Perlin et al. discuss recent findings in the field of zebrafish hematopoiesis, focusing on the transcriptional regulation of hematopoietic stem cell (HSC) induction and HSC–niche interactions. Manipulation of developmental signaling pathways may enhance HSC bioengineering, which would improve transplantation therapies.
Abstract
Hematopoietic stem cell transplantation (HSCT) is an important therapy for patients with a variety of hematological malignancies. HSCT would be greatly improved if patient-specific hematopoietic stem cells (HSCs) could be generated from induced pluripotent stem cells in vitro. There is an incomplete understanding of the genes and signals involved in HSC induction, migration, maintenance, and niche engraftment. Recent studies in zebrafish have revealed novel genes that are required for HSC induction and niche regulation of HSC homeostasis. Manipulation of these signaling pathways and cell types may improve HSC bioengineering, which could significantly advance critical, lifesaving HSCT therapies.
Introduction
Each day humans require the production of about 100 billion blood cells. Rare hematopoietic stem cells (HSCs) are required to replenish multilineage progenitors, which can then differentiate into lineage-restricted cells (reviewed by Orkin and Zon, 2008). These unique HSCs reside in specialized niches and rely on critical niche factors to regulate their maintenance, self-renewal, differentiation, and regeneration after injury. There is currently a large effort to generate and engineer HSCs in vitro for research and therapy (reviewed by Rowe et al., 2016). HSC transplantation (HSCT) is a curative therapy to treat a wide variety of hematological malignancies, but numbers of immunologically matched HSCs are a limiting factor in such treatments (Gragert et al., 2014). More research is needed to understand cell-autonomous and non-cell-autonomous regulation of HSC induction, homing, and engraftment. If we can further our understanding of developmental hematopoiesis and identify key regulators of these processes in vivo, it may be possible to apply this knowledge to in vitro efforts to derive patient-specific HSCs from induced pluripotent stem cells (iPSCs). This would help overcome current issues of limiting cell number and graft-versus-host disease. Zebrafish are an ideal model to use to study hematopoiesis. The sites of blood development and the molecular signals regulating hematopoiesis are conserved with mammalian systems, and zebrafish embryos are transparent, allowing for noninvasive visualization of HSC emergence, migration, and engraftment. Zebrafish are also amenable to forward genetic and chemical screening, making them a useful tool to uncover novel regulators of blood development.
Overview of zebrafish hematopoiesis and techniques
The zebrafish has emerged as a powerful model for the study of hematopoiesis. There are many strengths that make this system ideally suited to the study of both hematopoietic development and disorders. Zebrafish embryos are fertilized externally, develop rapidly, and are amenable to genetic modification, which allows relatively straightforward creation of transgenic reporter lines labeling specific cell populations. In addition, the transparency of the embryos has enabled real-time visualization of the emergence, migration, and behavior of blood cells as they populate the embryo and adult, both during endogenous development and upon various chemical or genetic perturbations. These unique attributes have uncovered novel genes, cell types, and cellular behaviors that are required for normal vertebrate hematopoiesis.
Developmental hematopoiesis is conserved across species
Vertebrate hematopoiesis occurs during two waves of development. Hematopoietic cells that support early stages of development are formed during the primitive wave (reviewed by Orkin and Zon, 2008). Primitive erythroid cells, important for oxygenation of rapidly growing tissues, are formed in the intermediate cell mass (ICM), which is derived from the posterior lateral mesoderm (PLM; reviewed by Orkin and Zon, 2008). Primitive macrophages are formed in the anterior lateral mesoderm and initiate innate immunity (Travnickova et al., 2015). Before the onset of definitive hematopoiesis, a transient population of multipotent erythromyeloid progenitors emerges from the ICM and seeds the fetal liver in mammals, or the caudal hematopoietic tissue (CHT) in zebrafish (reviewed by Ciau-Uitz et al., 2014). The hallmark of definitive hematopoiesis is the induction of HSCs, which can self-renew and give rise to more committed progenitors, such as myeloid and lymphoid cells (reviewed by Orkin and Zon, 2008). Definitive HSCs bud off from specialized endothelial cells of the aorta-gonad mesonephros (AGM) region, known as hemogenic endothelium, and migrate to successive niche sites as hematopoiesis occurs sequentially (Sánchez et al., 1996; Bertrand et al., 2010b; Boisset et al., 2010; Kissa and Herbomel, 2010). In mammals, definitive HSCs migrate to and colonize the placenta, fetal liver, thymus, spleen, and finally the bone marrow, which is the adult hematopoietic niche (reviewed by Orkin and Zon, 2008). In zebrafish, HSCs are born in the AGM around 30 h post fertilization (hpf; Bertrand et al., 2010b; Kissa and Herbomel, 2010) and migrate to the CHT, a vascular plexus in the tail, starting around 36 hpf (Murayama et al., 2006; Tamplin et al., 2015). HSCs begin to populate the thymus and kidney, the adult stem cell niches in zebrafish, at 4 d post fertilization (dpf; Murayama et al., 2006; Orkin and Zon, 2008).
Understanding the origin of HSCs
Investigation into the origin of HSCs indicated that human endothelial cells isolated from the embryonic dorsal aorta have the potential to differentiate into myeloid and lymphoid progeny when cultured in vitro (Oberlin et al., 2002). Lineage tracing experiments in mouse and chick models also suggested that the intra-aortic clusters that give rise to HSCs are derived from endothelial cells (Jaffredo et al., 1998; Zovein et al., 2008). However, there remained some debate over whether the HSCs were born in the aortic floor itself or in the subaortic mesenchyme (Bertrand et al., 2005; reviewed by Dieterlen-Lièvre et al., 2006). The unique strengths of the zebrafish enabled the first real-time, in vivo visualization of the emergence of HSCs in their native environment, and live imaging of developing zebrafish embryos confirmed their endothelial origin (Bertrand et al., 2010b; Kissa and Herbomel, 2010). Transgenic reporter lines with fluorescently labeled hematopoietic stem and progenitor cells (HSPCs), such as cmyb:eGFP and cd41:GFP, combined with the endothelial marker kdrl:mCherry or lmo2:dsred, have enabled the direct observation of HSPCs as they first arise from specialized hemogenic endothelium in the AGM. Between 28 and 32 hpf, endothelial cells lining the ventral wall of the dorsal aorta begin to contract and change shape, becoming more spherical and forming buds that extend into the aortic lumen (Bertrand et al., 2010b; Kissa and Herbomel, 2010). At the same time, they begin to express the HSPC-specific genes cmyb and cd41. During this process, which has been termed the endothelial hematopoietic transition (EHT; Kissa and Herbomel, 2010), nascent HSPCs bud off into the subaortic space before entering circulation via the dorsal wall of the cardinal vein, to migrate to the intermediate stem cell niche in the CHT (Kissa et al., 2008).
Using genetic and chemical manipulation to understand hematopoietic development
Zebrafish are extremely amenable to genetic manipulations, including overexpression by microinjection of RNA or DNA; knockdown by morpholino, TALENs, or CRISPR; or unbiased approaches such as large-scale forward genetic screens (reviewed by Avagyan and Zon, 2016). Since the breakthrough genetic screens of the 1990s, forward genetics in zebrafish has been used to identify important and novel genes in hematopoietic development, many of which have roles in known human diseases (Driever et al., 1996; Haffter et al., 1996; Ransom et al., 1996; reviewed by de Jong and Zon, 2005). For example, the ferroportin 1 iron transporter was first isolated from a zebrafish mutant, and then similar mutations were found in human patients with iron-overload disorders (Donovan et al., 2000; Gordeuk et al., 2003). Insertional mutagenesis screens have also been used to probe hematopoiesis and have identified mutations associated with human diseases, such as Diamond-Blackfan anemia (Taylor et al., 2012; Mirabello et al., 2014).
Chemical screening in zebrafish has uncovered essential hematopoietic signaling that is conserved in mammals and is a powerful model for drug discovery. Zebrafish are an ideal model for screening both new small molecules and libraries of US Food and Drug Administration–approved drugs to examine how they modulate a specific phenotype of interest or disease model (reviewed by Avagyan and Zon, 2016). Thousands of zebrafish embryos can easily be obtained and screened as intact embryos with temporal control. One of the best examples of chemical screening success is that of prostaglandin E2 (PGE2), which increases the number of HSPCs in the zebrafish AGM (North et al., 2007). A 2-h pulse treatment of PGE2 is also sufficient to increase the long-term repopulation activity of mouse HSPCs, demonstrating conservation. Currently, a human clinical trial is under way to determine whether PGE2 treatment of umbilical cord blood enhances HSCT. In the phase I trial, PGE2-treated cells preferentially engrafted in 10 of 12 patients (Cutler et al., 2013). Competitive HSPC transplantation can also be done in adult zebrafish. Such transplantation was combined with a chemical screen to determine that 11,12-epoxyeicosatrienoic acid enhances HSPC specification, homing, and engraftment (Li et al., 2015). Other emerging techniques in zebrafish include surgical parabiosis, in which embryos of two different genotypes or treatment can be conjoined with a shared circulation to examine the autonomous or nonautonomous role of various genes or factors in controlling HSPC migration and engraftment (Demy et al., 2013; Hagedorn et al., 2016).
Work in both mammals and zebrafish has shed light on the critical processes that regulate blood development and disease, but further efforts are needed to improve HSC transplant therapies. Recent studies in zebrafish, including time-lapse imaging and forward genetic screens, have revealed novel cell–cell interactions and signaling pathways that regulate HSPC induction, migration, and homing, and it may be possible to apply these new findings to strategies to develop HSCs in vitro for therapeutic purposes.
Transcriptional regulation of HSC specification
The specification and emergence of HSCs in the AGM are regulated by a plethora of molecular and biochemical signaling pathways, which are highly conserved between zebrafish and mammals. Many of these signaling pathways rely on the controlled and coordinated expression of transcription factors, which are fundamental drivers of cell fate. Recent studies have used transcription factor reprogramming approaches to try to generate HSCs in vitro, but these are not yet equivalent to bona fide HSCs (reviewed by Rowe et al., 2016). Furthering our understanding of the precise transcription factors that are required to regulate the emergence of definitive HSCs may reveal new molecular pathways that could be targeted to improve these HSC engineering strategies. Recent zebrafish studies have revealed new insights into the regulation of HSC induction by Notch1 and other transcription factors.
Regulation of HSC induction by Runx1 signaling
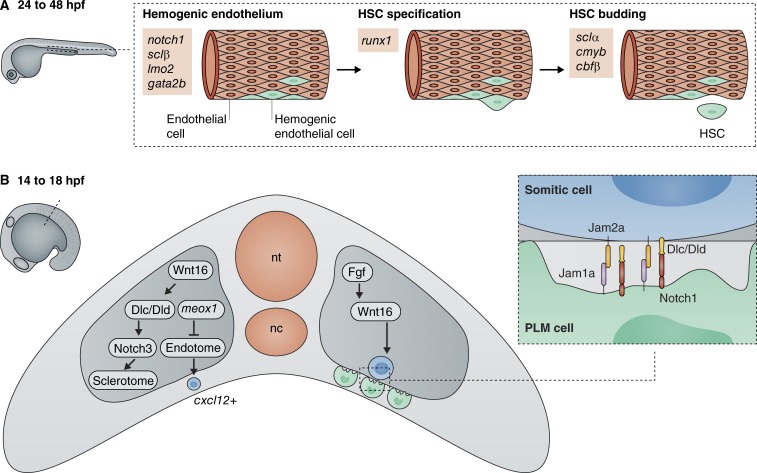
Runx1 is the master regulator of definitive HSC specification across species (North et al., 2002; Chen et al., 2009; Bertrand et al., 2010b; Kissa and Herbomel, 2010). Early studies in mice revealed that this key hematopoietic transcription factor is essential for definitive hematopoiesis (Okuda et al., 1996), as mice lacking Runx1 fail to initiate fetal liver hematopoiesis and are embryonic lethal. In zebrafish embryos lacking runx1, EHT events are aborted, and any cells that do emerge rapidly burst into pieces (Kissa and Herbomel, 2010). Runx1 expression is regulated by the transcription factors Gata2, Scl, and Lmo2 (Nottingham et al., 2007), which are required for both primitive and definitive hematopoiesis (Tsai et al., 1994; Shivdasani et al., 1995; Yamada et al., 1998). As in mammalian systems, Scl and Lmo2 are also fundamentally required for zebrafish hematopoiesis, and deletion of either factor results in loss of both primitive and definitive blood lineages (Dooley et al., 2005; Patterson et al., 2007). There are two isoforms of scl in zebrafish, which have distinct temporal requirements. The sclb isoform is expressed first in hemogenic endothelial cells before EHT, and the scla isoform is expressed later during HSC budding (Fig. 1 A; Zhen et al., 2013). Expression of sclb is dependent on adenosine signaling, which regulates hemogenic endothelium formation and enhances HSPC development (Jing et al., 2015). Zebrafish also express two Gata2 homologues, gata2a and gata2b, the latter of which is specifically expressed in hemogenic endothelial cells and is essential for HSC emergence (Butko et al., 2015).
Figure 1.
HSC specification is regulated by transcriptional signaling throughout development. (A) Zebrafish studies have revealed transcription factors that are important at different stages of definitive hematopoiesis: hemogenic endothelial formation, HSC specification in the AGM, and HSC budding. (B) Recent studies in zebrafish have identified Notch signaling pathways in the somite that influence HSC specification. Wnt16 signaling induces expression of Notch ligands DeltaC and DeltaD, which interact with Notch1 receptors on vascular precursors from the posterior lateral mesoderm (PLM) as they migrate over the somite surface. This interaction is dependent on the junctional adhesion molecules Jam1a and Jam2a. Notch3 is also expressed in the somite and may provide additional inductive cues to HSCs via the sclerotome. Endothelial precursor cells derived from the endotome are specified by the homeobox gene meox1 and migrate to the dorsal aorta, where they contribute to cxcl12-dependent HSC induction. nc, notochord; nt, neural tube.
Regulation of HSC induction by Notch signaling
Notch1 is a key transcriptional regulator required for the development of repopulating HSCs from hemogenic endothelium (Kumano et al., 2003; Hadland et al., 2004) and has been used in efforts to expand HSCs from endothelium in vitro with some success (Hadland et al., 2015). Notch1 is a conserved regulator of HSC emergence and acts in multiple cell-autonomous and non-cell-autonomous ways to regulate HSC emergence, but we still do not fully understand these mechanisms (reviewed by Butko et al., 2016). Canonical Notch signaling occurs when the extracellular Notch transmembrane receptor is activated by a transmembrane ligand on a neighboring cell (Nakagawa et al., 2006). This induces cleavage and release of the Notch intracellular domain, which translocates to the nucleus, where it functions as a transcription factor. Signaling pathways involving Bmp (Wilkinson et al., 2009), Wnt (Goessling et al., 2009; Clements et al., 2011), Shh (Lawson et al., 2002; Gering and Patient, 2005; Wilkinson et al., 2009), and Vegfa (Gering and Patient, 2005; Burns et al., 2009) are required for development of the arterial endothelium, activation of the Notch pathway, and specification of definitive HSCs. Notch signaling is required for successful EHT and HSC emergence and activates the expression of many hematopoietic genes, including scl, gata2, and runx1 (Burns et al., 2005; Nakagawa et al., 2006). The Notch intracellular domain activates Hes1, Hes5, Foxc2, and Gata 2 (Robert-Moreno et al., 2005; Guiu et al., 2013; Jang et al., 2015), and Notch signaling indirectly regulates Runx1 via this activation of Gata2 (Kumano et al., 2003; Hadland et al., 2004; Robert-Moreno et al., 2005), which interacts with Fli-1 and the Scl/Lmo2/Ldb1 complex to form a transcriptional signaling center that drives Runx1 expression (Nottingham et al., 2007). Studies show that the loss of Notch signaling pathway components results in reduced numbers of functional HSCs. For instance, the zebrafish mind bomb mutant, in which an E3 ligase required for Notch ligand ubiquitination and internalization is mutated, fails to specify definitive HSCs (Burns et al., 2005; Bertrand et al., 2010a). Primitive hematopoiesis remains unaffected, and the same phenotype is observed with this mutation in mammalian systems (Yoon et al., 2008). The Notch1 receptor is essential for specification of definitive HSCs in a cell-autonomous manner, and its deletion in mice and zebrafish results in reduced HSC emergence from the hemogenic endothelium (Kumano et al., 2003; Hadland et al., 2004; Kim et al., 2014). In contrast, the Notch2 receptor is dispensable (Kumano et al., 2003; Kim et al., 2014). Recently, Notch-mediated EHT has been shown to be dependent on the transcriptional regulator Evi1, which activates Notch via pAKT (Konantz et al., 2016).
Zebrafish studies have also revealed a novel requirement for Notch signaling in HSC development that occurs in a non-cell-autonomous manner (Clements et al., 2011). Fgf signaling in the somite from 14 to 17 hpf, before the onset of definitive hematopoiesis, activates Wnt16, which induces somitic expression of the Notch ligands DeltaC and DeltaD (Fig. 1 B; Clements et al., 2011; Lee et al., 2014). These ligands are required to induce hematopoietic potential in vascular precursors from the PLM as they migrate over the ventral surface of the somite. The transduction of Notch signals between the somite and precursor cells is dependent on close association of the two cell types, which is achieved via interaction of the junctional adhesion molecules jam1a (expressed on the PLM cells) and jam2a (expressed on the somite cells; Kobayashi et al., 2014). Knockdown of either of these molecules in zebrafish embryos results in a loss of Notch signaling and reduced HSC numbers. Non-cell-autonomous regulation of HSC specification is also mediated by notch3, which is expressed in the somites at 14 hpf (Kim et al., 2014). This receptor is activated by DeltaC and DeltaD, downstream of Wnt16. It is not yet clear how this somite-intrinsic signaling affects HSC specification, but it has been shown that both Notch3 and Wnt16 are essential for specification of the sclerotome, which is believed to provide additional inductive cues to HSCs (Kim et al., 2014). Recently, it was found that wnt16 expression is potentiated by the regulatory factor Rspo1, which also up-regulates vegfaa in the somite (Genthe and Clements, 2017). This activates TGFβ signaling in the hemogenic endothelium, which is required for HSC specification via up-regulation of the Notch ligand jag1a (Monteiro et al., 2016). Further studies are required to establish the precise interactions between signaling pathways in the somite and hemogenic endothelial cells over the course of their development, which would help determine the transcription factors that are required for HSC specification in a non-cell-autonomous manner. These, along with other signaling factors from the stem cell niche, are likely to be essential for the successful generation of HSCs in vitro for therapeutic purposes.
Regulation of HSC induction by HOX genes
Several homeodomain-containing transcription factors, including HOXA9, HOXB3, and HOXB4, are highly expressed in HSPCs, but the mechanisms by which they regulate hematopoiesis are not fully understood (reviewed by Alharbi et al., 2013). Knockdown of Hoxa9, Hoxb3, and Hoxb4 in mice leads to a reduction in the repopulating capacity of HSCs and a deficiency in myeloid and lymphoid precursors (Magnusson et al., 2007). An important role for hox genes has also been demonstrated in zebrafish HSC development. Mutations in the upstream activator of hox genes, cdx4, cause hematopoietic defects and aberrant hox gene expression (Davidson et al., 2003). These defects can be rescued by overexpression of hoxb7a or hoxa9a. The additional knockdown of cdx1 in the cdx4 mutant background results in a complete loss of blood cell specification, and overexpression of hoxa9 in this situation rescues both embryonic erythropoiesis and HSC formation in the AGM (Davidson and Zon, 2006). Overexpression of cdx4 leads to ectopic blood development, both in wildtype zebrafish embryos and mouse embryonic stem cells (ESCs; Davidson and Zon, 2004; Lengerke et al., 2007). These studies highlight the importance of hox genes as master regulators of HSC development, which supply key spatial and temporal information and may provide an important missing link to generate human HSCs from iPSCs. Early efforts to derive HSCs from mouse ESCs used overexpression of Hoxb4 to generate hematopoietic cells with some engraftment capacity (Kyba et al., 2002), and more recent transcriptional reprogramming strategies have used HOX genes in combination with other transcription factors (Doulatov et al., 2013; Sugimura et al., 2017). In the most recent study, HOXA5, HOXA9, and HOXA10 were used alongside ERG, LCOR, RUNX1, and SPI1 to convert hemogenic endothelium to HSPCs with multilineage engraftment potential (Sugimura et al., 2017). Further studies are necessary to precisely define the function of homeotic genes in HSC development.
Transcriptional regulation of budding HSCs
In addition to advancing our understanding of hemogenic endothelium development and HSC fate specification, zebrafish studies have also revealed insight into several additional transcriptional regulators that are critical for the release of nascent HSCs from the hemogenic endothelium. In zebrafish embryos with deletion of the Runx1-binding cofactor cbfb, HSCs are unable to extravasate from the dorsal aorta and definitive hematopoiesis does not occur (Bresciani et al., 2014). Deletion of Cbfb in mice blocks fetal liver hematopoiesis and causes embryonic lethality (Wang et al., 1996). HSC release is also inhibited in zebrafish with mutations in the cmyb gene, which encodes another transcription factor required for HSC maintenance (Zhang et al., 2011). Cmyb function is conserved in mammalian systems; in Cmyb mutant mice, impaired mobilization of HSCs from the AGM is indicated by reduced fetal liver hematopoiesis (Mucenski et al., 1991). Further research is necessary to broaden our understanding of the mechanisms that allow release of HSCs from the endothelium, which will be important for establishing efficient ways to generate large numbers of HSCs in vitro.
Understanding HSC–niche interactions
HSPCs intimately interact with niche cells throughout their development from specification to migration to engraftment (reviewed by Morrison and Scadden, 2014). HSC development relies on intimate interactions with niche cells of the microenvironment, which provide important signaling factors and physical properties. The ability to successfully engineer HSCs will likely rely on the incorporation of niche-derived factors and cues, and reprogramming studies have illustrated the importance of the vascular niche for the emergence of definitive hematopoietic cells (Ledran et al., 2008; Choi et al., 2012; Sturgeon et al., 2013; Elcheva et al., 2014; Lis et al., 2017). There is currently much that is not understood about niche ontogeny, gene expression, and function. Recent studies in zebrafish have elucidated new roles for niche cells in regulating HSC induction, migration, maintenance, and differentiation.
Niche interactions and signaling in the mammalian bone marrow niche
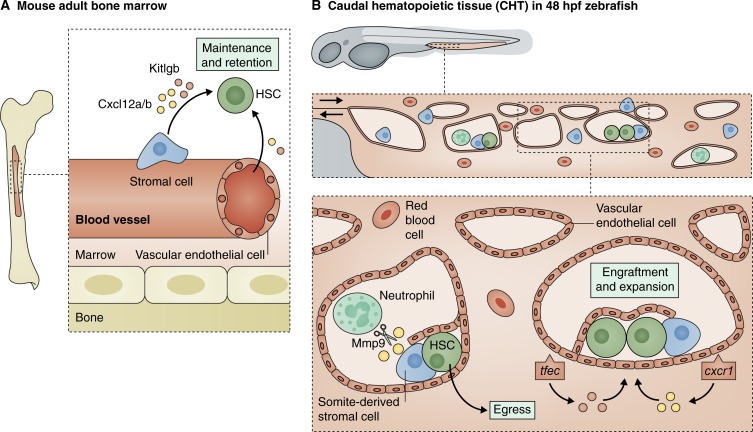
After HSCs bud off from the hemogenic endothelium, they enter circulation and home to successive sites of hematopoiesis (reviewed by Orkin and Zon, 2008). Homing to the niche and lodgment within that niche are critical processes that must occur both in development and in transplant (Heazlewood et al., 2014). The cellular and chemical composition of the adult mammalian bone marrow niche is made up of multiple types of stromal cells, endothelial cells, osteoblasts and osteoclasts, sympathetic nerves, megakaryocytes, and macrophages, among others, and is the subject of a large body of work (reviewed by Mendelson and Frenette, 2014; Morrison and Scadden, 2014; Birbrair and Frenette, 2016). For the past decade researchers have been working to elucidate the cell types and signals in the bone marrow niche that are essential for HSC homeostasis. Stromal cells and endothelial cells of the vascular niche are important sources of the two main niche factors, chemokine CXCL12 (also known as stromal derived factor-1) and cytokine KIT ligand (KITLG; also known as stem cell factor; Fig. 2 A). CXCL12 and KITLG promote HSC localization to, and maintenance within, the perivascular niche (reviewed by Kisanuki et al., 2001; Ding et al., 2012; Ding and Morrison, 2013; Greenbaum et al., 2013; Morrison and Scadden, 2014). These factors are expressed at much higher levels by stromal cells but are required in both cell types for niche function. Both endothelial and stromal cells maintain HSCs through KITLG and CXCL12 signaling, and efforts are under way to mimic this niche in vitro. Hematopoietic progenitors can be obtained when ESC- or AGM-derived cells are cultured with stromal cells, but the efficiency of this process needs to be improved (Ledran et al., 2008; Choi et al., 2012; Sturgeon et al., 2013). In an alternative approach, nonhematopoietic endothelial cells can be reprogrammed into engraftable hematopoietic cells but only if they are in contact with a vascular niche, indicating a requirement for inductive and supportive signals from associated endothelial cells (Sandler et al., 2014).
Figure 2.
Stromal and endothelial cells in the vascular niche modulate Cxcl12 and Kitlg signaling to regulate HSCs. (A) The mammalian bone marrow contains multiple types of perivascular stromal cells and endothelial cells in the vascular niche. Stromal and endothelial cells are both required to express niche factors Cxcl12 and Kitlg to influence HSC localization, maintenance, retention, and quiescence. Stromal cells express these factors at ∼1,000× greater levels. (B) HSPCs colonize the caudal hematopoietic tissue in the 48 hpf zebrafish. Within the niche, multiple factors influence the expression of niche factors Cxcl12 and Kitlg in the CHT. Vascular endothelial cells require expression of transcription factor tfec and chemokine receptor cxcr1 to regulate expression of kitlgb and cxcl12a, respectively, to control HSPC engraftment and expansion. mmp9 expressed by neutrophils in the niche cleaves cxcl12 to promote HSPC egress and colonization of secondary niches.
Novel niche interactions and signaling required for HSC homeostasis in zebrafish
HSPCs also interact with multiple cell types in the zebrafish niche, including endothelial cells, stromal cells, macrophages, and neutrophils, among others (Tamplin et al., 2015). Purified cell lines from the AGM, CHT, and kidney stroma have been shown to support hematopoiesis (Stachura et al., 2009; Campbell et al., 2015; Wolf et al., 2017). Recent work in zebrafish has revealed novel cell–cell interactions that are important for HSPC migration and engraftment. The transparency of the zebrafish embryo makes it ideally suited for live imaging of intimate HSC-niche interactions. Although recent studies in the mouse have optimized imaging in the endosteal niche in the calvarium of the skull (Lo Celso et al., 2009; Lassailly et al., 2013), zebrafish offers unparalleled opportunities for long-term and continuous noninvasive imaging of endogenous processes. Live imaging was used to track HSCs from their birthplace in the AGM to the fetal CHT niche and to the adult thymus and kidney niches (Murayama et al., 2006; Kissa et al., 2008). Recently our laboratory developed a novel transgenic zebrafish line to more specifically label HSPCs (Tamplin et al., 2015). Tamplin et al. (2015) cloned a mouse regulatory region located in the first intron of the transcription factor Runx1 upstream of GFP to mark definitive HSPCs in zebrafish. This transgenic line allowed for high-resolution mapping of the dynamic cell–cell interactions that occur during HSPC homing and niche engraftment in unperturbed embryos. Multiple HSPC-endothelial cell interactions were observed during HSPC engraftment. As described in mammals, HSPCs migrate through circulation to the niche, adhere to the endothelium, and extravasate across the endothelial cell wall. High-resolution imaging revealed a novel behavior where the arrival of the HSPC triggers a remodeling of the endothelial cells to surround the HSPC in a niche pocket, termed “endothelial cell cuddling” (Fig. 2 B; Tamplin et al., 2015). Within the pocket, HSPCs are anchored to mesenchymal stromal cells, which orient the division axis of the stem cell. Imaging in dissected mouse fetal livers showed that endothelial cells similarly surround HSPCs in the mammalian fetal niche, indicating it is a conserved cell–cell interaction (Tamplin et al., 2015). Future studies will reveal the functional effect of these endothelial and stromal cell interactions on the fate of the HSC.
Two recent studies identified novel somite-derived cell types that are required in the AGM for HSC induction, and in the CHT for HSPC retention. Nguyen et al. (2014) found that a subset of somite-derived cells, known as endotome, form endothelial cells in the dorsal aorta of the AGM, express niche factor cxcl12b, and induce HSC formation. Homeobox gene meox1 was found to be expressed in a subset of somitic muscle cells and is required to specify somitic cell fates. In choker mutants, which have a null mutation in meox1, endotome cells are expanded, increasing Cxcl12b signaling and thus HSC induction (Nguyen et al., 2014). The authors suggested that to be able to generate HSCs in vitro, somite-derived endothelial cells might be required to serve as niche cells and promote induction. Murayama et al. (2015) identified a distinct population of somite-derived stromal niche cells that are required in the fetal CHT niche for HSPC maintenance, expansion, and differentiation. The gene nascent polypeptide-associated complex, which is mutated in oloca mutants, is required cell autonomously for stromal cell maturation and survival in the developing CHT (Murayama et al., 2015). HSPCs can home to the CHT in the absence of these stromal cells, but naca is required nonautonomously in stromal cells for HSPC retention in the niche and subsequent expansion and differentiation of the stem cells (Fig. 2 B). In the absence of naca, these somite-derived stromal cells apoptose because of activation of the unfolded protein response (Murayama et al., 2015). These studies revealed a new role for somite-derived cells in HSC specification in the AGM, and in HSC expansion and differentiation in the fetal niche. These novel genes and cell types critical in HSC development were both identified through zebrafish forward genetic screens. Further studies will determine whether these cells and signals are important in mammalian HSC development.
Additional studies have uncovered new ways that niche signals Kitlg and Cxcl12 are modulated to regulate HSC retention, expansion, and egress from the niche (Fig. 2 B). The transcription factor Tfec was recently found to enhance HSC expansion in mouse HSCs nonautonomously through unknown mechanisms (Deneault et al., 2013). Studies in zebrafish determined that tfec is expressed in caudal endothelial cells, the region where the CHT niche will form, and is required for HSPC maintenance (Mahony et al., 2016). Mahoney et al. (2016) found that tfec acts nonautonomously in the vascular niche to regulate HSC expansion by controlling expression of cytokines, including kitlgb. Overexpressing kitlgb is sufficient to rescue HSPC expansion in mutants lacking tfec. Endothelial cell-derived niche signals are critical for proper HSPC development. In complementary experiments, gene expression profiling identified that niche-derived cxcl8/cxcr1 signaling enhances HSPC niche colonization by remodeling the vascular CHT niche (Blaser et al., 2017). Chemokine cxcl8 and its receptor cxcr1 are expressed in endothelial cells in the CHT, and this signaling positively regulates the amount of time HSPCs spend in the niche and the number of divisions and cuddling events that take place there. Increasing cxcr1 expression induces cxcl12a expression and increases the volume of the vascular niche. Through these mechanisms, cxcr1 acts nonautonomously to increase HSPC niche engraftment (Blaser et al., 2017). CXCL12 regulation of HSC migration is also regulated by matrix metalloproteinase 9 (MMP9) signaling in the adult bone marrow (Jin et al., 2008). MMP9 can degrade CXCL12 in plasma from human mobilized bone marrow to promote migration of CD34+ HSCs (Jin et al., 2008). In the zebrafish fetal niche, mmp9 expressed by neutrophils was recently shown to regulate HSPC homeostasis through regulation of cxcl12 expression (Theodore et al., 2017). Mmp9 activity was shown to be required for HSPC egress from the CHT and is required for colonization of secondary niches. Absence of mmp9 signaling led to vascular changes in the niche and retention of HSPCs in the CHT. These migration defects were blocked by knocking down cxcl12a signaling or phenocopied by overexpressing cxcl12b, indicating that Mmp9 is required to modulate this signaling axis to regulate HSPC homeostasis (Theodore et al., 2017). Together these studies have demonstrated that vascular-derived tfec and cxcr1 signaling, and neutrophil-derived mmp9 are important for regulating the vascular HSC niche, while expression of kitlgb and cxcl12 is important for controlling HSPC maintenance and migration. Modulation of these cell–cell interactions or signaling molecules could be of potential therapeutic benefit to patients receiving HSCT.
Concluding thoughts
Recent studies in zebrafish have capitalized on the unique experimental strengths of the system to probe the mechanisms of HSPC specification and niche interactions during HSPC emergence, migration, and engraftment. Continued efforts using emerging technologies in the zebrafish will reveal new insights into important aspects of hematopoiesis. Single-cell RNA sequencing will allow researchers to identify cell-specific gene expression, which can be used to more precisely mark distinct hematopoietic populations. Cell type–specific promoters will allow more detailed time-lapse imaging, lineage tracing, and gene knockout experiments, when combined with CRISPR/Cas9 technology. Multicolor fate mapping methods, such as Zebrabow, can be used to track HSC clonal dynamics in steady-state and in disease (Henninger et al., 2017). The unique strengths of the zebrafish have illuminated critical signaling pathways and cell–cell interactions that are conserved in mammalian hematopoiesis. New insights into the transcriptional regulation of HSC induction by factors such as Notch1, Runx1, and Cbfβ have highlighted the complexity of these signaling networks and the dynamic interactions that occur between HSC precursors and other cell types (reviewed by Bresciani et al., 2014; Butko et al., 2016). Zebrafish studies have also uncovered novel cell–cell interactions, such as endothelial cell cuddling (Tamplin et al., 2015), novel cell types, such as somite-derived niche cells (Nguyen et al., 2014; Murayama et al., 2015), and novel regulators of niche signals, such as Tfec and Cxcr1 (Mahony et al., 2016; Blaser et al., 2017). It may be important to recapitulate these processes and signals to generate definitive HSCs that can be used in the clinic. The ability to bioengineer patient-compatible HSCs would greatly enhance lifesaving bone marrow transplantation therapies used to treat patients with a variety of leukemias, lymphomas, anemias, and immunodeficiencies. Autologous transplantation of patient-derived HSCs would reduce the risks associated with allogeneic cell transplantation, such as immune rejection and graft-versus-host disease. Continued efforts to further characterize both the autonomous transcription factors that specify HSCs and the nonautonomous cell types and signals that are essential for HSC homeostasis are required to robustly engineer HSCs for therapeutic use.
Acknowledgments
This work was supported by National Institutes of Health grants R01HL04880, P01HL032262, P30DK049216, R01DK53298, U01HL10001, and R24DK092760. In addition, L.I. Zon is a Howard Hughes Medical Institute Investigator, and J.R. Perlin is an American Cancer Society Postdoctoral Fellow.
L.I. Zon is a founder and stock holder of Fate Therapeutics, Marauder Therapeutics, and Scholar Rock. The authors declare no additional competing financial interests.
Footnotes
Abbreviations used:
- AGM
- aorta-gonad mesonephros
- CHT
- caudal hematopoietic tissue
- dpf
- days post fertilization
- EHT
- endothelial hematopoietic transition
- ESC
- embryonic stem cell
- hpf
- hours post fertilization
- HSC
- hematopoietic stem cell
- HSCT
- HSC transplantation
- HSPC
- hematopoietic stem and progenitor cell
- ICM
- intermediate cell mass
- iPSC
- induced pluripotent stem cell
- PLM
- posterior lateral mesoderm
References
- Alharbi R.A., Pettengell R., Pandha H.S., and Morgan R.. 2013. The role of HOX genes in normal hematopoiesis and acute leukemia. Leukemia. 27:1000–1008. 10.1038/leu.2012.356 [DOI] [PubMed] [Google Scholar]
- Avagyan S., and Zon L.I.. 2016. Fish to learn: insights into blood development and blood disorders from zebrafish hematopoiesis. Hum. Gene Ther. 27:287–294. 10.1089/hum.2016.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Giroux S., Golub R., Klaine M., Jalil A., Boucontet L., Godin I., and Cumano A.. 2005. Characterization of purified intraembryonic hematopoietic stem cells as a tool to define their site of origin. Proc. Natl. Acad. Sci. USA. 102:134–139. 10.1073/pnas.0402270102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Chi N.C., Santoso B., Teng S., Stainier D.Y.R., and Traver D.. 2010b Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 464:108–111. 10.1038/nature08738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand J.Y., Cisson J.L., Stachura D.L., and Traver D.. 2010a Notch signaling distinguishes 2 waves of definitive hematopoiesis in the zebrafish embryo. Blood. 115:2777–2783. 10.1182/blood-2009-09-244590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birbrair A., and Frenette P.S.. 2016. Niche heterogeneity in the bone marrow. Ann. N. Y. Acad. Sci. 1370:82–96. 10.1111/nyas.13016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser B.W., Moore J.L., Hagedorn E.J., Li B., Riquelme R., Lichtig A., Yang S., Zhou Y., Tamplin O.J., Binder V., and Zon L.I.. 2017. CXCR1 remodels the vascular niche to promote hematopoietic stem and progenitor cell engraftment. J. Exp. Med. 214:1011–1027. 10.1084/jem.20161616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J.-C., van Cappellen W., Andrieu-Soler C., Galjart N., Dzierzak E., and Robin C.. 2010. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 464:116–120. 10.1038/nature08764 [DOI] [PubMed] [Google Scholar]
- Bresciani E., Carrington B., Wincovitch S., Jones M., Gore A.V., Weinstein B.M., Sood R., and Liu P.P.. 2014. CBFβ and RUNX1 are required at 2 different steps during the development of hematopoietic stem cells in zebrafish. Blood. 124:70–78. 10.1182/blood-2013-10-531988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C.E., Traver D., Mayhall E., Shepard J.L., and Zon L.I.. 2005. Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev. 19:2331–2342. 10.1101/gad.1337005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns C.E., Galloway J.L., Smith A.C.H., Keefe M.D., Cashman T.J., Paik E.J., Mayhall E.A., Amsterdam A.H., and Zon L.I.. 2009. A genetic screen in zebrafish defines a hierarchical network of pathways required for hematopoietic stem cell emergence. Blood. 113:5776–5782. 10.1182/blood-2008-12-193607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butko E., Distel M., Pouget C., Weijts B., Kobayashi I., Ng K., Mosimann C., Poulain F.E., McPherson A., Ni C.-W., et al. . 2015. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development. 142:1050–1061. 10.1242/dev.119180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butko E., Pouget C., and Traver D.. 2016. Complex regulation of HSC emergence by the Notch signaling pathway. Dev. Biol. 409:129–138. 10.1016/j.ydbio.2015.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C., Su T., Lau R.P., Shah A., Laurie P.C., Avalos B., Aggio J., Harris E., Traver D., and Stachura D.L.. 2015. Zebrafish embryonic stromal trunk (ZEST) cells support hematopoietic stem and progenitor cell (HSPC) proliferation, survival, and differentiation. Exp. Hematol. 43:1047–1061. 10.1016/j.exphem.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.J., Yokomizo T., Zeigler B.M., Dzierzak E., and Speck N.A.. 2009. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 457:887–891. 10.1038/nature07619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K.-D., Vodyanik M.A., Togarrati P.P., Suknuntha K., Kumar A., Samarjeet F., Probasco M.D., Tian S., Stewart R., Thomson J.A., and Slukvin I.I.. 2012. Identification of the hemogenic endothelial progenitor and its direct precursor in human pluripotent stem cell differentiation cultures. Cell Reports. 2:553–567. 10.1016/j.celrep.2012.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciau-Uitz A., Monteiro R., Kirmizitas A., and Patient R.. 2014. Developmental hematopoiesis: ontogeny, genetic programming and conservation. Exp. Hematol. 42:669–683. 10.1016/j.exphem.2014.06.001 [DOI] [PubMed] [Google Scholar]
- Clements W.K., Kim A.D., Ong K.G., Moore J.C., Lawson N.D., and Traver D.. 2011. A somitic Wnt16/Notch pathway specifies haematopoietic stem cells. Nature. 474:220–224. 10.1038/nature10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler C., Multani P., Robbins D., Kim H.T., Le T., Hoggatt J., Pelus L.M., Desponts C., Chen Y.-B., Rezner B., et al. . 2013. Prostaglandin-modulated umbilical cord blood hematopoietic stem cell transplantation. Blood. 122:3074–3081. 10.1182/blood-2013-05-503177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson A.J., and Zon L.I.. 2004. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 23:7233–7246. 10.1038/sj.onc.1207943 [DOI] [PubMed] [Google Scholar]
- Davidson A.J., and Zon L.I.. 2006. The caudal-related homeobox genes cdx1a and cdx4 act redundantly to regulate hox gene expression and the formation of putative hematopoietic stem cells during zebrafish embryogenesis. Dev. Biol. 292:506–518. 10.1016/j.ydbio.2006.01.003 [DOI] [PubMed] [Google Scholar]
- Davidson A.J., Ernst P., Wang Y., Dekens M.P.S., Kingsley P.D., Palis J., Korsmeyer S.J., Daley G.Q., and Zon L.I.. 2003. cdx4 mutants fail to specify blood progenitors and can be rescued by multiple hox genes. Nature. 425:300–306. 10.1038/nature01973 [DOI] [PubMed] [Google Scholar]
- de Jong J.L.O., and Zon L.I.. 2005. Use of the zebrafish system to study primitive and definitive hematopoiesis. Annu. Rev. Genet. 39:481–501. 10.1146/annurev.genet.39.073003.095931 [DOI] [PubMed] [Google Scholar]
- Demy D.L., Ranta Z., Giorgi J.-M., Gonzalez M., Herbomel P., and Kissa K.. 2013. Generating parabiotic zebrafish embryos for cell migration and homing studies. Nat. Methods. 10:256–258. 10.1038/nmeth.2362 [DOI] [PubMed] [Google Scholar]
- Deneault E., Wilhelm B.T., Bergeron A., Barabé F., and Sauvageau G.. 2013. Identification of non-cell-autonomous networks from engineered feeder cells that enhance murine hematopoietic stem cell activity. Exp. Hematol. 41:470–478.e4. 10.1016/j.exphem.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Dieterlen-Lièvre F., Pouget C., Bollérot K., and Jaffredo T.. 2006. Are intra-aortic hemopoietic cells derived from endothelial cells during ontogeny? Trends Cardiovasc. Med. 16:128–139. 10.1016/j.tcm.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Ding L., and Morrison S.J.. 2013. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 495:231–235. 10.1038/nature11885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., and Morrison S.J.. 2012. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 481:457–462. 10.1038/nature10783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan A., Brownlie A., Zhou Y., Shepard J., Pratt S.J., Moynihan J., Paw B.H., Drejer A., Barut B., Zapata A., et al. . 2000. Positional cloning of zebrafish ferroportin1 identifies a conserved vertebrate iron exporter. Nature. 403:776–781. 10.1038/35001596 [DOI] [PubMed] [Google Scholar]
- Dooley K.A., Davidson A.J., and Zon L.I.. 2005. Zebrafish scl functions independently in hematopoietic and endothelial development. Dev. Biol. 277:522–536. 10.1016/j.ydbio.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Doulatov S., Vo L.T., Chou S.S., Kim P.G., Arora N., Li H., Hadland B.K., Bernstein I.D., Collins J.J., Zon L.I., and Daley G.Q.. 2013. Induction of multipotential hematopoietic progenitors from human pluripotent stem cells via respecification of lineage-restricted precursors. Cell Stem Cell. 13:459–470. 10.1016/j.stem.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever W., Solnica-Krezel L., Schier A.F., Neuhauss S.C., Malicki J., Stemple D.L., Stainier D.Y., Zwartkruis F., Abdelilah S., Rangini Z., et al. . 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development. 123:37–46. [DOI] [PubMed] [Google Scholar]
- Elcheva I., Brok-Volchanskaya V., Kumar A., Liu P., Lee J.-H., Tong L., Vodyanik M., Swanson S., Stewart R., Kyba M., et al. . 2014. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat. Commun. 5:4372 10.1038/ncomms5372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genthe J.R., and Clements W.K.. 2017. R-spondin 1 is required for specification of hematopoietic stem cells through Wnt16 and Vegfa signaling pathways. Development. 144:590–600. 10.1242/dev.139956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gering M., and Patient R.. 2005. Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev. Cell. 8:389–400. 10.1016/j.devcel.2005.01.010 [DOI] [PubMed] [Google Scholar]
- Goessling W., North T.E., Loewer S., Lord A.M., Lee S., Stoick-Cooper C.L., Weidinger G., Puder M., Daley G.Q., Moon R.T., and Zon L.I.. 2009. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 136:1136–1147. 10.1016/j.cell.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordeuk V.R., Caleffi A., Corradini E., Ferrara F., Jones R.A., Castro O., Onyekwere O., Kittles R., Pignatti E., Montosi G., et al. . 2003. Iron overload in Africans and African-Americans and a common mutation in the SCL40A1 (ferroportin 1) gene. Blood Cells Mol. Dis. 31:299–304. 10.1016/S1079-9796(03)00164-5 [DOI] [PubMed] [Google Scholar]
- Gragert L., Eapen M., Williams E., Freeman J., Spellman S., Baitty R., Hartzman R., Rizzo J.D., Horowitz M., Confer D., and Maiers M.. 2014. HLA match likelihoods for hematopoietic stem-cell grafts in the U.S. registry. N. Engl. J. Med. 371:339–348. 10.1056/NEJMsa1311707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A., Hsu Y.-M.S., Day R.B., Schuettpelz L.G., Christopher M.J., Borgerding J.N., Nagasawa T., and Link D.C.. 2013. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 495:227–230. 10.1038/nature11926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiu J., Shimizu R., D’Altri T., Fraser S.T., Hatakeyama J., Bresnick E.H., Kageyama R., Dzierzak E., Yamamoto M., Espinosa L., and Bigas A.. 2013. Hes repressors are essential regulators of hematopoietic stem cell development downstream of Notch signaling. J. Exp. Med. 210:71–84. 10.1084/jem.20120993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B.K., Huppert S.S., Kanungo J., Xue Y., Jiang R., Gridley T., Conlon R.A., Cheng A.M., Kopan R., and Longmore G.D.. 2004. A requirement for Notch1 distinguishes 2 phases of definitive hematopoiesis during development. Blood. 104:3097–3105. 10.1182/blood-2004-03-1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadland B.K., Varnum-Finney B., Poulos M.G., Moon R.T., Butler J.M., Rafii S., and Bernstein I.D.. 2015. Endothelium and NOTCH specify and amplify aorta-gonad-mesonephros-derived hematopoietic stem cells. J. Clin. Invest. 125:2032–2045. 10.1172/JCI80137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P., Granato M., Brand M., Mullins M.C., Hammerschmidt M., Kane D.A., Odenthal J., van Eeden F.J., Jiang Y.J., Heisenberg C.P., et al. . 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 123:1–36. [DOI] [PubMed] [Google Scholar]
- Hagedorn E.J., Cillis J.L., Curley C.R., Patch T.C., Li B., Blaser B.W., Riquelme R., Zon L.I., and Shah D.I.. 2016. Generation of parabiotic zebrafish embryos by surgical fusion of developing blastulae. J. Vis. Exp. e54168 10.3791/54168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood S.Y., Oteiza A., Cao H., and Nilsson S.K.. 2014. Analyzing hematopoietic stem cell homing, lodgment, and engraftment to better understand the bone marrow niche. Ann. N. Y. Acad. Sci. 1310:119–128. 10.1111/nyas.12329 [DOI] [PubMed] [Google Scholar]
- Henninger J., Santoso B., Hans S., Durand E., Moore J., Mosimann C., Brand M., Traver D., and Zon L.. 2017. Clonal fate mapping quantifies the number of haematopoietic stem cells that arise during development. Nat. Cell Biol. 19:17–27. 10.1038/ncb3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffredo T., Gautier R., Eichmann A., and Dieterlen-Lièvre F.. 1998. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development. 125:4575–4583. [DOI] [PubMed] [Google Scholar]
- Jang I.-H., Lu Y.-F., Zhao L., Wenzel P.L., Kume T., Datta S.M., Arora N., Guiu J., Lagha M., Kim P.G., et al. . 2015. Notch1 acts via Foxc2 to promote definitive hematopoiesis via effects on hemogenic endothelium. Blood. 125:1418–1426. 10.1182/blood-2014-04-568170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin F., Zhai Q., Qiu L., Meng H., Zou D., Wang Y., Li Q., Yu Z., Han J., Li Q., and Zhou B.. 2008. Degradation of BM SDF-1 by MMP-9: the role in G-CSF-induced hematopoietic stem/progenitor cell mobilization. Bone Marrow Transplant. 42:581–588. 10.1038/bmt.2008.222 [DOI] [PubMed] [Google Scholar]
- Jing L., Tamplin O.J., Chen M.J., Deng Q., Patterson S., Kim P.G., Durand E.M., McNeil A., Green J.M., Matsuura S., et al. . 2015. Adenosine signaling promotes hematopoietic stem and progenitor cell emergence. J. Exp. Med. 212:649–663. 10.1084/jem.20141528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A.D., Melick C.H., Clements W.K., Stachura D.L., Distel M., Panáková D., MacRae C., Mork L.A., Crump J.G., and Traver D.. 2014. Discrete Notch signaling requirements in the specification of hematopoietic stem cells. EMBO J. 33:2363–2373. 10.15252/embj.201488784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisanuki Y.Y., Hammer R.E., Miyazaki J., Williams S.C., Richardson J.A., and Yanagisawa M.. 2001. Tie2-Cre transgenic mice: a new model for endothelial cell-lineage analysis in vivo. Dev. Biol. 230:230–242. 10.1006/dbio.2000.0106 [DOI] [PubMed] [Google Scholar]
- Kissa K., and Herbomel P.. 2010. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 464:112–115. 10.1038/nature08761 [DOI] [PubMed] [Google Scholar]
- Kissa K., Murayama E., Zapata A., Cortés A., Perret E., Machu C., and Herbomel P.. 2008. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 111:1147–1156. 10.1182/blood-2007-07-099499 [DOI] [PubMed] [Google Scholar]
- Kobayashi I., Kobayashi-Sun J., Kim A.D., Pouget C., Fujita N., Suda T., and Traver D.. 2014. Jam1a-Jam2a interactions regulate haematopoietic stem cell fate through Notch signalling. Nature. 512:319–323. 10.1038/nature13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konantz M., Alghisi E., Müller J.S., Lenard A., Esain V., Carroll K.J., Kanz L., North T.E., and Lengerke C.. 2016. Evi1 regulates Notch activation to induce zebrafish hematopoietic stem cell emergence. EMBO J. 35:2315–2331. 10.15252/embj.201593454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumano K., Chiba S., Kunisato A., Sata M., Saito T., Nakagami-Yamaguchi E., Yamaguchi T., Masuda S., Shimizu K., Takahashi T., et al. . 2003. Notch1 but not Notch2 is essential for generating hematopoietic stem cells from endothelial cells. Immunity. 18:699–711. 10.1016/S1074-7613(03)00117-1 [DOI] [PubMed] [Google Scholar]
- Kyba M., Perlingeiro R.C.R., and Daley G.Q.. 2002. HoxB4 confers definitive lymphoid-myeloid engraftment potential on embryonic stem cell and yolk sac hematopoietic progenitors. Cell. 109:29–37. 10.1016/S0092-8674(02)00680-3 [DOI] [PubMed] [Google Scholar]
- Lassailly F., Foster K., Lopez-Onieva L., Currie E., and Bonnet D.. 2013. Multimodal imaging reveals structural and functional heterogeneity in different bone marrow compartments: Functional implications on hematopoietic stem cells. Blood. 122:1730–1740. 10.1182/blood-2012-11-467498 [DOI] [PubMed] [Google Scholar]
- Lawson N.D., Vogel A.M., and Weinstein B.M.. 2002. Sonic hedgehog and vascular endothelial growth factor act upstream of the Notch pathway during arterial endothelial differentiation. Dev. Cell. 3:127–136. 10.1016/S1534-5807(02)00198-3 [DOI] [PubMed] [Google Scholar]
- Ledran M.H., Krassowska A., Armstrong L., Dimmick I., Renström J., Lang R., Yung S., Santibanez-Coref M., Dzierzak E., Stojkovic M., et al. . 2008. Efficient hematopoietic differentiation of human embryonic stem cells on stromal cells derived from hematopoietic niches. Cell Stem Cell. 3:85–98. 10.1016/j.stem.2008.06.001 [DOI] [PubMed] [Google Scholar]
- Lee Y., Manegold J.E., Kim A.D., Pouget C., Stachura D.L., Clements W.K., and Traver D.. 2014. FGF signalling specifies haematopoietic stem cells through its regulation of somitic Notch signalling. Nat. Commun. 5:5583 10.1038/ncomms6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengerke C., McKinney-Freeman S., Naveiras O., Yates F., Wang Y., Bansal D., and Daley G.Q.. 2007. The cdx-hox pathway in hematopoietic stem cell formation from embryonic stem cells. Ann. N. Y. Acad. Sci. 1106:197–208. 10.1196/annals.1392.006 [DOI] [PubMed] [Google Scholar]
- Li P., Lahvic J.L., Binder V., Pugach E.K., Riley E.B., Tamplin O.J., Panigrahy D., Bowman T.V., Barrett F.G., Heffner G.C., et al. . 2015. Epoxyeicosatrienoic acids enhance embryonic haematopoiesis and adult marrow engraftment. Nature. 523:468–471. 10.1038/nature14569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis R., Karrasch C.C., Poulos M.G., Kunar B., Redmond D., Duran J.G.B., Badwe C.R., Schachterle W., Ginsberg M., Xiang J., et al. . 2017. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature. 545:439–445. 10.1038/nature22326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C., Fleming H.E., Wu J.W., Zhao C.X., Miake-Lye S., Fujisaki J., Côté D., Rowe D.W., Lin C.P., and Scadden D.T.. 2009. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 457:92–96. 10.1038/nature07434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnusson M., Brun A.C.M., Lawrence H.J., and Karlsson S.. 2007. Hoxa9/hoxb3/hoxb4 compound null mice display severe hematopoietic defects. Exp. Hematol. 35:1421.e1–1428.e9. 10.1016/j.exphem.2007.05.011 [DOI] [PubMed] [Google Scholar]
- Mahony C.B., Fish R.J., Pasche C., and Bertrand J.Y.. 2016. tfec controls the hematopoietic stem cell vascular niche during zebrafish embryogenesis. Blood. 128:1336–1345. 10.1182/blood-2016-04-710137 [DOI] [PubMed] [Google Scholar]
- Mendelson A., and Frenette P.S.. 2014. Hematopoietic stem cell niche maintenance during homeostasis and regeneration. Nat. Med. 20:833–846. 10.1038/nm.3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirabello L., Macari E.R., Jessop L., Ellis S.R., Myers T., Giri N., Taylor A.M., McGrath K.E., Humphries J.M., Ballew B.J., et al. . 2014. Whole-exome sequencing and functional studies identify RPS29 as a novel gene mutated in multicase Diamond-Blackfan anemia families. Blood. 124:24–32. 10.1182/blood-2013-11-540278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro R., Pinheiro P., Joseph N., Peterkin T., Koth J., Repapi E., Bonkhofer F., Kirmizitas A., and Patient R.. 2016. Transforming growth factor β drives hemogenic endothelium programming and the transition to hematopoietic stem cells. Dev. Cell. 38:358–370. 10.1016/j.devcel.2016.06.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison S.J., and Scadden D.T.. 2014. The bone marrow niche for haematopoietic stem cells. Nature. 505:327–334. 10.1038/nature12984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucenski M.L., McLain K., Kier A.B., Swerdlow S.H., Schreiner C.M., Miller T.A., Pietryga D.W., Scott W.J. Jr., and Potter S.S.. 1991. A functional c-myb gene is required for normal murine fetal hepatic hematopoiesis. Cell. 65:677–689. 10.1016/0092-8674(91)90099-K [DOI] [PubMed] [Google Scholar]
- Murayama E., Kissa K., Zapata A., Mordelet E., Briolat V., Lin H.-F., Handin R.I., and Herbomel P.. 2006. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 25:963–975. 10.1016/j.immuni.2006.10.015 [DOI] [PubMed] [Google Scholar]
- Murayama E., Sarris M., Redd M., Le Guyader D., Vivier C., Horsley W., Trede N., and Herbomel P.. 2015. NACA deficiency reveals the crucial role of somite-derived stromal cells in haematopoietic niche formation. Nat. Commun. 6:8375 10.1038/ncomms9375 [DOI] [PubMed] [Google Scholar]
- Nakagawa M., Ichikawa M., Kumano K., Goyama S., Kawazu M., Asai T., Ogawa S., Kurokawa M., and Chiba S.. 2006. AML1/Runx1 rescues Notch1-null mutation-induced deficiency of para-aortic splanchnopleural hematopoiesis. Blood. 108:3329–3334. 10.1182/blood-2006-04-019570 [DOI] [PubMed] [Google Scholar]
- Nguyen P.D., Hollway G.E., Sonntag C., Miles L.B., Hall T.E., Berger S., Fernandez K.J., Gurevich D.B., Cole N.J., Alaei S., et al. . 2014. Haematopoietic stem cell induction by somite-derived endothelial cells controlled by meox1. Nature. 512:314–318. 10.1038/nature13678 [DOI] [PubMed] [Google Scholar]
- North T.E., de Bruijn M.F.T.R., Stacy T., Talebian L., Lind E., Robin C., Binder M., Dzierzak E., and Speck N.A.. 2002. Runx1 expression marks long-term repopulating hematopoietic stem cells in the midgestation mouse embryo. Immunity. 16:661–672. 10.1016/S1074-7613(02)00296-0 [DOI] [PubMed] [Google Scholar]
- North T.E., Goessling W., Walkley C.R., Lengerke C., Kopani K.R., Lord A.M., Weber G.J., Bowman T.V., Jang I.-H., Grosser T., et al. . 2007. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 447:1007–1011. 10.1038/nature05883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottingham W.T., Jarratt A., Burgess M., Speck C.L., Cheng J.-F., Prabhakar S., Rubin E.M., Li P.-S., Sloane-Stanley J., Kong-A-San J., and de Bruijn M.F.T.R.. 2007. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 110:4188–4197. 10.1182/blood-2007-07-100883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberlin E., Tavian M., Blazsek I., and Péault B.. 2002. Blood-forming potential of vascular endothelium in the human embryo. Development. 129:4147–4157. [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen J., Hiebert S.W., Grosveld G., and Downing J.R.. 1996. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 84:321–330. 10.1016/S0092-8674(00)80986-1 [DOI] [PubMed] [Google Scholar]
- Orkin S.H., and Zon L.I.. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 132:631–644. 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson L.J., Gering M., Eckfeldt C.E., Green A.R., Verfaillie C.M., Ekker S.C., and Patient R.. 2007. The transcription factors Scl and Lmo2 act together during development of the hemangioblast in zebrafish. Blood. 109:2389–2398. 10.1182/blood-2006-02-003087 [DOI] [PubMed] [Google Scholar]
- Ransom D.G., Haffter P., Odenthal J., Brownlie A., Vogelsang E., Kelsh R.N., Brand M., van Eeden F.J., Furutani-Seiki M., Granato M., et al. . 1996. Characterization of zebrafish mutants with defects in embryonic hematopoiesis. Development. 123:311–319. [DOI] [PubMed] [Google Scholar]
- Robert-Moreno A., Espinosa L., de la Pompa J.L., and Bigas A.. 2005. RBPjκ-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development. 132:1117–1126. 10.1242/dev.01660 [DOI] [PubMed] [Google Scholar]
- Rowe R.G., Mandelbaum J., Zon L.I., and Daley G.Q.. 2016. Engineering hematopoietic stem cells: Lessons from development. Cell Stem Cell. 18:707–720. 10.1016/j.stem.2016.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M.J., Holmes A., Miles C., and Dzierzak E.. 1996. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity. 5:513–525. 10.1016/S1074-7613(00)80267-8 [DOI] [PubMed] [Google Scholar]
- Sandler V.M., Lis R., Liu Y., Kedem A., James D., Elemento O., Butler J.M., Scandura J.M., and Rafii S.. 2014. Reprogramming human endothelial cells to haematopoietic cells requires vascular induction. Nature. 511:312–318. 10.1038/nature13547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani R.A., Mayer E.L., and Orkin S.H.. 1995. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature. 373:432–434. 10.1038/373432a0 [DOI] [PubMed] [Google Scholar]
- Stachura D.L., Reyes J.R., Bartunek P., Paw B.H., Zon L.I., and Traver D.. 2009. Zebrafish kidney stromal cell lines support multilineage hematopoiesis. Blood. 114:279–289. 10.1182/blood-2009-02-203638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgeon C.M., Ditadi A., Clarke R.L., and Keller G.. 2013. Defining the path to hematopoietic stem cells. Nat. Biotechnol. 31:416–418. 10.1038/nbt.2571 [DOI] [PubMed] [Google Scholar]
- Sugimura R., Jha D.K., Han A., Soria-Valles C., da Rocha E.L., Lu Y.-F., Goettel J.A., Serrao E., Rowe R.G., Malleshaiah M., et al. . 2017. Haematopoietic stem and progenitor cells from human pluripotent stem cells. Nature. 545:432–438. 10.1038/nature22370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamplin O.J., Durand E.M., Carr L.A., Childs S.J., Hagedorn E.J., Li P., Yzaguirre A.D., Speck N.A., and Zon L.I.. 2015. Hematopoietic stem cell arrival triggers dynamic remodeling of the perivascular niche. Cell. 160:241–252. 10.1016/j.cell.2014.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Humphries J.M., White R.M., Murphey R.D., Burns C.E., and Zon L.I.. 2012. Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp. Hematol. 40:228–237.e5. 10.1016/j.exphem.2011.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodore L.N., Hagedorn E.J., Cortes M., Natsuhara K., Liu S.Y., Perlin J.R., Yang S., Daily M.L., Zon L.I., and North T.E.. 2017. Distinct roles for matrix metalloproteinases 2 and 9 in embryonic hematopoietic stem cell emergence, migration, and niche colonization. Stem Cell Reports. 8:1226–1241. 10.1016/j.stemcr.2017.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova J., Tran Chau V., Julien E., Mateos-Langerak J., Gonzalez C., Lelièvre E., Lutfalla G., Tavian M., and Kissa K.. 2015. Primitive macrophages control HSPC mobilization and definitive haematopoiesis. Nat. Commun. 6:6227 10.1038/ncomms7227 [DOI] [PubMed] [Google Scholar]
- Tsai F.Y., Keller G., Kuo F.C., Weiss M., Chen J., Rosenblatt M., Alt F.W., and Orkin S.H.. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 371:221–226. 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Wang Q., Stacy T., Miller J.D., Lewis A.F., Gu T.L., Huang X., Bushweller J.H., Bories J.C., Alt F.W., Ryan G., et al. . 1996. The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell. 87:697–708. 10.1016/S0092-8674(00)81389-6 [DOI] [PubMed] [Google Scholar]
- Wilkinson R.N., Pouget C., Gering M., Russell A.J., Davies S.G., Kimelman D., and Patient R.. 2009. Hedgehog and Bmp polarize hematopoietic stem cell emergence in the zebrafish dorsal aorta. Dev. Cell. 16:909–916. 10.1016/j.devcel.2009.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf A., Aggio J., Campbell C., Wright F., Marquez G., Traver D., and Stachura D.L.. 2017. Zebrafish caudal haematopoietic embryonic stromal tissue (CHEST) cells support haematopoiesis. Sci. Rep. 7:44644 10.1038/srep44644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Warren A.J., Dobson C., Forster A., Pannell R., and Rabbitts T.H.. 1998. The T cell leukemia LIM protein Lmo2 is necessary for adult mouse hematopoiesis. Proc. Natl. Acad. Sci. USA. 95:3890–3895. 10.1073/pnas.95.7.3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon M.-J., Koo B.-K., Song R., Jeong H.-W., Shin J., Kim Y.-W., Kong Y.-Y., and Suh P.-G.. 2008. Mind bomb-1 is essential for intraembryonic hematopoiesis in the aortic endothelium and the subaortic patches. Mol. Cell. Biol. 28:4794–4804. 10.1128/MCB.00436-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Jin H., Li L., Qin F.X.-F., and Wen Z.. 2011. cMyb regulates hematopoietic stem/progenitor cell mobilization during zebrafish hematopoiesis. Blood. 118:4093–4101. 10.1182/blood-2011-03-342501 [DOI] [PubMed] [Google Scholar]
- Zhen F., Lan Y., Yan B., Zhang W., and Wen Z.. 2013. Hemogenic endothelium specification and hematopoietic stem cell maintenance employ distinct Scl isoforms. Development. 140:3977–3985. 10.1242/dev.097071 [DOI] [PubMed] [Google Scholar]
- Zovein A.C., Hofmann J.J., Lynch M., French W.J., Turlo K.A., Yang Y., Becker M.S., Zanetta L., Dejana E., Gasson J.C., et al. . 2008. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell. 3:625–636. 10.1016/j.stem.2008.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]