Perilipins are abundant proteins on the surface of mammalian cytoplasmic lipid droplets that protect the organelle from adventitious lipolysis. Gao et al. report the discovery of a perilipin in yeast and show that it is important for droplet stability and biogenesis.
Abstract
Pet10p is a yeast lipid droplet protein of unknown function. We show that it binds specifically to and is stabilized by droplets containing triacylglycerol (TG). Droplets isolated from cells with a PET10 deletion strongly aggregate, appear fragile, and fuse in vivo when cells are cultured in oleic acid. Pet10p binds early to nascent droplets, and their rate of appearance is decreased in pet10Δ. Moreover, Pet10p functionally interacts with the endoplasmic reticulum droplet assembly factors seipin and Fit2 to maintain proper droplet morphology. The activity of Dga1p, a diacylglycerol acyltransferase, and TG accumulation were both 30–35% lower in the absence of Pet10p. Pet10p contains a PAT domain, a defining property of perilipins, which was not previously known to exist in yeast. We propose that the core functions of Pet10p and other perilipins extend beyond protection from lipases and include the preservation of droplet integrity as well as collaboration with seipin and Fit2 in droplet assembly and maintenance.
Introduction
Cytoplasmic lipid droplets are nearly ubiquitous intracellular structures that store fat for combustion on site or in distant tissues. Droplets, which bud from the ER and consist of a core of neutral lipids, typically triacylglycerol (TG) and steryl esters (SEs), are surrounded by a phospholipid monolayer and associated proteins.
The formation of droplets is not spontaneous but is facilitated by ER proteins and specific lipids. Seipin, a protein that resides at ER–droplet junctions, functions early to stabilize the nascent structures, ensures droplets of correct morphology, prevents the accumulation of phosphatidic acid at the ER–droplet junction, and prevents ectopic budding into the nucleus (Cartwright et al., 2015; Grippa et al., 2015; Wolinski et al., 2015; Wang et al., 2016). Fit2, fat storage–inducing transmembrane protein 2, binds to TG and ensures that the expansion of the neutral lipid core in nascent droplets is toward the cytosolic rather than the luminal surface (Gross et al., 2011; Choudhary et al., 2015). Proteins that bind to budding droplets from the cytosolic surface may also facilitate droplet formation: Plin3 and Plin4 bind early to droplets, suggesting that they may function early in droplet formation (Wolins et al., 2003; Skinner et al., 2009). Indeed, droplets fail to mature in the absence of Plin3 (Bulankina et al., 2009). In addition to these proteins, DAG has also been shown to be important for droplet budding (Skinner et al., 2009; Adeyo et al., 2011).
Plin3 and Plin4 are members of the perilipin class of abundant droplet proteins in animal cells (Bickel et al., 2009; Kimmel and Sztalryd, 2016). Perilipins are best known as protectors of droplets from adventitious lipolysis. Other functions specific to members of the class include regulation of lipases and interaction with mitochondria. The class generally shares three domains with varying conservation: the signature PAT domain, an 11-mer amphipathic repeat segment that is important for targeting (Rowe et al., 2016), and a more divergent four-helix bundle with varying effector functions (Subramanian et al., 2004; Granneman et al., 2011). Perilipins are found in insects, in Dictyostelium, and in a pathogenic fungus (Lu et al., 2001; Wang and St. Leger, 2007); they are believed not to exist in yeast.
The research described herein began with an analysis of proteins associated with droplets enriched in one of the core lipids, TG or SE. We report that Pet10p, a known droplet surface protein with unknown function (Athenstaedt et al., 1999), preferentially localizes to TG droplets, where it is protected from degradation. Pet10p is required for droplet integrity; in its absence, droplet structure is less stable, exhibiting severe aggregation in vitro. In addition, loss of the protein results in droplet fusion in vivo when cells are cultured with oleic acid (OA). PET10-null cells accumulate less TG, which for the acyltransferase Dga1p is accompanied by a loss of enzyme activity and movement from the ER to droplets. Pet10p localizes to droplets early in their assembly and promotes droplet formation. We identify a PAT domain in Pet10p and consider it a yeast perilipin, and we propose that the activities of Pet10p reflect important functions of the perilipin class.
Results
Pet10p binds to TG but not to SE droplets
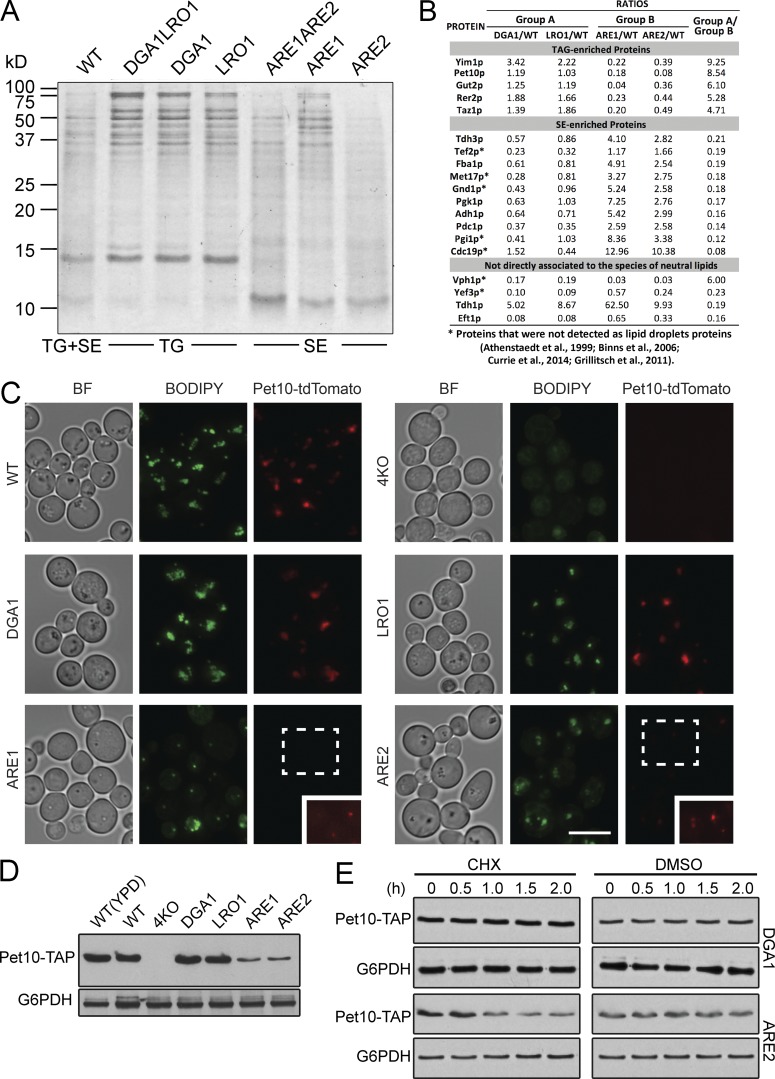
Two neutral lipids predominate in the cores of droplets from the yeast Saccharomyces cerevisiae: TGs, the product of Dga1p and Lro1p, and SEs, the product of Are1p and Are2p (Czabany et al., 2007). A previous study indicated that the protein components of droplets, assessed from a stained SDS gel, differed between droplets composed of TG and those composed of SE (Czabany et al., 2008). Consistent with this, specific mouse perilipins prefer to associate with TG or SE droplets (Hsieh et al., 2012). To further explore differences in protein composition between these droplets, we isolated droplets from triple knockout (3KO) cells expressing only one of the four acyltransferases (the strains are hereafter called by the remaining enzyme, such as DGA1) or double knockout (2KO) cells that lacked either the genes for the two DAG acyltransferases or the two steryl acyltransferases. As expected, the protein patterns of isolated droplets differed strikingly from strains producing TG droplets versus SE droplets on an SDS gel (Fig. 1 A). Proteomes from TG droplets were similar, as were those from SE droplets (Table S1). The major differences between the TG and SE droplet proteomes are listed in Fig. 1 B. We decided to focus on Pet10p, a protein of unknown function consistently observed in droplet proteomes (Athenstaedt et al., 1999; Binns et al., 2006; Grillitsch et al., 2011; Currie et al., 2014) that was greatly enriched on TG droplets. Pet10p had the highest spectral index (Griffin et al., 2010) among proteins associating with TG droplets (Table S1). Indeed, the abundant ∼14-kD protein of TG droplets (Fig. 1 A) is a fragment of Pet10p.
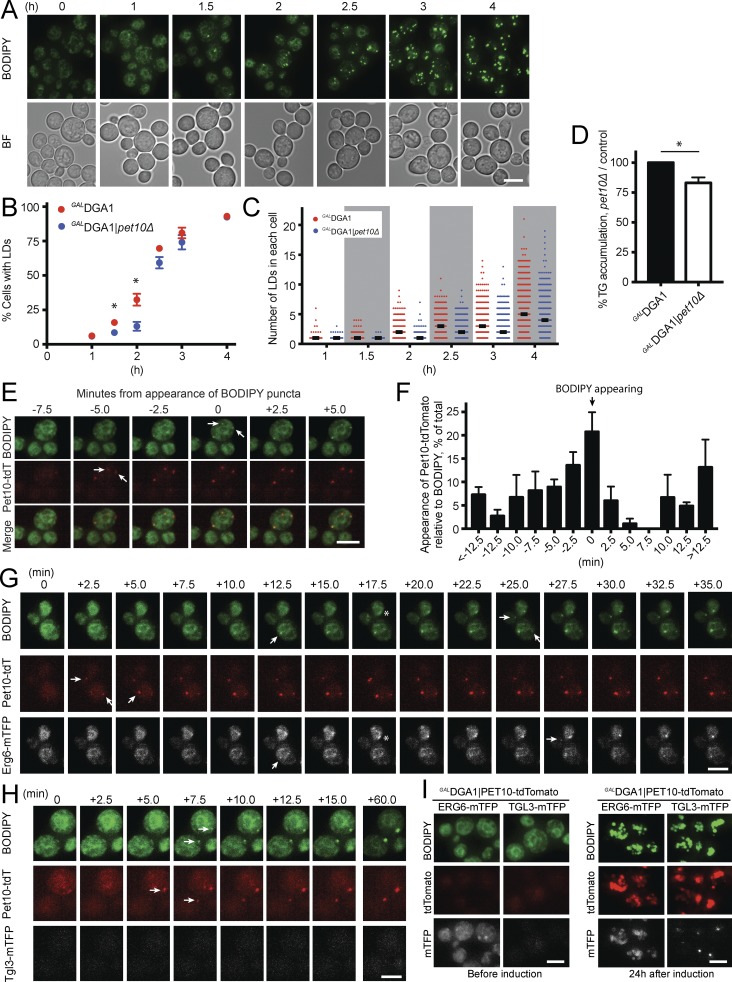
Figure 1.
Pet10p is an abundant protein specific to droplets containing TG. (A) Lipid droplets were purified from the indicated strains, subjected to SDS-PAGE (2 µg of protein per lane), and stained with Coomassie blue. The neutral lipid contained in the droplets is indicated below. The protein pattern is representative of three independent experiments. (B) Analysis of proteomics from TG and SE droplets (all from 3KO strains). The highest 100 hits overall in terms of spectral counts were considered, and their spectral indices were compared with that of the wild type. The last column expresses the ratio of signal from DGA1 and LRO1 droplets, termed group A, to that of ARE1 and ARE2 droplets, termed group B. Shown are those hits where the group A/group B ratios were >4 or <0.25. (B, bottom) Four hits where the spectral indices in all four 3KO strains were either >4 or <0.25 compared with the wild type. Asterisks indicate genes encoding proteins not previously identified in droplet proteomic studies (Athenstaedt et al., 1999; Binns et al., 2006; Grillitsch et al., 2011; Currie et al., 2014). (C) Cells from the indicated strains expressing chromosomal Pet10-tdTomato were stained with BODIPY to visualize droplets. (C, inset) The indicated area was enhanced in Photoshop to allow visualization of puncta. (D) Whole-cell extracts of the indicated strains were immunoblotted with anti-His (reacts with TAP tag) or anti-G6PDH antibody. (E) DGA1 or ARE2 cells expressing Pet10-TAP were subjected to CHX or DMSO vehicle for the indicated hours. Whole-cell extracts were subjected to SDS-PAGE and immunoblotting. For the ARE2 samples, the amount loaded on the gel for the anti-His blot was increased five times, and development time was increased 30 times to approximate the signal of the t0 point of the DGA1 sample. BF, brightfield. Bar, 5 µm.
PET10 was named in a meta-analysis of total yeast protein interactions in which it shared a subset of binding proteins with the product of PET9, the mitochondrial ATP transporter (Samanta and Liang, 2003). However, unlike other “petite” mutants (Ephrussi et al., 1949), the pet10Δ strain grew as wild type on nonfermentable carbon sources (Fig. S1 A), showing no defect in mitochondrial respiration.
To confirm the preference of Pet10p for TG droplets, we tagged Pet10p in the genome and monitored its behavior in these strains. In wild-type, DGA1, or LRO1 cells, Pet10-tdTomato unambiguously localized to lipid droplets as expected (Fig. 1 C). Pet10-tdTomato was difficult to detect in ARE1 and ARE2 cells unless exposure time was increased severalfold (Fig. 1 C, insets). In quadruple knockout cells (the strain cells lacking all four acyltransferases), Pet10-tdTomato could not be detected at all. Immunoblots of whole-cell extracts using Pet10-TAP (His6-biotinylation site-His6 [HBH] tag; Tagwerker et al., 2006) confirmed the expression of Pet10-tdTomato in the various strain backgrounds (Fig. 1 D).
To determine if Pet10p is degraded in the absence of TG droplets, we treated DGA1 and ARE2 cells expressing Pet10-TAP with the protein synthesis inhibitor cycloheximide (CHX). Pet10-TAP was stable over 2 h in the DGA1 strain (Fig. 1 E). In contrast, much of Pet10-TAP was degraded in the ARE2 strain. These results suggest that Pet10p is stabilized by TG; otherwise, the majority is degraded. Overall, our data indicate that the neutral lipid core in droplets likely plays a role in the localization of Pet10p to that organelle.
Lower TG synthesis in the absence of PET10
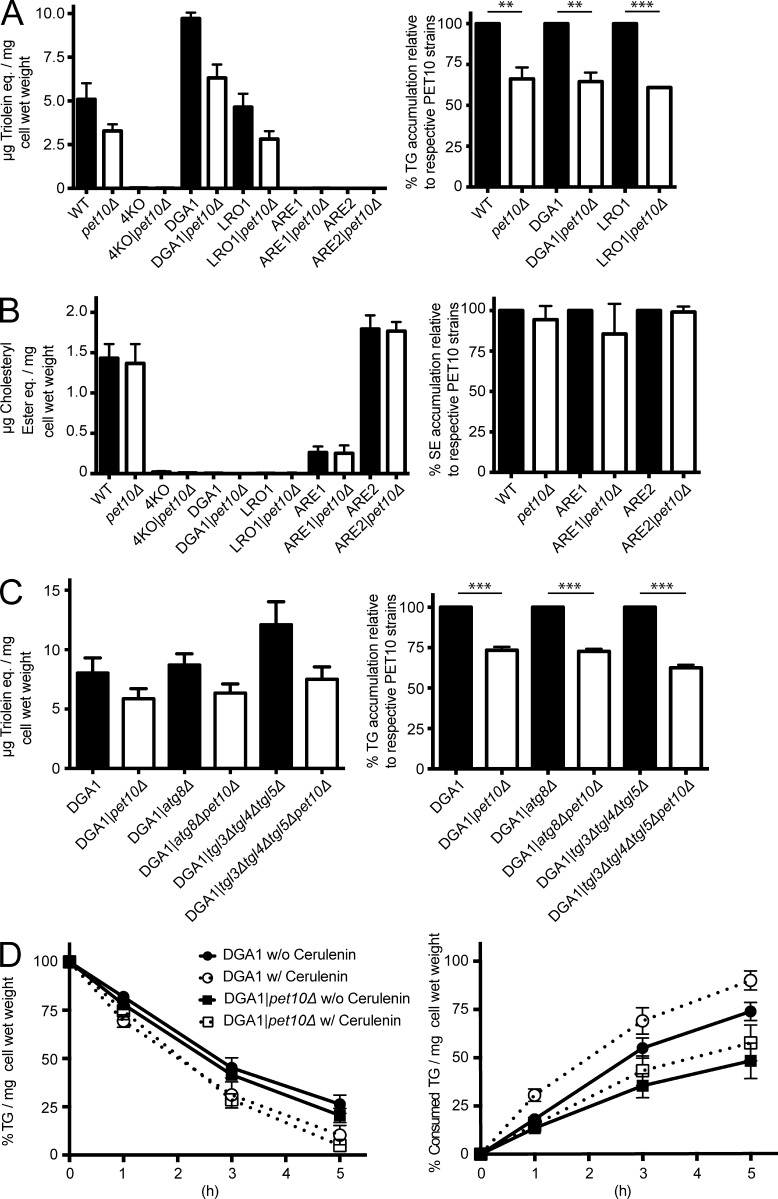
As Pet10p selectively binds to TG droplets, we asked whether it participates in TG homeostasis. Indeed, wild-type, DGA1, and LRO1 cells lacking PET10 contained ∼35% less TG (Fig. 2 A). In contrast, the absence of PET10 had no effect on SE accumulation in wild-type, ARE1, or ARE2 cells (Fig. 2 B).
Figure 2.
Pet10p promotes accumulation of TG, but not by decreased lipolysis. The amount of TG (A and C) or SE (B) was determined in whole-cell extracts of the indicated strains and plotted directly (left). The values in pet10Δ strains are plotted relative to their background strains, set at 100% (right). (D) The rate of lipolysis was determined after addition of cerulenin to the indicated strains. (D, left) TG level compared with t0 for each strain. (D, right) Calculated consumption of TG in all strains relative to DGA1, set at 100% at t0. For all panels, error bars represent SEM over all replicate experiments. **, P < 0.01; ***, P < 0.001.
To determine the basis of the drop in TG accumulation, we focused on the DGA1 strain. Dga1p, the homologue of mammalian DGAT2, is the enzyme mainly responsible for the accumulation of TG for cell storage in yeast (Oelkers et al., 2002). TG can strongly fluctuate during cell growth (Kurat et al., 2006), but the growth kinetics of DGA1 and DGA1|pet10Δ cells were identical (Fig. S1 B).
The decrease in TG accumulation in the absence of PET10 could be caused by an increase in lipolysis and/or a decrease in TG synthesis. We first considered an increase in lipolysis, which may occur through lipases on the surface of droplets or subsequent to lipophagy. The expression of the three principal TG lipases—Tgl3p, Tgl4p, and Tgl5p (Athenstaedt and Daum, 2003, 2005)—was not changed in DGA1|pet10Δ (Fig. S1 C), and the deletion of the corresponding genes did not alter the pet10Δ effect on TG accumulation (Fig. 2 C). Similarly, deletion of ATG8, a gene required for lipophagy (van Zutphen et al., 2014), had no significant effect (Fig. 2 C). To more directly measure lipolysis, we treated cells with cerulenin, which blocks TG synthesis by inhibiting production of acyl-CoA, and monitored the rate of decrease in TG (Fig. 2 D). The presence or absence of Pet10p made no difference in the rate when the initial TG amount was set to 100% for each strain (Fig. 2 D, left). Consumption of TG actually decreased in pet10Δ (Fig. 2 D, right).
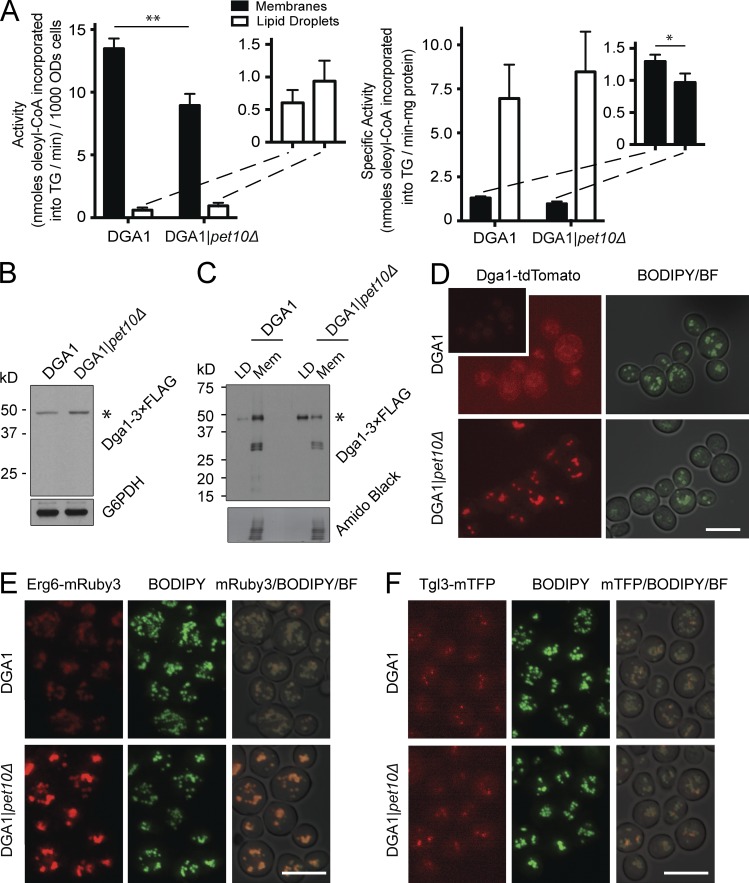
Having ruled out lipolysis as the cause of the decrease in TG accumulation in DGA1|pet10Δ, we turned to synthesis. Dga1p resides on the ER and on droplets (Jacquier et al., 2011; Markgraf et al., 2014). We observed a 34% decrease of DGAT activity in a crude membrane fraction of DGA1|pet10Δ cells compared with the DGA1 control strain, mirroring the drop in TG accumulation (Fig. 3 A, left). In contrast, activity in the droplet fraction was much lower, although it was slightly higher in pet10Δ. The addition of droplets to membranes in the assay did not inhibit activity, ruling out a negative factor (or substrate sink) in the droplet fraction. Although these values represent the partitioning of enzyme between ER and droplet, the specific activity (i.e., purity) was always severalfold higher on the droplets (Fig. 3 A, right), as previously reported (Sorger and Daum, 2002). The decrease in DGAT enzyme activity in DGA1|pet10Δ was not a result of a decrease in the Dga1p protein level, as shown by immunoblotting with a 3×FLAG-tagged version (Fig. 3 B); the addition of the FLAG epitope did not change the relative activities of the enzyme between the two strains (Fig. S1 D). The decrease of TG in the DGA1|pet10Δ strain was not caused by a decrease in the activities of the two yeast glycerol 3-phosphate acyltransferases (GPATs), Sct1p and Gpt2p, the enzymes responsible for the rate-limiting step in glycerolipid biosynthesis (Fig. S1 E). In summary, Pet10p ensures normal TG levels by maintaining DGAT activity.
Figure 3.
Pet10p promotes Dga1p activity and its localization in the ER. (A) Crude membranes and droplets were isolated from PNSs of the indicated strains, and DGAT activities in each were determined (A, left) Total activity expressed per 1,000 OD600 units of cells. (A, inset) Expands the scale for droplets. (A, right) Specific activity expressed per milligram of protein. Data are from four independent experiments. Paired t test was used to determine p-value. Error bars represent SEM over all replicate experiments. *, P < 0.05; **, P < 0.01. (B and C) Dga1–3×FLAG expression in whole-cell extracts (B) or droplets and crude membranes (C) from the indicated strains. Immunoblots are shown using anti-FLAG and anti-G6PDH antibodies. Asterisks show the predicted molecular weight of Dga1p-3xFLAG. Equal percentage of total membrane and droplet fractions analyzed (C). (C, top) Immunoblot using anti-FLAG antibody. (C, bottom) A collapsed view of the nitrocellulose stained with amido black as control for protein load. (D) Cells from the indicated strains containing chromosomal DGA1-tdTomato were stained with BODIPY. Note: The inset in the top left image is presented at the same settings as the others; the main image was enhanced in Photoshop to visualize ER rings. (E and F) Cells from the indicated strains containing Erg6-mRuby3 and Tgl3-mTFP were stained with BODIPY. Bars, 5 µm.
Dga1p accumulates on lipid droplets in the absence of PET10
We asked whether the decrease in DGAT activity in the membranes of the DGA1|pet10Δ strain reflected a change in the organellar localization of Dga1p. 3×FLAG- and tdTomato-tagged Dga1p in the DGA1 strain grown to the late log phase localized mainly to membranes, with a minor fraction associated with droplets (Fig. 3, C and D). In contrast, most of Dga1p associated with droplets in the DGA1|pet10Δ strain. The enzyme in the two locations behaves differently with respect to the effect of exogenous DAG to the in vitro assay: The activity of the membrane form is independent of exogenous DAG, whereas this substrate stimulates the droplet form (Fig. S1 F, compare left and right panels), suggesting that the enzyme has different conformations or access to substrates at the two sites. In addition to lower DGAT activity as a result of translocation, other factors are likely at play to explain the decrease in TG accumulation in DGA1|pet10Δ cells, as an ∼80% shift of Dga1p from membrane to droplet results in only an ∼35% drop in membrane DGAT activity.
Control of droplet localization by Pet10p is not specific to Dga1p. The absence of Pet10p also results in an increase in Erg6p on the droplet surface but no change in Tgl3p droplet localization (Fig. 3, E and F). Molecular crowding by Pet10p is therefore likely involved (Kory et al., 2015), but not all proteins are subjected to this effect.
The absence of Pet10p also lowers TG produced by Lro1p (Fig. 2 A). The other DGAT, Lro1p, appears punctate on the ER and can move to droplet contact sites (Wang and Lee, 2012). Unlike Dga1p, however, we observed no change in Lro1-tdTomato localization with respect to droplets with or without Pet10p (Fig. S1, G and H). In fact, the presence or absence of Lro1-tdTomato puncta had no effect on the droplet number at all (Fig. S1 I). As the deletion of PET10 results in an ∼35% decrease in TG accumulation in both DGA1 and LRO1 cells (Fig. 2 A), there may be a common mechanism to this decrease that we have yet to uncover.
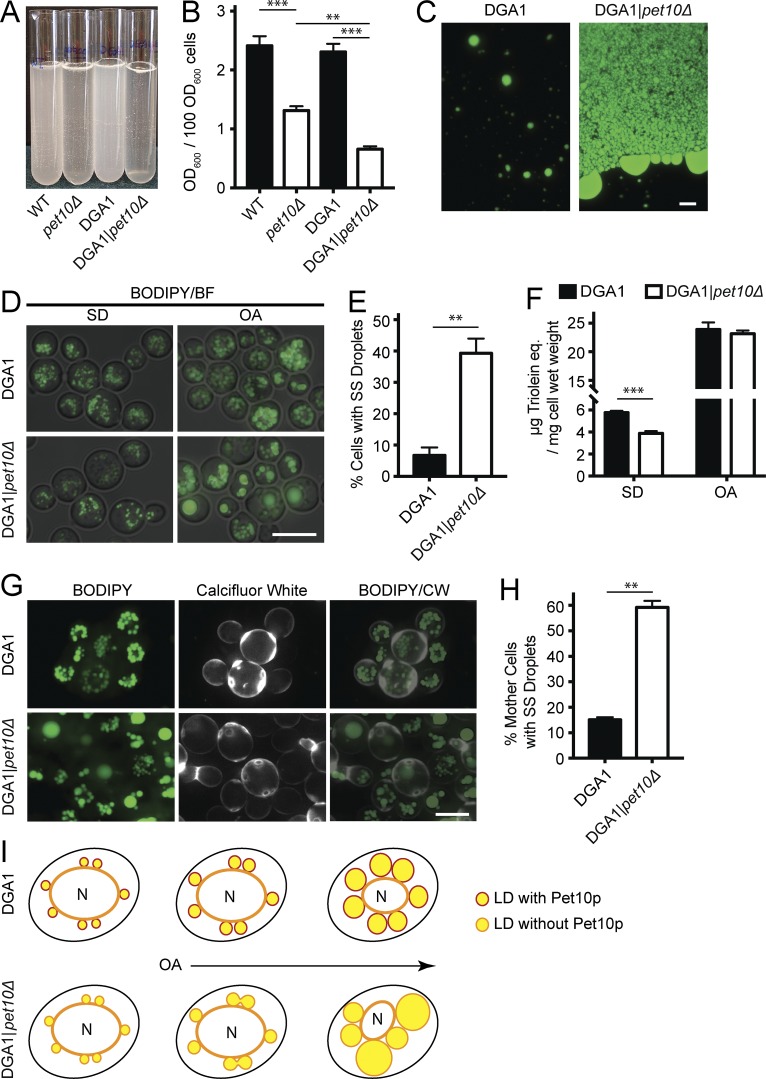
Lipid droplets aggregate in vitro and fuse in vivo in the absence of Pet10p
Lipid droplets are easily isolated by flotation during centrifugation of a cell lysate. Upon gentle resuspension, they form an emulsion of milky consistency (Fig. 4 A). Droplets from pet10Δ strains behaved differently, forming a more transparent suspension with visible white clumps. The optical density of the suspensions decreased by 45.5% in the wild-type background and by 71.5% in the DGA1 background (Fig. 4 B). The suspension of droplets from DGA1 cells, stained with BODIPY, revealed single droplets and small clusters of approximately five to six droplets (Fig. 4 C). In contrast, the aggregates in the DGA1|pet10Δ strain contained droplet structures much larger than those seen in situ, as though droplets had coalesced during centrifugation. The aggregates also contained globules of neutral lipid, especially at the periphery, as if released from damaged droplets. We conclude that droplets lacking Pet10p aggregate easily upon centrifugation and may be unstable.
Figure 4.
Pet10p promotes droplet stability. (A) Isolated droplets were gently resuspended in 3 ml flotation buffer. (B) The suspensions were subjected to light scattering at 600 nm. (C) BODIPY was added to an aliquot of the droplet suspensions from the indicated strains. (C, left) The suspension from DGA1 cells consists of single droplets and small aggregates. (C, right) An edge of a DGA1|pet10Δ aggregate. (D) Cells from the indicated strains were grown in SD or OA media for 24 h and stained with BODIPY. (E) The percentage of cells cultured in OA (experiment in D) with supersized (SS) droplets (>1.0 µm in diameter). (F) TG determined in whole-cell extracts from cells in the experiment shown in D and E. (G) Cells were cultured for 24 h in OA medium (as in D–F) and stained with BODIPY and Calcofluor White to identify mother cells. The Calcofluor White layer was pseudocolored white. The experiment was repeated once. (H) Quantification of mother cells with supersized droplets from experiment in G. (I) Diagram to illustrate possible fusion events in OA culture as lipid droplets (LDs) expand in the absence of Pet10p. BF, brightfield; CW, Calcofluor white. For B, E, F, and H, error bars represent SEM over all replicate experiments. **, P < 0.01; ***, P < 0.001. Bars, 5 µm.
Pet10p would be expected to comprise a significant amount of the droplet surface if it structurally stabilizes the organelle. We found this to be the case. We estimate that there are 8,000–34,000 Pet10p molecules per droplet (the range from four samples; see Materials and methods) and that 15–86% of the droplet surface may be comprised of copies of this protein.
Aggregated or coalesced droplets were not observed in intact DGA1|pet10Δ cells in glucose medium. However, when they were incubated in OA medium, a condition in which droplets expand with stored fat (Veenhuis et al., 1987), cells with single supersized droplets were frequently observed (Fig. 4 D). Such giant droplets were present in 39.3% of DGA1|pet10Δ cells compared with 6.7% of the DGA1 controls (Fig. 4 E), although the total TG in the two strains was virtually identical (Fig. 4 F).The giant droplets seen in the DGA1|pet10Δ strain growing in OA medium may reflect a biogenesis defect or fusion of preexisting droplets. To test for fusion, we took advantage of the slow growth of cells in OA; DGA1 cells divide once at most in 24 h in this medium. We reasoned that after 24 h of exposure to oleate, mother cells containing giant droplets at the expense of smaller ones probably generated them by fusion of the preexisting droplets. To test this hypothesis, we used Calcofluor White, which stains chitin in the cell wall and highlights bud scars (Hayashibe and Katohda, 1973). We scored mother cells with two or more bud scars and found that 59.2% of DGA1|pet10Δ mother cells had supersized droplets with few or no small ones compared with 15.1% of the DGA1 controls (Fig. 4, G and H), providing evidence for fusion. Why do pet10Δ cells fuse in oleate but not glucose medium? We reason that the small droplets in cells growing on glucose have little opportunity to physically interact compared with those enlarged with TG in oleate medium (Fig. 4 I). Moreover, the imported fatty acid itself, a chaotrope, could also facilitate fusion in the mutant. Pet10p is therefore an important droplet-stabilizing factor to ensure droplet integrity, as manifest in its absence by aggregation and fragility in vitro and fusion in vivo.
Pet10p stimulates droplet formation
We sought to determine whether Pet10p plays a direct role in droplet assembly in addition to its role in droplet integrity. Droplets can be induced to form in a (3KO) GALDGA1 strain upon switching growth medium from glucose to galactose, which drives Dga1p expression (Cartwright et al., 2015; Fig. 5 A). In the absence of Pet10p, the initial appearance of droplets (assessed by BODIPY staining) was delayed (Fig. 5 B). As expected, Pet10p expression was initially very low and increased with droplet formation (Fig. S2 A). After 2 h, droplets appeared in 32.4% of control cells but only in 13.1% of the GALDGA1|pet10Δ strain, accompanied by a relatively minor 17.0% drop in TG accumulation (Fig. 5 D). At later time points as Dga1p became overexpressed, these differences were attenuated. However, the number of droplets in cells with droplets was generally less in the GALDGA1|pet10Δ strain than in the control (Fig. 5 C).
Figure 5.
Pet10p binds early to nascent droplets and promotes biogenesis. Droplet formation was induced by galactose addition. (A–D) Cells were cultured in galactose medium and BODIPY for the indicated times. (A) The appearance of droplets (BODIPY stained) GALDGA1 cells. (B) Comparison of the number of cells with lipid droplets (LDs) in the GALDGA1 control versus the GALDGA1|pet10Δ strain. (C) The number of lipid droplets per cell in cells with droplets (thicker black bar represents median). At least 500 cells were scored at each time point per strain over three experiments. (D) Comparison of TG levels in cells after 2 h of induction. (E–H) After 40 min in galactose medium, cells containing BODIPY and fluorescent protein tags were transferred to a microscope slide to monitor appearance of nascent droplets (arrows). (E) An example of the appearance of two Pet10-tdTomato puncta 5 min before BODIPY staining. (F) Appearance of punctate Pet10-tdTomato with respect to BODIPY over time. Data represent 80 droplets appearing over three experiments. (G) An example of Pet10-tdTomato puncta appearing before Erg6-mTFP or BODIPY. Asterisks show a rare situation in which Erg6-mTFP and BODIPY appear before Pet10-tdTomato. (H) Lack of Tgl3-mTFP appearance during the time course. (I) Appearance of Erg6-mTFP or Tgl3-mTFP before and after 24 h in galactose medium. For B, D, and F, error bars represent SEM over all replicate experiments. *, P < 0.05. Bars, 5 µm.
Although the stimulation of droplet formation by Pet10p was much stronger than its effect on TG formation, it is possible that a threshold of TG was reached in the control that allowed the large observed increase in droplet numbers. To rule this out, we slightly modified the protocol to allow multiple simultaneous measurements of TG and droplet numbers. We observed that as droplets began to form (at the 1.5- and 2.0-h time points), there was consistently a 50–65% decrease in droplet number in the absence of Pet10p with only a 15% decrease in TG, regardless of the absolute amount of TG per cell (Fig. S2 B). Therefore, the slight increase of TG in the presence of Pet10p probably plays a small part in the stimulation of droplet formation.
To test for specificity, we performed the induction experiment in the absence of Tgl3p, the major TG lipase on droplets (Athenstaedt and Daum, 2003) that is not known to have a role in droplet formation. The absence of Tgl3p had no effect on droplet formation, in contrast to the absence of Pet10p (Fig. S2 C).
If Pet10p plays a direct role in droplet formation, it should appear on droplets early in the process. To test this, we tagged Pet10-tdTomato in the GALDGA1 strain and followed the appearance of Pet10p puncta with respect to BODIPY dots upon switching to galactose medium. In 47.4% of cases, Pet10-tdTomato dots preceded BODIPY staining (Fig. 5, E and F). In another 20.8% of cases, the two probes appeared simultaneously. Therefore, during biogenesis, most nascent droplets contain Pet10p.
For comparison, we simultaneously tracked Pet10p-tdTomato appearance with two other droplet proteins not known to have biogenesis activity: Erg6p and Tgl3p, both tagged with monomeric teal fluorescent protein (mTFP). Although Erg6p-mTFP was detected on the ER before Dga1p and droplets were induced, Erg6p-mTFP puncta appeared at the same time as BODIPY or close to it compared with Pet10p-tdTomato, which usually appeared earlier (Fig. S2 D). Tracking appearance on the same droplet, Pet10-tdTomato appeared earlier 58.6% of the time, and the two markers appeared simultaneously 13.4% of the time (Fig. 5 G and Fig. S2 D). Pet10p was not required for Erg6p droplet localization, however. In contrast, Tgl3-mTFP puncta were never observed during the first 2 h of droplet formation (Fig. 5 H illustrates the second hour). After 24 h of culture, both Erg6-mTFP and Tgl3-mTFP were easily observed on droplets, although Tgl3-mTFP puncta were localized in a small area on the droplet surface (Fig. 5 I).
Overall, the lag in droplet formation in the absence of Pet10p coupled with the rapid appearance of this protein even before detection of BODIPY suggest that Pet10p is an important accessory protein in droplet formation, although it is not an obligate player.
Functional interactions of Pet10p with seipin and Fit2 on droplet morphology
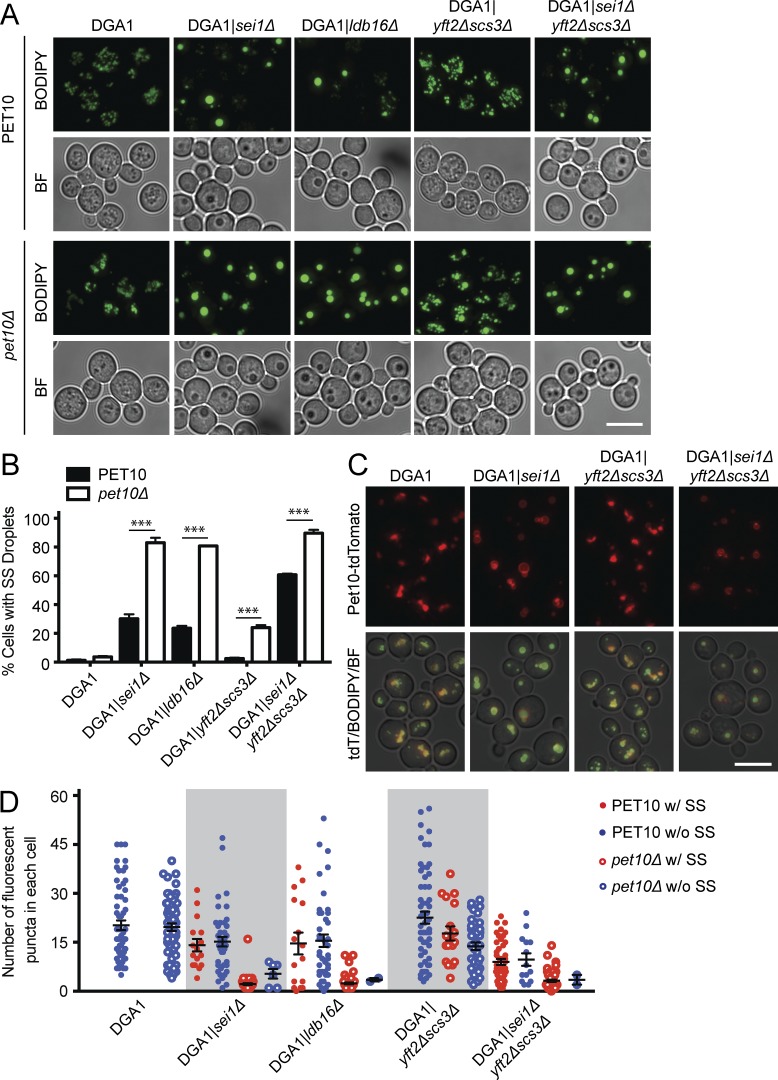
If Pet10p is indeed important for droplet formation, it may functionally interact with two other proteins that participate in this process: seipin (Sei1p and its binding partner in yeast, Ldb16p) and Fit2 (Yft2p and Scs3p; Szymanski et al., 2007; Fei et al., 2008; Kadereit et al., 2008; Wang et al., 2014). We wondered if the absence of Pet10p might contribute to the aberrant droplet morphology seen in the absence of seipin or Fit2, indicating a functional interaction and suggesting a collaborative role in biogenesis. The appearance of supersized droplets is one indicator that can be easily observed. Supersized droplets were very rare in the DGA1|pet10Δ strain (occurring in 3.7% of cells) cultured in glucose medium (Fig. 6, A and B). Deletion of seipin (SEI1) or its partner LDB16 alone in the DGA1 background resulted in the appearance of supersized droplets in 30.2% and 23.6% of cells, respectively. Remarkably, over 80% of cells had supersized droplets in the double-deletion mutants DGA1|sei1Δpet10Δ and DGA1|ldb16Δpet10Δ. Synergy was also strong when Fit2 deletions were coupled with that of PET10: 2.5% of cells contained supersized droplets in the absence of Fit2 proteins compared with 24.0% in the DGA1|yft2Δscs3Δpet10Δ strain, nearly a 10-fold increase. Further deleting SEI1 (in the DGA1|yft2Δscs3Δpet10Δ strain) resulted in supersized droplets in 89.7% of the population. Pet10p was able to bind to droplets in the absence of seipin or Fit2 (Fig. 6 C).
Figure 6.
Pet10p functionally interacts with seipin and Fit2 proteins to determine droplet number and size. (A) Cells were stained with BODIPY; the corresponding brightfield (BF) images are shown below. (B) The percentage of cells with supersized (SS) droplets (diameter > 0.5 µm) is plotted. (C) Pet10-tdTomato targets to seipin- and Fit2-null droplets. Shown are the red channel and a merge of the red, green, and brightfield channels. (D) Droplets in cells containing a supersized droplet (red) or lacking supersized droplets (blue) were counted in the indicated strains (closed circles) or corresponding ones that lacked PET10 (open circles). Droplets in 60 cells were scored in each strain. No cells with supersized droplets were seen in the DGA1 control strain. For B and D, error bars represent SEM over all replicate experiments. ***, P < 0.001. Bars, 5 µm.
An even stronger indication of a functional interaction between Pet10p and the other proteins is shown by the number of lipid droplets in these cells (Fig. S2, E and F). The DGA1 control strain and DGA1|pet10Δ had similar mean numbers of droplets per cell (20.2 and 19.7, respectively), whereas the number in DGA1|sei1Δ cells was slightly decreased (14.9 droplets). However, this number was diminished to 2.5 in the DGA1|sei1Δpet10Δ strain. Very similar results were obtained when LDB16 was deleted instead of SEI1. The functional interaction between PET10 and Fit2 deletions was considerably less dramatic. To test whether the decrease in droplet number was caused by fusion, we counted droplets separately in cells with and without supersized organelles. There was little or no difference in droplet number in all strains that contained cells in both populations (with or without supersized droplets; Fig. 6 D), although the nonsupersized droplets in cells were generally smaller and dimmer (Fig. S2 E). These results suggest that fusion is not responsible for the low number of droplets in the absence of seipin, Fit2, and Pet10p and that these proteins cooperate to ensure formation of droplets with regular morphology and adequate quantity (Fig. S2 G). Overall, we observed a profound interaction between seipin and Pet10p and a significant but weaker one between Fit2 and Pet10p.
Functional interactions of Pet10p with seipin and Fit2 in TG accumulation
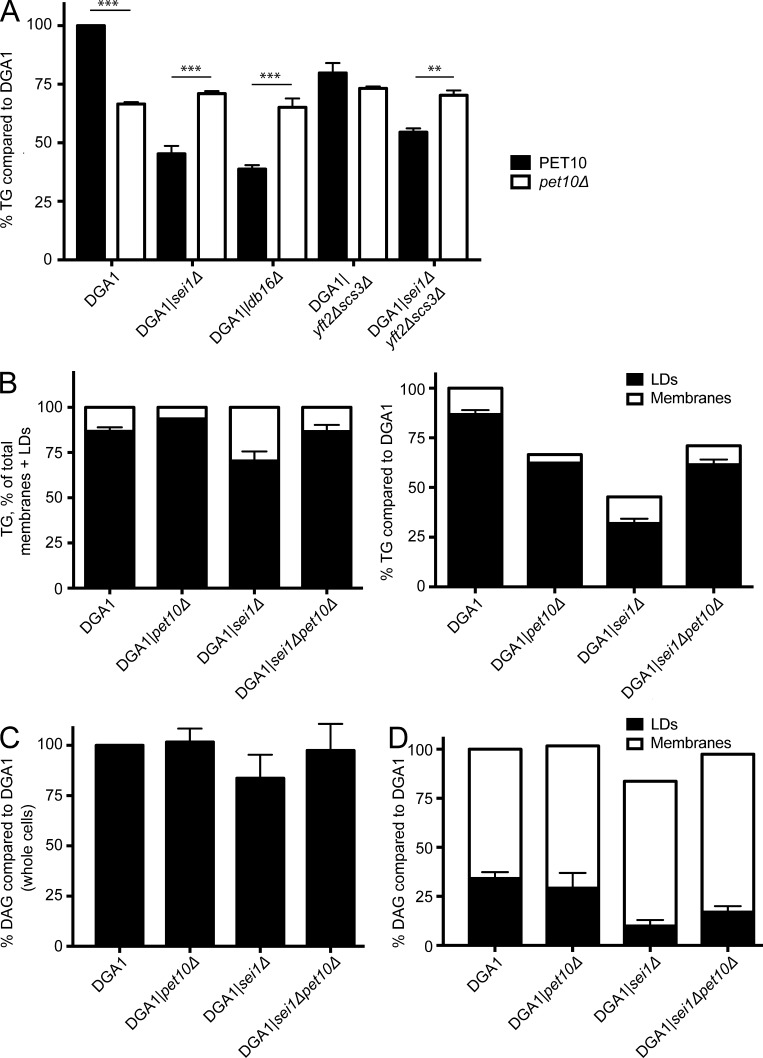
Similar to the effect in the W303 strain reported previously (Han et al., 2015), deletion of SEI1 or LDB16 in our derived DGA1 strain resulted in a 54.7% or 61.3% drop in TG, respectively, which was considerably more profound than the 33.5% drop in the DGA1|pet10Δ sample (Fig. 7 A). Deletion of the two yeast Fit2 genes had a minor effect compared with deletion of seipin, causing a 20.2% decrease in TG accumulation; deletion of Fit2 and seipin genes in combination was intermediate to both. However, in all cases, further deletion of PET10 was indistinguishable in TG accumulation from deletion of PET10 alone. In other words, Pet10p works upstream of seipin or Fit2p in TG accumulation. The genetic interaction between PET10 and seipin or Fit2 genes on TG levels, which is different from the effects on droplet morphology, may reflect independent functions of Pet10p in regulating droplet formation and neutral lipid homeostasis.
Figure 7.
Pet10p is epistatic to seipin and Fit2 in TG accumulation. (A) TG was determined in the indicated strains and compared with the DGA1 control. Note that strains are paired—the only difference being the presence or absence of PET10. (B) TG was determined in crude membrane and lipid droplet (LD) fractions and plotted relative to the total within each strain (left) or compared with the DGA1 control (right). (C and D) DAG in the strains was determined and plotted relative to the DGA1 strain. (C) Whole cells. (D) Membranes and droplets analyzed separately. Error bars represent SEM over all replicate experiments. **, P < 0.01; ***, P < 0.001.
Under conditions of Dga1p overexpression, TG accumulates in the ER in the absence of seipin, as droplet initiation is less efficient (Cartwright et al., 2015). Similarly, deletion of seipin in DGA1 resulted in more TG in a crude membrane fraction (Fig. 7 B). An opposite effect was seen in the pet10Δ strain: Only 6.4% of TG associated with membranes compared with 13.2% in the DGA1 control and 29.6% in the sei1Δ strain. One interpretation of these data is that seipin and Pet10p play complementary roles in droplet formation, whereby seipin helps to mobilize TG from the ER to droplets, whereas Pet10p controls this flux, perhaps by limiting elasticity of the nascent droplet surface. This presumption would explain the multiple small droplets often observed in the absence of seipin (Szymanski et al., 2007) but absent in pet10Δ. However, we cannot rule out that droplet fragility or altered droplet–organelle interactions upon lysis affects this distribution. The DGA1|pet10Δsei1Δ double-deletion strain yielded an intermediate distribution, similar to the DGA1 strain, consistent with the cancelation of these opposing forces.
Although TG accumulation varied in these mutants, whole-cell levels of the DAG precursor were similar among all strains tested (Fig. 7 C). The distribution of DAG between droplet and membrane fractions was altered in the SEI1 mutant but not in the PET10 mutant (Fig. 7 D), suggesting that this TG precursor is not limiting.
Pet10p is a yeast perilipin
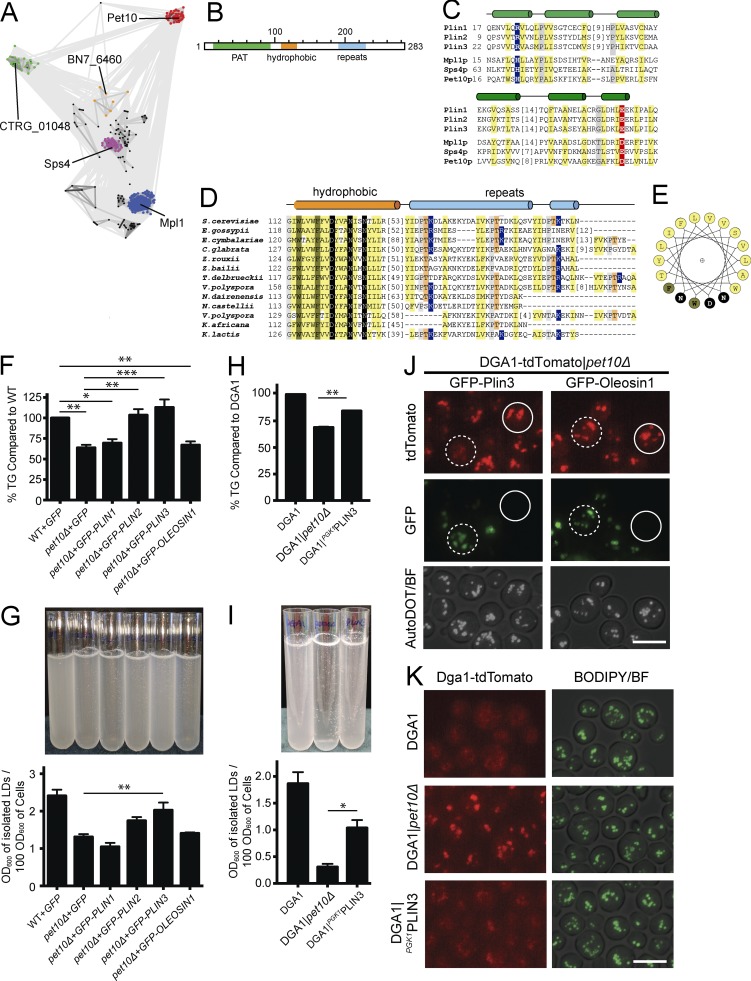
Using PSI-BLAST to probe for proteins homologous to Pet10p, we found numerous related sequence groups in fungi (Fig. 8 A), including the previously described perilipin homologue Mpl1p in Metarhizium anisopliae (Wang and St. Leger, 2007; Bickel et al., 2009), as well as yeast Sps4p. The Pet10p and Sps4p sequence clusters show a relatively divergent relationship to the Mpl1p perilipin group. SPS4 represents a gene whose expression is induced during sporulation (Garber and Segall, 1986), and its gene product also localizes to lipid droplets (Lam et al., 2014). The suggested link between Pet10p, Sps4p, and known fungal Mpl1p prompted us to further evaluate sequence relationships to perilipins using a sensitive sequence search. The results (Table 1) substantiate the sequence similarities identified between fungal groups and mammalian perilipin PAT domains (Plin1 to Plin5), with most relationships assigned by high confidence probabilities (>95%).
Figure 8.
Pet10p is one of two PAT-containing proteins in yeast. (A) Two-dimensional CLANS diagram (Frickey and Lupas, 2004) depicts sequence similarity (lines) between identified fungal sequences (nodes). Clustered nodes are labeled by gene name of representative and colored by family: MPL1 (blue), SPS4 (magenta), PET10 (red), sparse intermediate family (orange), family of hypothetical proteins (green), and others (black). (B) Proposed domain structure of Pet10p. (C) Alignment of known PAT domains from human perilipins Plin 1–Plin 3 and M. anisopliae Mpl1 with S. cerevisiae Sps4p and Pet10p highlights conserved positions: hydrophobic (yellow), small (gray), basic (blue), and acidic (red). Predicted secondary structure above alignment with helices as cylinders. Residue numbers of the first position in the alignment are on the left, and omitted residue counts are in brackets. (D) Alignment of fungal Pet10p family C-terminal sequence regions is depicted and colored as described previously, with additional conservations: aromatic (dark yellow), polar (black), and repeat threonines (orange). Secondary structures are colored according to sequence regions in B. (E) A helical wheel diagram of Pet10p conserved hydrophobic segment colored as in D. (F–I) Parallel cultures in three or four independent experiments were harvested for whole-cell TG analysis (F and H) or subjected to lipid droplet (LD) isolation, resuspension, and light scattering at 600 nm (G and I). Note that strains in the left panels are based on W303 wild type with plasmid expression of heterologous proteins, whereas those in the right panels are based on the DGA1 background with PLIN3 replacing PET10 in the chromosome as indicated. (J) Plasmids containing GFP-PLIN3 or GFP-OLEOSIN1 were expressed in the DGA1-tdTomato|pet10Δ strain. AutoDOT (pseudocolored white) tagged the droplets (Yang et al., 2012). (K) Localization of DGA1-tdTomato in the indicated strains. For panels F–I, error bars represent SEM over all replicate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Bars, 5 µm.
Table 1. Similarities in fungal and human sequences.
| Query | Hit | Query range | Hit range | Probability | E-value |
|---|---|---|---|---|---|
| A. Fungi | |||||
| pet10p | Pet10p | 1–283 | 1–283 | 100 | 8.0 × 10−121 |
| pet10p | Pet10pa | 203–238 | 188–231 | 81.5 | 3.4 × 1000 |
| pet10p | Sps4pb | 1–147 | 50–94 | 59 | 2.4 × 1001 |
| BN7_6460 | Sps4p | 10–238 | 53–312 | 99.9 | 9.9 × 10−24 |
| BN7_6460 | Pet10p | 1–242 | 1–266 | 97.9 | 5.2 × 10−04 |
| BN7_6460 | Perilipin | 20–211 | 11–255 | 94.3 | 2.7 × 1000 |
| sps4p | Sps4p | 1–338 | 1–338 | 100 | 2.0 × 10−111 |
| sps4p | Perilipin | 63–281 | 11–255 | 98.9 | 2.0 × 10−08 |
| sps4p | Pet10p | 50–321 | 3–271 | 95.1 | 3.4 × 10−01 |
| Mpl1 | Perilipin | 1–178 | 1–199 | 100 | 3.7 × 10−39 |
| Mpl1 | Sps4p | 3–179 | 55–240 | 99.2 | 1.1 × 10−11 |
| Mpl1 | Pet10p | 68–178 | 189–281 | 81.1 | 2.4 × 1000 |
| CTRG_01048 | Sps4p | 1–122 | 50–190 | 97.3 | 2.5 × 10−04 |
| CTRG_01048 | Pet10p | 1–111 | 1–139 | 91.1 | 3.9 × 1000 |
| B. Human | |||||
| PLIN1 | Perilipin | 7–402 | 1–390 | 100 | 2.0 × 10−94 |
| PLIN1 | Sps4p | 3–197 | 51–231 | 98.4 | 1.2 × 10−07 |
| PLIN2 | Perilipin | 2–396 | 4–390 | 100 | 6.0 × 10−107 |
| PLIN2 | Sps4p | 12–196 | 66–231 | 98.1 | 2.4 × 10−06 |
| PLIN3 | Perilipin | 12–412 | 1–390 | 100 | 2.0 × 10−108 |
| PLIN3 | Sps4p | 3–209 | 41–388 | 98.7 | 7.4 × 10−09 |
| PLIN4 | Perilipin | 925–1,341 | 78–390 | 100 | 2.2 × 10−70 |
| PLIN4 | Perilipin | 6–140 | 16–182 | 99.2 | 1.1 × 10−11 |
| PLIN5 | Perilipin | 25–396 | 2–390 | 100 | 6.4 × 10−95 |
| PLIN5 | Sps4p | 10–198 | 44–231 | 98.7 | 3.2 × 10−08 |
Low confidence hit.
Duplicated sequence region suggesting repeats.
Our confidently identified sequence relationships to the yeast proteins are limited to the PAT domain (Fig. 8, B and C; and Fig. S3 A) and are extended with less confidence to the more divergent carboxy-terminal helical segments (Fig. 8, D and E). The helical carboxy terminus of Pet10p beyond the PAT domain is distinguished by a unique conserved hydrophobic region not found in other PAT domain sequences, followed by a divergent repeat. The hydrophobic region contains several invariant residues, including two large aromatic and three polar residues; these should line one surface of the predicted helix (Fig. 8 E). Whether this segment plays the same role in droplet targeting as the 11-mer repeat amphipathic helix does in perilipins (Rowe et al., 2016) is not known. The repeat (hhxPTKxxh, with h representing any hydrophobic residue) differs in primary sequence from those identified in perilipins and diverges significantly even among the Pet10p family, both in sequence and in repeat number (Fig. 8 D). This suggests that it does not play a conserved role in Pet10p, let alone perilipin function.
The sequence conservation between Pet10p and mammalian perilipins suggests shared function. To test this idea, we first expressed plasmids (Jacquier et al., 2013) with human PLIN1-3 as well as Arabidopsis OLEOSIN1, all fused to GFP sequences in pet10Δ cells (W303 strain background), to determine if human perilipins could restore TG accumulation in the absence of Pet10p. Plin2, Plin3, and Oleosin1, but not Plin1, expressed well in our strains, regardless of the presence or absence of Pet10p (Fig. S3 B). They all targeted to droplets (Fig. S3 C), as was previously shown (Jacquier et al., 2013). We found that expression of Plin2 and Plin3 complemented the pet10Δ strain in TG accumulation, whereas Plin1 and Oleosin1 did not (Fig. 8 F). Plin3 also partially restored droplet stability (Fig. 8 G), and Dga1p was removed from the droplet surface in the DGA1-tdTomato strain (Fig. 8 J). These effects were not seen with Oleosin1. In addition to expression on a plasmid, replacing PET10 with PLIN3 driven by the PGK1 promoter in the yeast genome of the DGA1 strain showed similar results (Fig. 8, H, I, and K). The increase in TG accumulation with Plin3 was not caused by inhibition of lipolysis (Fig. S3 C). Interestingly, Plin3 was not very effective in restoring normal-sized droplets to the DGA1|pet10Δ strain grown on OA (Fig. S3 D), suggesting that this function may require a more specific interaction between Pet10p and other droplet components.
Discussion
We report herein that Pet10p, an established lipid droplet protein in yeast, which binds to TG-containing droplets where it is stabilized, is important for droplet integrity, influences protein distribution, facilitates droplet formation, contributes to TG reservoirs, and collaborates with seipin and Fit2 proteins to maintain normal droplet size. Pet10p and Sps4p both contain a PAT domain, and complementation of pet10Δ with human Plin2 and Plin3 indicates that Pet10p is indeed a yeast perilipin.
Pet10p prevents droplets from aggregating and bursting in vitro and from fusing in vivo in oleate cultures (Fig. 4). These observations argue that Pet10p, and by extension other perilipins, is responsible for the integrity of droplets. Although perilipins are well known to protect the droplet core from adventitious lipolysis (Bickel et al., 2009; Kimmel and Sztalryd, 2016), our data suggest that their stabilizing properties go much further. Perilipins may prevent the fragility and fusion activity that we observe by providing anchors of the phospholipid leaflet into the core, or they may shield the cytosol from the hydrophobic surface created when surface tension increases and local phospholipid concentration drops (Thiam et al., 2013). Although perilipins are less hydrophobic than plant oleosins, which bury deep into the core to stabilize the phospholipid–core interface (Tzen and Huang, 1992), they may still perform a similar function by extending below the phospholipid leaflet to bind to neutral lipids. A nuclear magnetic resonance study of Drosophila melanogaster Plin1 suggests that two helices of the protein extend deep into the droplet, beyond the phospholipid monolayer (Lin et al., 2014).
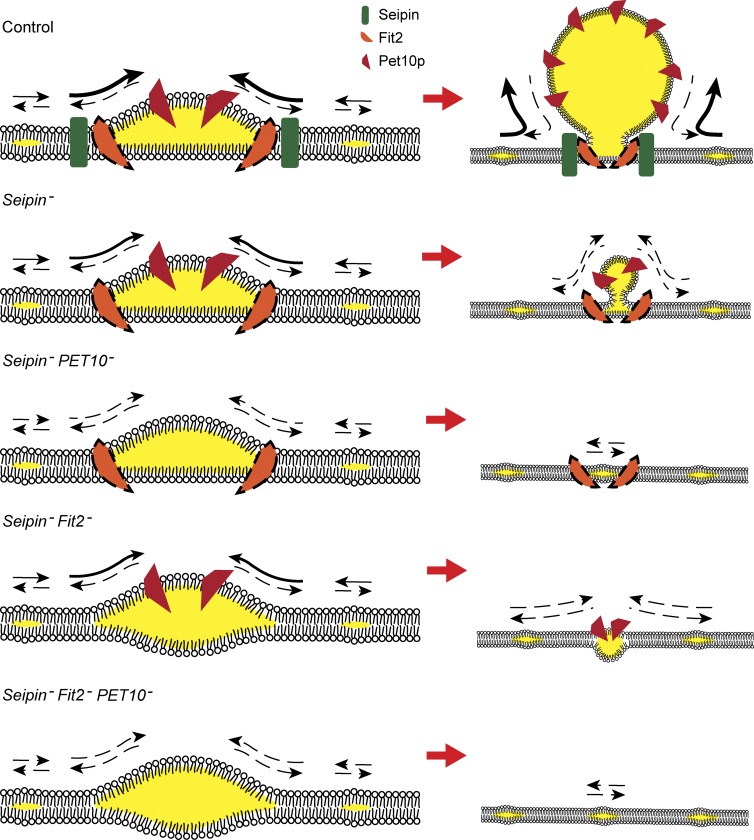
Three lines of evidence indicate that Pet10p contributes to the assembly of lipid droplets. First, there is a lag in droplet formation in the absence of this factor upon induction of DGA1 expression. This is manifest by the lower number of cells with droplets early on (Fig. 5 B and Fig. S2 B) and fewer droplets in cells with droplets (Fig. 5 C). There is precedence for this: Knockdown of Plin3 in HeLa cells leads to fewer mature droplets (Bulankina et al., 2009). Second, Pet10p arrives early to the nascent droplet, even before droplets can be visualized with BODIPY (Fig. 5, E and F) and before other droplet proteins (Fig. 5, G and H). Again, Plin3 arrives very early to nascent droplets in cultured cells (Skinner et al., 2009). Finally, Pet10p strongly interacts genetically with other droplet assembly proteins, seipin and Fit2p, in maintaining normal droplet size (Fig. 6). We hypothesize that prevention of supersized droplets by Pet10p, seipin, and Fit2p is an indirect measure of collaboration in droplet assembly. Early steps of assembly with various combinations of these factors are visualized in the working model shown in Fig. 9. We envision this process to occur on specific ER sites, dynamic in nature, where neutral lipids can pool. This model corresponds to the LiveDrop-positive, BODIPY-negative puncta that have recently been observed (Wang et al., 2016). Early pools are represented on the left side of Fig. 9. In the absence of assembly factors, such pools will be unstable, may fail to grow larger (Fig. 9, bottom right) or may coalesce with more lipids and become droplets of random size, even supersized organelles. If all assembly factors are present, nascent droplets will be stabilized and growth will be controlled both from the ER (by seipin and Fit2) and at the cytosolic side of the bud by Pet10p, yielding an appropriate number and size of droplets per cell. In the sole absence of Pet10p, although there is a lag in droplet formation, there is sufficient stabilization by seipin/Fit2 to yield droplets at near-normal numbers and size. However, in the absence of both Pet10p and seipin (or, to a lesser extent, Fit2, which prevents budding into the ER lumen [Choudhary et al., 2015]), stabilization of nascent droplets is lost, and the small pools of neutral lipids may not develop further or may continue to fuse with other pools, creating the supersized droplets shown in the combined deletions in Fig. 6.
Figure 9.
Working model of the contribution of Pet10p to droplet assembly. Membranes oriented with cytosol above ER lumen. (Left) Nascent droplets in the presence or absence of seipin, Fit2, or Pet10p, as indicated. These proteins are proposed to work cooperatively to control the rate of flux of TG and the final size of droplets. Arrows signify flux of TG relative to the droplet; dashed arrows represent less certain flux. Flux to droplet may depend on accompanying flux of phospholipids. The Fit2p icon has a dashed-line perimeter, as its early localization to droplets has not been demonstrated. Whether its localization is affected by the presence of seipin is also unknown. (Right) Growing droplets at a lower magnification derived from the nascent structures. Droplets occurring independently of these proteins as a result of TG saturation of the ER and membrane instability, forming droplets of varying sizes, including supersized, are not depicted. Morphology in Fit2 deletion mutants based on published work (Choudhary et al., 2015).
The accumulation of TG is lower in the absence of Pet10p, shown herein to be caused by a drop in Dga1p activity as well as a shift of this protein from the ER to droplets. Interestingly, suppression of Plin3 in HeLa cells also resulted in a significant drop in cellular TG (Bulankina et al., 2009), suggesting a conserved mechanism. Comparing the signals on immunoblots to enzyme activity (Fig. 3, A and C), it is clear that Dga1p is less active on droplets than it is on the ER. Analogous to Dga1p, squalene epoxidase (Erg1p) localizes to both the ER and droplets (Leber et al., 1998). The droplet form is inactive, and the authors concluded that an ER factor is required for activity.
However, although the drop we see in enzyme activity in pet10Δ cells corresponds to the drop in accumulated TG, this cannot be fully explained by the disappearance of Dga1p from the ER to droplets: Lack of Pet10p results in a loss of ∼80% of membrane Dga1p but only ∼35% of activity. Additionally, our data are not consistent with those of a recent study showing that Dga1p-dependent incorporation of fatty acids into TG in intact cells corresponds to relocalization of the enzyme to droplets (Markgraf et al., 2014). Other inputs toward Dga1p activity must be at play to reconcile these observations. Possible factors include a change in the availability of substrates or modifying factors, a conformational change in Dga1p (Kamisaka et al., 2010), or a posttranslational modification. A similar drop in TG accumulation in the absence of Pet10p occurs whether Dga1p or Lro1p is the sole synthetic enzyme, suggesting an upstream effector. Thus far, we have not been able to identify such a factor, although we can rule out three causes: (a) a GPAT enzyme does not appear to be responsible (Fig. S1 E); (b) the localization of Lro1p does not appear to change with or without Pet10p as it does for Dga1p (Fig. S1, G and H); and (c) there is no difference in the amount of DAG or its distribution between droplets and membranes (Fig. 7, C and D).
Also puzzling is the interaction among seipin, Fit2, and Pet10p with regard to TG levels (Fig. 7 A). Although these proteins clearly interact to synergistically suppress supersized droplets, suggesting action at a common step, the TG accumulation phenotypes suggest that Pet10p is upstream. The 35% drop in TG in pet10Δ is fixed, regardless of the additional deletion of seipin or Fit2p, as if Dga1p is altered in the absence of Pet10p to be nonresponsive to loss of seipin or Fit2. We suspect that these interactions to regulate TG levels are more indirect than those governing droplet assembly and size. As droplets are probably always covered with a perilipin, whether this protein physiologically regulates TG accumulation is uncertain.
Considering the abundance of Pet10p on the droplet surface and the PAT domain homology, we propose changing the corresponding gene name from PET10 to PLN1. The existence of a single perilipin in yeast that is normally expressed (the other PAT-containing protein, Sps4p, is expressed during sporulation; Garber and Segall, 1986) coupled with the ease of genetic manipulation are compelling reasons to study the conserved functions of perilipins in this simple eukaryote. We extrapolate from this work that physical stabilization of droplets, collaboration with ER factors to efficiently assemble droplets, and controlling protein accessibility to this organelle are three fundamental functions of perilipins. We further posit that well-described functions specific to particular perilipins, such as regulation of lipases or binding to organelles such as mitochondria, were the results of later evolution.
Materials and methods
Reagents
All chemicals were reagent grade and from Sigma-Aldrich unless noted otherwise. The following BD products were purchased from Thermo Fisher Scientific: Difco Yeast Nitrogen Base, Yeast Extract, Bacto-Peptone, Bacto-Agar, and Difco LB Broth. BODIPY 493/503 was purchased from Invitrogen. AUTOdot was purchased from Abgent. An antipeptide antibody against Pet10p was generated in rabbits using amino acids 264–283 by ProSci. Anti-GFP antibody was obtained from Roche, and anti-6xHis antibody was obtained from Thermo Fisher Scientific. Neutral lipid standards were obtained from Nu-Chek (no. TLC 18–5 C), and 1,2-dioleoyl-sn-glycerol was obtained from Avanti Polar Lipids. Oleoyl-1-14C-CoA was obtained from Perkin-Elmer Cetus, and Zymolyase T-100 was purchased from US Biological Life Sciences. The ECL kit for immunoblots was obtained from Amersham.
Strains and plasmids
The strains and plasmids used in this study are listed in Table S2. Unless noted, all strains are based on W303-1A (Thomas and Rothstein, 1989). The 2KO strains (DGA1LRO1 and ARE1ARE2 in the W303-1A background) were a gift from S. Sturley (Columbia University, New York, NY). The 3KO strains (DGA1, LRO1, ARE1, and ARE2) were constructed by replacement of markers for genes in the 2KO parents. The plasmids to express human perilipins and Arabidopsis thaliana Oleosin1 in yeast were gifts from R. Schneiter (University of Fribourg, Fribourg, Switzerland). The plasmids for generating C-terminal protein fusions with mRuby3 and mTFP were gifts from A. Doncic (University of Texas Southwestern Medical Center, Dallas, TX). For generating a purified Pet10p standard curve, Pet10p was overexpressed using a modified pRS316 plasmid (Sikorski and Hieter, 1989) in which the PET10 coding sequence was inserted between myc and FLAG sites in a cassette consisting of a PGK1 promoter, 2×myc, 2×FLAG, and a PGK1 terminator, generating pRS316-PGK1-2×myc-Pet10p-2×FLAG. The parent vector, pRS316-PGK1-2×myc-2×FLAG, was a gift from S. Han (University of Texas Southwestern Medical Center, Dallas, TX).
Unless indicated, all tags were inserted in the endogenous genes directly before the stop codon by homologous recombination, followed by the PGK1 terminator and the selectable marker. The TAP tag consists of an HBH tag and a KanMX6 marker (originally derived from plasmid; DQ407918; Tagwerker et al., 2006).
Growth conditions
Unless noted, all strains were precultured overnight in minimal synthetic dextrose (SD) medium (2% dextrose, Yeast Nitrogen Base, histidine, uracil, adenine, lysine, leucine, and tryptophan at 30°C in an Innova 43 rotary shaker; New Brunswick) at 210 rpm, diluted in SD to 0.1 OD600 unit/ml, and cultured for 24 h, by which time they had progressed to the late log-early stationary phase. For oleate cultures, cells were first precultured in SD overnight, washed and diluted to 0.1 OD600 units into SD containing 0.3% dextrose, grown for 24 h, washed and resuspended into OA medium (Binns et al., 2006), and grown for 24 h. For CHX experiments, two flasks of ARE2 or DGA1 cells expressing Pet10-TAP were precultured for 24 h and then subjected to 1 mg/ml CHX in DMSO or DMSO alone for the indicated hours before harvesting. For cerulenin experiments, two samples of 100 OD600 units from 24-h SD cultures (prepared as described earlier in this paragraph) were each resuspended in 200 ml of fresh SD. Either 200 µl of 10 mg/ml cerulenin (Sigma-Aldrich) in DMSO or DMSO alone was added to the flasks, and they were allowed to incubate at 30°C for the times indicated before harvesting.
For the droplet biogenesis experiments, a modification of a previous method (Cartwright et al., 2015) was used. In brief, lipid droplets were induced in the indicated strains by first preculturing cells in SD containing 0.2% dextrose overnight, washing them in distilled water, resuspending them in 10 ml SR (2% raffinose substituted for dextrose in SD) at 0.5 OD600 units, and culturing for 24 h. 20% galactose and 1 mg/ml BODIPY were then added to final concentrations of 2% and 1 µg/ml, respectively. Cells were observed at the indicated times. For the time-lapse experiments, cells were removed 40 min after galactose addition, concentrated 10-fold, placed on a microscope slide at ambient temperature, and imaged every 2.5 min for 1–1.5 h. For the experiment shown in Fig. 5 D, 50-ml cultures were used and BODIPY was omitted. For the experiment shown in Fig. S2 (B), 200-ml cultures were used, and BODIPY was added to small aliquots of cells at each time point before concentration and microscopy. 50 OD600 units were removed from the same cultures for TG quantification.
Protein extraction for immunoblotting
Whole-cell extracts for immunoblots were prepared by precipitating 50 OD600 units of cells in 10% trichloroacetic acid, resuspending in 4× Laemmli sample buffer (Laemmli, 1970) lacking bromophenol blue, neutralizing with NaOH, and vortexing with acid-washed glass beads on the MiniBeadBeater 16 (model 607; Biospec Products) for three 1-min pulses. Samples were boiled for 5 min and then microfuged for the same duration at 13,000 rpm in a microfuge to remove insoluble material. Bromophenol blue was then added to the supernatant, and the identical cell equivalents (typically 0.5–1.0 OD600 units) were subjected to SDS-PAGE and immunoblotting using ECL. For isolated organelles, 4× Laemmli buffer was added to cell fractions to yield 1× final concentration, and samples were subjected to SDS-PAGE and immunoblotting.
Purification of crude droplets and membranes
Postnuclear supernatant (PNS), lipid droplets, and crude membranes were prepared as previously described (Binns et al., 2010; Cartwright et al., 2015). Spheroplasts were made using Zymolyase 100T (8 mg/1,000 OD600 units) in the following medium (at 100 OD600 units per ml): 10 mM Tris-HCl, pH 7.4, 0.7 M sorbitol, 0.5% glucose, 0.75% yeast extract, 1.5% Bacto-Peptone, and 1 mM DTT, at 30°C, with gentle shaking. Cells were resuspended in lysis buffer at 500 OD600 units per ml (20 mM Hepes/KOH, pH 7.4, 50 mM potassium acetate, 0.2 M sorbitol, 2 mM EDTA, and 1 mM DTT with protease inhibitors). For all manipulations, protease inhibitors were from Sigmafast (Sigma-Aldrich), which contain 4-(2-aminoethyl)benzenesulfonyl fluoride, Bestatin hydrochloride, leupeptin, E-64, aprotinin, pepstatin A, and phosphoramidon disodium salt. Cells were flash frozen in liquid N2. Cells were thawed on ice with addition of ddH2O with 1 mM DTT and protease inhibitors (500 OD600 units/ml). Cells were homogenized by ten strokes in a Potter-Elvehjem tissue grinder (Wheaton; Thermo Fisher Scientific) at 4°C. Cells were centrifuged gently at 1,000 g for 5 min at 4°C. The pellet, which contains unbroken cells, was discarded. The resulting PNS was overlaid with cushion buffer (20 mM Hepes/KOH, pH 7.4, 100 mM KCl, and 2 mM MgCl2 with protease inhibitors) at ∼250 OD600 units of PNS in ∼1.0 ml volume with 3.0 ml of cushion buffer. The tubes were centrifuged at 120,000 g for 1 h at 4°C. The lipid droplet cake at the top was removed by aspiration. The pellet (crude membranes) was resuspended in PNS buffer.
Turbidity assay of isolated droplets
Droplet suspensions were diluted 1:20 (to be within the linear range of the spectrophotometer) in droplet isolation buffer, and the optical density was measured in a DU650 spectrophotometer (Beckman Coulter). The values shown in the figures represent extrapolation to the undiluted sample.
Extractions and analysis of lipids
For whole cells, 50–100 OD600 units were used. After a determination of cell pellet wet weight, cells were lysed in 400 µl H2O using acid-washed glass beads in a MiniBeadBeater using three 1-min pulses at 4°C. Lipids were extracted three times in 1 ml 2:1 chloroform/methanol, followed by two extractions in 3 ml chloroform:methanol. The lower phases were combined and washed twice with 1 ml 1.0 M KCl. The lower phase was dried under nitrogen stream and resuspended in the appropriate volume of 1:1 chloroform/methanol. For isolated organelles, samples were brought up to 1 ml in buffer and extracted three times in 3 ml chloroform:methanol, washed in KCl, and processed as described previously.
Samples and standards were separated by thin-layer chromatography on 20-cm TLC Silica Gel 60 plates (EMD Millipore) as previously described (Binns et al., 2006. Plates were charred and lipid bands were analyzed against an internal standard curve as described previously (Adeyo et al., 2011).
Estimation of Pet10p surface area on droplets
The determination of the Pet10p copy number was performed by first purifying overexpressed 2×myc-Pet10-2×FLAG to obtain a protein standard curve from glass-bead lysates using anti-myc beads (Binns et al., 2010). Before cell breakage of the protein cross-linker dithiobis(succinimidyl propionate) (DSP) was added, which we found prevented subsequent Pet10p degradation during affinity purification, the effect of the DSP treatment itself was found to decrease antigenicity by 32% based on immunoblotting on fresh lysates immediately boiled in SDS sample buffer. The resulting pure protein concentration was measured using the amido black method (Schaffner and Weissmann, 1973), and a standard curve of this material was applied to subsequent immunoblots to determine the amount of Pet10p in cells. For this purpose, TCA was added to DGA1 cells (with the undisturbed PET10 gene) grown to late log using the standard conditions, and proteins were extracted in sample buffer. We found that Pet10p was not fully solubilized even after four serial extractions; however, the fourth extraction usually yielded less Pet10p. For calculations, we assumed full recovery after four extractions. Immunoblots from gels containing these cell extracts as well as a purified Pet10p standard curve were performed using the anti-Pet10p antibody, and the values were interpolated from the Pet10p standard curve. TG was determined from chloroform-methanol extracts by TLC. To estimate the droplet surface area, we assumed that TG, the partial specific volume of which is 1.11 ml/g for triolein, comprised the entire core (as the DGA1 strain had no SE) and that all of this neutral lipid was in droplets visible with BODIPY; that is, we neglected any TG in the ER or other membranes. From the estimated volume, we calculated the surface area assuming that the droplets were spherical. Cells were counted in a hemocytometer to relate cell number to OD600 in our strain. The surface of Pet10p on the droplet assumed a partial specific volume of 0.73 cm3/g typical for a soluble protein (Erickson, 2009), a spherical shape, and its bisected cross section aligned with the droplet surface.
Fluorescence microscopy
Microscopy was performed as described previously (Cartwright et al., 2015), except the acquiring microscope was an Axiovert 200M (ZEISS), and the camera was from Q-Imaging (model 01-OPTIMOS-R-M-16-C). The objective used was a 100× 1.3 NA oil immersion lens (ZEISS; Plan-Neofluar). GFP and BODIPY images were acquired using the fluorescein isothiocyanate filter set, mRuby3 and tdTomato images were acquired using the CY3 filter set, Calcofluor White and autoDOT images were acquired with the DAPI filter set, and mTFP images were acquired using the CFP filter set (Chroma Technology Corp.). All images were obtained at room temperature. For BODIPY images, living cells were prestained for 10–15 min with BODIPY (1:1,000 dilution of a 1 mg/ml stock in DMSO) before pipetting onto a slide. A z-stack of 20 images 0.3 or 0.35 µm apart was collected with Slidebook software (v5.5.2 or 6.0.4; Intelligent Imaging Innovations), and maximal projections were generated with nearest-neighbor deconvolution using Slidebook commands. Unless otherwise noted in the figure legends, the microscope intensity settings and times of acquisition were identical for each sample in an experiment. Images for each channel were exported as monocolored 24-bit TIFF files, and merged images were produced in Adobe Photoshop Creative Cloud.
Mass spectrometry
Gel bands were digested overnight with trypsin (Promega) after reduction and alkylation with DTT and iodoacetamide. Digested samples were extracted from the gel with 50:50 water/acetonitrile (ACN), 5% trifluoroacetic acid, dried, and then cleaned by solid-phase extraction with Oasis HLB plates (Waters). The samples were then reconstituted and analyzed by liquid chromatography tandem mass spectrometry using a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled to an UltiMate 3000 RSLCnano liquid chromatography system (Dionex). Samples were injected onto a 75-µm i.d., 15-cm-long Easy Spray column (Thermo Fisher Scientific) and eluted with a gradient from 0–28% buffer B over 40 min. Buffer A contained 2% (vol/vol) ACN and 0.1% formic acid in water, and buffer B contained 80% (vol/vol) ACN, 10% (vol/vol) trifluoroethanol, and 0.08% formic acid in water. The mass spectrometer operated in positive ion mode with a source voltage of 2.4 kV and a capillary temperature of 250°C. Mass spectrometry scans were acquired at 70,000 resolution, and up to 20 tandem mass spectrometry spectra were obtained for each full spectrum acquired at 17,500 resolution using higher-energy collisional dissociation (HCD) for ions with a charge ≥2. Dynamic exclusion was set for 7 s.
Raw mass spectrometry data files were converted to a peak list format and analyzed using the central proteomics facilities pipeline (CPFP version 2.0.3; Trudgian et al., 2010; Trudgian and Mirzaei, 2012). Peptide identification was performed using the X!Tandem (Craig and Beavis, 2004) and open mass spectrometry search algorithm (Geer et al., 2004) search engines against the S. cerevisiae protein database from NCBI. Fragment and precursor tolerances of 20 ppm and 0.1 D, respectively, were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification, and oxidation of Met was set as a variable modification. Label-free quantitation of proteins across samples was performed using SINQ normalized spectral index software (Trudgian et al., 2011).
DAG acyltransferase assay
For analysis of DAG acyltransferase activity, crude membranes and lipid droplets were separated by centrifugation from PNS as described previously (Cartwright et al., 2015). The protein concentration of fractions was determined by amido black assay (Schaffner and Weissmann, 1973). Enzyme activity was measured generally as described previously (Cartwright et al., 2015) with modifications. For membrane fractions, 50 µg of membrane protein was used, which was within the linear range of the assay. For droplet fractions, 0.49 µg of droplet protein was used, which was supplemented with 10 µg of crude membrane protein from quadruple knockout cells. These membranes have no measurable DAG acyltransferase activity, but their addition was necessary for enzyme activity to be in the linear range with respect to droplet protein. Activities were performed in duplicate in each independent experiment. Lipids were visualized and activities calculated as described previously (Cartwright et al., 2015). Dependence on DAG was tested in some experiments by leaving out that exogenous substrate.
Identification of Pet10-sequence homologues
A PSI-BLAST (Altschul et al., 1997) search against the nonredundant database (July 9, 2015) with Pet10p query sequence (gi|398365145, range 1–283) identified fungal Pet10p-related sequences, followed by a more distantly related sequence encoded by BN7_6460 from Wickerhamomyces ciferrii (E-value 7 × 10–6, range 10–251). We further probed the nr database limited to fungi (taxid:4751) using this intermediate sequence (three iterations, E-value cutoff of 0.005) to identify additional Pet10p homologues. Identified sequences were clustered in two dimensions using CLANS (Frickey and Lupas, 2004) with a sequence linkage cutoff E-value of 0.0001. One of the identified groups of fungal sequences related to Pet10 included the previously described fungal perilipin homologue Mpl1 (Wang and St. Leger, 2007). To further confirm this suggested relationship, we submitted representative fungal sequences from the CLANS groupings as well as the five human perilipin sequences to the HHPRED server (Söding et al., 2005), searching profiles from the conserved domain database and the S. cerevisiae genome. Multiple-sequence alignments of Pet10, SpS4, and Mpl1 family sequences, as well as mammalian perilipin sequences, were generated using the MAFFT server (Katoh et al., 2002). Assembly of the individual multiple-sequence alignments into one global alignment of the PAT domain was guided by JPRED (Drozdetskiy et al., 2015) secondary structure predictions and HHPRED alignments.
Replications and statistical analyses
The mass spectroscopic analysis shown in Fig. 1 represents a single experiment. Unless otherwise mentioned, all other experiments were repeated at least twice (i.e., three independent experiments) with similar results. Micrographs represent one of these experiments. Quantified data include all the repetitions, unless stated otherwise. Two-sample comparisons were evaluated by an unpaired Student’s t test, except the data in Fig. 3 A, in which a paired t test was used. Multiple samples were first subjected to ANOVA, and pairs of samples were compared using Tukey’s test method. Unless otherwise noted, means ± SEM are shown throughout. Significance: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Analyses were performed using GraphPad Prism (v6 or 7) software.
Online supplemental material
Fig. S1 contains growth data of strains ± PET10, controls relating to Figs. 2 and 3, and Lro1p subcellular localization data. Fig. S2 contains controls relating to Figs. 5 and 6 and a diagram representing droplet morphology. Fig. S3 contains controls and additional data relating to Fig. 8. Table S1 is a dataset from mass spectrometry analysis of droplet proteins. Table S2 lists strains and plasmids used in this study.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health grant GM084210 (awarded to J.M. Goodman) and GM094575 (awarded to N.V. Grishin); American Diabetes Association Basic Science Award 7-13-BS-055 (awarded to J.M. Goodman); American Heart Association Grant-in-Aid 27540010 (awarded to J.M. Goodman); and Welch Foundation grant I-1505 (awarded to N.V. Grishin).
The authors declare no competing financial interests.
Author contributions: Q. Gao conceived and performed experiments, analyzed data, and wrote the paper. D.D. Binns, N. Ortiz, and X. Chen performed experiments and edited the paper. L.N. Kinch analyzed data and wrote the paper. N.V. Grishin analyzed data. J.M. Goodman conceived experiments, analyzed data, and wrote the paper.
Footnotes
Abbreviations used:
- 2KO
- double knockout
- 3KO
- triple knockout
- ACN
- acetonitrile
- CHX
- cycloheximide
- mTFP
- monomeric teal fluorescent protein
- OA
- oleic acid
- PNS
- postnuclear supernatant
- SD
- synthetic dextrose
- SE
- steryl ester
- TG
- triacylglycerol
References
- Adeyo O., Horn P.J., Lee S., Binns D.D., Chandrahas A., Chapman K.D., and Goodman J.M.. 2011. The yeast lipin orthologue Pah1p is important for biogenesis of lipid droplets. J. Cell Biol. 192:1043–1055. 10.1083/jcb.201010111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., and Lipman D.J.. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athenstaedt K., and Daum G.. 2003. YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278:23317–23323. 10.1074/jbc.M302577200 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., and Daum G.. 2005. Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280:37301–37309. 10.1074/jbc.M507261200 [DOI] [PubMed] [Google Scholar]
- Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S.D., and Daum G.. 1999. Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181:6441–6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel P.E., Tansey J.T., and Welte M.A.. 2009. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta. 1791:419–440. 10.1016/j.bbalip.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D., Januszewski T., Chen Y., Hill J., Markin V.S., Zhao Y., Gilpin C., Chapman K.D., Anderson R.G., and Goodman J.M.. 2006. An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173:719–731. 10.1083/jcb.200511125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns D., Lee S., Hilton C.L., Jiang Q.X., and Goodman J.M.. 2010. Seipin is a discrete homooligomer. Biochemistry. 49:10747–10755. 10.1021/bi1013003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulankina A.V., Deggerich A., Wenzel D., Mutenda K., Wittmann J.G., Rudolph M.G., Burger K.N., and Höning S.. 2009. TIP47 functions in the biogenesis of lipid droplets. J. Cell Biol. 185:641–655. 10.1083/jcb.200812042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartwright B.R., Binns D.D., Hilton C.L., Han S., Gao Q., and Goodman J.M.. 2015. Seipin performs dissectible functions in promoting lipid droplet biogenesis and regulating droplet morphology. Mol. Biol. Cell. 26:726–739. 10.1091/mbc.E14-08-1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V., Ojha N., Golden A., and Prinz W.A.. 2015. A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J. Cell Biol. 211:261–271. 10.1083/jcb.201505067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R., and Beavis R.C.. 2004. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics. 20:1466–1467. 10.1093/bioinformatics/bth092 [DOI] [PubMed] [Google Scholar]
- Currie E., Guo X., Christiano R., Chitraju C., Kory N., Harrison K., Haas J., Walther T.C., and Farese R.V. Jr. 2014. High confidence proteomic analysis of yeast LDs identifies additional droplet proteins and reveals connections to dolichol synthesis and sterol acetylation. J. Lipid Res. 55:1465–1477. 10.1194/jlr.M050229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabany T., Athenstaedt K., and Daum G.. 2007. Synthesis, storage and degradation of neutral lipids in yeast. Biochim. Biophys. Acta. 1771:299–309. 10.1016/j.bbalip.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Czabany T., Wagner A., Zweytick D., Lohner K., Leitner E., Ingolic E., and Daum G.. 2008. Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283:17065–17074. 10.1074/jbc.M800401200 [DOI] [PubMed] [Google Scholar]
- Drozdetskiy A., Cole C., Procter J., and Barton G.J.. 2015. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 43:W389–W394. 10.1093/nar/gkv332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ephrussi B., Hottintguer H., and Chimenes A.-M.. 1949. Action de l’acriflavine sur les levures. I. La mutation “petite colonie.” Ann. Inst. Pasteur (Paris). 76:351–367. [Google Scholar]
- Erickson H.P. 2009. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol. Proced. Online. 11:32–51. 10.1007/s12575-009-9008-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frickey T., and Lupas A.. 2004. CLANS: A Java application for visualizing protein families based on pairwise similarity. Bioinformatics. 20:3702–3704. 10.1093/bioinformatics/bth444 [DOI] [PubMed] [Google Scholar]
- Garber A.T., and Segall J.. 1986. The SPS4 gene of Saccharomyces cerevisiae encodes a major sporulation-specific mRNA. Mol. Cell. Biol. 6:4478–4485. 10.1128/MCB.6.12.4478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geer L.Y., Markey S.P., Kowalak J.A., Wagner L., Xu M., Maynard D.M., Yang X., Shi W., and Bryant S.H.. 2004. Open mass spectrometry search algorithm. J. Proteome Res. 3:958–964. 10.1021/pr0499491 [DOI] [PubMed] [Google Scholar]
- Granneman J.G., Moore H.P., Mottillo E.P., Zhu Z., and Zhou L.. 2011. Interactions of perilipin-5 (Plin5) with adipose triglyceride lipase. J. Biol. Chem. 286:5126–5135. 10.1074/jbc.M110.180711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin N.M., Yu J., Long F., Oh P., Shore S., Li Y., Koziol J.A., and Schnitzer J.E.. 2010. Label-free, normalized quantification of complex mass spectrometry data for proteomic analysis. Nat. Biotechnol. 28:83–89. 10.1038/nbt.1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grillitsch K., Connerth M., Köfeler H., Arrey T.N., Rietschel B., Wagner B., Karas M., and Daum G.. 2011. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: Lipidome meets proteome. Biochim. Biophys. Acta. 1811:1165–1176. 10.1016/j.bbalip.2011.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A., Buxó L., Mora G., Funaya C., Idrissi F.Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P.. 2015. The seipin complex Fld1/Ldb16 stabilizes ER–lipid droplet contact sites. J. Cell Biol. 211:829–844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross D.A., Zhan C., and Silver D.L.. 2011. Direct binding of triglyceride to fat storage-inducing transmembrane proteins 1 and 2 is important for lipid droplet formation. Proc. Natl. Acad. Sci. USA. 108:19581–19586. 10.1073/pnas.1110817108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Binns D.D., Chang Y.F., and Goodman J.M.. 2015. Dissecting seipin function: The localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(ΔNterm) only in combination with Ldb16p. BMC Cell Biol. 16:29 10.1186/s12860-015-0075-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashibe M., and Katohda S.. 1973. Initiation of budding and chitin-ring. J. Gen. Appl. Microbiol. 19:23–39. 10.2323/jgam.19.23 [DOI] [Google Scholar]
- Hsieh K., Lee Y.K., Londos C., Raaka B.M., Dalen K.T., and Kimmel A.R.. 2012. Perilipin family members preferentially sequester to either triacylglycerol-specific or cholesteryl-ester-specific intracellular lipid storage droplets. J. Cell Sci. 125:4067–4076. 10.1242/jcs.104943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N., Choudhary V., Mari M., Toulmay A., Reggiori F., and Schneiter R.. 2011. Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J. Cell Sci. 124:2424–2437. 10.1242/jcs.076836 [DOI] [PubMed] [Google Scholar]
- Jacquier N., Mishra S., Choudhary V., and Schneiter R.. 2013. Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J. Cell Sci. 126:5198–5209. 10.1242/jcs.131896 [DOI] [PubMed] [Google Scholar]
- Kadereit B., Kumar P., Wang W.J., Miranda D., Snapp E.L., Severina N., Torregroza I., Evans T., and Silver D.L.. 2008. Evolutionarily conserved gene family important for fat storage. Proc. Natl. Acad. Sci. USA. 105:94–99. 10.1073/pnas.0708579105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamisaka Y., Kimura K., Uemura H., and Shibakami M.. 2010. Activation of diacylglycerol acyltransferase expressed in Saccharomyces cerevisiae: Overexpression of Dga1p lacking the N-terminal region in the Δsnf2 disruptant produces a significant increase in its enzyme activity. Appl. Microbiol. Biotechnol. 88:105–115. 10.1007/s00253-010-2725-x [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., and Miyata T.. 2002. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel A.R., and Sztalryd C.. 2016. The perilipins: Major cytosolic lipid droplet–associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu. Rev. Nutr. 36:471–509. 10.1146/annurev-nutr-071813-105410 [DOI] [PubMed] [Google Scholar]
- Kory N., Thiam A.R., Farese R.V. Jr., and Walther T.C.. 2015. Protein crowding is a determinant of lipid droplet protein composition. Dev. Cell. 34:351–363. 10.1016/j.devcel.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat C.F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., and Kohlwein S.D.. 2006. Obese yeast: Triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281:491–500. 10.1074/jbc.M508414200 [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 227:680–685. 10.1038/227680a0 [DOI] [PubMed] [Google Scholar]
- Lam C., Santore E., Lavoie E., Needleman L., Fiacco N., Kim C., and Neiman A.M.. 2014. A visual screen of protein localization during sporulation identifies new components of prospore membrane-associated complexes in budding yeast. Eukaryot. Cell. 13:383–391. 10.1128/EC.00333-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leber R., Landl K., Zinser E., Ahorn H., Spök A., Kohlwein S.D., Turnowsky F., and Daum G.. 1998. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol. Biol. Cell. 9:375–386. 10.1091/mbc.9.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P., Chen X., Moktan H., Arrese E.L., Duan L., Wang L., Soulages J.L., and Zhou D.H.. 2014. Membrane attachment and structure models of lipid storage droplet protein 1. Biochim. Biophys. Acta. 1838:874–881. 10.1016/j.bbamem.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X., Gruia-Gray J., Copeland N.G., Gilbert D.J., Jenkins N.A., Londos C., and Kimmel A.R.. 2001. The murine perilipin gene: The lipid droplet-associated perilipins derive from tissue-specific, mRNA splice variants and define a gene family of ancient origin. Mamm. Genome. 12:741–749. 10.1007/s00335-01-2055-5 [DOI] [PubMed] [Google Scholar]
- Markgraf D.F., Klemm R.W., Junker M., Hannibal-Bach H.K., Ejsing C.S., and Rapoport T.A.. 2014. An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Reports. 6:44–55. 10.1016/j.celrep.2013.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelkers P., Cromley D., Padamsee M., Billheimer J.T., and Sturley S.L.. 2002. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J. Biol. Chem. 277:8877–8881. 10.1074/jbc.M111646200 [DOI] [PubMed] [Google Scholar]
- Rowe E.R., Mimmack M.L., Barbosa A.D., Haider A., Isaac I., Ouberai M.M., Thiam A.R., Patel S., Saudek V., Siniossoglou S., and Savage D.B.. 2016. Conserved amphipathic helices mediate lipid droplet targeting of perilipins 1–3. J. Biol. Chem. 291:6664–6678. 10.1074/jbc.M115.691048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta M.P., and Liang S.. 2003. Predicting protein functions from redundancies in large-scale protein interaction networks. Proc. Natl. Acad. Sci. USA. 100:12579–12583. 10.1073/pnas.2132527100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W., and Weissmann C.. 1973. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56:502–514. 10.1016/0003-2697(73)90217-0 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinner J.R., Shew T.M., Schwartz D.M., Tzekov A., Lepus C.M., Abumrad N.A., and Wolins N.E.. 2009. Diacylglycerol enrichment of endoplasmic reticulum or lipid droplets recruits perilipin 3/TIP47 during lipid storage and mobilization. J. Biol. Chem. 284:30941–30948. 10.1074/jbc.M109.013995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J., Biegert A., and Lupas A.N.. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33(Web Server, Suppl. 2):W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger D., and Daum G.. 2002. Synthesis of triacylglycerols by the acyl-coenzyme A:diacyl-glycerol acyltransferase Dga1p in lipid particles of the yeast Saccharomyces cerevisiae. J. Bacteriol. 184:519–524. 10.1128/JB.184.2.519-524.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian V., Rothenberg A., Gomez C., Cohen A.W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M.P., and Brasaemle D.L.. 2004. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J. Biol. Chem. 279:42062–42071. 10.1074/jbc.M407462200 [DOI] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagwerker C., Zhang H., Wang X., Larsen L.S., Lathrop R.H., Hatfield G.W., Auer B., Huang L., and Kaiser P.. 2006. HB tag modules for PCR-based gene tagging and tandem affinity purification in Saccharomyces cerevisiae. Yeast. 23:623–632. 10.1002/yea.1380 [DOI] [PubMed] [Google Scholar]
- Thiam A.R., Farese R.V. Jr., and Walther T.C.. 2013. The biophysics and cell biology of lipid droplets. Nat. Rev. Mol. Cell Biol. 14:775–786. 10.1038/nrm3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.J., and Rothstein R.. 1989. Elevated recombination rates in transcriptionally active DNA. Cell. 56:619–630. 10.1016/0092-8674(89)90584-9 [DOI] [PubMed] [Google Scholar]
- Trudgian D.C., and Mirzaei H.. 2012. Cloud CPFP: A shotgun proteomics data analysis pipeline using cloud and high performance computing. J. Proteome Res. 11:6282–6290. [DOI] [PubMed] [Google Scholar]
- Trudgian D.C., Thomas B., McGowan S.J., Kessler B.M., Salek M., and Acuto O.. 2010. CPFP: A central proteomics facilities pipeline. Bioinformatics. 26:1131–1132. 10.1093/bioinformatics/btq081 [DOI] [PubMed] [Google Scholar]
- Trudgian D.C., Ridlova G., Fischer R., Mackeen M.M., Ternette N., Acuto O., Kessler B.M., and Thomas B.. 2011. Comparative evaluation of label-free SINQ normalized spectral index quantitation in the central proteomics facilities pipeline. Proteomics. 11:2790–2797. 10.1002/pmic.201000800 [DOI] [PubMed] [Google Scholar]
- Tzen J.T., and Huang A.H.. 1992. Surface structure and properties of plant seed oil bodies. J. Cell Biol. 117:327–335. 10.1083/jcb.117.2.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T., Todde V., de Boer R., Kreim M., Hofbauer H.F., Wolinski H., Veenhuis M., van der Klei I.J., and Kohlwein S.D.. 2014. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 25:290–301. 10.1091/mbc.E13-08-0448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenhuis M., Mateblowski M., Kunau W.H., and Harder W.. 1987. Proliferation of microbodies in Saccharomyces cerevisiae. Yeast. 3:77–84. 10.1002/yea.320030204 [DOI] [PubMed] [Google Scholar]
- Wang C.W., and Lee S.C.. 2012. The ubiquitin-like (UBX)-domain-containing protein Ubx2/Ubxd8 regulates lipid droplet homeostasis. J. Cell Sci. 125:2930–2939. 10.1242/jcs.100230 [DOI] [PubMed] [Google Scholar]
- Wang C., and St. Leger R.J.. 2007. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J. Biol. Chem. 282:21110–21115. 10.1074/jbc.M609592200 [DOI] [PubMed] [Google Scholar]
- Wang C.W., Miao Y.H., and Chang Y.S.. 2014. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127:1214–1228. 10.1242/jcs.137737 [DOI] [PubMed] [Google Scholar]
- Wang H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. . 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5:e16582 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolins N.E., Skinner J.R., Schoenfish M.J., Tzekov A., Bensch K.G., and Bickel P.E.. 2003. Adipocyte protein S3-12 coats nascent lipid droplets. J. Biol. Chem. 278:37713–37721. 10.1074/jbc.M304025200 [DOI] [PubMed] [Google Scholar]
- Wolinski H., Hofbauer H.F., Hellauer K., Cristobal-Sarramian A., Kolb D., Radulovic M., Knittelfelder O.L., Rechberger G.N., and Kohlwein S.D.. 2015. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta. 1851:1450–1464. 10.1016/j.bbalip.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Yang H.J., Hsu C.L., Yang J.Y., and Yang W.Y.. 2012. Monodansylpentane as a blue-fluorescent lipid-droplet marker for multi-color live-cell imaging. PLoS One. 7:e32693 10.1371/journal.pone.0032693 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.