOI] [PubMed] [Google Scholar]
- Oxenius A., Bachmann M.F., Zinkernagel R.M., and Hengartner H.. 1998. Virus-specific MHC-class II-restricted TCR-transgenic mice: effects on humoral and cellular immune responses after viral infection. Eur. J. Immunol. 28:390–400. [DOI] [PubMed] [Google Scholar]
- Plantinga M., Guilliams M., Vanheerswynghels M., Deswarte K., Branco-Madeira F., Toussaint W., Vanhoutte L., Neyt K., Killeen N., Malissen B., et al. 2013. Conventional and monocyte-derived CD11b+ dendritic cells initiate and maintain T helper 2 cell-mediated immunity to house dust mite allergen. Immunity. 38:322–335. 10.1016/j.immuni.2012.10.016 [DOI] [PubMed] [Google Scholar]
- Robinette M.L., Fuchs A., Cortez V.S., Lee J.S., Wang Y., Durum S.K., Gilfillan S., Colonna M., and Immunological Genome C.. Immunological Genome Consortium . 2015. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat. Immunol. 16:306–317. 10.1038/ni.3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer S. 2015. Ligands for the nuclear peroxisome proliferator-activated receptor γ. Trends Pharmacol. Sci. 36:688–704 (published erratum appears in Trends Pharmacol. Sci. 2016. 37:167). 10.1016/j.tips.2015.06.010 [DOI] [PubMed] [Google Scholar]
- Schmitz J., Owyang A., Oldham E., Song Y., Murphy E., McClanahan T.K., Zurawski G., Moshrefi M., Qin J., Li X., et al. 2005. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 23:479–490. 10.1016/j.immuni.2005.09.015 [DOI] [PubMed] [Google Scholar]
- Schneider C., Nobs S.P., Heer A.K., Kurrer M., Klinke G., van Rooijen N., Vogel J., and Kopf M.. 2014a Alveolar macrophages are essential for protection from respiratory failure and associated morbidity following influenza virus infection. PLoS Pathog. 10:e1004053 10.1371/journal.ppat.1004053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C., Nobs S.P., Kurrer M., Rehrauer H., Thiele C., and Kopf M.. 2014b Induction of the nuclear receptor PPAR-γ by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat. Immunol. 15:1026–1037. 10.1038/ni.3005 [DOI] [PubMed] [Google Scholar]
- Soroosh P., Doherty T.A., Duan W., Mehta A.K., Choi H., Adams Y.F., Mikulski Z., Khorram N., Rosenthal P., Broide D.H., and Croft M.. 2013. Lung-resident tissue macrophages generate Foxp3+ regulatory T cells and promote airway tolerance. J. Exp. Med. 210:775–788. 10.1084/jem.20121849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szanto A., Balint B.L., Nagy Z.S., Barta E., Dezso B., Pap A., Szeles L., Poliska S., Oros M., Evans R.M., et al. 2010. STAT6 transcription factor is a facilitator of the nuclear receptor PPARγ-regulated gene expression in macrophages and dendritic cells. Immunity. 33:699–712. 10.1016/j.immuni.2010.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmari I., Töröcsik D., Agostini M., Nagy T., Gurnell M., Barta E., Chatterjee K., and Nagy L.. 2007. PPARγ regulates the function of human dendritic cells primarily by altering lipid metabolism. Blood. 110:3271–3280. 10.1182/blood-2007-06-096222 [DOI] [PubMed] [Google Scholar]
- Tan H.T., Sugita K., and Akdis C.A.. 2016. Novel biologicals for the treatment of allergic diseases and asthma. Curr. Allergy Asthma Rep. 16:70 10.1007/s11882-016-0650-5 [DOI] [PubMed] [Google Scholar]
- Thepen T., Van Rooijen N., and Kraal G.. 1989. Alveolar macrophage elimination in vivo is associated with an increase in pulmonary immune response in mice. J. Exp. Med. 170:499–509. 10.1084/jem.170.2.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend M.J., Fallon P.G., Matthews D.J., Jolin H.E., and McKenzie A.N.. 2000. T1/ST2-deficient mice demonstrate the importance of T1/ST2 in developing primary T helper cell type 2 responses. J. Exp. Med. 191:1069–1076. 10.1084/jem.191.6.1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifilieff A., Bench A., Hanley M., Bayley D., Campbell E., and Whittaker P.. 2003. PPAR-α and -γ but not -δ agonists inhibit airway inflammation in a murine model of asthma: In vitro evidence for an NF-κB-independent effect. Br. J. Pharmacol. 139:163–171. 10.1038/sj.bjp.0705232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rijt L.S., Jung S., Kleinjan A., Vos N., Willart M., Duez C., Hoogsteden H.C., and Lambrecht B.N.. 2005. In vivo depletion of lung CD11c+ dendritic cells during allergen challenge abrogates the characteristic features of asthma. J. Exp. Med. 201:981–991. 10.1084/jem.20042311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mutius E. 2009. Gene-environment interactions in asthma. J. Allergy Clin. Immunol. 123:3–11. 10.1016/j.jaci.2008.10.046 [DOI] [PubMed] [Google Scholar]
- Webb D.C., Cai Y., Matthaei K.I., and Foster P.S.. 2007. Comparative roles of IL-4, IL-13, and IL-4Rα in dendritic cell maturation and CD4+ Th2 cell function. J. Immunol. 178:219–227. 10.4049/jimmunol.178.1.219 [DOI] [PubMed] [Google Scholar]
- Willart M.A., Deswarte K., Pouliot P., Braun H., Beyaert R., Lambrecht B.N., and Hammad H.. 2012. Interleukin-1α controls allergic sensitization to inhaled house dust mite via the epithelial release of GM-CSF and IL-33. J. Exp. Med. 209:1505–1517. 10.1084/jem.20112691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerly G., Honda K., Loyens M., Papin J.P., Auwerx J., Staels B., Capron M., and Dombrowicz D.. 2003. Peroxisome proliferator-activated receptors α and γ down-regulate allergic inflammation and eosinophil activation. J. Exp. Med. 198:411–421. 10.1084/jem.20021384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlfert E.A., Grainger J.R., Bouladoux N., Konkel J.E., Oldenhove G., Ribeiro C.H., Hall J.A., Yagi R., Naik S., Bhairavabhotla R., et al. 2011. GATA3 controls Foxp3+ regulatory T cell fate during inflammation in mice. J. Clin. Invest. 121:4503–4515. 10.1172/JCI57456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfer A., Bakker T., Wilson A., Nicolas M., Ioannidis V., Littman D.R., Lee P.P., Wilson C.B., Held W., MacDonald H.R., and Radtke F.. 2001. Inactivation of Notch 1 in immature thymocytes does not perturb CD4 or CD8T cell development. Nat. Immunol. 2:235–241. 10.1038/85294 [DOI] [PubMed] [Google Scholar]
- Wynn T.A. 2015. Type 2 cytokines: Mechanisms and therapeutic strategies. Nat. Rev. Immunol. 15:271–282. 10.1038/nri3831 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Huang Y., He J., Li C., Deng W., Ran X., and Wang D.. 2014. Rosiglitazone, a peroxisome proliferator-activated receptor-γ agonist, attenuates airway inflammation by inhibiting the proliferation of effector T cells in a murine model of neutrophilic asthma. Immunol. Lett. 157:9–15. 10.1016/j.imlet.2013.11.004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.