Zhu et al. show that the Zika virus, which has a tropism for fetal and adult neuroprogenitor cells, targets and kills cancer stem cells while leaving differentiated tumor cells relatively unaffected, providing a new potential oncolytic virus therapy in neuro-oncology.
Abstract
Glioblastoma is a highly lethal brain cancer that frequently recurs in proximity to the original resection cavity. We explored the use of oncolytic virus therapy against glioblastoma with Zika virus (ZIKV), a flavivirus that induces cell death and differentiation of neural precursor cells in the developing fetus. ZIKV preferentially infected and killed glioblastoma stem cells (GSCs) relative to differentiated tumor progeny or normal neuronal cells. The effects against GSCs were not a general property of neurotropic flaviviruses, as West Nile virus indiscriminately killed both tumor and normal neural cells. ZIKV potently depleted patient-derived GSCs grown in culture and in organoids. Moreover, mice with glioblastoma survived substantially longer and at greater rates when the tumor was inoculated with a mouse-adapted strain of ZIKV. Our results suggest that ZIKV is an oncolytic virus that can preferentially target GSCs; thus, genetically modified strains that further optimize safety could have therapeutic efficacy for adult glioblastoma patients.
Introduction
Glioblastoma is the most prevalent primary intrinsic brain tumor. Despite surgery, radiation, and chemotherapy, glioblastomas remain lethal, with a median survival below two years (Stupp et al., 2009). Glioblastoma is a heterogeneous disease, with extensive contributions from nontransformed cells and a cellular hierarchy within the neoplastic compartment. Atop the hierarchy resides a self-renewing, tumorigenic, stem-like tumor cell population called glioblastoma stem cells (GSCs) or tumor-initiating cells (Chen et al., 2012). GSCs contribute to tumor malignancy due to sustained proliferation, promotion of angiogenesis, invasive potential, immune escape, and therapeutic resistance (Bao et al., 2006; Alvarado et al., 2017).
Unlike many lethal cancers, glioblastomas rarely metastasize out of the central nervous system (CNS), and a majority of patients suffer recurrence within 2–3 cm of the original resection cavity (Wallner et al., 1989); this behavior has prompted investigation of local therapies, including oncolytic viruses (Martuza et al., 1991; Alonso et al., 2012; Kaufmann and Chiocca, 2014; Miska et al., 2016; Cassady et al., 2017; Cattaneo and Russell, 2017). Efficacy of virotherapy against tumors depends on the ability to infect and kill tumor cells specifically (Cattaneo and Russell, 2017). Several oncolytic DNA viruses have been developed to achieve tumor cell killing with limited toxicity (Martuza et al., 1991; Alonso et al., 2012).
Zika virus (ZIKV) is a member of the flavivirus genus of RNA viruses, which includes dengue, West Nile virus (WNV), and yellow fever viruses. The recent outbreak of ZIKV-induced fetal microcephaly has spurred extensive research into its cell tropism (Garcez et al., 2016; Lazear et al., 2016; Li et al., 2016; Ming et al., 2016; Qian et al., 2016; Shan et al., 2016). ZIKV infects the developing CNS, with neural stem and progenitor cells prominently affected. Neural precursors infected with ZIKV undergo differentiation, loss of proliferation, and cell death (Gromeier et al., 2000; Li et al., 2016; Ming et al., 2016; Qian et al., 2016; Gabriel et al., 2017). In contrast, the effects of ZIKV in adults are generally less severe, with rare cases of meningoencephalitis, suggesting that ZIKV infection has fewer deleterious effects in the adult brain (Parra et al., 2016). We hypothesized that the tropism of ZIKV for neural precursor cells could be leveraged against glioblastomas.
Results and discussion
ZIKV infects human GSCs and inhibits proliferation in vitro
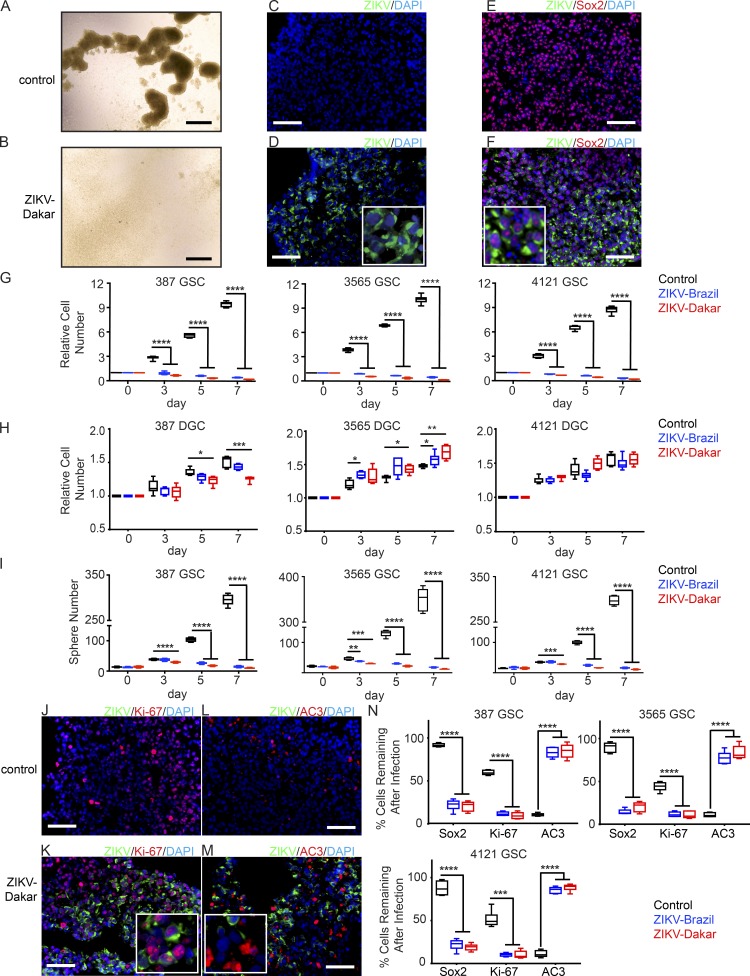
To interrogate the effects of ZIKV on glioblastoma, we used patient-derived GSCs that express stem cell markers, self-renew, have differentiation potential, and form tumors upon xenotransplantation, as well as differentiated glioma cells (DGCs; Bao et al., 2006; Wang et al., 2017). We selected four GSC models representing the major transcriptional glioblastoma subtypes—proneural, classical, and mesenchymal—and induced cellular differentiation through serum exposure (Bao et al., 2006). We infected GSCs (Fig. 1 A; multiplicity of infection [MOI] of 5) with representative African (Dakar 1984) and American (Brazil 2015) ZIKV strains. 7 d later, spheres were obliterated (Fig. 1 B). Immunofluorescence microscopy demonstrated that greater than 60% of GSCs were infected by either strain at 48 h after infection (Fig. 1, C and D). We analyzed the fraction of ZIKV-infected cells that expressed a GSC marker (SOX2); greater than 90% of infected cells were SOX2+ (Fig. 1, E and F; and Fig. S1 A). Flow cytometry results were consistent with the microscopy data (Fig. S1, B–G) and demonstrated that the percentage of infected GSCs increased over time, consistent with virus spread. We next determined the impact of ZIKV on matched GSCs and DGCs. ZIKV could infect DGCs, but at a significantly lower rate than GSCs (Fig. S1, H and I). Infectious yield assays corroborated higher ZIKV levels from GSCs than from DGCs (Fig. S1, J and K). Whereas GSC proliferation was abolished by either ZIKV strain (Fig. 1, B and G), DGCs were nearly unaffected (Fig. 1 H). Sphere formation in serum-free conditions has been used as a surrogate for self-renewal (Reynolds and Weiss, 1992). Consistent with its preferential targeting of GSCs, ZIKV reduced GSC sphere formation (Fig. 1 I). These effects on cell number and sphere formation were also associated with reductions in SOX2 expression and proliferation (measured by Ki-67) and increases in apoptosis (measured by activated caspase-3 [AC3]; Fig. 1, J–N).
Figure 1.
ZIKV causes loss of GSC self-renewal and proliferation. (A and B) GSCs were uninfected (A) or infected (B) with ZIKV-Dakar (7 dpi). (C–F) GSCs uninfected (C and E) or infected (D and F) with ZIKV-Dakar for 48 h underwent immunofluorescence staining for ZIKV envelope protein (green) and DAPI (blue; C–F) with Sox2 (red; E and F). Brightfield and immunofluorescence images in A–F are representative of three independent experiments. (G and H) Relative cell number of paired GSCs (387, 3565, and 4121; G) and DGCs (H), infected with ZIKV-Dakar or ZIKV-Brazil, at an MOI of 5 for 7 d; all data were normalized to day 0. Data from G and H are from three independent experiments. (I) Sphere formation capacity of 387, 3565, and 4121 GSCs infected with indicated ZIKV strains or control. Data are from three independent experiments. (J–M) GSCs uninfected (J and L) or infected (K and M) with ZIKV-Dakar for 48 h underwent staining for ZIKV (green) and DAPI (blue) with Ki-67 (red; J and K) or AC3 (red; L and M). J–M are representative of three independent experiments. (N) At day 4, the frequency of Sox2+, Ki-67+, and AC3+ cells was measured by visual quantification in the three GSC lines with or without ZIKV infection. Data are derived from experiments performed in duplicate and pooled from three independent experiments. Error bars indicate SDs. Significance was analyzed with one-way ANOVA with Tukey’s multiple comparison test (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Bars: (A and B) 725 µm; (C–F and J–M) 100 µm.
The selective effects of ZIKV on GSCs are not observed with WNV
More than 60 yr ago, WNV was tested for its oncolytic efficacy but displayed substantial toxicity (Southam and Moore, 1952; Moore, 1954). We compared the effects of WNV (New York 1999) to those of ZIKV in our models. WNV infected both GSCs and DGCs to high levels (Fig. S2, A–G), inducing death in both cell types (Fig. S2, A and B). WNV also infected normal human neural cells in culture and brain slices from freshly resected epilepsy tissues and targeted NeuN+ neurons and GFAP+ astrocytes (Fig. S2, H–K). WNV continued to infect cells over time (Fig. S2 L), and this was associated with significant reductions in cell number (Fig. S2 M). Thus, the GSC specificity of ZIKV is not a general property of related neurotropic flaviviruses.
ZIKV causes loss of self-renewal and proliferation in glioblastoma organoids
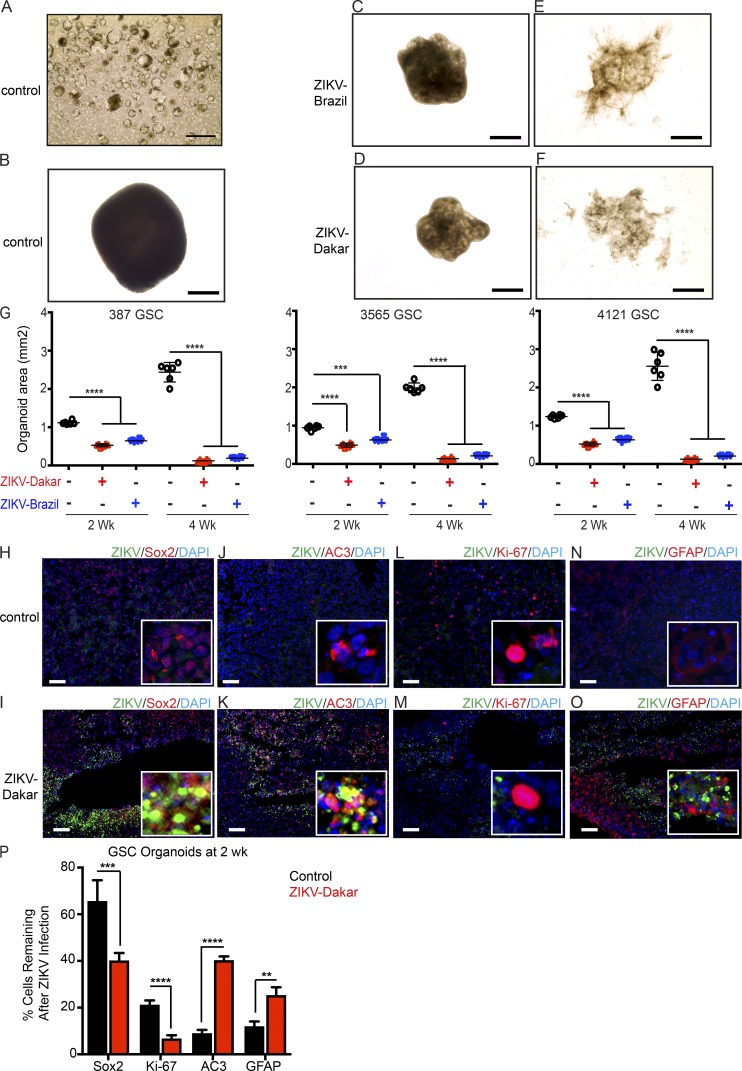
To test the specificity of ZIKV for GSCs in the context of the cellular heterogeneity that exists in patients, we used an in vitro human glioblastoma organoid model (Hubert et al., 2016). Three GSC models (387, 3565, and 4121) were used that form small organoids by 3 d (Fig. 2 A) and mature organoids within 3 wk (Fig. 2 B). Infection with ZIKV-Brazil or ZIKV-Dakar slowed organoid growth at 2 wk (Fig. 2, C and D) and 4 wk (Fig. 2, E and F), respectively, as assessed by organoid area (Fig. 2 G). ZIKV infected the glioblastoma organoids, with preference for cells expressing the GSC marker SOX2 (Fig. 2, H and I). Colocalization of ZIKV-infected cells and the apoptotic marker, AC3, confirmed that ZIKV induced tumor cell death (Fig. 2, J and K). However, ZIKV did not efficiently infect the proliferating tumor cells, as marked by Ki-67 (Fig. 2, L and M), or differentiated tumor cells (Fig. 2, N and O). ZIKV infection reduced undifferentiated GSCs in glioblastoma organoids, as shown by reduction in SOX2 and Ki-67 staining, and increased apoptosis, which resulted in a relative increase in DGCs (marked by GFAP) compared with the uninfected control (Fig. 2 P).
Figure 2.
ZIKV infection causes loss of self-renewal and proliferation of human glioblastoma-derived organoids. (A–F) Brightfield images of GSC organoids after infection with ZIKV. GSCs were incubated in Matrigel for 3 d (A) or 3 wk (B). Organoids were infected with ZIKV-Brazil (C and E) or ZIKV-Dakar (D and F) 2 (C and D) or 4 (E and F) wk after infection. Images in A–F are representative of two independent experiments. (G) Organoid areas at 2 or 4 wk after ZIKV infection were determined for three GSC organoid models (387, 3565, and 4121). Data represent two independent experiments. (H–O) Representative images of uninfected control and ZIKV-Dakar-infected GSC organoids 2 wk after infection stained for ZIKV (green) and DAPI (blue; H–O) with Sox2 (red; H and I), AC3 (red; J and K), Ki-67 (red; L and M), or GFAP (red; N and O). Images are representative of three independent experiments. (P) Quantification of Sox2+, Ki-67+, AC3+, and GFAP+ subpopulations of DAPI+ cells; n = 6 organoids for each condition from two independent experiments. Values represent mean ± SD. Significance was analyzed with one-way ANOVA with Tukey’s multiple comparison test (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Bars: (A–F) 725 µm; (H–O) 200 µm.
In human tissue specimens, ZIKV targets GSCs, with fewer effects on DGCs and normal neural cells
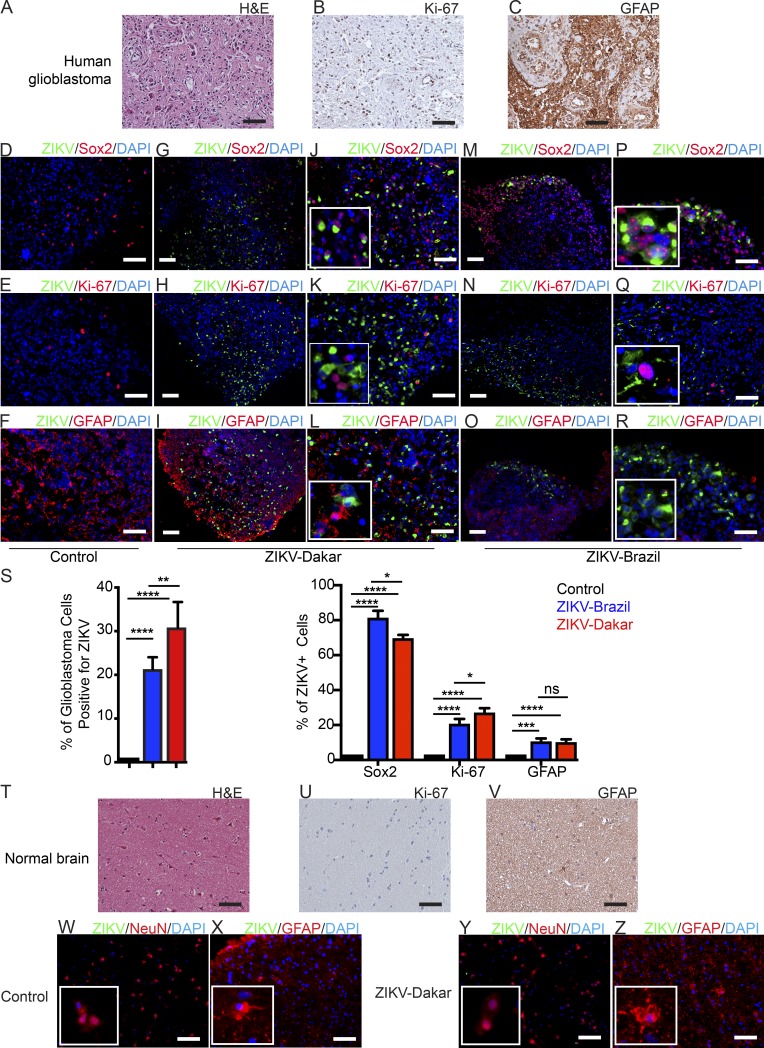
To confirm these results in the absence of culture, we collected glioblastoma specimens (Fig. 3, A–C) immediately after surgical resection and inoculated them with the two ZIKV strains. Over a 1-wk period, ZIKV progressively infected the tumors (Fig. 3, D–S; and Fig. S2 N). Costaining of sections revealed that the majority of ZIKV-infected cells expressed SOX2 (Fig. 3, D, G, J, M, P, and S). Unlike in organoids in patient specimens, which have different growth dynamics, ZIKV did infect proliferating cells (Fig. 3, E, H, K, N, Q, and S) but still rarely infected DGCs (Fig. 3, F, I, L, O, R, and S). These results support the hypothesis that ZIKV preferentially targets and kills GSCs.
Figure 3.
ZIKV infects isolated human glioblastoma but not normal brain tissue slices. (A–C) Representative images from three independent experiments showing freshly resected glioblastoma after staining with H&E (A) or for Ki-67 (B) or GFAP (C). (D–R) Immunofluorescence staining of glioblastoma tissue uninfected (D–F) or infected with ZIKV-Dakar (G–L) or ZIKV-Brazil (M–R) after 7 d for ZIKV E protein (green) and DAPI (blue) with Sox2 (red; D, G, J, M, and P), Ki-67 (red; E, H, K, N, and Q), or GFAP (red; F, I, L, O, and R). D–R are representative images from three independent experiments. (S) Quantification of tumor cells infected with ZIKV (left) and ZIKV-infected cells that co-stain for Sox2, Ki-67, or GFAP (right). Values represent mean ± SD and are pooled from three independent experiments. (T–Z) Representative images showing freshly resected normal brain after staining with H&E (T) or for Ki-67 (U) or GFAP (V). (W–Z) Normal brain tissue uninfected (W and X) or infected with ZIKV-Dakar (Y and Z) after 7 d stained for ZIKV (green) and DAPI (blue) with NeuN (red; W and Y) or GFAP (red; X and Z). Images in W–Z are representative of three independent experiments. Significance was analyzed with one-way ANOVA with Tukey’s multiple comparison test (*, P < 0.05; ***, P < 0.001; ****, P < 0.0001). Bars: (A–I and R–Z) 100 µm.
To test the effects of ZIKV on normal adult human neural cells, we inoculated nonmalignant neural tissues from adult epilepsy specimens (Fig. 3, T–V). ZIKV did not infect normal adult human brain tissues, including NeuN+ neurons (Fig. 3, W and Y) and GFAP+ glial cells (Fig. 3, X and Z), as limited viral replication was detected compared with glioblastomas (Fig. S2, N and O). In addition, compared with GSCs and DGCs, the human brain neural cell cultures that were derived from epilepsy patients (NM55 and NM177) or from differentiated human neural stem cells (Hu-DNCs) demonstrated limited ZIKV infection (Fig. S2, P–U). Limited toxicity in these neural cell models was confirmed using a cell viability assay over a 1-wk time course with two ZIKV strains (Fig. S2 V), and ZIKV replicated poorly in normal neural cells (Fig. S2, W and X).
ZIKV attenuates glioma growth, prolongs survival, and has marginal effects on normal neural cells
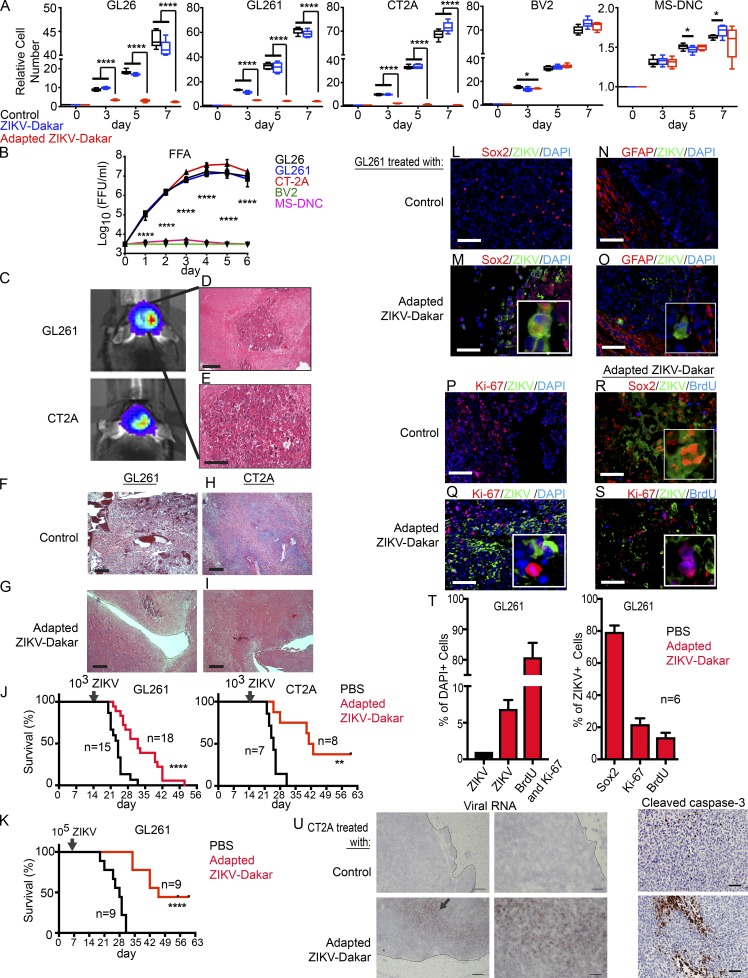
We recapitulated relevant conditions for human brain tumor therapy using mice. As mice are not natural hosts for ZIKV, pathogenesis studies have used animals with acquired or genetic deficiencies of type I IFN signaling (Lazear et al., 2016). To overcome this limitation, we used a mouse-adapted ZIKV-Dakar that had gained virulence through passage in a Rag1−/− host (Govero et al., 2016; Sapparapu et al., 2016). We first compared the efficacy of the parental and mouse-adapted ZIKV-Dakar strains against three mouse glioma models developed in the C57BL/6 background (GL261, GL26, and CT-2A; Wang et al., 2017) and two mouse CNS lines (BV2 and mouse-differentiated neural cells [MS-DNCs]) in vitro (Fig. 4, A and B). The mouse-adapted ZIKV-Dakar strain attenuated the growth of the mouse glioma cells, whereas the parental ZIKV strain was less effective (Fig. 4 A). In contrast, neither the parental nor the mouse-adapted ZIKV-Dakar inhibited the growth of other mouse CNS cells (BV2 and MS-DNCs; Fig. 4 A). These results were confirmed by virus titration at 1 wk with mouse glioma cells (GL26, GL261, and CT-2A) but not with BV2 cells and MS-DNCs (Fig. 4 B).
Figure 4.
Mouse-adapted ZIKV-Dakar attenuates growth of mouse glioma cells and prolongs survival of mice with glioma in vivo. (A) Mouse glioma cells (C57BL/6 background: GL26, GL261, and CT-2A), microglial cells (BV2), and MS-DNCs were infected with the parental or mouse-adapted ZIKV-Dakar, and relative cell number was assessed over 1 wk, normalized to day 0. (B) Viral titer from supernatants of ZIKV-Dakar-infected cells (GL26, GL261, CT-2A, BV2, and MS-DNCs) was measured at 1 wk by FFA. (C–I) Mouse glioma model with GL261 and CT-2A. 1 wk after implantation, bioluminescence imaging (BLI; C) and H&E staining (D and E) demonstrating glioma. 3 wk after GL261 (F and G) and CT-2A (H and I) implantation without (F and H) or with mouse-adapted ZIKV-Dakar treatment (G and I). (J, left) Mice bearing GL261 glioma were treated with PBS (n = 15) or 103 FFU of the mouse-adapted-ZIKV-Dakar (n = 18). (J, right) Mice bearing CT2A glioma were treated with PBS (n = 7) or 103 FFU of the mouse-adapted ZIKV-Dakar (n = 8). (K) Mice bearing GL261 glioma were treated with PBS (n = 9) or 105 FFU of the mouse-adapted ZIKV-Dakar (n = 9). (L–S) Immunofluorescence staining of GL261 glioma tumor–bearing mice at the endpoint after treatment with PBS control (L, N, and P) or 103-FFU–adapted ZIKV-Dakar (M, O, Q, R, and S) for ZIKV (green) with DAPI (blue; L–Q), Sox2 (red; L, M, and R), GFAP (red; N and O), Ki-67 (red; P, Q, and S), and BrdU (blue; R and S). (T) At survival endpoint (J, left), quantification of cells infected with ZIKV; cells positive for BrdU and Ki-67 (left); and ZIKV-infected cells positive for Sox2, Ki-67, or BrdU (right). (U, left) Representative low- and high-power images of in situ hybridization staining for viral RNA in mice with CT2A glioma 2 wk after treatment with ZIKV-Dakar or PBS (representative of two experiments). Arrow indicates positive staining. (U, right) Representative high-power images of cleaved caspase-3 staining on the same tumors. In vitro experiments were pooled from three independent experiments performed in duplicate. Animal survival data were pooled from two independent experiments. Quantification of immunostaining was derived from six mice. Values represent mean ± SD. Significance was analyzed by one-way ANOVA with Tukey’s multiple comparison test for A and B and the log-rank test for J and K (**, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Bars: (D, F–I) 200 µm; (E; L–S; U, left; and U, right) 100 µm; (U, middle) 10 µm.
To assess the oncolytic effects of ZIKV in vivo, we generated mouse gliomas from two different mouse cell lines (GL261 and CT-2A) grown in syngeneic hosts. Glioma cells were transduced with a luciferase reporter and permitted to form tumors, which were validated by bioluminescence imaging and histopathology (Fig. 4, C–E). Mice with tumors were randomized into two groups and treated 2 wk after implantation with either PBS control or mouse-adapted ZIKV-Dakar (103 focus-forming units [FFU]). Histological examination at 1 wk after tumor treatment demonstrated that the ZIKV-treated tumors were smaller in size compared with PBS-treated tumors (Fig. 4, F–I). Notably, ZIKV infection extended the life spans of tumor-bearing mice (Fig. 4 J). To test whether tumor-bearing mice could benefit from a higher dose of virus, we inoculated 105 FFU of the mouse-adapted ZIKV-Dakar at 1 wk after implantation with the GL261 model. The survival time of tumor-bearing mice was greater compared with that of the control or the 103-FFU dose (Fig. 4 K). To determine the specificity of cell targeting, we stained for ZIKV antigen and markers of stem cell proliferation and differentiation (Fig. 4, L–T). ZIKV infected ∼6% of glioma cells at the endpoint (Fig. 4 T), with the majority of these cells expressing the precursor marker SOX2 (Fig. 4, L, M, and T). In contrast, GFAP+ tumor cells were less infected (Fig. 4, N and O). Effects on proliferating cell populations were measured by Ki-67 staining and BrdU treatment and staining. The majority of ZIKV+ cells were negative for Ki-67 (>70%; Fig. 4, P, Q, and T) or BrdU (>80%; Fig. 4, R–T). These results support the efficacy of ZIKV in vivo against nonproliferating, stem-like cells (Li et al., 2016; Qian et al., 2016). 2 wk after treatment, viral RNA remained localized to the tumor, and ZIKV-treated tumors had extensive cell death as assessed by caspase-3 staining (Fig. 4 U).
IFN signaling is one determinant of differential sensitivity of GSCs to ZIKV
Although the mechanism by which ZIKV preferentially targets GSCs for infection and killing remains unknown, GSCs can suppress antitumor immune responses (Sarkar et al., 2014; Alvarado et al., 2017). To address the possible target specificity of ZIKV, we performed RNA sequencing (RNA-seq) comparing uninfected GSCs and DGCs and defined a group of differentially expressed immune genes, including type I IFN–stimulated genes (ISGs; Fig. S3). Gene set enrichment analysis revealed that many ISGs were up-regulated in DGCs (Fig. S3, A and B). To further elucidate the signaling pathways that regulate ZIKV targeting of GSCs, we infected three GSC models (387, 3565, and 4121) with the ZIKV-Dakar strain for 36–48 h and then performed RNA-seq. IFN signaling was the top gene ontology pathway activated by ZIKV infection (Fig. S3, C and D). The RNA-seq data were validated by qPCR (Fig. S3 E). As ZIKV cannot fully antagonize ISGs, IFN responses may contribute to the specificity of ZIKV inhibition of GSC growth but more limited killing of DGCs or normal brain neurons and glial cells. Consistent with this hypothesis, treatment of DGCs with a blocking antibody against IFN-α/β receptor 2 (IFNAR2) resulted in increased ZIKV infection (Fig. S3 F) and decreased relative cell number (Fig. S3 G).
An attenuated ZIKV maintains effectiveness against GSCs and has additive effects with temozolomide chemotherapy
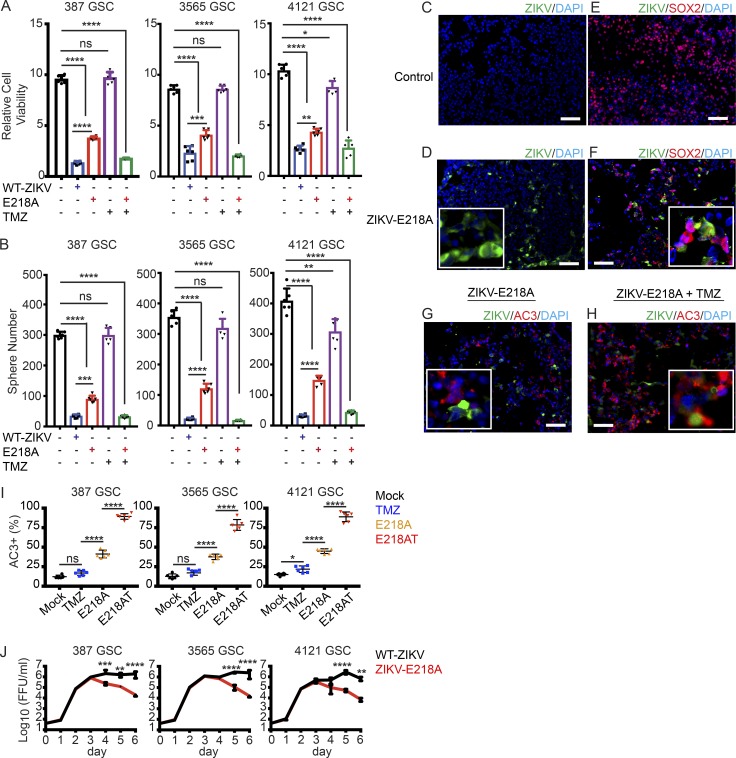
We previously reported a mutation of the flavivirus NS5 gene (E218A) that sensitizes the virus to translational inhibition by type I IFN and IFIT1 (Daffis et al., 2010) resulting in attenuation in cells responsive to type I IFNs. To begin to enhance the safety features of a potential oncolytic ZIKV, we compared the tumoricidal effects of a wild-type ZIKV (FSS13025, Cambodia 2010; Shan et al., 2016) and its recombinant derivative ZIKV-E218A against three GSC models (Fig. 5). Both the parental and E218A mutant ZIKV strains displayed anti-GSC activity, as measured by cell viability and sphere formation (Fig. 5, A and B). Although the parental ZIKV strain was more potent in reducing GSC growth, both strains were effective. Both the parental strain and the E218A-attenuated strain preferentially infected SOX2+ tumor cells and induced apoptosis (Fig. 5, C–H). Although tumor cells have the potential to activate the IFN response pathway, we still observed significant death of GSCs treated with ZIKV-E218A. As GSCs often display resistance to chemotherapy, including the standard-of-care temozolomide (TMZ; Stupp et al., 2009; Chen et al., 2012), we evaluated the combinatorial efficacy of TMZ and ZIKV E218A. Whereas TMZ alone had limited effects on GSCs, ZIKV-E218A combined with TMZ for 1 wk showed greater antitumor efficacy (Fig. 5, A and B) and induction of apoptosis (Fig. 5, G–I). We tested ZIKV-E218A infection capacity over a 1-wk time course. ZIKV-E218A had self-limited replication capacity in three GSC models (387, 3565, and 4121) relative to that of the parental ZIKV strain (Fig. 5 J). These data suggest that engineered mutant ZIKV strains may promote infection and lysis of GSCs with less toxicity to surrounding differentiated neural cells.
Figure 5.
ZIKV-E218A inhibits the growth of GSCs and has additive effects with TMZ. (A and B) GSCs were mock treated or incubated with parental ZIKV (MOI of 5), ZIKV-E218A (MOI of 5), TMZ (250 µM), or ZIKV-E218A (MOI of 5) and TMZ (250 µM) combined (E218AT). After 1 wk, GSC lines (387, 3565, and 4121) were assayed for relative cell number normalized to day 0 (A) and sphere formation (B). (C–F) Immunofluorescence staining of uninfected control (C and E) and ZIKV-E218A treated (D and F) GSCs on day 7 for Sox2 (red), DAPI (blue), and ZIKV-E218A (green). (G and H) Immunofluorescence staining of ZIKV-E218A-infected GSCs without (G) and with TMZ (250 µM; H) on day 7 for AC3 (red), DAPI (blue), and ZIKV-E218A (green). (I) Quantification of AC3+ apoptotic cells in three GSC lines treated with TMZ, ZIKV-E218A, or ZIKV-E218A combined with TMZ (E218AT). (J) Viral titer from supernatants of parental ZIKV-infected and ZIKV-E218A-infected GSCs over 1 wk measured by FFA. For every panel, data were pooled from three independent experiments performed in duplicate. Values represent mean ± SD. Significance was analyzed with one-way ANOVA with Tukey’s multiple comparison test (A, B, and I) and two-way ANOVA with the Bonferroni multiple comparison test (J; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001). Bars: (C–H) 100 µm.
Engineering of ZIKV may provide a therapeutic modality against GSCs
Our findings suggest that because of its tropism for neuroprogenitor cells, ZIKV may offer a tailored therapy that could be used in combination with conventional therapies (e.g., cytotoxic chemotherapy) that target bulk tumor cell populations. The reason for its tropism for specific cells is uncertain, as no definitive ZIKV receptor has been established. Our study is a first step in the development of engineered ZIKV as a glioblastoma therapy. Safety remains a paramount concern. The E218A mutant virus has two nucleotide changes in the same codon that abrogate the 2'-O methyltransferase activity in the NS5 protein (Zhou et al., 2007) and attenuates its replication in cells responsive to type I IFNs (Daffis et al., 2010). Regression to pathogenic virus requires a low-probability event of concurrent nucleotide changes causing the exact amino acid reversion. Notwithstanding this, the E218A mutant virus represents a framework for further genetic modification to ensure safety and maintain efficacy. For example, second-site mutations in the 3′ untranslated region that affect production of a subgenomic RNA (Akiyama et al., 2016; Donald et al., 2016) could further sensitize the virus to type I IFN. Public health concerns must be addressed with additional preclinical testing, including assays that evaluate for dissemination and genetic reversion.
Although we observed significant effects on mouse high-grade glioma models in vivo and on patient-derived GSCs in vitro, it remains to be determined how ZIKV strains perform in patient-derived GSCs in vivo. This will require overcoming the technical challenges of creating patient-derived tumor models in more immunocompetent mice. We envision possible therapeutic use of modified ZIKV strains to target GSCs through orthotopic injection into the tumor beds of patients. Other modified viruses are under development to treat glioblastoma, including measles (Bach et al., 2013; Hardcastle et al., 2017), polio (Dobrikova et al., 2008), and herpes viruses (Cheema et al., 2013). Our work serves as a foundation for further mechanistic studies and the genetic engineering of a safe and effective ZIKV, which could become an important tool in neuro-oncology.
Materials and methods
Ethics statement
This study was performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH). The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance no. A338101). Inoculations were performed under anesthesia induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering.
Isolation and culture of GSCs, differentiated tumor cells, and nonmalignant brain cultures
Glioblastoma or nonmalignant epilepsy resection tissues were obtained from excess surgical materials from patients at the Cleveland Clinic after neuropathology review with appropriate informed consent in accordance with a Cleveland Clinic Foundation Institutional Review Board–approved protocol (2559). To prevent culture-induced drift in glioblastoma models, patient-derived subcutaneous xenografts were generated in NOD-scid IL2Rgnull mice (Jackson Laboratory) and maintained as a recurrent source of tumor cells for study. Upon xenograft removal, a papain dissociation system (Worthington Biochemical) was used to dissociate tumors according to the manufacturer’s instructions. Cells were then cultured in Neurobasal complete media (Neurobasal Medium; Life Technologies) supplemented with 1× B27 without vitamin A (Thermo Fisher), 2 mM l-glutamine (Thermo Fisher), 1 mM sodium pyruvate (Thermo Fisher), 10 ng/ml basic fibroblast growth factor (bFGF), and 10 ng/ml epidermal growth factor (EGF; R&D Systems). The GSC phenotype was validated by Olig2 and Sox2 expression, functional assays of self-renewal (serial neurosphere passage), and tumor propagation using in vivo limiting dilution.
Proliferation and sphere formation assay
Cell viability was measured using CellTiter-Glo (Promega). After addition of ZIKV, all data were normalized to day 0 and expressed as a relative cell number. Neurosphere formation was measured as previously described (Wang et al., 2017). In brief, GSCs (1,000 cells) were plated into 96-well plates. The presence and number of neurospheres in each well were recorded on days 0, 3, 5, and 7.
ZIKV strains
ZIKV Dakar 41519 strain (Senegal 1984) and Brazil (Paraiba 2015) were provided by the World Reference Center for Emerging Viruses and Arboviruses (University of Texas Medical Branch) and S. Whitehead (NIH, Bethesda, MD). Parental and ZIKV-E218A (mutation in NS5 gene) were generated from an infectious cDNA clone of the Cambodian strain FSS13025 (2010) using site-directed mutagenesis as described previously (Shan et al., 2016). This Cambodian strain of ZIKV was chosen for mutagenesis, as opposed to the Dakar strain, because of the availability of an infectious cDNA clone (Shan et al., 2016). ZIKV stocks were propagated in Vero cells after inoculating at an MOI of 0.01 and incubating for 72 h. Viral titers were quantified by plaque assay as described previously (Govero et al., 2016), and stocks were stored at −80°C in single-use aliquots. ZIKV strain Dakar 41519 was passaged four times in Rag1−/− mice to create a mouse-adapted, more pathogenic variant of ZIKV-Dakar (Govero et al., 2016; Sapparapu et al., 2016). This variant has a single mutation in the NS4B gene that is associated with increased infectivity.
Cells
Vero (African green monkey kidney epithelial, ATCCCCL-81) cells, BV2 cells (microglia), GL261 (mouse glioma), GL26 (mouse glioma; a gift from M. Castro, University of Michigan, Ann Arbor, MI), CT2A (Oh et al., 2014; mouse glioma; a gift from T. Seyfried, Boston College, Boston, MA), NM55 and NM177 nonmalignant epilepsy cultures, or DGCs were maintained in DMEM supplemented with 10% FBS (Atlas). MS-DNCs were generated by isolating cells from the lateral ventricles of 4–6-wk-old C57BL/6 mice and placing them in DMEM with 10% FBS. For animal studies, GL261 and CT2A cells were virally transduced with a luciferase construct and selected with puromycin (1 µg/ml). GSCs, epilepsy tissues, and glioblastoma tissues were maintained in Neurobasal complete media. All cells were incubated at 37°C in humidified incubators supplemented with 5% CO2. All cell lines were negative for mycoplasma.
In vitro viral infection and drug treatment experiments
GSCs were plated at 1,000 cells/well in 96-well tissue culture treated plates (TPP) and allowed to attach overnight. For viral infection and growth inhibition assays, wild-type ZIKV-Dakar, ZIKV-Brazil, or mouse-adapted ZIKV-Dakar were used at an MOI of 5. For combined drug and virus therapy experiments, the wild-type parental ZIKV-Cambodia and its derivative ZIKV-E218A were used for infection. TMZ (Sigma-Aldrich) was dissolved in PBS and diluted in Neurobasal complete media. ZIKV was added at an MOI of 5 by itself or 4 h before TMZ (250 µM) addition. Cell supernatants were stored at −80°C for subsequent analysis. For IFNAR antibody blockade experiments, DGCs were treated with 20 µg/ml of anti–human IFNAR2 neutralizing monoclonal antibody (clone MMHAR-2; 21385-1; PBL Assay Science) for 12 h before the addition of indicated ZIKV strains at an MOI of 5.
Infectious virus titration
Focus-forming assays (FFAs) were performed with Vero cells as described previously (Govero et al., 2016). Supernatant samples containing ZIKV were serially diluted and added to Vero cell monolayers in 96-well plates. The virus was allowed to infect for 2–4 h, and then 100 µl of a 1:1 solution of 2× DMEM with 8% FBS and 2% methylcellulose was added to cells. Plates were incubated for 48 h and then fixed by the addition of 2% paraformaldehyde (PFA). Cells were then incubated with 500 ng/ml of the flavivirus cross-reactive mouse monoclonal antibody E60 (Oliphant et al., 2006) for 2 h at room temperature. After incubation for 1 h with a 1:5,000 dilution of horseradish peroxidase (HRP)–conjugated goat anti–mouse IgG (Sigma-Aldrich), foci were detected by addition of TrueBlue substrate (KPL). Foci were analyzed with a CTL Immunospot instrument.
Histology
5-µm-thick sections of paraffin-embedded tissues were analyzed for hematoxylin and eosin (H&E; Thermo Fisher), Picro-Sirius Red (Sigma-Aldrich), and Masson’s Trichrome (Diagnostic Biosystems) according to the manufacturer’s instructions. 4×, 10×, and 20× images were captured on an Eclipse 80i brightfield microscope (Nikon). Image analysis was performed by thresholding for positive staining and normalizing to total tissue area using ImageJ (NIH) and MetaMorph v7.7.0.0 (Molecular Devices) software (Jiang et al., 2016).
Immunofluorescence staining and microscopy
8-µm-thick cryosections were air dried and fixed in 4% PFA for 15 min before being washed twice with PBS. Tissues were permeabilized by incubating the slides with 1% Triton X-100 in PBS for 15 min at room temperature and peroxidase quenched by incubating them in 1% H2O2 (Invitrogen) for 10 min at room temperature. After blocking for 1 h at room temperature in blocking buffer (5% goat serum, 2.5% BSA in 1× PBS), slides were incubated overnight in a humidified chamber at 4°C with primary antibodies ZIKV (Millipore; AB10216; working dilution 1:1,000), Sox2 (Millipore; AB5603; stock: 1 mg/ml; working dilution 1:400), Ki-67 (Millipore; AB9620; working dilution 1:400), GFAP (Sigma-Aldrich; G9269; working dilution 1:1,000), PAX6 (Abcam; AB5790; stock: 1 mg/ml; working dilution 1:200), NeuN (Abcam; AB177487; working dilution 1:500), Stat1 (Abcam; AB31369; stock: 1 mg/ml; working dilution 1:1,000), BrdU (Abcam; AB6326; stock: 1 mg/ml; working dilution 1:1,000), and Ifnar1 (Sino Biological; 50469; stock: 1 mg/ml; working dilution 1:1,000). After PBST (1× PBS with 0.05% Tween 20) washes, slides were incubated with Alexa Fluor 488–, 594–, or 647–conjugated anti–mouse, anti–rat, or anti–rabbit secondary antibodies (Thermo Fisher). Slides were subsequently washed and mounted using VECTASHIELD with DAPI (Vector Labs). For cell immunofluorescence staining, 105 cells were seeded into a 12-well chamber slide (Thermo Fisher) and cultured overnight. Slides were then processed as described previously for tissue staining. 10×, 20×, and 40× images were collected at room temperature on an Eclipse 80i epifluorescence microscope (Nikon; Jiang et al., 2016). The cells were identified based on DAPI. Image analysis was performed by thresholding for positive staining and normalizing to total tissue area using ImageJ and MetaMorph v7.7.0.0 software. Quantitation was initially performed in an unblinded manner. However, many of the key results were requantitated by a second individual in a blinded manner to eliminate bias.
Immunohistochemistry
Tissues were fixed in 10% formalin, embedded in paraffin, and incubated with antibodies as previously described (Jiang et al., 2016). In brief, 6-µm-thick sections were deparaffinized in xylene, rehydrated in graded ethanol, and subjected to antigen retrieval by steam heating in Citra antigen retrieval solution (BioGenex). After blocking for 1 h at room temperature in blocking buffer (5% goat serum, 2.5% BSA in 1× PBS), slides were incubated overnight in a humidified chamber at 4°C with primary antibodies Ki-67 (Millipore; AB9620; working dilution 1:400), GFAP (Sigma-Aldrich; G9269; working dilution 1:1,000), and cleaved caspase-3 (Cell Signaling; 9661; working dilution 1:100). Slides were then incubated at room temperature for 30 min with anti–rabbit (EnVision+ System HRP Labeled Polymer; Dako) or anti–mouse (VECTASTAIN ABC Rabbit IgG detection kit; Vector Laboratories) secondary antibodies. Staining was detected using 3,3-diaminobenzidine (DAB). Image acquisition and analysis were similar to those of immunofluorescence imaging.
RNA in situ hybridization (ISH) was performed using RNAscope 2.5 HD (Brown; Advanced Cell Diagnostics) according to the manufacturer’s instructions and as previously described (Govero et al., 2016). PFA-fixed, paraffin-embedded tissue sections (from ZIKV-infected or uninfected tumors) were deparaffinized by incubating them for 60 min at 60°C, and endogenous peroxidases were quenched with H2O2 for 10 min at room temperature. Slides were then boiled for 15 min in RNAscope Target Retrieval reagents and incubated for 30 min in RNAscope Protease Plus reagent before ZIKV RNA probe (Advanced Cell Diagnostics; 467771). Sections were counterstained with Gill’s hematoxylin and visualized by brightfield microscopy.
Organoids
Organoids were formed by suspending tumor cells in Matrigel and forming 20-ml pearls on Parafilm molds before culture (Hubert et al., 2016). Organoids were cultured in 6-well or 10-cm plates, shaking in Neurobasal complete media. Images of growing organoids were acquired using an EVOS FL Cell Imaging System (Invitrogen) for microscopic imaging. Organoids were grown until 35 d under these conditions. Organoids were infected with ZIKV-Dakar or ZIKV-Brazil at 106 FFU for 2 h, and then the media was removed. Organoids were subsequently washed three times with PBS, and fresh Neurobasal complete media was added. Images were acquired using EVOS Cell Imaging System 3 (Thermo Fisher). Areas of individual organoids were measured with ImageJ software.
Animal experiments
Mice and tumor implantation
Mouse glioblastoma cells (GL261 and CT2A) transduced with luciferase were grown in DMEM supplemented with 10% serum. Cells were harvested by trypsinization and then washed and resuspended in PBS. A total of 2 × 104 cells were implanted into 6-wk-old C57BL/6 female mice (Jackson Laboratory). In brief, animals were anesthetized by intraperitoneal injection of ketamine (10 mg/kg) and xylazine (100 mg/kg). The animals were placed in a stereotactic apparatus (Stoelting), and an incision was made over the cranial midline. A burr hole was made 1.5 mm anterior to the lambda and 2.5 mm right of the midline. A 29.5-gauge Hamilton syringe was inserted to a depth of 3 mm and withdrawn 0.5 mm to a depth of 2.5 mm. 3 µl of GL261 or CT-2A-luc2 cells were injected over the course of 5 min. The incision site was closed by Vetbond (3M).
Treatment and animal monitoring
1 (GL261) or 2 wk (GL261 or CT2A) after tumor implantation, animals were placed into two groups for mouse-adapted ZIKV-Dakar inoculation or saline injection. There was no formal animal randomization process; animals were taken from serial cages and treated with the control or virus. 103 or 105 FFU of the mouse-adapted ZIKV-Dakar was diluted in 10-µl volume. The same coordinates from surgery were used for this treatment. Animals were monitored daily for signs of neurological impairment. The monitor was not blinded to the treatment received.
Flow cytometry
At different time points after ZIKV infection, cells were fixed with 2% PFA diluted in PBS for 10 min at room temperature and permeabilized with HBSS buffer (10 mM Hepes), 0.1% (wt/vol) saponin (Sigma-Aldrich), and 0.025% NaN3 for 10 min at room temperature. GSCs were transferred to a V-bottom plate (Costar) and incubated for 1 h at 4°C with 2 µg/ml of ZV-64 mAb (Zhao et al., 2016). After washing, cells were incubated with an Alexa Fluor 647–conjugated goat anti–mouse IgG (Invitrogen) for 30 min at 4°C, washed twice with HBSS buffer, processed on a FACS Array (BD Biosciences), and analyzed using FlowJo software (Tree Star).
Bioluminescence imaging
Beginning 1 wk after tumor cell implantation, brain tumor formation was detected using bioluminescence imaging. Mice, under isoflurane anesthesia (2% vaporized in O2), were injected intraperitoneally with d-luciferin (150 mg/kg in PBS; Gold Biotechnology) and imaged using an IVIS50 imaging system (PerkinElmer). Exposure times were 10 or 60 s, and software-defined contour regions of interest were used to measure total photon flux (photons/s) using Living Image 2.6 (PerkinElmer).
Quantitative RT-PCR (qRT-PCR) for ZIKV RNA
ZIKV RNA levels were determined by one-step qRT-PCR (Thermo Fisher) on an ABI 7500 Fast Instrument (Applied Biolsystems) using standard cycling conditions. Viral burden was expressed on a log10 scale as viral RNA equivalents per g after comparison with a standard curve produced using serial 10-fold dilutions of ZIKV RNA. For ZIKV, the following primer sets were used: For, 5′-CCACCAATGTTCTCTTGCAGACATATTG-3′; Rev, 5′-TTCGGACAGCCGTTGTCCAACACAAG-3′; and Probe, 5′-56-FAM/AGCCTACCT TGACAAGCAGTC/3IABkFQ-3′.
qRT-PCR for GSCs and DGCs
Total cellular RNA was isolated using TRIzol reagent (Sigma-Aldrich), followed by RT into cDNA using the qScript cDNA Synthesis kit (Quanta BioSciences). Real-time PCR was performed using an Applied Biosystems 7900HT cycler using SYBR Green PCR Master Mix (Thermo Fisher). Sequences for gene-specific primer sets were as follows: human IFNAR1 forward 5′-AACAGGAGCGATGAGTCTGTC-3′ and reverse 5′-TGCGAAATGGTGTAAATGAGTCA-3′; human STAT1 forward 5′-CAGCTTGACTCAAAATTCCTGGA-3′ and reverse 5′-TGAAGATTACGCTTGCTTTTCCT-3′; human IRF1 forward 5′-ATGCCCATCACTCGGATGC-3′ and reverse 5′-CCCTGCTTTGTATCGGCCTG-3′; human IFIT1 forward 5′-ATGACGATGAAATGCCTGA-3′ and reverse 5′-CAGGTCACCAGACTCCTCAC-3′; human OAS2-1 forward 5′-CTCAGAAGCTGGGTTGGTTTAT-3′ and reverse 5′-ACCATCTCGTCGATCAGTGTC-3′; human IFIH1 forward 5′-TCGAATGGGTATTCCACAGACG-3′ and reverse 5′-GTGGCGACTGTCCTCTGAA-3′; 18S RNA forward 5′-AACCCGTTGAACCCCATT-3′ and reverse 5′-CCATCCAATCGGTAGTAGCG-3′; and GAPDH forward 5′-CCTGTTCGACAGTCAGCCG-3′ and reverse 5′-CGACCAAATCCGTTGACTCC-3′.
RNA-seq data acquisition, quality control, and processing
RNA was obtained from GSCs infected with ZIKV-Dakar for 36–48 h. Total cellular RNA was isolated using the RNeasy kit (Qiagen). RNA-seq reads were aligned to the Ensembl release 76 assembly with STAR v. 2.5.1a. Gene counts were derived from the number of uniquely aligned unambiguous reads by Subread:feature Count v. 1.4.6p5. Transcript counts were produced by Sailfish v. 0.6.3. Sequencing performance was assessed for total number of aligned reads, total number of uniquely aligned reads, genes and transcripts detected, ribosomal fraction, known junction saturation, and read distribution over known gene models with RSeQC v. 2.6.2.
All gene-level and transcript counts were imported into the R/Bioconductor package EdgeR, and TMM normalization size factors were calculated to adjust for samples for differences in library size. Genes or transcripts not expressed in any sample were excluded from further analysis. The TMM size factors and the matrix of counts were imported into R/Bioconductor package Limma, and weighted likelihoods based on the observed mean–variance relationship of every gene/transcript were calculated for all samples with the Voom function. Performance of the samples was assessed with a Spearman correlation matrix and multidimensional scaling plots. Gene/transcript performance was assessed with plots of residual standard deviation of every gene to their mean log count with a robustly fitted trend line of the residuals. Generalized linear models were created to test for gene- or transcript-level differential expression. Differentially expressed genes and transcripts were then filtered for FDR-adjusted P values less than or equal to 0.05.
To enhance the biological interpretation of the large set of transcripts, grouping of genes/transcripts based on functional similarity was achieved using the R/Bioconductor packages GAGE and Pathview. GAGE and Pathview were also used to generate pathway maps on known signaling and metabolism pathways curated by KEGG.
For the matched GSC and DGC lines, RNA-seq data were evaluated for type I and II IFN signatures between GSCs and DGCs using gene set enrichment analysis and were validated using RNA-seq data on additional matched cell lines derived from Suvà et al. (2014). IFN signatures were derived from the Molecular Signature Database curated by the Broad Institute (Subramanian et al., 2005; http://www.broad.mit.edu/gsea/). Unsupervised hierarchical clustering was performed using FPKM values from matched TPC and DGC lines. All RNA-seq data are available through the Gene Expression Omnibus (GEO) data repository under accession nos. GSE102244 and GSE102924.
Statistical analysis
All statistical analysis was performed using Prism 7.0 software (GraphPad). The number of animals and replicate experiments is specified in each figure legend. No statistical methods were used to predetermine sample sizes, but our choice of sample sizes is similar to those reported in previous publications (Jiang et al., 2016; Wang et al., 2017). All animals that survived tumor implantation and virus injection surgeries were included in the analyses. All grouped data were presented as mean ± SD or SEM as indicated in the figure legends. Student’s t tests, one-way ANOVA with multiple comparison correction, and two-way ANOVA with multiple comparison correction were used to assess the significance of differences between groups. These tests were performed when the sample size was large enough to assume that the means were normally distributed or that the distribution of residuals was normal, respectively. For groups being statistically compared, variances in data were similar. For animal survival analysis, Kaplan–Meier curves were generated and the log-rank test was performed to assess statistical significance between groups.
Online supplemental material
Fig. S1 demonstrates that ZIKV infection efficiency is lower in DGCs compared with GSCs. Fig. S2 demonstrates that WNV has global toxicity on normal cells, GSCs, and DGCs, whereas ZIKV has minimal impact on the normal adult brain compared with GSCs and DGCs. Fig. S3 demonstrates that IFN signaling is one determinant of the differential effects of ZIKV on GSCs compared with DGCs.
Supplementary Material
Acknowledgments
We thank Julie Prior, Samuel Achilefu, and the Molecular Imaging Center and Oncologic Imaging Program of the Siteman Cancer Center for animal imaging; Jennifer Govero for preparation of virus stocks; Edward Oates for help with animal experiments; David G. DeNardo for use of his microscope; and Qi Xie for providing RNA sequencing data. We thank the Genome Technology Access Center in the Department of Genetics at the Washington University School of Medicine for assistance with RNA sequencing and analysis.
National Institutes of Health (NIH) grants (R01 AI073755 and R01 AI104972 awarded to M.S. Diamond and CA197718, CA154130, CA169117, CA171652, NS087913, and NS089272 awarded to J.N. Rich) and grants from the Elsa U. Pardee Foundation, the Concern Foundation, the Cancer Research Foundation, and the McDonnell Center for Cellular and Molecular Neurobiology of Washington University (awarded to M.G. Chheda) supported this work. The Genome Technology Access Center of Washington University in St. Louis is supported by National Center for Advancing Translational Sciences (NCATS) ICTS/CTSA grant UL1 TR002345 and NCI Cancer Center support grant P30 CA91842. The Imaging Program is supported by NCI Cancer Center support grant P50 CA094056. The Center is partially supported by NCI Cancer Center support grant P30 CA91842 to the Siteman Cancer Center and by National Center for Research Resources (NCRR) ICTS/CTSA grant UL1TR000448, a component of the NIH, and the NIH Roadmap for Medical Research.
M.S. Diamond is a consultant for Inbios and Visterra, is on the Scientific Advisory Boards of Moderna and OvaGene, and is a recipient of grants from Moderna and Visterra. The authors declare no additional competing conflicts of interest. This publication is solely the responsibility of the authors and does not necessarily represent the official views of the NCRR or NIH.
Author contributions: Z. Zhu, M.J. Gorman, L.D. McKenzie, J.N. Chai, C.G. Hubert, B.C. Prager, E. Fernandez, J.M. Richner, R. Zhang, X. Wang, and M.G. Chheda performed the experiments. C. Shan and P.-Y. Shi provided key reagents. E. Tycksen provided RNA-seq analysis and methods. Z. Zhu, J.N. Chai, M.S. Diamond, M.G. Chheda, and J.N. Rich wrote the initial draft of the manuscript, with the other authors contributing to editing the manuscript into its final form.
Footnotes
Abbreviations used:
- CNS
- central nervous system
- DGC
- differentiated glioma cell
- FFA
- focus-forming assay
- FFU
- focus-forming unit
- GSC
- glioblastoma stem cell
- Hu-DNC
- differentiated human neural stem cell
- ISG
- type I IFN–stimulated gene
- MOI
- multiplicity of infection
- MS-DNC
- mouse-differentiated neural cell
- PFA
- paraformaldehyde
- qRT-PCR
- quantitative RT-PCR
- RNA-seq
- RNA sequencing
- TMZ
- temozolomide
- WNV
- West Nile virus
- ZIKV
- Zika virus
References
- Akiyama B.M., Laurence H.M., Massey A.R., Costantino D.A., Xie X., Yang Y., Shi P.-Y., Nix J.C., Beckham J.D., and Kieft J.S.. 2016. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science. 354:1148–1152. 10.1126/science.aah3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso M.M., Jiang H., Gomez-Manzano C., and Fueyo J.. 2012. Targeting brain tumor stem cells with oncolytic adenoviruses. Methods Mol. Biol. 797:111–125. 10.1007/978-1-61779-340-0_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarado A.G., Thiagarajan P.S., Mulkearns-Hubert E.E., Silver D.J., Hale J.S., Alban T.J., Turaga S.M., Jarrar A., Reizes O., Longworth M.S., et al. 2017. Glioblastoma cancer stem cells evade innate immune suppression of self-renewal through reduced TLR4 expression. Cell Stem Cell. 20:450–461.e4. 10.1016/j.stem.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach P., Abel T., Hoffmann C., Gal Z., Braun G., Voelker I., Ball C.R., Johnston I.C., Lauer U.M., Herold-Mende C., et al. 2013. Specific elimination of CD133+ tumor cells with targeted oncolytic measles virus. Cancer Res. 73:865–874. 10.1158/0008-5472.CAN-12-2221 [DOI] [PubMed] [Google Scholar]
- Bao S., Wu Q., McLendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., and Rich J.N.. 2006. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 444:756–760. 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- Cassady K.A., Bauer D.F., Roth J., Chambers M.R., Shoeb T., Coleman J., Prichard M., Gillespie G.Y., and Markert J.M.. 2017. Pre-clinical assessment of C134, a chimeric oncolytic herpes simplex virus, in mice and non-human primates. Mol. Ther. Oncolytics. 5:1–10. 10.1016/j.omto.2017.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R., and Russell S.J.. 2017. How to develop viruses into anticancer weapons. PLoS Pathog. 13:e1006190 10.1371/journal.ppat.1006190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheema T.A., Wakimoto H., Fecci P.E., Ning J., Kuroda T., Jeyaretna D.S., Martuza R.L., and Rabkin S.D.. 2013. Multifaceted oncolytic virus therapy for glioblastoma in an immunocompetent cancer stem cell model. Proc. Natl. Acad. Sci. USA. 110:12006–12011. 10.1073/pnas.1307935110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li Y., Yu T.S., McKay R.M., Burns D.K., Kernie S.G., and Parada L.F.. 2012. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 488:522–526. 10.1038/nature11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S., Szretter K.J., Schriewer J., Li J., Youn S., Errett J., Lin T.Y., Schneller S., Zust R., Dong H., et al. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature. 468:452–456. 10.1038/nature09489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova E.Y., Broadt T., Poiley-Nelson J., Yang X., Soman G., Giardina S., Harris R., and Gromeier M.. 2008. Recombinant oncolytic poliovirus eliminates glioma in vivo without genetic adaptation to a pathogenic phenotype. Mol. Ther. 16:1865–1872. 10.1038/mt.2008.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donald C.L., Brennan B., Cumberworth S.L., Rezelj V.V., Clark J.J., Cordeiro M.T., Freitas de Oliveira França R., Pena L.J., Wilkie G.S., Da Silva Filipe A., et al. 2016. Full genome sequence and sfRNA interferon antagonist activity of Zika virus from Recife, Brazil. PLoS Negl. Trop. Dis. 10:e0005048 10.1371/journal.pntd.0005048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel E., Ramani A., Karow U., Gottardo M., Natarajan K., Gooi L.M., Goranci-Buzhala G., Krut O., Peters F., Nikolic M., et al. 2017. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell. 20:397–406.e5. 10.1016/j.stem.2016.12.005 [DOI] [PubMed] [Google Scholar]
- Garcez P.P., Loiola E.C., Madeiro da Costa R., Higa L.M., Trindade P., Delvecchio R., Nascimento J.M., Brindeiro R., Tanuri A., and Rehen S.K.. 2016. Zika virus impairs growth in human neurospheres and brain organoids. Science. 352:816–818. 10.1126/science.aaf6116 [DOI] [PubMed] [Google Scholar]
- Govero J., Esakky P., Scheaffer S.M., Fernandez E., Drury A., Platt D.J., Gorman M.J., Richner J.M., Caine E.A., Salazar V., et al. 2016. Zika virus infection damages the testes in mice. Nature. 540:438–442. 10.1038/nature20556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromeier M., Lachmann S., Rosenfeld M.R., Gutin P.H., and Wimmer E.. 2000. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. USA. 97:6803–6808. 10.1073/pnas.97.12.6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle J., Mills L., Malo C.S., Jin F., Kurokawa C., Geekiyanage H., Schroeder M., Sarkaria J., Johnson A.J., and Galanis E.. 2017. Immunovirotherapy with measles virus strains in combination with anti–PD-1 antibody blockade enhances antitumor activity in glioblastoma treatment. Neuro Oncol. 19:493–502. 10.1093/neuonc/now179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert C.G., Rivera M., Spangler L.C., Wu Q., Mack S.C., Prager B.C., Couce M., McLendon R.E., Sloan A.E., and Rich J.N.. 2016. A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 76:2465–2477. 10.1158/0008-5472.CAN-15-2402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Hegde S., Knolhoff B.L., Zhu Y., Herndon J.M., Meyer M.A., Nywening T.M., Hawkins W.G., Shapiro I.M., Weaver D.T., et al. 2016. Targeting focal adhesion kinase renders pancreatic cancers responsive to checkpoint immunotherapy. Nat. Med. 22:851–860. 10.1038/nm.4123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann J.K., and Chiocca E.A.. 2014. Glioma virus therapies between bench and bedside. Neuro-oncol. 16:334–351. 10.1093/neuonc/not310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazear H.M., Govero J., Smith A.M., Platt D.J., Fernandez E., Miner J.J., and Diamond M.S.. 2016. A mouse model of Zika virus pathogenesis. Cell Host Microbe. 19:720–730. 10.1016/j.chom.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Saucedo-Cuevas L., Regla-Nava J.A., Chai G., Sheets N., Tang W., Terskikh A.V., Shresta S., and Gleeson J.G.. 2016. Zika virus infects neural progenitors in the adult mouse brain and alters proliferation. Cell Stem Cell. 19:593–598. 10.1016/j.stem.2016.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martuza R.L., Malick A., Markert J.M., Ruffner K.L., and Coen D.M.. 1991. Experimental therapy of human glioma by means of a genetically engineered virus mutant. Science. 252:854–856. 10.1126/science.1851332 [DOI] [PubMed] [Google Scholar]
- Ming G.L., Tang H., and Song H.. 2016. Advances in Zika virus research: Stem cell models, challenges, and opportunities. Cell Stem Cell. 19:690–702. 10.1016/j.stem.2016.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska J., Rashidi A., Chang A.L., Muroski M.E., Han Y., Zhang L., and Lesniak M.S.. 2016. Anti-GITR therapy promotes immunity against malignant glioma in a murine model. Cancer Immunol. Immunother. 65:1555–1567. 10.1007/s00262-016-1912-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore A.E. 1954. Effects of viruses on tumors. Annu. Rev. Microbiol. 8:393–410. 10.1146/annurev.mi.08.100154.002141 [DOI] [PubMed] [Google Scholar]
- Oh T., Fakurnejad S., Sayegh E.T., Clark A.J., Ivan M.E., Sun M.Z., Safaee M., Bloch O., James C.D., and Parsa A.T.. 2014. Immunocompetent murine models for the study of glioblastoma immunotherapy. J. Transl. Med. 12:107 10.1186/1479-5876-12-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant T., Nybakken G.E., Engle M., Xu Q., Nelson C.A., Sukupolvi-Petty S., Marri A., Lachmi B.E., Olshevsky U., Fremont D.H., et al. 2006. Antibody recognition and neutralization determinants on domains I and II of West Nile virus envelope protein. J. Virol. 80:12149–12159. 10.1128/JVI.01732-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra B., Lizarazo J., Jiménez-Arango J.A., Zea-Vera A.F., González-Manrique G., Vargas J., Angarita J.A., Zuñiga G., Lopez-Gonzalez R., Beltran C.L., et al. 2016. Guillain-Barré syndrome associated with Zika virus infection in Colombia. N. Engl. J. Med. 375:1513–1523. 10.1056/NEJMoa1605564 [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. 2016. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 165:1238–1254. 10.1016/j.cell.2016.04.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds B.A., and Weiss S.. 1992. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 255:1707–1710. 10.1126/science.1553558 [DOI] [PubMed] [Google Scholar]
- Sapparapu G., Fernandez E., Kose N., Cao B., Fox J.M., Bombardi R.G., Zhao H., Nelson C.A., Bryan A.L., Barnes T., et al. 2016. Neutralizing human antibodies prevent Zika virus replication and fetal disease in mice. Nature. 540:443–447. 10.1038/nature20564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Döring A., Zemp F.J., Silva C., Lun X., Wang X., Kelly J., Hader W., Hamilton M., Mercier P., et al. 2014. Therapeutic activation of macrophages and microglia to suppress brain tumor-initiating cells. Nat. Neurosci. 17:46–55. 10.1038/nn.3597 [DOI] [PubMed] [Google Scholar]
- Shan C., Xie X., Muruato A.E., Rossi S.L., Roundy C.M., Azar S.R., Yang Y., Tesh R.B., Bourne N., Barrett A.D., et al. 2016. An infectious cDNA clone of Zika virus to study viral virulence, mosquito transmission, and antiviral inhibitors. Cell Host Microbe. 19:891–900. 10.1016/j.chom.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southam C.M., and Moore A.E.. 1952. Clinical studies of viruses as antineoplastic agents with particular reference to Egypt 101 virus. Cancer. 5:1025–1034. [DOI] [PubMed] [Google Scholar]
- Stupp R., Hegi M.E., Mason W.P., van den Bent M.J., Taphoorn M.J., Janzer R.C., Ludwin S.K., Allgeier A., Fisher B., Belanger K., et al. National Cancer Institute of Canada Clinical Trials Group . 2009. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 10:459–466. 10.1016/S1470-2045(09)70025-7 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suvà M.L., Rheinbay E., Gillespie S.M., Patel A.P., Wakimoto H., Rabkin S.D., Riggi N., Chi A.S., Cahill D.P., Nahed B.V., et al. 2014. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 157:580–594. 10.1016/j.cell.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner K.E., Galicich J.H., Krol G., Arbit E., and Malkin M.G.. 1989. Patterns of failure following treatment for glioblastoma multiforme and anaplastic astrocytoma. Int. J. Radiat. Oncol. Biol. Phys. 16:1405–1409. 10.1016/0360-3016(89)90941-3 [DOI] [PubMed] [Google Scholar]
- Wang X., Yang K., Xie Q., Wu Q., Mack S.C., Shi Y., Kim L.J.Y., Prager B.C., Flavahan W.A., Liu X., et al. 2017. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat. Neurosci. 20:661–673. 10.1038/nn.4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Fernandez E., Dowd K.A., Speer S.D., Platt D.J., Gorman M.J., Govero J., Nelson C.A., Pierson T.C., Diamond M.S., and Fremont D.H.. 2016. Structural basis of Zika virus-specific antibody protection. Cell. 166:1016–1027. 10.1016/j.cell.2016.07.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Ray D., Zhao Y., Dong H., Ren S., Li Z., Guo Y., Bernard K.A., Shi P.Y., and Li H.. 2007. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 81:3891–3903. 10.1128/JVI.02704-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.