Abstract
Background
Chagas disease is a debilitating often fatal disease resulting from infection by the protozoan parasite Trypanosoma cruzi. Chagas disease is endemic in 21 countries of the Americas, and it is an emerging disease in other countries as a result of migration. Given the chronic nature of the infection where intracellular parasites persist for years, the diagnosis of T. cruzi by direct detection is difficult, whereas serologic tests though sensitive may yield false-positive results. The development of new rapid test based on the identification of soluble parasitic antigens in serum would be a real innovation in the diagnosis of Chagas disease.
Methods
To identify new soluble biomarkers that may improve diagnostic tests, we investigated the proteins secreted by T. cruzi using mass spectrometric analyses of conditioned culture media devoid of serum collected during the emergence of trypomastigotes from infected Vero cells. In addition, we compared the secretomes of two T. cruzi strains from DTU Tc VI (VD and CL Brener).
Results
Analysis of the secretome collected during the emergence of trypomastigotes from Vero cells led to the identification of 591 T. cruzi proteins. Three hundred sixty three proteins are common to both strains and most belong to different multigenic super families (i.e. TcS, GP63, MASP, and DGF1). Ultimately we have established a list of 94 secreted proteins, common to both DTU Tc VI strains that do not belong to members of multigene families.
Conclusions
This study provides the first comparative analysis of the secretomes from two distinct T. cruzi strains of DTU TcVI. This led us to identify a subset of common secreted proteins that could potentially serve as serum markers for T. cruzi infection. Their potential could now be evaluated, with specific antibodies using sera collected from patients and residents from endemic regions.
Introduction
Trypanosoma cruzi is a protozoan parasite of the order Kinetoplastide and the aetiological agent of Chagas disease a vector-borne infection with a high prevalence in Central and South America. According to recent estimates, nearly 6 million people are infected with this neglected disease in Latin American countries [1]. Moreover, Chagas disease is now an increasing global health problem because of increased migration of infected persons to non-endemic regions [2].
T. cruzi reproduction is mostly clonal, with occasional events of genetic exchange leading to the emergence of hybrid genotypes [3]. These features led to a complex population structure, showing remarkable genetic diversity [4]. Biochemical and genetic typing schemes developed throughout the last decades converged in the delineation of six major T. cruzi evolutionary lineages or discrete typing units (DTUs) termed TcI to TcVI [5]. However the remarkable genetic heterogeneity of T.cruzi could partially account for their wide range of biological features, eco-epidemiological traits, and the large spectrum of clinical manifestations of Chagas disease [6]. No clear correlation between serodiagnostic test reactivity and molecular diversity of T. cruzi has been observed. Instead, discrepancies between serologic test’s sensitivity may reflect adaptive immune responses to parasite antigens [7]. In human parasite transmission occurs by vectorial route when metacyclic trypomastigotes present in blood-sucking triatomine bug faeces penetrate the skin or mucous membranes. However, humans can also become infected via blood transfusion or organ transplantation [8], through the ingestion of tainted food and fluids [9], or via vertical transmission from mother-to-child during pregnancy or delivery [10][11].
The clinical course of the T. cruzi infectionhas three phases: acute, chronic without symptoms, and chronic with symptoms [12]. The initial acute phase, lasting for about 2 months, is characterised by high levels of trypomastigotes (the circulating form of T. cruzi) in the blood. This is followed by a chronic phase (with or without symptoms) characterized by low trypomastigotes levels but with many intra-cellular parasites (amastigotes) present in target tissues (cardiac and smooth muscle) that can lead to severe morbidity. During this stage diagnosis is essentially based on the detection for classical (not lytic) anti-T cruzi antibodies in patient serum. At present, there are no diagnostic criteria for treatment response because anti-T. cruzi antibodies can persist many years after parasitological cure. Therefore, there is an urgent need to find a means to improve diagnosis in order to evaluate treatment efficacy.
A biomarker is defined as a parameter that can be objectively measured as an indicator of normal or pathogenic biological processes, or as an indicator of pharmacological responses to therapeutic interventions[13] [14]. In infectious diseases, there are two main types of markers: markers from the host and markers from the pathogen[15,16]. Only the latter markers provide evidence for the presence of the pathogen. Such markers can be of different types (carbohydrate, peptide and proteins or chemical product, lipids, sugar or nucleic acid) and from different locations (secreted in the serum of infected patients or present inside infected cells). The detection of circulating parasite excreted-secreted antigens (TESA) in chronically infected persons could serve as suitable biomarkers for the infection and treatment follow-up [17,18]. Proteomic approaches present a unique opportunity for identifying new parasite proteins that are potentially detectable in infected patients. We were particularly interested in extracellular proteins (secreted or released by the parasite), as these are often key mediators of host–parasite interactions involved in immune regulation, signalling, or invasion. Many proteomic analysis of T. cruzi secretome are already published [19,20]. These analyses made it possible to obtain a better understanding of the diverse pathways of secretion used by different parasite stages (epimastigote or metacyclic forms) to release many proteins into the extracellular medium and many secreted proteins could be identified. However, these studies have been carried out with parasite stages in the absence of vertebrate host cells, and where secretion was artificially induced. We propose here to complement these studies by analysing the secretome of Vero cells infected with two strains of T. cruzi DTU TcVI [18, 19] CL Brener, the strain used for the first genomic sequence of T. cruzi [21], with preferential tropism for heart and muscle cells [19] and VD, a strain isolated from a case of congenital Chagas disease, with preferential tropism for plancenta [18].
The main objective was to characterize as comprehensively as possible parasitic proteins secreted by T. cruzi (TcVI)-infected cells. The ultimate aim of this first step is to establish a list of antigens with potential usefulness as antigenic markers in the diagnosis and treatment follow-up of Chagas disease.
Materials and methods
Vero cells culture
Vero cells (normal kidney epithelial cells of Cercopithecus aethiops) were obtained from the Virology Laboratory of the Pitié Salpêtrière Hospital (Paris, France). At late exponential growth phase, trypsin-treated Vero cells were subcultured every seven days in RPMI-1640 medium (Life technologies) supplemented with streptomycin/penicillin (Life technologies) and 5% heat-inactivated foetal bovine serum (FBS) (Life technologies). Subcultures were maintained at 37°C in a humidified atmosphere of 5% CO2.
Trypanosoma cruzi culture and stocks
CL Brener strain (collection number: MNHN-CEU- 2016–0159) was a gift from Pr. P. Grellier of the Muséum National d’Histoire Naturelle (Paris, France). VD strain (Tc VI) was isolated from a congenital case of Chagas disease diagnosed at the Parasitology and Chagas Disease Service of the “Dr. Ricardo Gutiérrez Children’s’ Hospital” (Buenos Aires, Argentina.) Both strains were maintained in CF-1 (Non “Swiss” albino mice from Charles Rivers laboratory) mice prior to their use.
Large-scale trypomastigote production (CL Brener or VD strains) was achieved as follow. At the end of the exponential phase, Vero cells were harvested from subcultures and seeded into 75 cm2 flasks with 104 cells per cm2, followed by 3 days of incubation at 37°C in 5% CO2 in air. The cells were then infected with T. cruzi trypomastigotes at a parasite-cell-ratio 1:5. After 24 hours, the flasks were washed with HBSS buffer in order to remove non-attached cells and free parasites, and were subsequently incubated in RPMI-1640 medium. On day 4 (VD strain) or day 5 (CL Brener strain) post-infection, trypomastigotes were released from the cells. The culture medium was removed and transferred to a centrifuge tube. Attached infected cells were washed with 15 mL of HBSS buffer. The culture medium and wash containing trypomastigotes were mixed and centrifuged at 200 g for 10 minutes at room temperature to remove host cells and their debris. Subsequently, trypomastigotes were collected by supernatant centrifugation at 2000 g for 10 minutes, resuspended in 10 mL and counted in a haemocytometer using a light microscope. The recovered parasites served for the secretome analysis.
Determining the secretome
The parasite suspension was added to each flask containing Vero cells at 7.104 cells per cm2, in a 5:1 parasite-cell ratio. After 24 hours, the cells were washed once with HBSS buffer (without Ca2+ and Mg2+) to remove free parasites from the culture media and they were then incubated for 2 days (VD strain) or 3 days (CL Brener strain) in RPMI-1640 medium-5%SBF. After this, the cells were washed five times with HBSS buffer (with Ca2+ and Mg 2+) to remove all traces of foetal bovine serum (FBS) and then incubated for 24 hours in a medium used for hybridoma culture (PFHM II),.a synthetic culture medium that contains no proteins. After the release of the trypomastigotes into the culture supernatant, the medium was transferred to a 50 mL centrifuge tube and centrifuged at 2000 g for 10 minutes at room temperature to remove any contamination with host cells, T. cruzi and other debris. The supernatant was collected and filtered on a 0.22 μm membrane to remove any residual cells or parasites, and then dialyzed and concentrated 200 -fold on a Vivaspin® 15R centrifugal concentrator (Vivaspin) at 4°C. The resultant concentrate was conserved at -80°C until its use.
One-dimensional electrophoretic analysis
Equal aliquots (40 µg) of proteins obtained from infected Vero cells supernatant were submitted to a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (12% SDS-PAGE). Proteins were denaturized by incubating samples with Laemlli buffer (0.5 M Tris–HCl, pH 6.8, 50% glycerol, 10% SDS, 5% β-mercaptoethanol, and 0.05% bromophenol blue) at room temperature for 16 hours before electrophoresis. Electrophoresis was carried out running buffer (50 mM Tris, 192 mM glycine and 0.1% SDS). After protein separation, gels were stained with Instant Blue (Expedeon Ltd., Harston, U.K.) for 20 minutes and transferred to distilled water for direct use.
In-gel digestion
Each track was cut into 9 gel bands using a sterile scalpel and these were individually transferred to 1.5 mL Eppendorf sterile tubes. The gel was covered with 200 μL of destaining solution (50% acetonitrile, 25 mM ammonium bicarbonate in Milli-Q-H2O) and incubated under stirring at room temperature for 10 minutes. This step was repeated until the blue colour was removed. The gel pieces were then dehydrated using 200 μL of acetonitrile for 5 min at room temperature and air dried for 30 min.
Subsequently, 100 μL of buffer (10mM dithiothreitol (DTT), 100 mM ammonium bicarbonate) were added to reduce proteins for 30 minutes at 56°C. Next, 100 μL of 50 mM iodoacetamide buffer was added at room temperature for 15 min. Excess solution was removed and 200 μL of acetonitrile were added to dehydrate gel pieces for 5 min at room temperature until white coloration appeared. Excess acetonitrile was removed and the gel was further air dried until complete dryness (30 min, in general).
Trypsin reagent was prepared by adding 1 mL of ice-cold 50mM ammonium bicarbonate to 20 μg of trypsin (Promega) (final concentration 20 ng/ μL) and the solution was kept on ice. Then, 30 μL of trypsin solution was added to samples to rehydrate gel pieces for 10 min on ice while gently mixing. A total of 30 μL of 50 mM ammonium bicarbonate was added to gel pieces and trypsin digestion was carried out overnight at 37°C. Excess solution was removed and transferred into a new 1.5 mL tube.
30 μL of extraction buffer (50% acetonitrile, 1% tri-fluoro-acetic acid (TFA) in Milli-Q-H2O), was sequentially added to gel pieces for incubation for 10 minutes and transferred into the same tube. After a last incubation with extraction buffer, the samples were completely dried in a vacuum centrifuge and then resuspended in 30 μL of buffer A (98% Milli-Q-H2O, 2% acetonitrile, 0.1% TFA).
Mass spectrometry analysis
Nano LC–MS/MS
Protein digests were analyzed by a Q-TOF mass spectrometer (Impact II, Bruker Daltonik GmbH; Bremen, Germany) using a Captive Spray source, interfaced with a nano-HPLC RSLC System (Ultimate 3000 Thermo Scientific). Samples were concentrated on a pre-column (Thermo Scientific, C18 PepMap100, 2 cm × 100 μm ID, 5 μm particle size, 100 A) at a flow rate of 10 μL/min using 0.1% tri-fluoro acetic acid. After pre-concentration, peptides were separated on a Thermo Scientific C18 PepMap100 UHPLC column (50 cm x 75 μm ID, 2 μm particles, 100A) at a flow rate of 400 nL/ min using a 90 min linear gradient (buffer A: 2% ACN in 0.1% FA; buffer B 100% ACN in 0.1% FA). Acquisition is data dependent InstantExpertise™ mode (Bruker Daltonik GmbH; Bremen, Germany) for precursor selection based on fixed time (1 MS every 2 sec at 0.25 sec/scan) for MS and as many as possible MSMS in between (1.75 sec) at acquisition time from 0.0625s to 0.25 s/scan based on precursor intensity.
Protein identification with reference to databases
MS/MS raw data were processed using the Data Analysis software (Bruker Daltonik GmbH; Bremen, Germany) to generate the peak lists. The resulting mgf (Mascot Generic File Format) were submitted to Mascot server version 2.5 (MatrixScience–London, UK) through Proteinscape platform version 4.0 (Bruker Daltonics). Files were then searched against a home-built database (38685 entries) made of compiled Chlorocebus sabeus protein database from UniProtKb (19441 entries) and T. cruzi (strain CL Brener) protein databases from UniProtKb (19244 entries) using trypsin as hydrolysis enzyme. Mass tolerances were set at 10 ppm and 0.01 Da for precursor and product ions, respectively. Two missed cleavages per peptide were allowed considering both tryptic and semi-tryptic cleavages, while the search included fixed carbamidomethylation of Cysteine, and the following variable modifications: N-terminal acetylation, deamidation of asparagines and glutamines and, methionine oxidation. Searches were filtered using both a minimum peptide ion score of 20 and a false discovery rate (FDR) <1% calculated using Percolator.
Results
We opted to analyse all the proteins secreted from Vero cells infected with trypomastigotes of either one of two strains, CL Brener and VD, as a means to derive a list of proteins that could be potential biomarkers for a T. cruzi infection.
In our in vitro infection model, following the multiplication of the intracellular amastigotes and their differentiation into trypomastigotes, the parasites are released from the cells in the culture medium. The culture supernatant is defined in this study as the "secretome", and it includes: 1) soluble proteins from Vero cells, 2) parasite proteins secreted into Vero cells cytoplasm that are then released into the culture media, and 3) all the trypomastigote proteins that have been directly secreted into the culture supernatant.
Comparing the secretome of two T. cruzi strains
To increase the validity of the results we selected either proteins whose LC-MSMS score was over 35 and with at least two peptides, or proteins with a score over 50 but with only one peptide identified. In this way, we identified 441 parasites proteins (Additional File 1) from the secretome derived from Vero cells infected with the CL Brener strain, and, a total of 515 proteins (Additional File 2) from Vero cells infected with the VD strain. Since Vero lineage was isolated from kidney epithelial cells extracted from an African green monkey and this study focuses on parasite proteins, those derived from the Vero cells were excluded from further analyses.
A total of the 591 T. cruzi proteins were found in the secretomes of the 2 strains, 363 were common to both strains (additional file 3), while 78 were exclusive to CL Brener strain and 151 were detected only from the VD strain (see Fig 1).
Fig 1. Overlap between secretomes of two different T. cruzi strains DTU Tc VI.
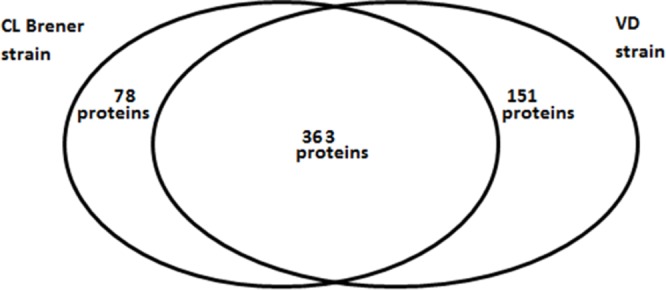
A total of 591 proteins of T. cruzi were identified. We note that 78 proteins are specific to the CL Brener strain whereas 151 proteins are specific to VD strain. However, 363 proteins are common to both strains.
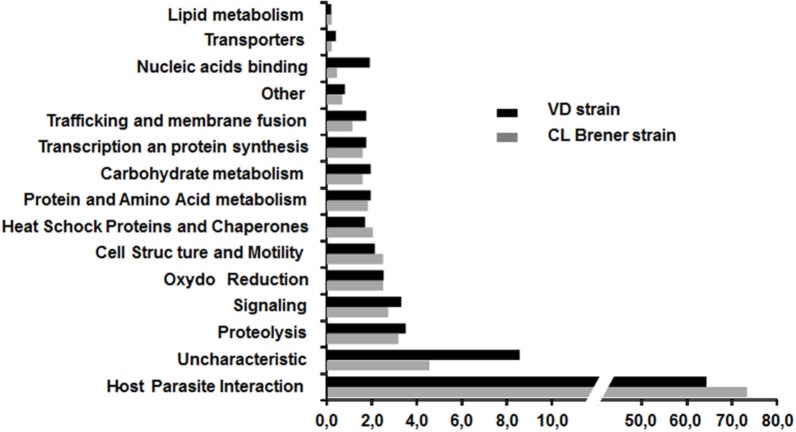
In order to ascertain the possible role of these secreted proteins, the 363 specific proteins were assigned to 15 functional groups based on information from the literature (Mainly the work of E. Bayer santos et al) and UniprotKB annotation (see Fig 2).
Fig 2. Functional categories classification of T. cruzi proteins from CL Brener and VD strains.
Proteins were classified into 15 functional categories using literature and UniprotKB annotation. Y-axis, categories are indicated. X-axis shows the percentage of each category for each strain.
For both strains, approximately 70% of the proteins belong to the host-parasite interaction category that plays a very important role in virulence. A significant difference is remarkable; the number of identified proteins in the secretome with unknown function for the VD strain (44 proteins) is slightly more twice that for the CL Brener strain (20 proteins). For the VD strain, this subset represents more than 8% of the total proteins identified. A similar difference was noticed for the proteins involved in nucleic acid binding, but with a much lower number of proteins (10 proteins versus 2 proteins, respectively). For the other categories, there was no significant difference between the two strains. The set of T. cruzi proteins identified for each of the strains is provided in Additional Files 1 and 2.
Multigenic superfamilies identified in the secretomes
Among the 591 proteins identified in both T. cruzi secretomes, 379 (64%) correspond to those encoded by different multigenic superfamilies. Trans-sialidase proteins (TcS) and trans-sialidase like proteins are the major families founded in the secretomes of the two strains (315 proteins for TcS, 18 proteins for DGF1, 15 proteins for Masps and 31 proteins for GP63 surface proteins). A total of 315 TcS proteins from the T. cruzi secretomes have been identified for the 2 strains. Among these proteins, 46 are fragments of TcS. We can remark that 184 complete TcS proteins were common to both strains. This implies that 42 proteins are exclusively found in CL Brener strain and 43 for VD strain.
Since, previous studies have shown the presence of 508 complete TcS genes that can be classified into 8 Groups [22], we analysed the expression profile of these TcS in the two secretomes and classified these TcS proteins into groups I to VIII, according to previously described classes. For a better description of the expression of different TcS groups, we derived the ratio between the number of TcS identified in our analysis, and that in each group (see Fig 3).
Fig 3. Analyse of trans-sialidase (TcS) proteins found in secretome of 2 strains.
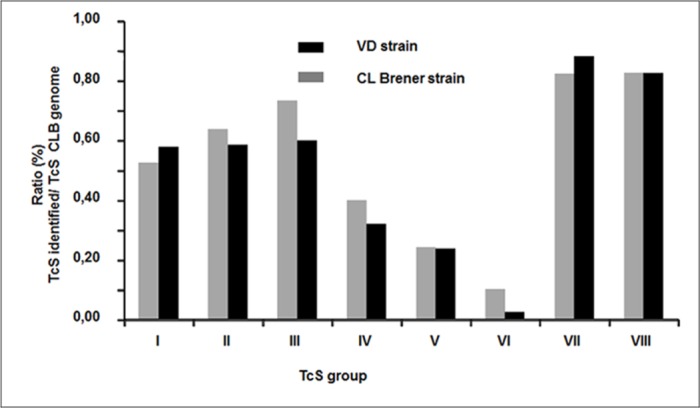
Classification of TcS proteins for each strain into 8 group previously described [22]. Groups IV, V and VI are less than I, II, III groups VII and VIII for two strains. On the x-axis, the number of group is indicated. The Y-axis shows the percentage of each TcS identified in our analyses.
Discussion
A reliable biomarker for the T. cruzi infection could be of different nature (protein, lipid, sugar or nucleid acid) but it must be specific to T. cruzi and ubiquitous in the different T. cruzi genotypes. The objective of this work was to obtain and compare the secretomes of two T. cruzi strains infecting Vero celIs, as a way to focus on protein biomarkers for Chagas disease.
For this study we chose to work with two T. cruzi strains of the same DTU (TcVI). The genetic intra-DTU variability in the parasite was poorly studied. The T. cruzi CL Brener genome (TcVI) was published in 2005 [21]. Other genomes, such as that of the Sylvio X10/1 strain and DM28c (TcI), have also been published [23,24]. The VD strain was isolated from a case of congenital Chagas disease and its genome is not sequenced. The comparison of two strains of TcVI genome but with different tropism increases the degree of selection of the most important secreted proteins and eliminated the strain-dependent proteins.
Secretome comparison
We have identified 515 proteins for VD strain compared to 441 proteins for the CL Brener strain. The result of the comparison shows a high proportion (more than 60%) of protein secreted common to both T. cruzi strains. It would seem that gene expression in both T. cruzi strain of the same DTU is relatively homogeneous. This high degree of homology makes possible to identify proteins secreted by all strains of T. cruzi and thus potential markers. This result is very different from that obtained by Geiger et al. who analyzed the secretome of T. brucei and found a small number of proteins common to the two strains of the same group (36 proteins common to the Feo strain and the strain OK out of a total of 270 identified proteins [25].
After functional classification of the parasite proteins, we noted that the proportion of identified proteins involved in proteolysis, signalling, oxydo-reduction, cell structure and motilities was quite different for the two strains. This result suggests that proteins involved in metabolic pathways of stress response, protein folding, proteolysis, and oxidoreductase activity, along with proteins that can interact with different signalling pathways, are important for T. cruzi’s capacity to infect its host cell, and that these are probably more conserved in all T. cruzi strains. Furthermore, proteins involved in DNA binding are also more numerous in the secretome of the VD strain as compared to that of the CL Brener strain. This class of proteins plays an essential role in the control of gene expression in T. cruzi, modulating RNA processing stability, turnover, and translation [26].
The VD strain secretome contained more proteins with unknown functions than the CL Brener strain. The VD strain was isolated from a pediatric patient with congenital infection. As previously noted from in vivo investigations [27,28], in our study the VD strain seemed more virulent than the CL Brener strain due the faster emergence from infected Vero cells. The hypothetical proteins of unknown function could be involved in virulence, and also be a source of biomarkers candidates for strain discrimination and for further studies to understand the underlying mechanisms of pathogenicity.
However, more than 70% of the proteins that have been identified in the secretome of the two strains belong to the host-parasite interaction category, and these play a very important role as virulence factors. In this class there are essentially four major multigenic proteins families. A remarkable feature of the T. cruzi genome is the massive expansion of these multigenes family that encodes polymorphic surface proteins such as trans-sialidase (TcS), mucin and mucin-associated surface proteins (MASPs), dispersed gene family 1 (DGF-1), and gp63 peptidases. The gene families coding for these proteins represent about 10% to 30% of the T. cruzi genome [29]. The high sequence variability of these different gene families could confer upon the parasite the ability to invade a significant number of cell types, as well as to have pleiotropic tissue tropisms, as described previously in in vitro, and in in vivo and clinical studies [30] where a wide variety of syndromes associated with T. cruzi infection were described.
The trans-sialidase and tran-sialidase like gene (TcS) super family represents the largest T. cruzi gene family, with more than 1,400 genes. However, only half of these genes are apparently functional [21]. The TcS superfamily is composed of glycosylphosphatidylinositol proteins anchored on T. cruzi surface, but they are also released to the extracellular space. The TcS gene family is highly polymorphic and only a few members have critical residues necessary for catalytic activity [31]. The very large number of different TcS proteins found in both secretomes is not unexpected. It has been shown that TcS expression is highly induced when cells become full of parasites and when amastigotes transform to trypomastigotes [32]. Thus, it would be possible to correlate the presence of large amounts of TcS into culture medium with cell host rupture. Finally, Affranchino JL et al. reported that Vero cells infected with trypomastigotes release vesicles carrying some protein members of the trans-sialidase multigene family [33]. However, our study provides a more detailed description (number and identification) of the TcS members expressed during the life cycle of T. cruzi (DTU TcVI). Recently, a comparative analysis of TcS sequences (508 complete genes) in the T. cruzi CL Brener strain genome led to the differentiation of eight different groups [22]. These authors further showed that the majority of TcS transcripts of all groups are present both in trypomastigotes and amastigotes forms. However, some transcripts are highly expressed in trypomastigote stage whereas other transcripts are expressed in amastigotes. It is interesting to note that Freitas LM and their colleagues found a very low level of expression for two genes from TcS group V in all the developmental stages [22]. The protein structure of the TcS groups V and VI are very similar. Trans-sialidases of these two groups have one signal peptide, a single asp box, a canonical VTV motif and a GPI anchor [22]. Our study appears to confirm these observations since we observed that more than half of the TcS proteins belonging to groups I, II and III, are expressed (19, 117 and 15 proteins, respectively). This percentage increases up to 80% for the VII and VIII groups (17 and 46 sequences, respectively). While for group IV the expression of TcS is less than 40%, and for groups V and VI less than 25% (227 and 39, respectively). This observation is noteworthy, because groups V and VI represent more than 60% of the trans-sialidases (TcS).
Finally, antibodies against two repeated epitopes, SAPA epitope (PVDSSAHG/STPST) and TcD epitope (PKPAE) of trans-sialidases were found in the sera of patients with Chagas' disease in acute and chronic phase [34]. Our sequence analysis shows trans-sialidases that possess the SAPA epitope (Group I) and the TcD epitope (Group IV) in the secretome of both analysed strains. There is a high homology in the TcS group expressed in both strains from same DTU (TcVI), we could suppose that some variations in their expression might be related to the genetic difference between strains. Numerous proteomic studies have revealed that the T. cruzi secretomes contain many proteins. The first secretome analysis has been made from ‘reservosome’ fraction belonging to Dm28c strain (TcI) epimastigotes [19], suggesting that there are several possible pathways for the secretion of proteins from T. cruzi. In another study [25] the authors identified T. brucei proteins from purified microvesicles extracted from infected rats sera.
In the latest and most comprehensive study [20], the authors identified two types of microvesicles released by epimastigotes and metacyclic forms of Dm28c T. cruzi strain. All these studies pointed to the use of microvesicles to deliver cargo of many proteins into host cells. The role of the proteins released from microvesicles is fundamental in Chagas disease. For example, Cestari et al. observed microvesicles release from infected cells into bloodstream that could allow immune evasion by inhibiting C3 convertase [35]. The protein content of the microvesicles was found to be ubiquitous, contributing to virulence, infectivity and immunity in many pathogens such as Trypanosoma spp. and other related parasites such as Leishmania spp. [36] and Plasmodium spp. [37], but also from bacteria [38] or virus [39].
Nevertheless, all these studies were carried out without the host cell context. In the present study, we analyzed the secretome of T. cruzi in the presence of host cells and particularly at the time when trypomastigotes are released from the cell.
In our model we have identified a set of parasite proteins that have been released from the host cells by secretion, by microvesicles, or directly from the cell cytoplasm to the culture medium. These proteins are linked to biochemical processes that enable the transformation of amastigotes in trypomastigotes, the multiplication of amastigotes, the release of trypomastigotes from host cells and finally invasion of new cells. In the culture medium, we had access to all the proteins secreted or excreted by the parasite.
Our data show that the majority of the proteins identified for both strains were found in the microvesicles of the epimastigote and the metacyclic forms of T.cruzi. However, we have also identified some proteins that were previously found only in microvesicles from epimastigotes, such as the 10 KDa heat shock protein [20]. This result suggests that there are slight differences in the secretome of trypomastigote and amastigote forms of T. cruzi. Although these parasitic proteins are not required for cell invasion (metacyclic form) they may have a role in survival within host cell cytoplasm.
Short list of parasite proteins as a potential marker of infection with T. cruzi
Although the protein superfamilies (TcS, GP63, MASP, and DGF1) are highly represented in the secretome of both strains, they cannot be used as biomarkers for T. cruzi infection. The sequence identity of the TcS is low and is often limited to common motifs (Asp-box, GPI anchor, VTV and FRIP box). These motifs include a maximum of ten amino acids. In addition it should be noted that these proteins have sequences that are fairly variable between the various T. cruzi strains. Pattern searches by the protein BLAST algorithm from National Center for Biotechnology Information (NCBI)show that the TcD epitope is repeated 430 times in the genome of T. cruzi strain CL Brener while it is identified only 5 times in strain DM28C. Similarly we did not find the SAPA epitope In the DM28C strain whereas in the strain CL Brener this pattern is repeated 80 times. So we propose a selected list of proteins (see Table 1) that could potentially serve as markers of an infection with T. cruzi. Indeed, these proteins are secreted, clearly produced abundantly, and are common to at least two strains of TcVI.
Table 1. List of secreted proteins that are common in both T.cruzi strains without of multigene proteins.
| Proteins | UNIPROT Accession number | MW (Kda) | ||
|---|---|---|---|---|
| 10 kDa heat shock protein, putative | Q4DFA8 | 10.7 | ||
| 40S ribosomal protein S15a, putative | Q4E0N6 | 14.7 | ||
| 40S ribosomal protein S18, putative | Q4E093 | 17.5 | ||
| Actin, putative | Q4D7A6 | 38.1 | ||
| Adenosylhomocysteinase | Q4D455 | 48.4 | ||
| ADP-ribosylation factor 1, putative | Q4D7Y8 | 20.7 | ||
| Alpha tubulin, putative | Q4CLA1 | 49.8 | ||
| Arginine kinase, putative | Q4CWA5 | 40.2 | ||
| ATP synthase subunit beta | Q4DTX7 | 55.7 | ||
| Beta tubulin, putative | Q4DQP2 | 49.7 | ||
| Calcium-binding protein, putative | Q4D1Q2 | 19.6 | ||
| Calmodulin, putative (Fragment) | Q4D2S5 | 9.5 | ||
| Calpain-like cysteine peptidase, putative | Q4D066 | 12.8 | ||
| Calreticulin, putative | Q4DDX3 | 46.2 | ||
| Chaperonin HSP60, mitochondrial | Q4DYP5 | 59.1 | ||
| Complement regulatory protein, putative | Q4DQ07 | 113.7 | ||
| Cysteine peptidase C | Q4DQB0 | 36.7 | ||
| Dynein light chain, putative | Q4E4N7 | 10.4 | ||
| Elongation factor 1-alpha (Fragment) | Q4CXI2 | 42.8 | ||
| Elongation factor 2, putative | Q4D3T1 | 94.1 | ||
| Enolase, putative | Q4DZ98 | 46.4 | ||
| Glucose-regulated protein 78, putative | Q4D620 | 71.3 | ||
| Glutamate dehydrogenase | Q4D5C2 | 45 | ||
| Glutathione peroxidase | Q4DEJ5 | 19.7 | ||
| Glyceraldehyde 3-phosphate dehydrogenase, putative | Q4D3Y9 | 14.7 | ||
| Glyceraldehyde 3-phosphate dehydrogenase, putative | Q4DHF0 | 39 | ||
| Heat shock 70 kDa protein, mitochondrial, putative | Q4CVR9 | 70.9 | ||
| Heat shock 70 kDa protein, putative (Fragment) | Q4CU95 | 40.8 | ||
| Heat shock 70 kDa protein, putative (Fragment) | Q4DAZ6 | 30.1 | ||
| Heat shock protein 70 (HSP70), putative | Q4DTM9 | 70.9 | ||
| Heat shock protein 85, putative | Q4CQS6 | 80.7 | ||
| IgE-dependent histamine-releasing factor, putative | Q4CW52 | 19.6 | ||
| Isocitrate dehydrogenase [NADP] | Q4E4L7 | 46.8 | ||
| Lysosomal alpha-mannosidase, putative | Q4DXL4 | 111.2 | ||
| Malate dehydrogenase (Fragment) | Q4D4A0 | 31.5 | ||
| Microtubule-associated protein, putative (Fragment) | Q4CMT2 | 85.2 | ||
| NAD/FAD dependent dehydrogenase, putative | Q4CVH0 | 43 | ||
| Neutral sphingomyelinase activation associated protein | Q4DSD4 | 90.5 | ||
| Nucleoside diphosphate kinase | Q4E256 | 16.9 | ||
| Peptidyl-prolyl cis-trans isomerase | Q4E4L9 | 18.8 | ||
| Peptidyl-prolyl cis-trans isomerase | Q4DPB9 | 21.9 | ||
| Phosphoglycerate kinase | Q4D193 | 44.4 | ||
| Proteasome regulatory ATPase subunit 1, putative | Q4D9J1 | 48.5 | ||
| Proteasome regulatory ATPase subunit 2, putative | Q4D0B9 | 49 | ||
| Rab7 GTP binding protein, putative | Q4E4T4 | 23.9 | ||
| Ras-related protein rab-2a, putative (Fragment) | Q4DM40 | 10.5 | ||
| Ras-related protein rab-5, putative (Fragment) | Q4D504 | 20.4 | ||
| S-adenosylmethionine synthase | Q4CSC4 | 43.5 | ||
| Serine carboxypeptidase S28, putative | Q4DM56 | 72.1 | ||
| Serine/threonine-protein phosphatase | Q4D9Y4 | 35 | ||
| Seryl-tRNA synthetase, putative (Fragment) | Q4CW46 | 25.7 | ||
| Small GTP-binding protein Rab1, putative | Q4CZR0 | 22.8 | ||
| Superoxide dismutase | Q4D5A6 | 21.9 | ||
| Transitional endoplasmic reticulum ATPase, putative | Q4DWB5 | 86.1 | ||
| tRNA synthetase, putative (Fragment) | Q4E397 | 78.8 | ||
| Tryparedoxin peroxidase, putative | Q4CM56 | 22.4 | ||
| Tryparedoxin peroxidase, putative | Q4CX87 | 25.5 | ||
| Ubiquitin-activating enzyme E1, putative | Q4DYM1 | 114.3 | ||
| Ubiquitin-conjugating enzyme E2, putative | Q4CTN0 | 17.5 | ||
| Cofilin/actin depolymerizing factor, putative | Q4D8D3 | Q4CVE9 | 15.7 | 15.7 |
| 14-3-3 protein, putative | Q4DJB6 | Q4DRH6 | 29.9 | 29.9 |
| Cysteine peptidase inhibitor | Q4DH32 | Q4DY71 | 12 | 12.1 |
| Cysteine peptidase, putative | Q4DW02 | Q4E0J7 | 49,9 | 49.8 |
| Cytochrome c, putative | Q4D480 | Q4CV48 | 12.2 | 12.2 |
| Fructose-bisphosphate aldolase | Q4D0Q0 | Q4D4R9 | 40.8 | 40.8 |
| Serine carboxypeptidase (CBP1), putative | Q4CMQ4 | Q4DTP7 | 59,5 | 59.5 |
| Uncharacterized protein | Q4D3H5 | Q4DNJ6 | 16.7 | 16.8 |
| Uncharacterized protein | Q4CNH1 | 66 | ||
| Uncharacterized protein | Q4CPM9 | 112.1 | ||
| Uncharacterized protein | Q4CPX4 | 23.1 | ||
| Uncharacterized protein | Q4CUB2 | 36.5 | ||
| Uncharacterized protein | Q4CUQ4 | 23 | ||
| Uncharacterized protein | Q4CVJ1 | 16.4 | ||
| Uncharacterized protein | Q4CW22 | 13 | ||
| Uncharacterized protein | Q4CW23 | 13.5 | ||
| Uncharacterized protein | Q4D1D9 | 323 | ||
| Uncharacterized protein | Q4D6D8 | 21.1 | ||
| Uncharacterized protein | Q4DPV6 | 24.4 | ||
| Uncharacterized protein | Q4DT54 | 23.6 | ||
| Uncharacterized protein | Q4DUX0 | 22.3 | ||
| Uncharacterized protein (Fragment) | Q4CQX1 | 41.3 | ||
| Uncharacterized protein (Fragment) | Q4CTF0 | 21.3 | ||
| Uncharacterized protein (Fragment) | Q4DHN4 | 38.1 | ||
The list includes 94 proteins of which 8 are represented by two isoforms and 18 have no known function. These various proteins might also be future therapeutic targets and/or markers of Chagas disease. The protein Q4CVJ1 was not identified in the various secretomes published for T.cruzi, but it has been found in high-throughput screening and is highly expressed and immunogenic; moreover, 68% of sera from patients with Chagas disease recognize this antigen [40]. Finally, the proteins with unknown function, Q4CTF0 and Q4CUB2, have been described as part of the last multigenic family (about 40 members in Cl Brener) subdivided into 3 subfamilies TcTASV-A, B and C. Q4CTF0 is part of the subgroup family TcTASV-A while Q4CUB2 is part of the TcTASV-C family. This subfamily (8 proteins) was identified on the surface of trypomastigotes of T. cruzi and in (strain CL Brener, Sylvio and RA). About 30% of human sera infected by T. cruzi reacted with TcTASV-C [41].
We note that for some proteins in this list, Microtubule-associated protein, Heat shock proteins (HSP70), Complement regulatory protein and Flagellar Calcium-binding protein, specific antibodies can be found in the acute and chronic phase sera of patients with Chagas' disease [38]–[42].
The EMBRARIO laboratory (Brazil) offers a confirmatory test (HBK 740 Imunoblot Linhas) based on a multi-epitope recombinant peptide, including the TC-24 epitope (Flagellar calcium-binding protein) and the MAP repeated epitope (Microtubule-associated protein) [43,44].
Some proteins have already been identified as potential markers in other infections. For example, the fructose biphosphate aldolase enzyme, a key enzyme in the glycolytic pathway, was found in plasma of patients infected with P. falciparum [45]).
The abundance of the parasite proteins and their relative stability in serum are two criteria to help in the identification of suitable markers for T. cruzi infection. In our study the strategy adopted to obtain the T. cruzi secretome may eliminate the most labile proteins since the presence of proteases in the culture supernatant is likely to degrade many of the secreted proteins.
Conclusion
The development and availability of a simple, inexpensive, easy to handle, sensitive and specific method to effectively demonstrate the presence of T. cruzi from a blood sample is a major challenge for the diagnosis of Chagas disease. Identification of one or more proteins (biomarkers) in the patient's serum will be clear evidence for the presence of the parasite.
This study provides the first comparative overview of the secretomes of cells infected by the T. cruzi CL Brenner and the VD strains. Not surprisingly, the majority of excreted proteins belong to the four multigenic gene families (TcS, MASPs, DGF1, and gp63 surface protein), that unfortunately cannot be used as infection marker because of their very high, inter-species, degree of variability.
Some of the proteins identified had not been studied previously, particularly from the VD strain, and some may be new potential therapeutic targets. We have thus established a list of 94 secreted proteins, common to both strains and that do not belong to members of multigene families. These proteins are secreted in large quantities and are relatively stable in presence of many proteases. These conditions are necessary to identify good serum markers. This descriptive study provides the first step toward the identification of potential diagnostic markers in the sera of T. cruzi-infected persons.
Supporting information
The proteins secreted have selected with LCMSMS score was over 35 and with at least two peptides identified, or with a score over 50 but with only one peptide identified. For each protein, the number of matched proteins and peptides and the highest score are described.
(PDF)
The proteins secreted have selected with LCMSMS score was over 35 and with at least two peptides identified, or with a score over 50 but with only one peptide identified. For each protein, the number of matched proteins and peptides and the highest score are described.
(PDF)
For each protein, the number of matched proteins and peptides and the highest score for CL Brener (white box) and VD (gray box) strain are described.
(PDF)
Acknowledgments
We would to like to thanks Georges Snounou and Angelita Rebollo for fruitful scientific discussion.
We thank Dr. María Elisa Solana (Facultad de Medicina, Instituto de Investigaciones en Microbiología y Parasitología Médica (IMPAM),UBA-CONICET, Universidad de Buenos Aires) to maintain of the VD strain of T. cruzi".
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The Bruker Society provided support in the form of salary for M. Chapelle, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. The specific role of M. Chapelle is articulated in the ‘author contributions’ section. The study was also supported by Ecos-Sud (Grant number: A13S03; URL: www.univ-paris13.fr/cofecub-ecos/ecos-sud). Ecos-Sud had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO | 6 February 2015, vol. 90, 6 (pp. 33–44) [Internet]. WHO. [cited 2017 Jan 4]. Available from: http://www.who.int/wer/2015/wer9006/en/
- 2.Steverding D. The history of Chagas disease. Parasit Vectors. 2014. July 10;7:317 doi: 10.1186/1756-3305-7-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Messenger LA, Miles MA. Evidence and importance of genetic exchange among field populations of Trypanosoma cruzi. Acta Trop. 2015. November;151:150–5. doi: 10.1016/j.actatropica.2015.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tibayrenc M, Ayala FJ. The population genetics of Trypanosoma cruzi revisited in the light of the predominant clonal evolution model. Acta Trop. 2015. November;151:156–65. doi: 10.1016/j.actatropica.2015.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zingales B, Andrade SG, Briones MRS, Campbell DA, Chiari E, Fernandes O, et al. A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz. 2009. November;104(7):1051–4. [DOI] [PubMed] [Google Scholar]
- 6.Zingales B, Miles MA, Campbell DA, Tibayrenc M, Macedo AM, Teixeira MMG, et al. The revised Trypanosoma cruzi subspecific nomenclature: rationale, epidemiological relevance and research applications. Infect Genet Evol J Mol Epidemiol Evol Genet Infect Dis. 2012. March;12(2):240–53. [DOI] [PubMed] [Google Scholar]
- 7.Martin DL, Marks M, Galdos-Cardenas G, Gilman RH, Goodhew B, Ferrufino L, et al. Regional variation in the correlation of antibody and T-cell responses to Trypanosoma cruzi. Am J Trop Med Hyg. 2014. June;90(6):1074–81. doi: 10.4269/ajtmh.13-0391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gascon J, Bern C, Pinazo M-J. Chagas disease in Spain, the United States and other non-endemic countries. Acta Trop. 2010. August;115(1–2):22–7. doi: 10.1016/j.actatropica.2009.07.019 [DOI] [PubMed] [Google Scholar]
- 9.Segovia M, Carrasco HJ, Martínez CE, Messenger LA, Nessi A, Londoño JC, et al. Molecular epidemiologic source tracking of orally transmitted Chagas disease, Venezuela. Emerg Infect Dis. 2013. July;19(7):1098–101. doi: 10.3201/eid1907.121576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlier Y, Torrico F, Sosa-Estani S, Russomando G, Luquetti A, Freilij H, et al. Congenital Chagas disease: recommendations for diagnosis, treatment and control of newborns, siblings and pregnant women. PLoS Negl Trop Dis. 2011. October;5(10):e1250 doi: 10.1371/journal.pntd.0001250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freilij H, Altcheh J. Congenital Chagas’ disease: diagnostic and clinical aspects. Clin Infect Dis Off Publ Infect Dis Soc Am. 1995. September;21(3):551–5. [DOI] [PubMed] [Google Scholar]
- 12.Chagas’ Disease—NEJM [Internet]. [cited 2017 Apr 28]. Available from: http://www.nejm.org/doi/full/10.1056/NEJMra1410150
- 13.Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001. March;69(3):89–95. doi: 10.1067/mcp.2001.113989 [DOI] [PubMed] [Google Scholar]
- 14.Hulka BS. ASPO Distinguished Achievement Award Lecture. Epidemiological studies using biological markers: issues for epidemiologists. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 1991. December;1(1):13–9. [PubMed] [Google Scholar]
- 15.McNerney R, Maeurer M, Abubakar I, Marais B, McHugh TD, Ford N, et al. Tuberculosis diagnostics and biomarkers: needs, challenges, recent advances, and opportunities. J Infect Dis. 2012. May 15;205 Suppl 2:S147–158. [DOI] [PubMed] [Google Scholar]
- 16.Jain P, Chakma B, Patra S, Goswami P. Potential Biomarkers and Their Applications for Rapid and Reliable Detection of Malaria. BioMed Res Int [Internet]. 2014. April 2 [cited 2017 Apr 26];2014. Available from: https://www.hindawi.com/journals/bmri/2014/852645/abs/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Araujo FF, Nagarkatti R, Gupta C, Marino AP, Debrabant A. Aptamer-based detection of disease biomarkers in mouse models for chagas drug discovery. PLoS Negl Trop Dis. 2015. January;9(1):e3451 doi: 10.1371/journal.pntd.0003451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umezawa ES, Nascimento MS, Stolf AM. Enzyme-linked immunosorbent assay with Trypanosoma cruzi excreted-secreted antigens (TESA-ELISA) for serodiagnosis of acute and chronic Chagas’ disease. Diagn Microbiol Infect Dis. 2001. March;39(3):169–76. [DOI] [PubMed] [Google Scholar]
- 19.Sant’Anna C, Nakayasu ES, Pereira MG, Lourenço D, de Souza W, Almeida IC, et al. Subcellular proteomics of Trypanosoma cruzi reservosomes. Proteomics. 2009. April;9(7):1782–94. doi: 10.1002/pmic.200800730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayer-Santos E, Aguilar-Bonavides C, Rodrigues SP, Cordero EM, Marques AF, Varela-Ramirez A, et al. Proteomic analysis of Trypanosoma cruzi secretome: characterization of two populations of extracellular vesicles and soluble proteins. J Proteome Res. 2013. February 1;12(2):883–97. doi: 10.1021/pr300947g [DOI] [PubMed] [Google Scholar]
- 21.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran A-N, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005. July 15;309(5733):409–15. doi: 10.1126/science.1112631 [DOI] [PubMed] [Google Scholar]
- 22.Freitas LM, Santos SL dos, Rodrigues-Luiz GF, Mendes TAO, Rodrigues TS, Gazzinelli RT, et al. Genomic Analyses, Gene Expression and Antigenic Profile of the Trans-Sialidase Superfamily of Trypanosoma cruzi Reveal an Undetected Level of Complexity. PLOS ONE. 2011. October 19;6(10):e25914 doi: 10.1371/journal.pone.0025914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Franzén O, Ochaya S, Sherwood E, Lewis MD, Llewellyn MS, Miles MA, et al. Shotgun sequencing analysis of Trypanosoma cruzi I Sylvio X10/1 and comparison with T. cruzi VI CL Brener. PLoS Negl Trop Dis. 2011. March 8;5(3):e984 doi: 10.1371/journal.pntd.0000984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grisard EC, Teixeira SMR, de Almeida LGP, Stoco PH, Gerber AL, Talavera-López C, et al. Trypanosoma cruzi Clone Dm28c Draft Genome Sequence. Genome Announc. 2014. January 30;2(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiger A, Hirtz C, Bécue T, Bellard E, Centeno D, Gargani D, et al. Exocytosis and protein secretion in Trypanosoma. BMC Microbiol. 2010. January 26;10:20 doi: 10.1186/1471-2180-10-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.De Gaudenzi JG, Noé G, Campo VA, Frasch AC, Cassola A. Gene expression regulation in trypanosomatids. Essays Biochem. 2011;51:31–46. doi: 10.1042/bse0510031 [DOI] [PubMed] [Google Scholar]
- 27.Gulin JEN, Eagleson MA, Postan M, Cutrullis RA, Freilij H, Bournissen FG, et al. Efficacy of voriconazole in a murine model of acute Trypanosoma cruzi infection. J Antimicrob Chemother. 2013. April;68(4):888–94. doi: 10.1093/jac/dks478 [DOI] [PubMed] [Google Scholar]
- 28.Risso MG, Garbarino GB, Mocetti E, Campetella O, Gonzalez Cappa SM, Buscaglia CA, et al. Differential expression of a virulence factor, the trans-sialidase, by the main Trypanosoma cruzi phylogenetic lineages. J Infect Dis. 2004. June 15;189(12):2250–9. doi: 10.1086/420831 [DOI] [PubMed] [Google Scholar]
- 29.De Pablos LM, Osuna A. Multigene families in Trypanosoma cruzi and their role in infectivity. Infect Immun. 2012. July;80(7):2258–64. doi: 10.1128/IAI.06225-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tonelli RR, Giordano RJ, Barbu EM, Torrecilhas AC, Kobayashi GS, Langley RR, et al. Role of the gp85/trans-sialidases in Trypanosoma cruzi tissue tropism: preferential binding of a conserved peptide motif to the vasculature in vivo. PLoS Negl Trop Dis. 2010. November 2;4(11):e864 doi: 10.1371/journal.pntd.0000864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schenkman S, Eichinger D, Pereira ME, Nussenzweig V. Structural and functional properties of Trypanosoma trans-sialidase. Annu Rev Microbiol. 1994;48:499–523. doi: 10.1146/annurev.mi.48.100194.002435 [DOI] [PubMed] [Google Scholar]
- 32.Frevert U, Schenkman S, Nussenzweig V. Stage-specific expression and intracellular shedding of the cell surface trans-sialidase of Trypanosoma cruzi. Infect Immun. 1992. June;60(6):2349–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Affranchino JL, Ibañez CF, Luquetti AO, Rassi A, Reyes MB, Macina RA, et al. Identification of a Trypanosoma cruzi antigen that is shed during the acute phase of Chagas’ disease. Mol Biochem Parasitol. 1989. May 15;34(3):221–8. [DOI] [PubMed] [Google Scholar]
- 34.Frasch AC, Cazzulo JJ, Aslund L, Pettersson U. Comparison of genes encoding Trypanosoma cruzi antigens. Parasitol Today Pers Ed. 1991. June;7(6):148–51. [DOI] [PubMed] [Google Scholar]
- 35.Cestari I, Ansa-Addo E, Deolindo P, Inal JM, Ramirez MI. Trypanosoma cruzi immune evasion mediated by host cell-derived microvesicles. J Immunol Baltim Md 1950. 2012. February 15;188(4):1942–52. [DOI] [PubMed] [Google Scholar]
- 36.Silverman JM, Chan SK, Robinson DP, Dwyer DM, Nandan D, Foster LJ, et al. Proteomic analysis of the secretome of Leishmania donovani. Genome Biol. 2008;9(2):R35 doi: 10.1186/gb-2008-9-2-r35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mantel P-Y, Hoang AN, Goldowitz I, Potashnikova D, Hamza B, Vorobjev I, et al. Malaria-infected erythrocyte-derived microvesicles mediate cellular communication within the parasite population and with the host immune system. Cell Host Microbe. 2013. May 15;13(5):521–34. doi: 10.1016/j.chom.2013.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ståhl A, Arvidsson I, Johansson KE, Chromek M, Rebetz J, Loos S, et al. A novel mechanism of bacterial toxin transfer within host blood cell-derived microvesicles. PLoS Pathog. 2015. February;11(2):e1004619 doi: 10.1371/journal.ppat.1004619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mercier SK, Donaghy H, Botting RA, Turville SG, Harman AN, Nasr N, et al. The microvesicle component of HIV-1 inocula modulates dendritic cell infection and maturation and enhances adhesion to and activation of T lymphocytes. PLoS Pathog. 2013. October;9(10):e1003700 doi: 10.1371/journal.ppat.1003700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooley G, Etheridge RD, Boehlke C, Bundy B, Weatherly DB, Minning T, et al. High throughput selection of effective serodiagnostics for Trypanosoma cruzi infection. PLoS Negl Trop Dis. 2008. October 8;2(10):e316 doi: 10.1371/journal.pntd.0000316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bernabó G, Levy G, Ziliani M, Caeiro LD, Sánchez DO, Tekiel V. TcTASV-C, a Protein Family in Trypanosoma cruzi that Is Predominantly Trypomastigote-Stage Specific and Secreted to the Medium. PLOS ONE. 2013. Juil;8(7):e71192 doi: 10.1371/journal.pone.0071192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meira WSF, Galvão LMC, Gontijo ED, Machado-Coelho GLL, Norris KA, Chiari E. Use of the Trypanosoma cruzi recombinant complement regulatory protein to evaluate therapeutic efficacy following treatment of chronic chagasic patients. J Clin Microbiol. 2004. February;42(2):707–12. doi: 10.1128/JCM.42.2.707-712.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krautz GM, Galvão LM, Cançado JR, Guevara-Espinoza A, Ouaissi A, Krettli AU. Use of a 24-kilodalton Trypanosoma cruzi recombinant protein to monitor cure of human Chagas’ disease. J Clin Microbiol. 1995. August;33(8):2086–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernández-Villegas A, Pinazo MJ, Marañón C, Thomas MC, Posada E, Carrilero B, et al. Short-term follow-up of chagasic patients after benzonidazole treatment using multiple serological markers. BMC Infect Dis. 2011. July 31;11:206 doi: 10.1186/1471-2334-11-206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thézénas ML, Huang H, Njie M, Ramaprasad A, Nwakanma DC, Fischer R, et al. PfHPRT: a new biomarker candidate of acute Plasmodium falciparum infection. J Proteome Res. 2013. March 1;12(3):1211–22. doi: 10.1021/pr300858g [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The proteins secreted have selected with LCMSMS score was over 35 and with at least two peptides identified, or with a score over 50 but with only one peptide identified. For each protein, the number of matched proteins and peptides and the highest score are described.
(PDF)
The proteins secreted have selected with LCMSMS score was over 35 and with at least two peptides identified, or with a score over 50 but with only one peptide identified. For each protein, the number of matched proteins and peptides and the highest score are described.
(PDF)
For each protein, the number of matched proteins and peptides and the highest score for CL Brener (white box) and VD (gray box) strain are described.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.